Abstract
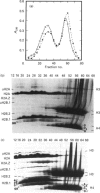
The properties of the nucleosomes of a salt-soluble, transcriptionally active gene-enriched fraction of chicken erythrocyte chromatin were evaluated by hydroxyapatite dissociation chromatography. We have demonstrated previously that the salt-soluble, transcriptionally active gene-enriched polynucleosomes are enriched in dynamically acetylated and ubiquitinated histones, and in an atypical U-shaped nucleosome that possessed about 20% less protein than a typical nucleosome. Further, newly synthesized histones H2A and H2B exchange preferentially with the nucleosomal histones H2A and H2B of this salt-soluble chromatin fraction. Analysis of the histones eluting from the hydroxyapatite-bound chromatin demonstrated that hyperacetylated and ubiquitinated (u), including multi-ubiquitinated, H2A-H2B.1 dimers dissociated at lower concentrations of NaCl than unmodified dimers or dimers with histone variants H2A.Z and/or H2B.2. Cross-linking studies revealed that at least 50% of uH2B.1 was paired with uH2A. uH2A-uH2B.1 dimers dissociated at lower NaCl concentrations than H2A-uH2B.1 dimers. Hyperacetylated histone (H3-H4)2 tetramers also eluted at lower concentrations of NaCl than unmodified tetramers. Our results support the idea that acetylation and ubiquitination of histones H2A and H2B.1 increase the lability of H2A-H2B.1 dimers in transcriptionally active nucleosomes. In contrast, our observations suggest that histone variants H2A.Z and H2B.2. stabilize the association of the H2A-H2B dimer in nucleosomes. The elevated lability of the H2A-H2B dimer may facilitate processes such as the exchange of these dimers with newly synthesized histones, the elongation process of transcription and transcription factor binding.
Full text
PDF

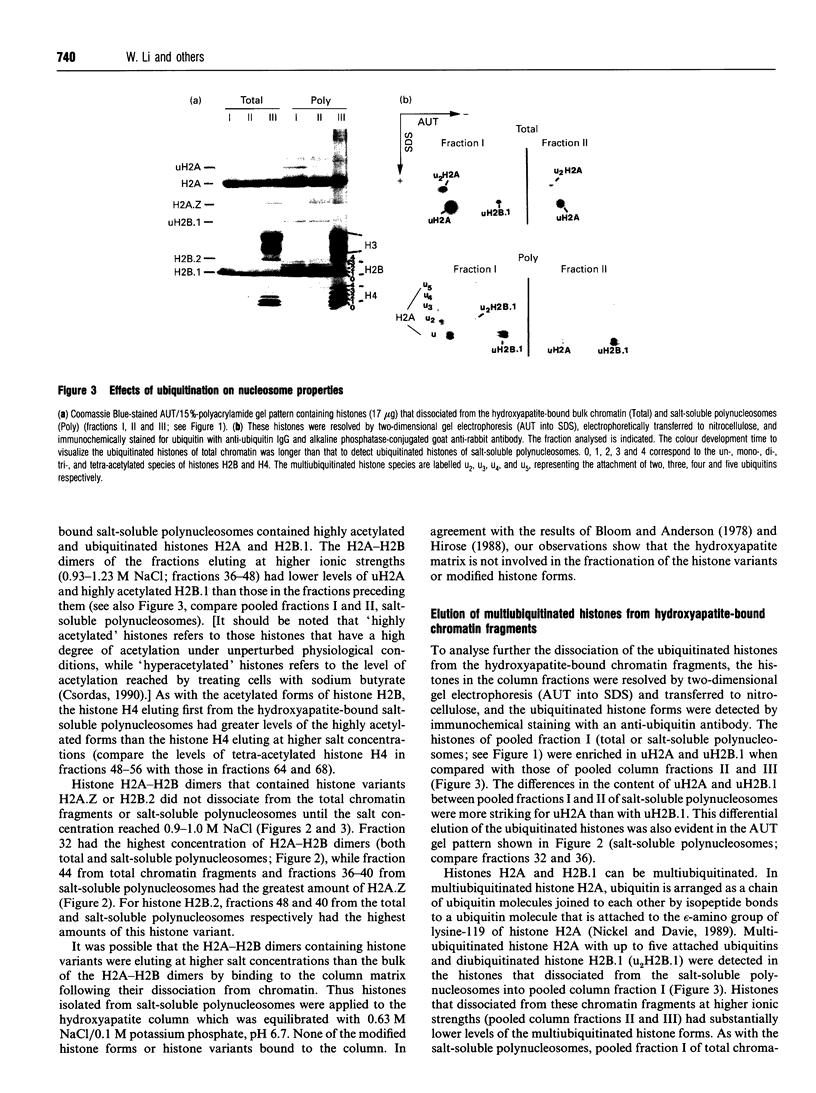

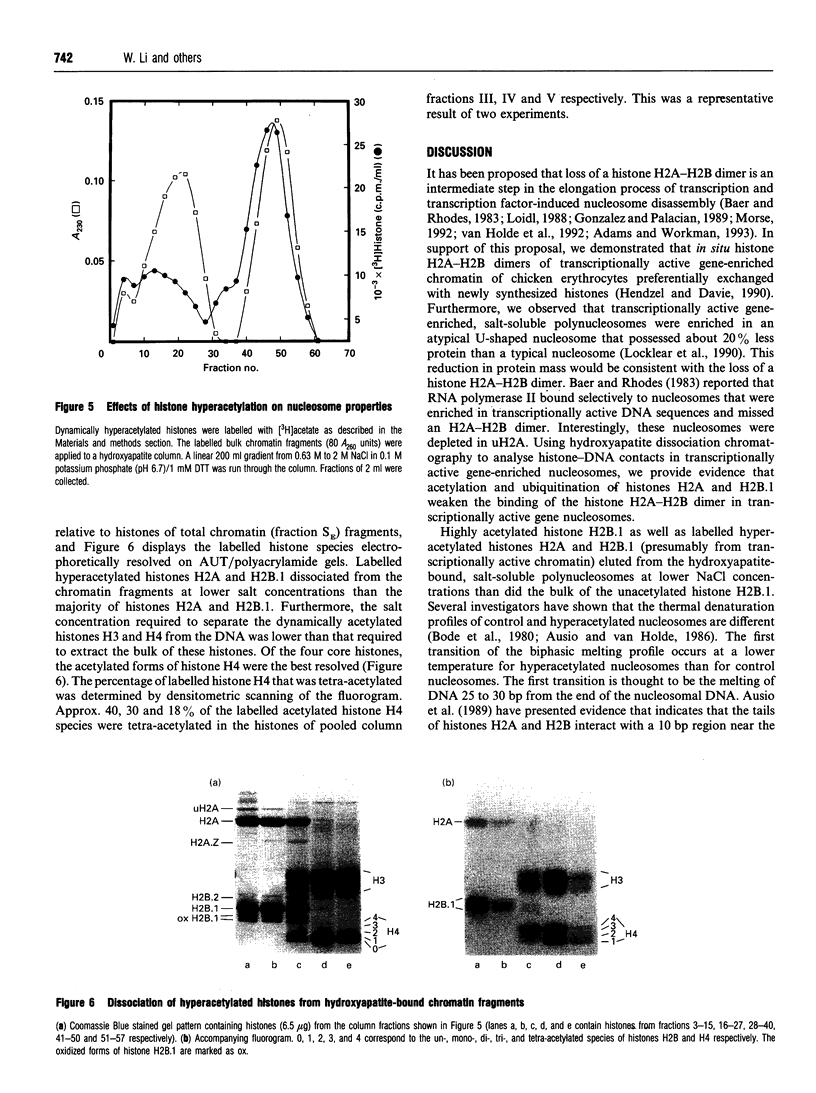


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams C. C., Workman J. L. Nucleosome displacement in transcription. Cell. 1993 Feb 12;72(3):305–308. doi: 10.1016/0092-8674(93)90109-4. [DOI] [PubMed] [Google Scholar]
- Ausio J., Dong F., van Holde K. E. Use of selectively trypsinized nucleosome core particles to analyze the role of the histone "tails" in the stabilization of the nucleosome. J Mol Biol. 1989 Apr 5;206(3):451–463. doi: 10.1016/0022-2836(89)90493-2. [DOI] [PubMed] [Google Scholar]
- Ausio J., van Holde K. E. Histone hyperacetylation: its effects on nucleosome conformation and stability. Biochemistry. 1986 Mar 25;25(6):1421–1428. doi: 10.1021/bi00354a035. [DOI] [PubMed] [Google Scholar]
- Baer B. W., Rhodes D. Eukaryotic RNA polymerase II binds to nucleosome cores from transcribed genes. Nature. 1983 Feb 10;301(5900):482–488. doi: 10.1038/301482a0. [DOI] [PubMed] [Google Scholar]
- Bloom K. S., Anderson J. N. Fractionation and characterization of chromosomal proteins by the hydroxyapatite dissociation method. J Biol Chem. 1978 Jun 25;253(12):4446–4450. [PubMed] [Google Scholar]
- Bode J., Henco K., Wingender E. Modulation of the nucleosome structure by histone acetylation. Eur J Biochem. 1980 Sep;110(1):143–152. doi: 10.1111/j.1432-1033.1980.tb04849.x. [DOI] [PubMed] [Google Scholar]
- Boffa L. C., Walker J., Chen T. A., Sterner R., Mariani M. R., Allfrey V. G. Factors affecting nucleosome structure in transcriptionally active chromatin. Histone acetylation, nascent RNA and inhibitors of RNA synthesis. Eur J Biochem. 1990 Dec 27;194(3):811–823. doi: 10.1111/j.1432-1033.1990.tb19474.x. [DOI] [PubMed] [Google Scholar]
- Burlingame R. W., Love W. E., Wang B. C., Hamlin R., Nguyen H. X., Moudrianakis E. N. Crystallographic structure of the octameric histone core of the nucleosome at a resolution of 3.3 A. Science. 1985 May 3;228(4699):546–553. doi: 10.1126/science.3983639. [DOI] [PubMed] [Google Scholar]
- Chen T. A., Sterner R., Cozzolino A., Allfrey V. G. Reversible and irreversible changes in nucleosome structure along the c-fos and c-myc oncogenes following inhibition of transcription. J Mol Biol. 1990 Apr 5;212(3):481–493. doi: 10.1016/0022-2836(90)90327-I. [DOI] [PubMed] [Google Scholar]
- Csordas A. On the biological role of histone acetylation. Biochem J. 1990 Jan 1;265(1):23–38. doi: 10.1042/bj2650023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davie J. R., Lin R., Allis C. D. Timing of the appearance of ubiquitinated histones in developing new macronuclei of Tetrahymena thermophila. Biochem Cell Biol. 1991 Jan;69(1):66–71. doi: 10.1139/o91-009. [DOI] [PubMed] [Google Scholar]
- Davie J. R., Murphy L. C. Level of ubiquitinated histone H2B in chromatin is coupled to ongoing transcription. Biochemistry. 1990 May 22;29(20):4752–4757. doi: 10.1021/bi00472a002. [DOI] [PubMed] [Google Scholar]
- Davie J. R. Two-dimensional gel systems for rapid histone analysis for use in minislab polyacrylamide gel electrophoresis. Anal Biochem. 1982 Mar 1;120(2):276–281. doi: 10.1016/0003-2697(82)90348-7. [DOI] [PubMed] [Google Scholar]
- Delcuve G. P., Davie J. R. Chromatin structure of erythroid-specific genes of immature and mature chicken erythrocytes. Biochem J. 1989 Oct 1;263(1):179–186. doi: 10.1042/bj2630179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcuve G. P., Davie J. R. Western blotting and immunochemical detection of histones electrophoretically resolved on acid-urea-triton- and sodium dodecyl sulfate-polyacrylamide gels. Anal Biochem. 1992 Feb 1;200(2):339–341. doi: 10.1016/0003-2697(92)90475-m. [DOI] [PubMed] [Google Scholar]
- Eickbush T. H., Godfrey J. E., Elia M. C., Moudrianakis E. N. H2a-specific proteolysis as a unique probe in the analysis of the histone octamer. J Biol Chem. 1988 Dec 15;263(35):18972–18978. [PubMed] [Google Scholar]
- Eickbush T. H., Moudrianakis E. N. The histone core complex: an octamer assembled by two sets of protein-protein interactions. Biochemistry. 1978 Nov 14;17(23):4955–4964. doi: 10.1021/bi00616a016. [DOI] [PubMed] [Google Scholar]
- Elgin S. C. Chromatin structure and gene activity. Curr Opin Cell Biol. 1990 Jun;2(3):437–445. doi: 10.1016/0955-0674(90)90125-x. [DOI] [PubMed] [Google Scholar]
- González P. J., Palacián E. Interaction of RNA polymerase II with structurally altered nucleosomal particles. Transcription is facilitated by loss of one H2A.H2B dimer. J Biol Chem. 1989 Nov 5;264(31):18457–18462. [PubMed] [Google Scholar]
- Grandy D. K., Dodgson J. B. Structure and organization of the chicken H2B histone gene family. Nucleic Acids Res. 1987 Feb 11;15(3):1063–1080. doi: 10.1093/nar/15.3.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein M. Nucleosomes: regulators of transcription. Trends Genet. 1990 Dec;6(12):395–400. doi: 10.1016/0168-9525(90)90299-l. [DOI] [PubMed] [Google Scholar]
- Harvey R. P., Whiting J. A., Coles L. S., Krieg P. A., Wells J. R. H2A.F: an extremely variant histone H2A sequence expressed in the chicken embryo. Proc Natl Acad Sci U S A. 1983 May;80(10):2819–2823. doi: 10.1073/pnas.80.10.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch C. L., Bonner W. M., Moudrianakis E. N. Minor histone 2A variants and ubiquinated forms in the native H2A:H2B dimer. Science. 1983 Jul 29;221(4609):468–470. doi: 10.1126/science.6306766. [DOI] [PubMed] [Google Scholar]
- Hatch C. L., Bonner W. M. Sequence of cDNAs for mammalian H2A.Z, an evolutionarily diverged but highly conserved basal histone H2A isoprotein species. Nucleic Acids Res. 1988 Feb 11;16(3):1113–1124. doi: 10.1093/nar/16.3.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendzel M. J., Davie J. R. Distribution of methylated histones and histone methyltransferases in chicken erythrocyte chromatin. J Biol Chem. 1989 Nov 15;264(32):19208–19214. [PubMed] [Google Scholar]
- Hendzel M. J., Davie J. R. Nucleosomal histones of transcriptionally active/competent chromatin preferentially exchange with newly synthesized histones in quiescent chicken erythrocytes. Biochem J. 1990 Oct 1;271(1):67–73. doi: 10.1042/bj2710067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendzel M. J., Delcuve G. P., Davie J. R. Histone deacetylase is a component of the internal nuclear matrix. J Biol Chem. 1991 Nov 15;266(32):21936–21942. [PubMed] [Google Scholar]
- Hirose M. Effects of histone acetylation on nucleosome properties as evaluated by polyacrylamide gel electrophoresis and hydroxylapatite dissociation chromatography. J Biochem. 1988 Jan;103(1):31–35. doi: 10.1093/oxfordjournals.jbchem.a122234. [DOI] [PubMed] [Google Scholar]
- Ip Y. T., Jackson V., Meier J., Chalkley R. The separation of transcriptionally engaged genes. J Biol Chem. 1988 Oct 5;263(28):14044–14052. [PubMed] [Google Scholar]
- Lee D. Y., Hayes J. J., Pruss D., Wolffe A. P. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell. 1993 Jan 15;72(1):73–84. doi: 10.1016/0092-8674(93)90051-q. [DOI] [PubMed] [Google Scholar]
- Locklear L., Jr, Ridsdale J. A., Bazett-Jones D. P., Davie J. R. Ultrastructure of transcriptionally competent chromatin. Nucleic Acids Res. 1990 Dec 11;18(23):7015–7024. doi: 10.1093/nar/18.23.7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loidl P. Towards an understanding of the biological function of histone acetylation. FEBS Lett. 1988 Jan 25;227(2):91–95. doi: 10.1016/0014-5793(88)80874-3. [DOI] [PubMed] [Google Scholar]
- Moehs C. P., Baxevanis A. D., Moudrianakis E. N., Spiker S. Enhanced stability of histone octamers from plant nucleosomes: role of H2A and H2B histones. Biochemistry. 1992 Nov 10;31(44):10844–10851. doi: 10.1021/bi00159a027. [DOI] [PubMed] [Google Scholar]
- Morse R. H. Transcribed chromatin. Trends Biochem Sci. 1992 Jan;17(1):23–26. doi: 10.1016/0968-0004(92)90422-6. [DOI] [PubMed] [Google Scholar]
- Nickel B. E., Davie J. R. Structure of polyubiquitinated histone H2A. Biochemistry. 1989 Feb 7;28(3):964–968. doi: 10.1021/bi00429a007. [DOI] [PubMed] [Google Scholar]
- Nickel B. E., Roth S. Y., Cook R. G., Allis C. D., Davie J. R. Changes in the histone H2A variant H2A.Z and polyubiquitinated histone species in developing trout testis. Biochemistry. 1987 Jul 14;26(14):4417–4421. doi: 10.1021/bi00388a034. [DOI] [PubMed] [Google Scholar]
- Ridsdale J. A., Davie J. R. Chicken erythrocyte polynucleosomes which are soluble at physiological ionic strength and contain linker histones are highly enriched in beta-globin gene sequences. Nucleic Acids Res. 1987 Feb 11;15(3):1081–1096. doi: 10.1093/nar/15.3.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridsdale J. A., Hendzel M. J., Delcuve G. P., Davie J. R. Histone acetylation alters the capacity of the H1 histones to condense transcriptionally active/competent chromatin. J Biol Chem. 1990 Mar 25;265(9):5150–5156. [PubMed] [Google Scholar]
- Simon R. H., Felsenfeld G. A new procedure for purifying histone pairs H2A + H2B and H3 + H4 from chromatin using hydroxylapatite. Nucleic Acids Res. 1979 Feb;6(2):689–696. doi: 10.1093/nar/6.2.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. T. Structure of chromatin containing extensively acetylated H3 and H4. Cell. 1978 Apr;13(4):691–699. doi: 10.1016/0092-8674(78)90219-2. [DOI] [PubMed] [Google Scholar]
- Stefanovsky VYu, Dimitrov S. I., Russanova V. R., Angelov D., Pashev I. G. Laser-induced crosslinking of histones to DNA in chromatin and core particles: implications in studying histone-DNA interactions. Nucleic Acids Res. 1989 Dec 11;17(23):10069–10081. doi: 10.1093/nar/17.23.10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickland M., Strickland W. N., Von Holt C. The occurrence of sperm in isohistones H2B in single sea urchins. FEBS Lett. 1981 Nov 30;135(1):86–88. doi: 10.1016/0014-5793(81)80949-0. [DOI] [PubMed] [Google Scholar]
- Svaren J., Chalkley R. The structure and assembly of active chromatin. Trends Genet. 1990 Feb;6(2):52–56. doi: 10.1016/0168-9525(90)90074-g. [DOI] [PubMed] [Google Scholar]
- Urban M. K., Franklin S. G., Zweidler A. Isolation and characterization of the histone variants in chicken erythrocytes. Biochemistry. 1979 Sep 4;18(18):3952–3960. doi: 10.1021/bi00585a017. [DOI] [PubMed] [Google Scholar]
- Walker I. O. Differential dissociation of histone tails from core chromatin. Biochemistry. 1984 Nov 6;23(23):5622–5628. doi: 10.1021/bi00318a037. [DOI] [PubMed] [Google Scholar]
- White E. M., Shapiro D. L., Allis C. D., Gorovsky M. A. Sequence and properties of the message encoding Tetrahymena hv1, a highly evolutionarily conserved histone H2A variant that is associated with active genes. Nucleic Acids Res. 1988 Jan 11;16(1):179–198. doi: 10.1093/nar/16.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Nelson D. A. Histone acetylation in chicken erythrocytes. Estimation of the percentage of sites actively modified. Biochem J. 1986 Dec 15;240(3):857–862. doi: 10.1042/bj2400857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Daal A., Elgin S. C. A histone variant, H2AvD, is essential in Drosophila melanogaster. Mol Biol Cell. 1992 Jun;3(6):593–602. doi: 10.1091/mbc.3.6.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Daal A., White E. M., Gorovsky M. A., Elgin S. C. Drosophila has a single copy of the gene encoding a highly conserved histone H2A variant of the H2A.F/Z type. Nucleic Acids Res. 1988 Aug 11;16(15):7487–7497. doi: 10.1093/nar/16.15.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Holde K. E., Lohr D. E., Robert C. What happens to nucleosomes during transcription? J Biol Chem. 1992 Feb 15;267(5):2837–2840. [PubMed] [Google Scholar]