Abstract
The objective of this study is to develop a multimodal neural network (MMNN) model that analyzes clinical variables and MRI images of a soft tissue sarcoma (STS) patient, to predict overall survival and risk of distant metastases. We compare the performance of this MMNN to models based on clinical variables alone, radiomics models, and an unimodal neural network. We include patients aged 18 or older with biopsy-proven STS who underwent primary resection between January 1st, 2005, and December 31st, 2020 with complete outcome data and a pre-treatment MRI with both a T1 post-contrast sequence and a T2 fat-sat sequence available. A total of 9380 MRI slices containing sarcomas from 287 patients are available. Our MMNN accepts the entire 3D sarcoma volume from T1 and T2 MRIs and clinical variables. Gradient blending allows the clinical and image sub-networks to optimally converge without overfitting. Heat maps were generated to visualize the salient image features. Our MMNN outperformed all other models in predicting overall survival and the risk of distant metastases. The C-Index of our MMNN for overall survival is 0.77 and the C-Index for risk of distant metastases is 0.70. The provided heat maps demonstrate areas of sarcomas deemed most salient for predictions. Our multimodal neural network with gradient blending improves predictions of overall survival and risk of distant metastases in patients with soft tissue sarcoma. Future work enabling accurate subtype-specific predictions will likely utilize similar end-to-end multimodal neural network architecture and require prospective curation of high-quality data, the inclusion of genomic data, and the involvement of multiple centers through federated learning.
Subject terms: Metastasis, Sarcoma
Introduction
Soft tissue sarcoma (STS) accounts for 1% of adult malignancies but comprises over 70 distinct subtypes with varying biology, genetics, clinical characteristics, and disease prognoses1. Over time, the treatment of patients with STS has started to shift from a relatively homogenous approach to therapy that is more tailored to the relevant subtype2. However, 5-year survival has largely plateaued over the past 3 decades at around 65–70% and new innovations in matching patients to optimal treatments are needed to further improve outcomes3.
An example of tailored therapy in STS is the use of systemic chemotherapy in addition to wide surgical resection and radiation therapy2. While not indicated for all STS patients, the decision by a multidisciplinary tumor board to use chemotherapy is influenced by the predicted aggressivity of a patient’s sarcoma, and thus the patient’s risk of developing metastases1. Furthermore, the postoperative surveillance of STS patients, commonly with CT scans of the chest, is more intensive in the first two years after surgery when the average risk of developing a metastasis is highest4,5. A current randomized trial is investigating if intensive surveillance regimens increase patient survival by detecting metastases sooner and offering the opportunity for earlier intervention4. However, the ideal surveillance regimen may be one tailored to a patient’s perspectives and individualized risk of metastases6.
Some nomograms, such as the ‘Sarculator’7, have been developed to help guide management by predicting the risk of metastases and overall survival in patients with extremity or retroperitoneal STS. It is externally validated8 but is based on only 4 clinical variables (patient age, tumor size, tumor grade, and tumor subtype). Previous nomograms from Memorial Sloan Kettering Cancer Center (MSKCC) which predict local recurrence9 and overall survival10 have been similarly based on only clinical variables. These tools are increasingly used in practice, yet they capture aggregate median data and not individual granular data. The creator of Sarculator himself stated in an editorial that nomograms such as his “represent the mean outcome of patients but individual variability cannot be caught”11. Importantly, data from other modalities are not incorporated into these nomograms. 3D magnetic resonance imaging (MRI) scans of sarcoma are a rich source of data that can be used in modelling but have not been routinely incorporated into sarcoma nomograms. MRIs can provide individualized data for each patient including important tumor features such as vascularity, necrosis, peritumoral edema, and heterogeneity. The inclusion of rich data like MRIs in prediction models – thus advancing from unimodal to multimodal models, may result in predictions accurate enough to enable individualized treatment of sarcoma patients.
Radiomics is one method of extracting features from image data. With radiomics, a predetermined set of human-engineered features such as texture, heterogeneity, and size are extracted from the image. The best combination of these features is then related to the outcome in question12,13. In contrast, deep learning methods using convolutional neural networks (CNNs) are able to learn the optimal features from the images directly, they are not limited by a pre-specified set of features which may not capture all of the salient information13,14. Recent comparisons of the performance of radiomics and CNNs on the same datasets have shown that CNNs are not only more accurate but achieve better results on external validation15,16. Thus, while advantages of radiomics include easier implementation, being computationally inexpensive, and standardization based on the Image Biomarker Standardization Initiative (IBSI)17, neural network-based techniques are likely to be superior in capturing important individualized features from MRI data which then generalize to patients from other institutions.
Recently, neural network methods have been used to predict the grade of sarcoma based on imaging alone18, predict the risk of metastases based on imaging alone14, or provide histologic diagnosis based on imaging19. However, none of these models were multimodal, their sample sizes were generally small (N = 51) and lacked a hold-out test set14, or they predict a diagnosis among a limited set of possibilities, as opposed to the entire breadth of sarcoma subtypes20. Whereas multimodal AI is of greater importance in soft tissue sarcoma due to the low prevalence and many subtypes21, to our knowledge there is no multimodal AI model that analyzes MRI data and clinical variables to predict the patient-important outcomes of overall survival or the risk of distant metastases.
The objective of this study is to develop a multimodal neural network (MMNN) model that analyzes clinical variables as well as MRI images of a sarcoma, to predict an STS patient’s overall survival and risk of distant metastases. We compare the performance of this multimodal model to other models based on clinical variables alone, radiomics models, and a unimodal neural network model.
Results and Discussion
Our cohort includes 287 patients with biopsy-confirmed soft tissue sarcoma from ages 18-91 (IQR 45–68) and 121/287 (42%) of patients are female [Table 1]. The list of included subtypes is Table 2. The median follow-up time was 4.1 years / 1511 days (IQR 2.2–7.3 years). In our cohort, 86/287 patients (30%) died from their disease during the follow-up period while 111/287 (38.7%) developed distant metastases. Not every patient who developed metastases died during their follow-up period.
Table 1.
Patient demographics in entire cohort and in the training, test and validation sets
| Missing | Overall | TRAIN SET | VALID SET | TEST SET | ||
|---|---|---|---|---|---|---|
| n | 287 | 199 | 44 | 44 | ||
| Age, mean (SD) | 0 | 55.7 (17.4) | 55.2 (17.1) | 59.0 (19.3) | 54.7 (16.7) | |
| Sex, n (%) | F | 0 | 122 (42.5) | 81 (40.7) | 25 (56.8) | 16 (36.4) |
| M | 165 (57.5) | 118 (59.3) | 19 (43.2) | 28 (63.6) | ||
| Tumor Size, median [Q1,Q3] | 0 | 9.0 [6.1,12.0] | 9.0 [6.2,11.8] | 9.0 [6.7,12.1] | 8.4 [5.7,13.0] | |
| Tumor Depth, n (%) | Superficial | 0 | 28 (9.8) | 23 (11.6) | 2 (4.5) | 3 (6.8) |
| Deep to fascia | 259 (90.2) | 176 (88.4) | 42 (95.5) | 41 (93.2) | ||
| Tumor Grade, n (%) | Low Grade | 0 | 21 (7.3) | 16 (8.0) | 2 (4.5) | 3 (6.8) |
| High Grade | 266 (92.7) | 183 (92.0) | 42 (95.5) | 41 (93.2) | ||
| Tumor Volume (cm^3), median [Q1,Q3] | 0 | 284.1 [82.4,694.3] | 292.5 [84.1,668.3] | 284.6 [81.8,830.6] | 250.3 [77.3,757.9] | |
| Follow up Time, median [Q1,Q3] | 0 | 1511.0 [819.0,2680.0] | 1577.0 [821.5,2741.0] | 1413.5 [729.2,2307.2] | 1423.0 [835.0,2529.5] | |
| Location, n (%) | Axial | 0 | 40 (13.9) | 27 (13.6) | 8 (18.2) | 5 (11.4) |
| Lower Extremity | 194 (67.6) | 136 (68.3) | 31 (70.5) | 27 (61.4) | ||
| Upper Extremity | 53 (18.5) | 36 (18.1) | 5 (11.4) | 12 (27.3) | ||
| Chemo (Neoadjuvant), n (%) | False | 0 | 193 (67.2) | 131 (65.8) | 30 (68.2) | 32 (72.7) |
| True | 94 (32.8) | 68 (34.2) | 14 (31.8) | 12 (27.3) | ||
| RT Type, n (%) | Pre-op | 0 | 132 (46.0) | 96 (48.2) | 16 (36.4) | 20 (45.5) |
| Post-op | 155 (54.0) | 103 (51.8) | 28 (63.6) | 24 (54.5) | ||
| Margin, n (%) | Negative (R0) | 1 | 259 (90.6) | 181 (91.0) | 41 (93.2) | 37 (86.0) |
| R1 | 26 (9.1) | 17 (8.5) | 3 (6.8) | 6 (14.0) | ||
| R2 | 1 (0.3) | 1 (0.5) | ||||
| Length of Surgery, median [Q1,Q3] | 0 | 3.5 [2.5,5.5] | 3.5 [2.5,5.8] | 3.4 [2.5,5.1] | 3.2 [2.0,5.5] | |
| Necrosis, median [Q1,Q3] | 13 | 0.3 [0.1,0.7] | 0.3 [0.1,0.7] | 0.3 [0.1,0.8] | 0.3 [0.1,0.7] | |
| Vital Status, n (%) | Dead | 0 | 201 (70.0) | 139 (69.8) | 32 (72.7) | 30 (68.2) |
| Alive | 86 (30.0) | 60 (30.2) | 12 (27.3) | 14 (31.8) | ||
| Local Recurrence, n (%) | No | 0 | 259 (90.2) | 180 (90.5) | 39 (88.6) | 40 (90.9) |
| Yes | 28 (9.8) | 19 (9.5) | 5 (11.4) | 4 (9.1) | ||
| Distant Mets, n (%) | No | 0 | 176 (61.3) | 118 (59.3) | 28 (63.6) | 30 (68.2) |
| Yes | 111 (38.7) | 81 (40.7) | 16 (36.4) | 14 (31.8) |
Breakdown of train, test and val sets. Tumor Grade: Grade is based on the final pathology report. Low Grade = Grade 1. High Grade = Grade 2, 3, or dedifferentiated. The 21 Low Grade sarcomas include 17 myxoid liposarcomas with round cell component <5%, two extraskeletal myxoid chondrosarcomas and two solitary fibrous tumors.
Table 2.
Included sarcoma histologic subtypes
| OVERALL n (%) | TRAIN SET n (%) | VALID SET n (%) | TEST SET n (%) | |
|---|---|---|---|---|
| HISTOLOGIC SUBTYPE | 287 | 199 | 44 | 44 |
| Undifferentiated Pleomorphic Sarcoma | 87 (30.5) | 61 (30.7) | 15 (35.7) | 11 (25.0) |
| Myxofibrosarcoma | 63 (22.1) | 40 (20.1) | 13 (31.0) | 10 (22.7) |
| Myxoid Liposarcoma | 33 (11.6) | 24 (12.1) | 3 (7.1) | 6 (13.6) |
| Synovial sarcoma | 35 (12.3) | 23 (11.6) | 3 (7.1) | 9 (20.5) |
| Liposarcoma | 22 (7.7) | 16 (8.0) | 2 (4.8) | 4 (9.1) |
| Leiomyosarcoma | 8 (2.8) | 5 (2.5) | 1 (2.4) | 2 (4.5) |
| Rhabdomyosarcoma | 6 (2.1) | 5 (2.5) | 1 (2.4) | |
| Myxoinflammatory Fibroblastic sarcoma | 6 (2.1) | 4 (2.0) | 2 (4.8) | |
| MPNST | 5 (1.8) | 2 (1.0) | 3 (7.1) | |
| Ewings of soft tissue | 4 (1.4) | 4 (2.0) | ||
| Solitary Fibrous Tumor | 4 (1.4) | 4 (2.0) | ||
| Extraskeletal Myxoid Chondrosarcoma | 4 (1.4) | 2 (1.0) | 2 (4.5) | |
| Extraskeletal Osteosarcoma | 3 (1.1) | 3 (1.0) | ||
| ASPS | 2 (0.7) | 2 (1.0) | ||
| Spindle cell sarcoma | 2 (0.7) | 2 (1.0) | ||
| Clear cell sarcoma | 1 (0.3) | 1 (2.4) | ||
| Epithelioid sarcoma | 1 (0.3) | 1 (1.5) | ||
| Fibrosarcoma | 1 (0.3) | 1 (1.0) |
Subtype is based on the final pathology report.
Predictions
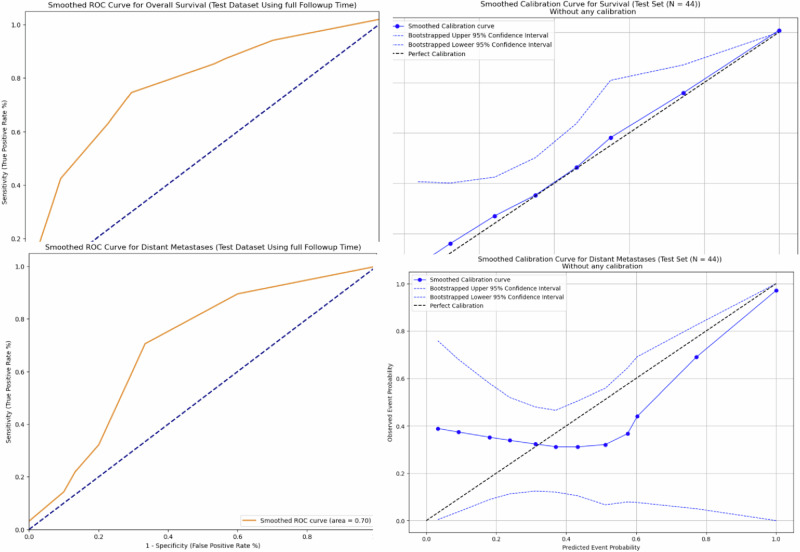
Our end-to-end MMNN, which combines images features from T1 and T2 MRIs and clinical features, outperformed all other models in predicting both outcomes, overall survival and the risk of distant metastases [Table 3]. The C-Index and standard deviation (SD) of our MMNN for overall survival is 0.769 (SD 0.126), with a Brier score of 0.30. This represents an absolute increase of 9% in AUC compared to the next best-performing model. The C-Index and SD of our MMNN for predicting the risk of distant metastases is 0.699 (SD 0.092), with a Brier score of 0.27, which represents an absolute increase of 6% in AUC compared to the next best-performing model. A lift in performance was seen when clinical variables were added to unimodal image features in both the neural network and radiomics models. Gradient blending resulted in the optimal use of specific losses across epochs [Fig. 1]. Without the implementation of gradient blending, the predictive performance of our MMNN was comparable to the other models we evaluated. Smoothed ROC curves and calibration plots are provided in Fig. 2. The larger clinical model only performed slightly better than the model using Sarculator variables.
Table 3.
Performance of different models on predicting overall survival and risk of distant metastases
| Model | C-Index (SD) for predicting Overall Survival | C-Index (SD) for predicting Distant Metastases | Number of Trainable Parameters | Floating Point Operations (FLOPs) |
|---|---|---|---|---|
| Sarculator7 variables (RSF) | 0.654 (0.109) | 0.618 (0.106) | 4 | – |
| Sarculator7 variables (CoxPH) | 0.614 (0.117) | 0.631 (0.097) | – | – |
| Larger clinical model (RSF) | 0.655 (0.111) | 0.639 (0.108) | 11 | – |
| Radiomics model (RSF) | 0.532 (0.116) | 0.541 (0.099) | 200 | – |
| Radiomics + Clinical (RSF) | 0.582 (0.112) | 0.558 (0.108) | 211 | – |
| Radiomics + Clinical (CoxPH with ElasticNet penalty) | 0.707 (0.095) | 0.658 (0.085) | 21 (Overall survival) 30 (Distant Metastases) | |
| Unimodal DenseNet | 0.553 (0.081) | 0.562 (0.068) | 4194,304 (2 x 128 x 128 × 128) | 97,538,861,056 |
| Multimodal Neural Network | 0.769 (0.126) | 0.699 (0.092) | 4194,315 (2 x 128 x 128 × 128) + 11 | 97,538,863,848 |
Fig. 1. Gradient Blending.
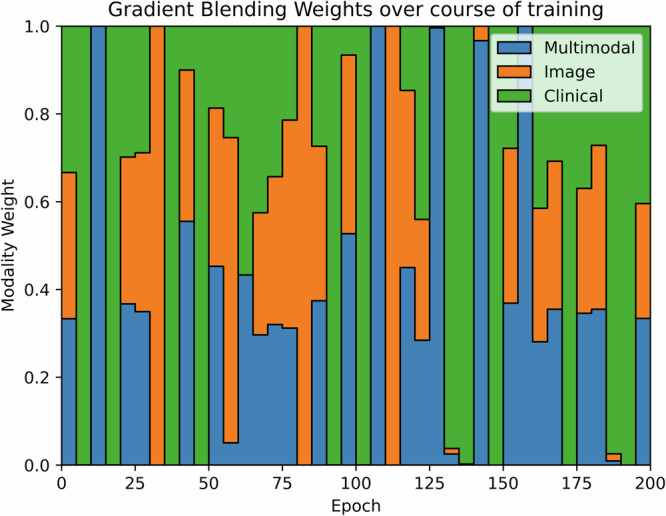
This diagram indicates the relative weighting of modality losses that are contributed to the overall loss over the course of training. The relative weightings are initially uniform and are adjusted every 5 epochs according to their relative overfitting to generalization ratios.
Fig. 2.
Smoothed ROC curves and calibration plots for our MMNN in predicting overall survival and the risk of distant metastases.
Model Interpretability
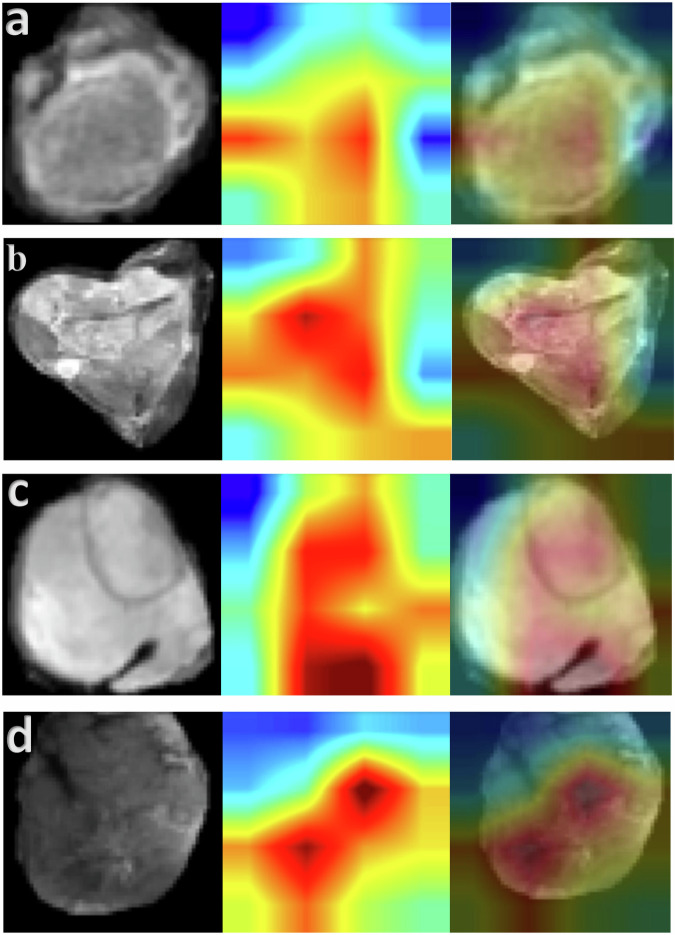
Representative examples of the heatmaps obtained from our test set are depicted in Fig. 3. We provide examples of correctly predicted low-risk and high-risk patients, as well as examples of when the model was wrong. In all cases, the model deemed pixels within the tumor volume as most relevant. There were no cases where the model considered the empty padded pixels on the periphery as the most salient in its predictions.
Fig. 3. Heat maps using the GradCAM method on our test set.
Representative T2 fat sat axial slices of four test set patients in our study (patients which were never encountered during model training) are displayed. The corresponding heat map from the same patient was pulled from the image subnetwork of the MMNN model. The merged image is provided. In all cases, the model deemed pixels within the tumor volume as most relevant. a Patient predicted to have very low risk of death and metastases, survived with no development of metastases over a 10-year follow-up period. (Low predicted risk, model correct). b Patient predicted to have high risk of death and metastases, perished shortly after developing metastases 1.2 years after surgery. (High predicted risk, model correct). c Patient predicted to have high risk of metastases, did not develop metastases in 3.8 years of follow-up (High predicted risk, model wrong). d Among patients who developed metastases in our test set, this patient had the lowest predicted risk. The model was correct in all other predictions indicating a lower risk of distant metastases. (Low-intermediate predicted risk, model incorrect since patient developed metastases two years after surgery).
Discussion
Our results demonstrate the increased predictive capability of multimodal models that utilize image data in addition to previously validated clinical variables. This is the first end-to-end multimodal neural network in the field of soft tissue sarcoma that predicts overall survival and the risk of distant metastases. Given the rarity of sarcoma, the use of multimodal data in prediction algorithms is essential to overcoming the limits of small sample sizes compared to other types of cancer.
Our model includes several notable advances in neural network methodology. Firstly, we implemented gradient blending between our image and clinical variable subnetworks. This ensures that one subnetwork does not overfit while the other subnetworks are still converging. Furthermore, we are among the first to acquire heat maps from the image subnetwork of a multimodal model trained with gradient blending22.
Performance compared to other models without image features
Our MMNN outperformed the Sarculator feature model for both predictions of overall survival (C-index 0.769 vs 0.654) and distant metastases (C-Index 0.699 vs 0.618). Another available nomogram to predict overall survival in sarcoma patients, PERSARC, is based on a Cox Proportional hazards model using six clinical variables23. Despite a larger patient population (766), their C-Index for overall survival is lower at 0.67723. While subsequent studies demonstrated that a multidisciplinary tumor board can change their systemic treatment recommendations based on PERSARC predictions24, and that the patients predicted to be at highest risk by the PERSARC model benefitted from the addition of chemotherapy (11% increase in 5 year survival)25, it is unlikely that the ultimate prediction model used to guide the management of sarcoma patients and enable precision medicine will be based on clinical variables alone or features extracted by radiomics. Our work demonstrates the added benefit of including image data and the superiority of neural networks over radiomics in extracting relevant image features. It is worth noting that our unimodal DenseNet model performed less accurately than a simpler model based on the four Sarculator clinical variables. This is likely due to our sample size. The demonstrated advances in prediction capability are due to the multimodal nature of our work, and the implementation of gradient blending which was crucial to properly combining the inputs from different modalities.
Performance compared to other models incorporating image features
While Sarculator and PERSARC are among the most well-known, validated and utilized prediction models in sarcoma, recently, several studies using radiomics for image feature extraction have been published and are well summarized in a review by Crombé et al. 12. Despite some radiomics studies reporting C-indices >0.9, many of the studies were not IBSI compliant26–29, had a sample size of 50 patients or less27,29,30, and lacked a hold-out or independent test-set26,29,31,32. For instance, Crombe et al. used radiomics features to predict metastasis-free survival in myxoid liposarcomas29. While they report a C-index of 0.94, their sample size was 35 patients, they did not have an independent test set and were not IBSI compliant29,33. Moreover, despite many radiomics studies adhering to IBSI standards, recent research has shown that these studies often struggle to generalize across different centers, even with IBSI compliance34.
However, an excellent use of combined radiomics features and deep learning features was demonstrated by Liu et al. in predicting local recurrence risk in sarcoma patients35. While only 40% of their patients had contrast-enhanced MRIs, thus necessitating two different test sets, Liu et al. achieved impressive performance in predicting local recurrence (C-Index of 0.766 for non-enhanced MRIs and 0.722 for enhanced MRIs). However, the presence of ki67, a cellular marker of proliferation in sarcoma, outperformed both of their models in predicting local recurrence35,36. In contrast, there is no known single marker able to predict overall survival and the risk of distant metastases with such accuracy.
Compared to previous studies extracting image features in sarcoma, our study contains one harmonized cohort where all patients have the same MRI sequences. Moreover, our sample size is larger, our deep learning MMNN analyzes 3D MRI data and does not rely on handcrafted features, and we have a hold-out test set. We are also the first to describe the use of gradient blending, which is crucial for multimodal AI. These features render models such as ours more likely to generalize well to patients from other institutions. Moreover, our model is the first end-to-end multimodal neural network in sarcoma. Previously, Esteva et al. published a multimodal model to predict the risk of metastases and overall survival in patients with prostate cancer37. However, they only used deep learning to extract image features and then predicted their outcomes with a separate classical machine algorithm (CatBoost)37,38. Because their model was not end-to-end deep learning, their deep learning subnetwork that extracted image features did not undergo further supervised learning while predicting outcomes. In contrast, given our end-to-end neural network architecture, the image subnetwork continues to learn throughout the entire model training process. Moreover, this end-to-end neural network architecture permitted model explainability through heat maps and allowed for gradient blending.
Ability to influence personalized medicine and future directions
Given that almost every sarcoma patient obtains an MRI prior to treatment and the clinical variables we used are validated and routinely collected, our MMNN has low barriers to external validation and adoption. Currently, there is significant variation between centers in North America and internationally in which chemotherapy is offered to STS patients39, if at all, and how STS patients are followed after surgery40.
Our MMNN is not designed to predict which chemotherapy regimen is most suitable for a patient. However, patients with a higher predicted risk of metastases are more likely to be treated with chemotherapy and survival has been demonstrated to be better, in some but not all studies, if treatment with chemotherapy is initiated when the lung metastases are smaller41. Notably, the ideal chemotherapy regimen for a particular STS subtype can be difficult to determine2, but several systematic reviews42,43 and a recent network meta-analysis44 have demonstrated increased overall survival in certain STS populations when chemotherapy is added to surgery and radiation therapy. What is missing from all previous studies is an individualized predicted risk of metastases that can guide the decision of a multidisciplinary tumor board to use chemotherapy.
Regarding ideal post-operative surveillance for early identification of metastases, a single-center trial from India showed no survival benefit to more intensive surveillance (3 months compared to every 6 months)45, and a multicenter trial is underway to investigate this question with a more generalizable population4. However, other outcomes such as patient anxiety from intensive surveillance, radiation exposure from CT scans, and patient and hospital expenses must be weighed as well. When STS patients were surveyed on their surveillance preferences, many expressed concern about radiation exposure, cost, and anxiety related to intensive surveillance6. Rather than determining one optimal surveillance regimen for all STS patients, there is likely a role for individualized surveillance regimens based on individualized risk of metastases.
In terms of limitations, our deep learning model required manual segmentation masks to determine a region of interest (ROI). Only the pixels in the ROI, representing the tumor volume and immediate periphery, were analyzed by the model. Important contextual information that is not contained in the ROI includes the proximity of important neurovascular structures46, the full extent of peritumoral edema47, nearby bones, or the location of a tumor deep in the pelvis rather than the extremity (although the location is captured as a clinical variable). Future versions of our MMNN will explore including the entire MRI slice as this contextual information may provide more accurate risk predictions for other important outcomes such as local recurrence or surgical complications48.
While our sample size is larger than almost all previous studies using image features to predict outcomes in sarcoma patients, it is smaller than previous models which used only clinical variables; we were limited by including patients with specific MRI sequences available. We compensate for the smaller sample size by using richer data sources21. The inclusion of MRI data in our prediction algorithm renders it more difficult to provide a web-based app similar to those based just on clinical variables such as Sarculator and PERSARC, as users would have to upload T1 and T2 MRIs.
Given the rapid rate of advances in AI algorithms and the rare prevalence of sarcomas, the most likely factor limiting the predictive accuracy of multimodal models in STS is the availability of high-quality prospective data. While current sarcoma prediction models require the inclusion of many STS subtypes in order to obtain an adequate sample size, subtype-specific prediction models would be ideal1. Ultimately, to achieve the goal of the most accurate predictions to guide patient management, multi-center international collaboration and prospective data collection is required. Currently, only around 2% of machine learning research uses prospective data49. Federated learning would allow multicenter collaboration while preserving patient privacy50. Furthermore, future multimodal models could also incorporate genomic information in addition to image data51. Genomic information is being collected more routinely in sarcoma patients and is likely to explain some of the remaining variability in the outcomes of our patients.
In conclusion, our multimodal neural network with gradient blending improves predictions of overall survival and the risk of distant metastases in patients with soft tissue sarcoma. Future work should focus on enabling accurate subtype-specific predictions to further individualize sarcoma patient management. The future deep learning AI models that guide the management of our patients are likely to incorporate end-to-end neural networks, gradient blending, prospective curation of high-quality data, the inclusion of genomic data, and the involvement of multiple centers through federated learning.
Methods
Patients
We retrospectively analyzed our prospectively maintained database of sarcoma patients at Memorial Sloan Kettering Cancer Center. All patients aged 18 or older with biopsy-confirmed non-retroperitoneal malignant STS who underwent primary resection at MSKCC between January 1st, 2005, and December 31st, 2020 were reviewed. We included all patients with complete outcome data and an MRI with T1 post-contrast fast-sat sequence and a T2 fat-sat sequences acquired prior to initiation of radiation therapy or chemotherapy. Radiation therapy (RT) and MRI protocols were standardized at MSKCC as of 2002, which limits heterogeneity52. The data set includes information on patient characteristics (demographics, medical history, and smoking status), tumor characteristics (sarcoma subtype, tumor size and volume, location, grade, stage, margin status, and margin size), and adjuvant therapy variables (overall RT dose and fractions, and the use tumor-directed chemotherapy or immunotherapy clinical trials or novel agents to control for confounding factors). The grade of every sarcoma included in this study was determined by histopathological analysis of the resection specimen and ranged from low grade (1) to dedifferentiated (4). Grade 2, 3, and 4 tumors are grouped together as “High Grade”, consistent with previous work from our institution and American Joint Committee on Cancer (AJCC) guidelines10,53,54. We obtained approval for this project from the Institutional Review Board of MSKCC (16-1123).
Image acquisition, definition of volumes of interest, and preprocessing
All T1 post-contrast and T2 fatsat MRIs were deidentified and stored on XNAT v1.8 for training of our models. Information on MRI parameters is included in Supplementary Material. Segmentation of the entire sarcoma volume of interest (VOI) was performed by AB through the Open Health Imaging Foundation (OHIF) viewer v 3.3 (Cancer Research UK, London, England). The entire 3D sarcoma volume was included for each MRI of each patient. A fellowship-trained attending musculoskeletal oncology radiologist (SS) segmented a portion of the MRIs to calculate intraclass correlation coefficients (0.92, 95%CI 0.912-0.928) and Dice coefficients55 (0.911, 95% CI 0.898-0.922). Preprocessing of the MRI data included N4 bias correction as well as z-score normalization to control for differences in the acquisition parameters of these sequences56. A total of 9380 MRI slices containing sarcomas in 287 patients are available for analysis. The same set of preprocessed images were used in the radiomics models, the unimodal DenseNet and the MMNN.
Clinical variable models
A model using the same variables (features) as Sarculator7 (patient age, tumor size, tumor grade, and histological subtype) was constructed. We also developed a larger clinical model based on 11 total features including patient sex, tumor location (axial, upper extremity, or lower extremity), tumor volume, depth (above or below fascia), the use of neo-adjuvant chemo, the presence of metastases at presentation, and whether radiation therapy was delivered in the neoadjuvant or adjuvant setting, in addition to the four Sarculator features. The clinical models were built using Random Survival Forest (RSF) and CoxPH. RSF was used because it is fully non-parametric, handles interaction effects between features automatically, and is very effective in high-dimensional settings such as this.
Radiomics models
A total of 100 radiomics features were extracted with PyRadiomics 3.0.1 for each T1 and T2 MRI57, of which 98 obtained an ICC > 0.9 and the others were >0.8. The T1 and T2 features of each patient were combined using concatenation, a side-to-side merging, for a total of 200 radiomics features for each patient. We first used RSF to predict our outcomes based on the extracted features. For the Radiomics + Clinical model, the radiomics features were combined with the larger clinical model variables using concatenation, for a total of 211 features for each patient. Subsequently, a regularized CoxPH model using the clinical and radiomics features was also constructed. Dimensionality reduction was trialed with different methods. The Elastic Net model, which combines the subset selection property of LASSO with the regularization strength of the Ridge model by using a weighted combination of L1 and L2 regularization, performed the best. The strength of the regularization was determined by using 5-fold CV and optimizing the concordance index. Standard deviation values for the C-indices were estimated using Bootstrap on the predicted values. The CLEAR and CLAIM checklists are available in Supplementary Material.
Neural network models
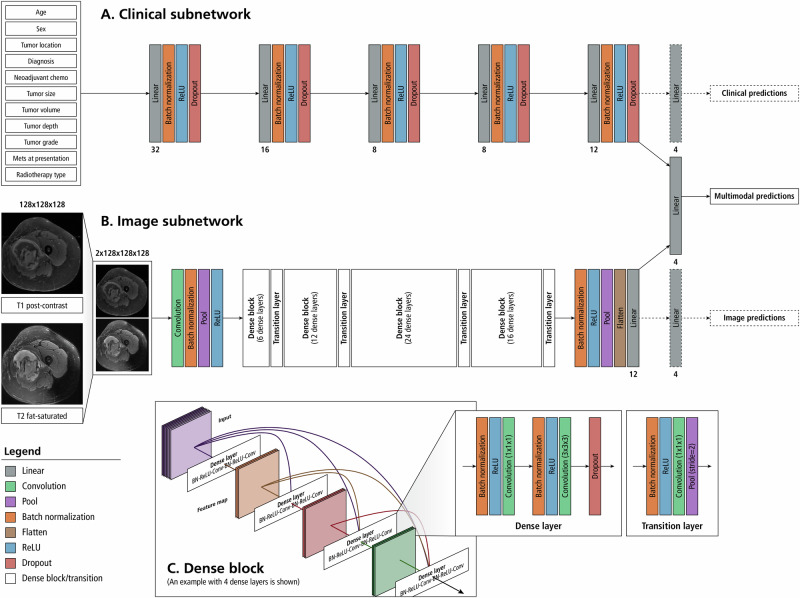
Our MMNN [Fig. 4] accepts the T1 and T2 MRIs in their native resolution through an image subnetwork consisting of a 2-channel DenseNet-12158,59 [Fig. 4B]. The T1 and T2 sequences are masked and cropped to contain the tumor volume and immediate perilesional area, resized to a predetermined spatial resolution, and concatenated along the channel dimension to be fed through the image subnetwork. Early fusion is used to combine data from T1 and T2 image modalities, but is not used to combine image and clinical modalities due to the differing dimensionalities of the data. In the MMNN, late blending of the image features with the clinical features occurs through concatenation. Clinical variables are analyzed in a parallel deep neural network [Fig. 4A] and this information is concatenated with the image features. A fully connected layer analyzes the combined multimodal features before outputting the predicted risk of each of our outcomes. This architecture represents end-to-end deep learning, without the use of radiomics or classical machine learning. Image augmentation was applied at training time and included vertical and horizontal flip, random rotation, and random zoom. Augmentations were jointly applied to the 2-channel T1 + T2 image, rather than separately to T1 and T2 images. This is to preserve the colocation of spatial features in each channel under spatial transformation. No augmentation or perturbation is applied to clinical variables. Because all MRI slices are integrated into a 3D volumetric representation in preprocessing, and because T1 and T2 images are stacked prior to being provided to the neural network, the neural network sees all image data for a patient simultaneously. Therefore, the model makes a single prediction per patient, and no aggregation strategies are needed. A separate unimodal DenseNet-121, equivalent to the image subnetwork of the MMNN, was also constructed. Models were constructed using Python version 3.960.
Fig. 4. Architecture of our multimodal neural network model.
A deep neural network (A) will interpret the 11 clinical variables and a 2-channel convolutional neural network (DenseNet-121) analyzes the MRI input (B). Image features from T1 and T2 MRI sequences are extracted by the convolutional neural network and this information is concatenated along with the features extracted from the clinical variables. Analysis of the combined feature set is used to predict the risk of distant metastases and overall survival. Gradient blending is used to moderate the weight updates between modalities. Dashed lines are used to indicate connections that are only present during training to facilitate Gradient Blending. 1A: Clinical Subnetwork Model. A deep neural network is implemented to extract features from a vector of clinical variables corresponding to the patient. Numbers under the linear layers correspond to the number of output features for those linear layers. The clinical model extracts 12 features that will be used for the multimodal prediction. 1B: Image Subnetwork Model. T1 post contrast and T2 fat-sat MRI sequences are concatenated along the channel dimension prior to being fed through a 2-channel DenseNet-121 model. Twelve features are extracted for use in the multimodal prediction. The numbers in each dense block correspond to the number of dense layers within that dense block. The architecture presented is representative of a 3-dimensional, 2-channel densenet-121 with 12 output neurons. Because the model is being used as a feature extractor rather than a classifier, the size of the output layer is a tunable parameter and not limited to the number of predictions made by the multimodal output head. 1C: Dense Block– Dense blocks consists of a series of dense layers. Within each dense block, the resolution of the feature map is constant. This allows all dense layers within a dense block to contain feed-forward bypass connections to every other dense layer in that dense block. These features are concatenated at the input of each dense layer. Transition layers are placed between dense blocks. Transition layers use 1x1x1 convolutions to act as channel pooling layers, reducing the number of feature maps by a factor of 2. In addition, stride 2 average pooling layers are used which reduce the resolution in all spatial dimensions by a factor of 2.
Gradient blending
Gradient blending61 is used to moderate the loss contributions of the different modalities during the training of multimodal neural networks. During training, gradient blending uses the overfitting to generalization ratios of each modality to promote losses from subnetworks that are generalizing well to the validation set, while downweighting losses from subnetworks that are overfitting the training set. This enables the simultaneous, end-to-end training of two separate model architectures that would typically converge and overfit at different rates. Model selection during training is performed considering only the unweighted contribution of the multimodal head to the loss function. The model checkpoint with the lowest value for the unweighted multimodal component of the loss function over the course of training is selected for evaluation.
Model training
The image model parameters for both the MMNN and the unimodal DenseNet model are initialized using a pre-trained self-supervised DenseNet-121 trained on the BHB-10k dataset consisting of neurological MRIs62. Parameters in the clinical subnetwork and all output heads are randomly initialized. A CoxPH loss function63 was used. This controls for the different follow-up times of our patients and allows for predictions at any time point for future patients being evaluated by our model. The training was performed for 200 epochs, with a batch size of 8, OneCycle learning rate scheduler64, and Stochastic Gradient Descent with Nesterov Momentum65.
Model interpretation
Visualization of the features extracted by the image model was obtained using the Grad-CAM methodology66. We extract the heat maps from the final convolutional layer in the image subnetwork of the multimodal neural network model. The heat maps show the areas of the original image that most contribute to the features extracted by the image model.
Statistical analysis and model evaluation
We used a 70/15/15 train/test/val split stratified by tumor volume, location, and whether radiation was administered preoperatively or postoperatively. The predictive ability of the model is assessed by the C-Index, which corresponds to the time-dependent area under the ROC curve67,68. The C-Index represents the proportion of included patients whose survival times can be ordered such that the subject with higher predicted survival actually survived longer. It is a better measure than raw accuracy for unbalanced classes. The performance of eight models – three models with clinical variables only, a unimodal radiomics model, two multimodal radiomics models, a unimodal neural network model, and a multimodal neural network model, were compared.
Supplementary information
Acknowledgements
Anthony Bozzo was supported by the Cedars Cancer Foundation, the Montreal General Hospital Foundation, the Richard H. Tomlinson Doctoral Fellowship award, the Major Family Orthopaedic Oncology Education Fund, and the MSTS Sarcoma Strong Mentored Research award. The authors thank Sarah Kulig for assistance in figure preparation.
Author contributions
A.B. is responsible for the project conception, organization of the team, curation of tabular and image data, segmentation of the MRIs, neural network design and implementation, and is the lead writer and editor of the manuscript. A.H. is responsible for the development of the first-of-its-kind end-to-end multimodal neural network, implementation of gradient blending, and model explainability using heat maps. He constructed the deep learning pipeline and performance analysis of the deep learning models. He is responsible for revising and editing the manuscript. S.C. is responsible for the development of the radiomics models, the development of performance metrics, and the verification of all results and model comparisons. He is responsible for revising and editing the manuscript. A.A. is responsible for data cleaning and preprocessing of image data, and editing of the manuscript. J.D. is responsible for figure generation, data interpretation, and editing of the manuscript. S.S. is responsible for the segmentation of the MRIs, clinical input related to radiology, and editing of the manuscript. W.T. is responsible for project design, clinical input relating to medical oncology, and editing of the manuscript. A.A. is responsible for project design, clinical input, and editing of the manuscript. S.B. is responsible for project design, statistical input, and editing of the manuscript. JHH is responsible for overall project coordination, project design, and significant revising and editing of the manuscript. All authors read and approved the final manuscript.
Data availability
The data that support the findings of this study are not openly available due to reasons of sensitivity and patient privacy. They are available from the corresponding author upon reasonable request and institutional review. Data are located in controlled access data storage at Memorial Sloan Kettering Cancer Center.
Code availability
The code that supports the multimodal neural network with gradient blending is openly available on github at https://github.com/DigITs-AIML/MMNN_STS. The terms of use are included in the repository.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41698-024-00695-7.
References
- 1.Gamboa, A. C., Gronchi, A. & Cardona, K. Soft‐tissue sarcoma in adults: An update on the current state of histiotype‐specific management in an era of personalized medicine. CA: Cancer J. Clin.70, 200–229 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Gronchi, A. et al. Histotype-tailored neoadjuvant chemotherapy versus standard chemotherapy in patients with high-risk soft-tissue sarcomas (ISG-STS 1001): an international, open-label, randomised, controlled, phase 3, multicentre trial. Lancet Oncol.18, 812–822 (2017). 10.1016/S1470-2045(17)30334-0 [DOI] [PubMed] [Google Scholar]
- 3.Weitz, J. R., Antonescu, C. R. & Brennan, M. F. Localized extremity soft tissue sarcoma: improved knowledge with unchanged survival over time. J. Clin. Oncol.21, 2719–2725 (2003). 10.1200/JCO.2003.02.026 [DOI] [PubMed] [Google Scholar]
- 4.Schneider, P. & Ghert, M. Surveillance AFter Extremity Tumor surgerY (SAFETY): A Protocol for an International Randomized Controlled Trial. (2018).
- 5.Wilson, D. A. et al. Designing a rational follow-up schedule for patients with extremity soft tissue sarcoma. Ann. Surg. Oncol.27, 2033–2041 (2020). 10.1245/s10434-020-08240-z [DOI] [PubMed] [Google Scholar]
- 6.Tepper, S. C. et al. Examining patient perspectives on sarcoma surveillance: The Sarcoma Surveillance Survey. Surg. Oncol.45, 101861 (2022). 10.1016/j.suronc.2022.101861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Callegaro, D. et al. Development and external validation of two nomograms to predict overall survival and occurrence of distant metastases in adults after surgical resection of localised soft-tissue sarcomas of the extremities: a retrospective analysis. Lancet Oncol.17, 671–680 (2016). 10.1016/S1470-2045(16)00010-3 [DOI] [PubMed] [Google Scholar]
- 8.Voss, R. K. et al. Sarculator is a good model to predict survival in resected extremity and trunk sarcomas in US patients. Ann. Surg. Oncol.29, 4376–4385 (2022). 10.1245/s10434-022-11442-2 [DOI] [PubMed] [Google Scholar]
- 9.Cahlon, O. et al. A postoperative nomogram for local recurrence risk in extremity soft tissue sarcomas after limb-sparing surgery without adjuvant radiation. Ann. Surg.255, 343 (2012). 10.1097/SLA.0b013e3182367aa7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mariani, L. et al. Validation and adaptation of a nomogram for predicting the survival of patients with extremity soft tissue sarcoma using a three‐grade system. Cancer103, 402–408 (2005). 10.1002/cncr.20778 [DOI] [PubMed] [Google Scholar]
- 11.Callegaro, D., Miceli, R. & Gronchi, A. Sarcoma nomograms: a light over the darkness. Oncoscience4, 15 (2017). 10.18632/oncoscience.341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crombé, A. et al. Systematic review of sarcomas radiomics studies: Bridging the gap between concepts and clinical applications? Eur. J. Radiol.132, 109283 (2020). 10.1016/j.ejrad.2020.109283 [DOI] [PubMed] [Google Scholar]
- 13.Whitney, H. M., Li, H., Ji, Y., Liu, P. & Giger, M. L. Comparison of breast MRI tumor classification using human-engineered radiomics, transfer learning from deep convolutional neural networks, and fusion methods. Proc. IEEE108, 163–177 (2019). 10.1109/JPROC.2019.2950187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang, L. & Ren, Z. Comparison of CT and MRI images for the prediction of soft-tissue sarcoma grading and lung metastasis via a convolutional neural networks model. Clin. Radiol.75, 64–69 (2020). 10.1016/j.crad.2019.08.008 [DOI] [PubMed] [Google Scholar]
- 15.Ziegelmayer, S. et al. Feature robustness and diagnostic capabilities of convolutional neural networks against radiomics features in computed tomography imaging. Investig. Radiol.57, 171–177 (2022). 10.1097/RLI.0000000000000827 [DOI] [PubMed] [Google Scholar]
- 16.Nyflot, M. J., Thammasorn, P., Wootton, L. S., Ford, E. C. & Chaovalitwongse, W. A. Deep learning for patient‐specific quality assurance: Identifying errors in radiotherapy delivery by radiomic analysis of gamma images with convolutional neural networks. Med. Phys.46, 456–464 (2019). 10.1002/mp.13338 [DOI] [PubMed] [Google Scholar]
- 17.Hatt, M., Vallieres, M., Visvikis, D. & Zwanenburg, A. (Soc Nuclear Med, 2018). [DOI] [PMC free article] [PubMed]
- 18.Navarro, F. et al. Development and external validation of deep-learning-based tumor grading models in soft-tissue sarcoma patients using MR imaging. Cancers13, 2866 (2021). 10.3390/cancers13122866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kathavate, P. N. & Amudhavel, J. Optimized convolutional neural network for soft tissue sarcoma diagnosis. Multimed. Tools Appl.82, 4497–4515 (2023). 10.1007/s11042-022-13429-3 [DOI] [Google Scholar]
- 20.Foersch, S. et al. Deep learning for diagnosis and survival prediction in soft tissue sarcoma. Ann. Oncol.32, 1178–1187 (2021). 10.1016/j.annonc.2021.06.007 [DOI] [PubMed] [Google Scholar]
- 21.Acosta, J. N., Falcone, G. J., Rajpurkar, P. & Topol, E. J. Multimodal biomedical AI. Nat. Med.28, 1773–1784 (2022). 10.1038/s41591-022-01981-2 [DOI] [PubMed] [Google Scholar]
- 22.Hu, W. et al. Interpretable multimodal fusion networks reveal mechanisms of brain cognition. IEEE Trans. Med. imaging40, 1474–1483 (2021). 10.1109/TMI.2021.3057635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Praag, V. M. et al. A prediction model for treatment decisions in high-grade extremity soft-tissue sarcomas: Personalised sarcoma care (PERSARC). Eur. J. Cancer83, 313–323 (2017). 10.1016/j.ejca.2017.06.032 [DOI] [PubMed] [Google Scholar]
- 24.Hagenmaier, H. et al. The influence of Personalised Sarcoma Care (PERSARC) prediction modelling on clinical decision making in a multidisciplinary setting. Sarcoma2021 (2021). [DOI] [PMC free article] [PubMed]
- 25.Acem, I. et al. The role of perioperative chemotherapy in primary high-grade extremity soft tissue sarcoma: a risk-stratified analysis using PERSARC. Eur. J. Cancer165, 71–80 (2022). 10.1016/j.ejca.2022.01.013 [DOI] [PubMed] [Google Scholar]
- 26.Vallières, M., Freeman, C. R., Skamene, S. R. & El Naqa, I. A radiomics model from joint FDG-PET and MRI texture features for the prediction of lung metastases in soft-tissue sarcomas of the extremities. Phys. Med. Biol.60, 5471 (2015). 10.1088/0031-9155/60/14/5471 [DOI] [PubMed] [Google Scholar]
- 27.Gao, Y. et al. Prediction of soft tissue sarcoma response to radiotherapy using longitudinal diffusion MRI and a deep neural network with generative adversarial network‐based data augmentation. Med. Phys.48, 3262–3372 (2021). 10.1002/mp.14897 [DOI] [PubMed] [Google Scholar]
- 28.Yang, Y., Ma, X., Wang, Y. & Ding, X. Prognosis prediction of extremity and trunk wall soft-tissue sarcomas treated with surgical resection with radiomic analysis based on random survival forest. Updates Surg.74, 355–365 (2022). 10.1007/s13304-021-01074-8 [DOI] [PubMed] [Google Scholar]
- 29.Crombé, A. et al. Can radiomics improve the prediction of metastatic relapse of myxoid/round cell liposarcomas? Eur. Radiol.30, 2413–2424 (2020). 10.1007/s00330-019-06562-5 [DOI] [PubMed] [Google Scholar]
- 30.Fields, B. K. et al. Predicting soft tissue sarcoma response to neoadjuvant chemotherapy using an MRI-based delta-radiomics approach. Mol. Imaging Biol.25, 776–787 (2023). 10.1007/s11307-023-01803-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miao, L. et al. Predicting pathological complete response of neoadjuvant radiotherapy and targeted therapy for soft tissue sarcoma by whole-tumor texture analysis of multisequence MRI imaging. Eur. Radiol.33, 3984–3994 (2023). 10.1007/s00330-022-09362-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fadli, D., Kind, M., Michot, A., Le Loarer, F. & Crombé, A. Natural changes in radiological and radiomics features on MRIs of soft‐tissue sarcomas naïve of treatment: correlations with histology and patients’ outcomes. J. Magn. Reson. Imaging56, 77–96 (2022). 10.1002/jmri.28021 [DOI] [PubMed] [Google Scholar]
- 33.Crombé, A. et al. Radiomics and artificial intelligence for soft-tissue sarcomas: Current status and perspectives. Diagn. Interven. Imaging104, 567–583 (2023). [DOI] [PubMed]
- 34.Paquier, Z. et al. Radiomics software comparison using digital phantom and patient data: IBSI-compliance does not guarantee concordance of feature values. Biomed. Phys. Eng. Express8, 065008 (2022). 10.1088/2057-1976/ac8e6f [DOI] [PubMed] [Google Scholar]
- 35.Liu, S. et al. Deep learning radiomic nomogram to predict recurrence in soft tissue sarcoma: a multi-institutional study. Eur. Radiol.32, 793–805 (2022). 10.1007/s00330-021-08221-0 [DOI] [PubMed] [Google Scholar]
- 36.Campos, M. et al. Ki‑67 and CD100 immunohistochemical expression is associated with local recurrence and poor prognosis in soft tissue sarcomas, respectively. Oncol. Lett.5, 1527–1535 (2013). 10.3892/ol.2013.1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Esteva, A. et al. Prostate cancer therapy personalization via multi-modal deep learning on randomized phase III clinical trials. NPJ Digital Med.5, 71 (2022). 10.1038/s41746-022-00613-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hancock, J. T. & Khoshgoftaar, T. M. CatBoost for big data: an interdisciplinary review. J. big data7, 94 (2020). 10.1186/s40537-020-00369-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Villano, A. M. et al. National trends in treatment for retroperitoneal soft tissue sarcoma: a modern appraisal of variability in therapeutic strategies. Ann. Surg. Oncol.29, 1–11 (2021). [DOI] [PubMed]
- 40.Greenberg, D. D. & Crawford, B. Surveillance strategies for sarcoma: results of a survey of members of the musculoskeletal tumor society. Sarcoma2016, 1–5 (2016). [DOI] [PMC free article] [PubMed]
- 41.Lindner, L. H. et al. Prognostic factors for soft tissue sarcoma patients with lung metastases only who are receiving first‐line chemotherapy: An exploratory, retrospective analysis of the European Organization for Research and Treatment of Cancer‐Soft Tissue and Bone Sarcoma Group (EORTC‐STBSG). Int. J. cancer142, 2610–2620 (2018). 10.1002/ijc.31286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Collaboration, S. M.-A. Adjuvant chemotherapy for localised resectable soft-tissue sarcoma of adults: meta-analysis of individual data. Lancet350, 1647–1654 (1997). 10.1016/S0140-6736(97)08165-8 [DOI] [PubMed] [Google Scholar]
- 43.Pervaiz, N. et al. A systematic meta‐analysis of randomized controlled trials of adjuvant chemotherapy for localized resectable soft‐tissue sarcoma. Cancer: Interdiscip. Int. J. Am. Cancer Soc.113, 573–581 (2008). 10.1002/cncr.23592 [DOI] [PubMed] [Google Scholar]
- 44.Haussmann, J. et al. Comparison of different systemic therapeutic regimes in resectable soft-tissue sarcoma—results of a network meta-analysis. Cancers13, 5631 (2021). 10.3390/cancers13225631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Puri, A. et al. Does a less intensive surveillance protocol affect the survival of patients after treatment of a sarcoma of the limb? Updated results of the randomized TOSS study. bone Jt. J.100, 262–268 (2018). 10.1302/0301-620X.100B2.BJJ-2017-0789.R1 [DOI] [PubMed] [Google Scholar]
- 46.Gerrand, C. et al. Classification of positive margins after resection of soft-tissue sarcoma of the limb predicts the risk of local recurrence. J. Bone Jt. Surg. Br.83, 1149–1155 (2001). 10.1302/0301-620X.83B8.0831149 [DOI] [PubMed] [Google Scholar]
- 47.Zhao, F. et al. Can MR imaging be used to predict tumor grade in soft-tissue sarcoma? Radiology272, 192–201 (2014). 10.1148/radiol.14131871 [DOI] [PubMed] [Google Scholar]
- 48.Casale, R., De Angelis, R., Coquelet, N., Mokhtari, A. & Bali, M. A. The impact of Edema on MRI radiomics for the prediction of lung metastasis in soft tissue sarcoma. Diagnostics13, 3134 (2023). 10.3390/diagnostics13193134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ben-Israel, D. et al. The impact of machine learning on patient care: a systematic review. Artif. Intell. Med.103, 101785 (2020). 10.1016/j.artmed.2019.101785 [DOI] [PubMed] [Google Scholar]
- 50.Rieke, N. et al. The future of digital health with federated learning. NPJ digital Med.3, 119 (2020). 10.1038/s41746-020-00323-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barretina, J. et al. Subtype-specific genomic alterations define new targets for soft-tissue sarcoma therapy. Nat. Genet.42, 715 (2010). 10.1038/ng.619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hong, L., Alektiar, K. M., Hunt, M., Venkatraman, E. & Leibel, S. A. Intensity-modulated radiotherapy for soft tissue sarcoma of the thigh. Int. J. Radiat. Oncol. * Biol. * Phys.59, 752–759 (2004). 10.1016/j.ijrobp.2003.11.037 [DOI] [PubMed] [Google Scholar]
- 53.Yoon, S. S. The New American Joint Commission on Cancer Staging system for soft tissue sarcomas: splitting versus lumping. Ann. Surg. Oncol.25, 1101–1102 (2018). 10.1245/s10434-018-6419-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Amin, M., Edge, S. & Byrd, D. AJCC Cancer Staging Manual. 8th Ed. 1–45 (2017).
- 55.Dice, L. R. Measures of the amount of ecologic association between species. Ecology26, 297–302 (1945). 10.2307/1932409 [DOI] [Google Scholar]
- 56.Carré, A. et al. Standardization of brain MR images across machines and protocols: bridging the gap for MRI-based radiomics. Sci. Rep.10, 1–15 (2020). 10.1038/s41598-020-69298-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Van Griethuysen, J. J. et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res.77, e104–e107 (2017). 10.1158/0008-5472.CAN-17-0339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu, X., Lin, J., Tao, Y. & Wang, X. In 2018 7th International Conference on Digital Home (ICDH). 137–140 (IEEE).
- 59.Wang, S.-H. & Zhang, Y.-D. DenseNet-201-based deep neural network with composite learning factor and precomputation for multiple sclerosis classification. ACM Trans. Multimed. Comput., Commun., Appl. (TOMM)16, 1–19 (2020). [Google Scholar]
- 60.Van Rossum, G. USENIX annual technical conference. Python programming language (2007).
- 61.Wang, W., Tran, D. & Feiszli, M. What makes training multi-modal classification networks hard?. In Proceedings of the IEEE/CVF conference on computer vision and pattern recognition 12695–12705 (2020).
- 62.Dufumier, B. et al. Openbhb: a large-scale multi-site brain mri data-set for age prediction and debiasing. NeuroImage, 263, 119637 (2022). [DOI] [PubMed]
- 63.Kvamme, H., Borgan, Ø. & Scheel, I. Time-to-event prediction with neural networks and Cox regression. J mach lear res. 20, 1–30 (2019).
- 64.Smith, L. N. Cyclical learning rates for training neural networks. In 2017 IEEE winter conference on applications of computer vision (WACV) 464–472 (IEEE, 2017).
- 65.Nesterov, Y. A method for solving the convex programming problem with convergence rate O (1/k2). Dokl akad nauk Sssr 269, 543–547 (1983).
- 66.Selvaraju, R. R. et al. Grad-CAM: visual explanations from deep networks via gradient-based localization. Int. J. Comput. Vis. 128, 336–359 (2020).
- 67.Uno, H., Cai, T., Pencina, M. J., D’Agostino, R. B. & Wei, L.-J. On the C‐statistics for evaluating overall adequacy of risk prediction procedures with censored survival data. Stat. Med.30, 1105–1117 (2011). 10.1002/sim.4154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Heagerty, P. J., Lumley, T. & Pepe, M. S. Time‐dependent ROC curves for censored survival data and a diagnostic marker. Biometrics56, 337–344 (2000). 10.1111/j.0006-341X.2000.00337.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are not openly available due to reasons of sensitivity and patient privacy. They are available from the corresponding author upon reasonable request and institutional review. Data are located in controlled access data storage at Memorial Sloan Kettering Cancer Center.
The code that supports the multimodal neural network with gradient blending is openly available on github at https://github.com/DigITs-AIML/MMNN_STS. The terms of use are included in the repository.