Abstract
Pharmacologically active small organic molecules derived from natural resources are prominent drug candidates due to their inherent structural diversity. Herein, we explored one such bioactive molecule, niloticin, which is a tirucallane-type triterpenoid isolated from the stem barks of Aphanamixis polystachya (Wall.) Parker. After initial screening with other isolated compounds from the same plant, niloticin demonstrated selective cytotoxicity against cervical cancer cells (HeLa) with an IC50 value of 11.64 μM. Whereas the compound exhibited minimal cytotoxicity in normal epithelial cell line MCF-10A, with an IC50 value of 83.31 μM. Subsequently, in silico molecular docking studies of niloticin based on key apoptotic proteins such as p53, Fas, FasL, and TNF β revealed striking binding affinity, reflecting docking scores of −7.2, −7.1, −6.8, and −7.2. Thus, the binding stability was evaluated through molecular dynamic simulation. In a downstream process, the apoptotic capability of niloticin was effectively validated through in vitro fluorimetric assays, encompassing nuclear fragmentation. Additionally, an insightful approach involving surface-enhanced Raman spectroscopy (SERS) re-establishes the occurrence of DNA cleavage during cellular apoptosis. Furthermore, niloticin was observed to induce apoptosis through both intrinsic and extrinsic pathways. This was evidenced by the upregulation of upstream regulatory molecules such as CD40 and TNF, which facilitate the activation of caspase 8. Concurrently, niloticin-induced p53 activation augmented the expression of proapoptotic proteins Bax and Bcl-2 and downregulation of IAPs, leading to the release of cytochrome C and subsequent activation of caspase 9. Therefore, the reflection of mitochondrial-mediated apoptosis is in good agreement with molecular docking studies. Furthermore, the anti-metastatic potential was evidenced by wound area closure and Ki67 expression patterns. This pivotal in vitro assessment confirms the possibility of niloticin being a potent anti-cancer drug candidate, and to the best of our knowledge, this is the first comprehensive anticancer assessment of niloticin in HeLa cells.
Niloticin, a triterpenoid from Aphanamixis polystachya, induces apoptosis in HeLa cells which is primarily validated through in silico analysis and subsequently in vitro apoptotic evaluation by modulating the apoptotic proteins.
Introduction
Throughout history, humans have leveraged the therapeutic potential of nature, which provides a vast repository of bioactive compounds. Nature is often esteemed as the quintessential healer, with its components playing a critical role in managing numerous chronic diseases. Medicinal plants have been integral to healthcare from ancient traditional medicine to contemporary medical systems owing to their remarkable efficacy, safety, and cost-effectiveness. The extracts and isolated compounds from medicinal plants are preferred treatments for a variety of ailments. The relevance of natural product-based drug discovery persists as some of these compounds are indispensable even in the era of modern synthetic medicines due to their diverse physicochemical properties, including high bioactivity, biocompatibility, and bioavailability.1,2 Although many modern therapeutic interventions have been practiced in recent decades, cancer continues to be a leading cause of death worldwide. Intensive research aims to discover a cure for this lethal affliction as existing treatments such as chemotherapy, radiation, and surgery pose risks to healthy cells and tissues. The escalating cancer incidence highlights the pressing need for an effective and affordable treatment with minimal side effects. In addition to the challenges associated with the disease itself, the development of chemoresistance has emerged as a major concern.3–5 Within the spectrum of malignancies, cervical cancer holds the fourth position in terms of incidence, emerging as the predominant cause of mortality among women.6 In this preview, natural products exert their actions by regulating multiple signaling pathways involved in cell growth and apoptosis, consequently impeding cancer cell progression and possessing immune activation, which results in tumor cell death.7,8 Currently, there is a growing interest in the search for naturally occurring active molecules with high immunomodulation for cancer treatment.9,10
Aphanamixis polystachya (Wall.) Parker, also known as Amoora rohituka, a member of the Meliaceae family, is distinguished for its extensive medicinal properties.11 Owing to its therapeutic potential, this plant has traditionally been used to address various health issues, including spleen and liver diseases, abdominal problems, tumors, eye disorders, rheumatoid arthritis, ulcers, and jaundice.12,13 It exhibits pharmacological properties, such as antibacterial, antifungal, and insecticidal activities.14 Furthermore, it possesses CNS depressant, analgesic, and clot lysis properties.15 The ethyl acetate fraction of A. polystachya has demonstrated its ability to safeguard bone marrow cells against chromosomal abnormalities induced by gamma radiation in Swiss albino mice. This finding suggests potential applications in radiation therapy for cancer treatment.16 Niloticin, a tirucallane-type triterpenoid isolated from A. polystachya, has demonstrated significant biological activities. One of the promising reported activities of niloticin is its ability to inhibit osteoclastogenesis by suppression of the RANKL-induced activation of the AKT, MAPK and NF-κB signaling pathways.17 Niloticin functions as an antagonist of myeloid differentiation factor-2 (MD-2) and possesses anti-inflammatory activity.18 The literature also hints at the insecticidal properties of niloticin, which is considered a prominent feature of this compound. Additionally, anti-plasmodial and anti-respiratory syncytial virus (RSV) activities have been reported for niloticin.19,20 The cytotoxicity of niloticin is identified in several cancer cell lines, including gastric (BGC-823, KE-97), liver (Huh-7), breast (MDA), prostate (PC3, LNCaP), fibrosarcoma (HT-1080), and hepatoma (Hep3B).21–24 However, comprehensive anti-cancer studies specifically targeting cervical cancer cell proliferation have not been reported, which constitutes a primary focus of the current investigation.
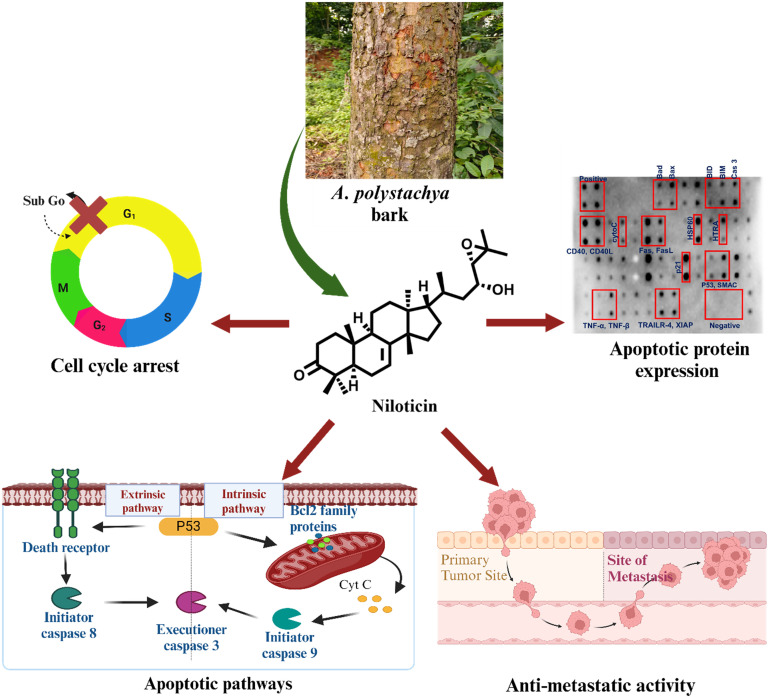
Considering these facets, our objective is to meticulously investigate the anticancer properties of the phytomolecule niloticin against cervical cancer, which is prevalent in the female reproductive system. In this process, the extraction and isolation of the desired phytomolecules have been carried out from the stem bark of Aphanamixis polystachya. The antiproliferative effect of the acetone extract was evaluated in cervical cancer (HeLa) cells, which demonstrates significant activity. Hence, we extended our studies with the isolation of key phytomolecules from acetone extract. The isolation yielded five distinct phytomolecules, falling into the category of tirucallane-type triterpenoids, which are characteristic of plants within the meliaceae family. The anticancer potential of the isolated compounds was then scrutinized in HeLa cells, resulting in niloticin being the most potent among them. This eventually led us to focus on the triterpenoid compound, niloticin, for an in-depth anticancer profiling investigation. In silico screening of niloticin with major protein targets of the apoptotic pathway show reasonably good binding affinity with key proteins involved in the signaling cascade, including p53, Fas receptor, Fas L, Bax, CDK2, BCL2 and TNF β. Therefore, to complement the in silico results, an in vitro downstream assessment was carried out to elucidate the apoptotic mechanisms by which niloticin operates, employing a variety of cell-based assays and analyzed DNA fragmentation by agarose gel electrophoresis, which was fully complemented by surface-enhanced Raman scattering (SERS) as an ultrasensitive spectroscopic modality. Furthermore, niloticin exerts its anti-proliferative potential by sub-G0 cell cycle arrest, confirmed by cell cycle assay and expression analysis of cell cycle regulatory proteins. To evaluate the apoptotic pathway, the expression of caspases was analyzed by fluorometric assays. Interestingly, it was observed that the niloticin followed both extrinsic and intrinsic modes of apoptosis, which is well supported by the molecular docking and protein expression studies of the involved key factors. Additionally, wound healing properties and the ability of clonogenic inhibition were evaluated, aiming to establish the anti-metastatic potential of niloticin backed by the expression analysis of Ki67 by immunofluorescence study. Finally, the pathway of the molecule's action was well confirmed by examining the expression of signaling proteins, thus solidifying niloticin's potential for further studies as a promising hit compound against cervical cancer. To date, this is the first detailed investigation of the anticancer potential of niloticin in cervical cancer using the HeLa cell line (Scheme 1).
Scheme 1. Cell death induced by niloticin through intrinsic and extrinsic modes of apoptosis.
Results and discussion
Extraction, isolation, and characterization of phyto entities from Aphanamixis polystachya
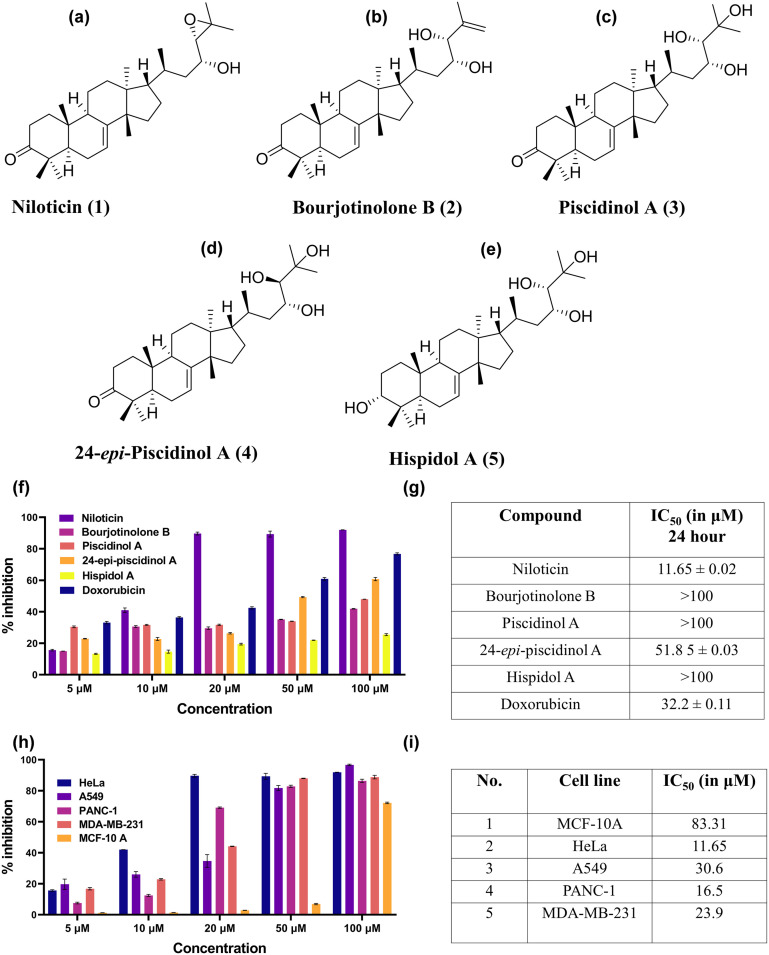
The acetone extract of the bark of Aphanamixis polystachya (Wall.) Parker was prepared by percolation, and its antiproliferative potential was primarily checked in HeLa (cervical cancer) cells using the MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide) assay (Fig. S1, Table S1†), with IC50 values of 39.96 and 17.18 μg mL−1 at 24 and 48 hours of incubation, respectively. This led to the isolation and purification of phytomolecules from the acetone extract. The acetone extract, subjected to repeated column chromatographic separation, yielded five distinct molecules: niloticin (15 mg), bourjotinolone B (32 mg), piscidinol A (10 mg), 24-epi-piscidinol A (21 mg), and hispidol B (18 mg) (Fig. 1a–e). All molecules belong to tirucallane type-triterpenoid, and were characterized by 1D NMR, 2D NMR and HRMS, which were in accordance with the reported data (Fig. S3–S340†).25–28
Fig. 1. Structure of a) niloticin, b) bourjotinolone, c) piscidinol A, d) 24-epi-piscidinol, and e) hispidol A. f) Graph showing the percentage toxicity of isolated molecules in HeLa cells, g) and their corresponding IC50 values. h) Comparative study of the percentage inhibition of niloticin in different cancer cell lines, i) and their IC50 values using the MTT assay.
Preliminary cytotoxic screening of isolated phytomolecules
A preliminary cytotoxic analysis of tirucallane type-triterpenoid against cervical cancer cells was conducted using the MTT assay in HeLa cells at concentrations ranging from 5 to 100 μM, and the IC50 values are shown in Fig. 1g. Among the phytomolecules, niloticin was found to be the most potent with an IC50 value of 11.64 μM at an incubation period of 24 hours. To check its anticancer potential in other cancer cells, we specifically conducted the MTT assay of niloticin on A549 (lung), PANC-1 (pancreatic), and MDA-MB-231 (breast) cancer cell lines by keeping the same concentrations of niloticin as in HeLa cells. The minimal inhibitory concentration (IC50) was determined, which turned out to be 30.6 μM (A549), 16.5 μM (PANC-1) and 23.9 μM (MDA-MB-231), respectively, at 24 hours of incubation. This infers that niloticin is most active against cervical cancer (HeLa cell line) (Fig. 1i). As a better cytotoxicity was observed in HeLa cells, we also evaluated the activity of niloticin in SiHa (squamous cell carcinoma) with an IC50 value of 16.23 μM, which further proved the anti-proliferative potential of niloticin in cervical cancer cells. Since niloticin exhibited a pronounced inhibitory potential against the cervical cancer cell line, we evaluated its potential in MCF 10A, a non-tumorigenic epithelial cell line, and the observed IC50 was approximately 83.31 μM. This finding of the selective anti-cancer potential of niloticin urges us to conduct an in-depth apoptotic assessment on HeLa cells.
Computational screening of niloticin
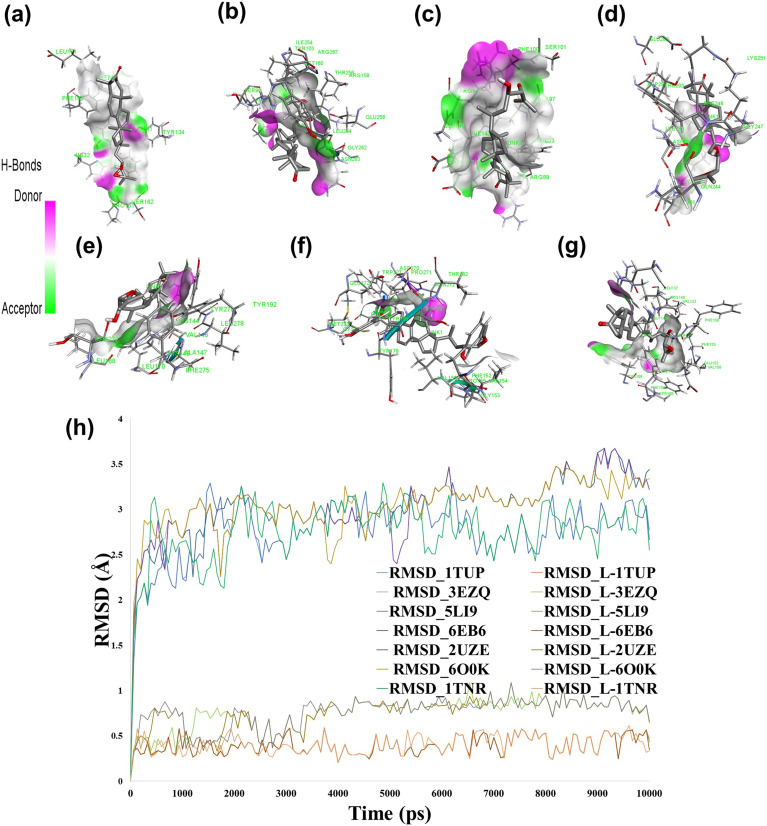
In the pursuit of understanding the pharmacokinetic properties of niloticin, our initial evaluations revealed promising characteristics. Niloticin exhibited high human intestinal absorption, accompanied by a favorable lipophilicity with an iLOGP value of 4.58. Furthermore, the absence of PAINS (pan assay interference compounds) was confirmed (Fig. S42†). To elucidate the intricate interactions between niloticin and key signaling proteins within the apoptotic pathway of cervical cancer, we conducted comprehensive molecular docking simulations. Seven pivotal proteins in the pathway were selected for this study: p53 (PDB ID: 1TUP), Fas receptor (PDB ID: 3EZQ), Fas L (PDB ID: 5L19), Bax (PDB ID: 6EB6), CDK2 (PDB ID: 2UZE), BCL2 (PDB ID: 6O0K), and TNF β (PDB ID: 1TNR). Notably, docking scores below −6.5 were observed, indicating robust interactions between niloticin and these proteins (Table S2†) (Fig. 2a–g).
Fig. 2. 3-D interaction of niloticin with a) TNF β, b) P53, c) Bax, d) Fas receptor, e) FasL, f) CDK2, and g) BCL2. h) RMSD plot of niloticin with different proteins.
Our analysis revealed the strong binding of niloticin with each protein, as evidenced by docking scores ranging from −6.8 to −9.0 kcal mol−1. For instance, with the tumor suppressor protein p53 (PDB ID: 1TUP), niloticin formed hydrogen bonds and hydrophobic interactions with residues such as ASN 263, GLY 262, and GLU 258, resulting in a docking score of −7.2 kcal mol−1. Similarly, interactions with the Fas receptor (PDB ID: 3EZQ) and Fas L (PDB ID: 5L19) yielded scores of −7.1 and −6.8 kcal mol−1, respectively, involving crucial residues such as GLY 247, PHE 248, and ARG 144. Moreover, niloticin demonstrated high affinity for proteins like Bax (PDB ID: 6EB6), CDK2 (PDB ID: 2UZE), BCL2 (PDB ID: 6O0K), and TNF β (PDB ID: 1TNR),29–34 with docking scores of −7.7, −7.2, −9.0, and −7.2 kcal mol−1, respectively. These findings underscore the efficacy of niloticin as a promising therapeutic agent in cervical cancer treatment, warranting further investigation into its mechanistic pathways and clinical applications.
Molecular dynamics simulation of protein–niloticin complex
Subsequently, the exploration of niloticin's interactions with the selected proteins delved even deeper through extensive molecular dynamics simulations. These simulations, conducted using the OPLS-2005 force field, extended over a meticulous time frame of 100 nanoseconds. The root mean square deviation (RMSD) plot emerged as a crucial analytical tool in this phase. This plot not only provided insights into the structural dynamics, but also served as a robust indicator of the stability exhibited by both the individual proteins and their respective complexes with niloticin. Remarkably, the RMSD analysis revealed that, throughout the simulation period, the proteins and their complexes maintained structural integrity, demonstrating a maximum RMSD value of 3.5 Å (Fig. 2h).
This meticulous assessment of structural fluctuations underscores the resilience of the protein–niloticin complexes, affirming the sustained stability of the binding interactions. The limited deviation observed in the RMSD values accentuates the consistency and endurance of the strong molecular bonds between niloticin and the crucial proteins in the apoptotic pathway. This substantiates the premise that niloticin's impact extends beyond mere binding affinity, indicating a profound and enduring influence on the structural stability of the targeted proteins, further fortifying its potential as a key player in inducing apoptosis in human cervical cancer cells.
Apoptotic evaluation of niloticin
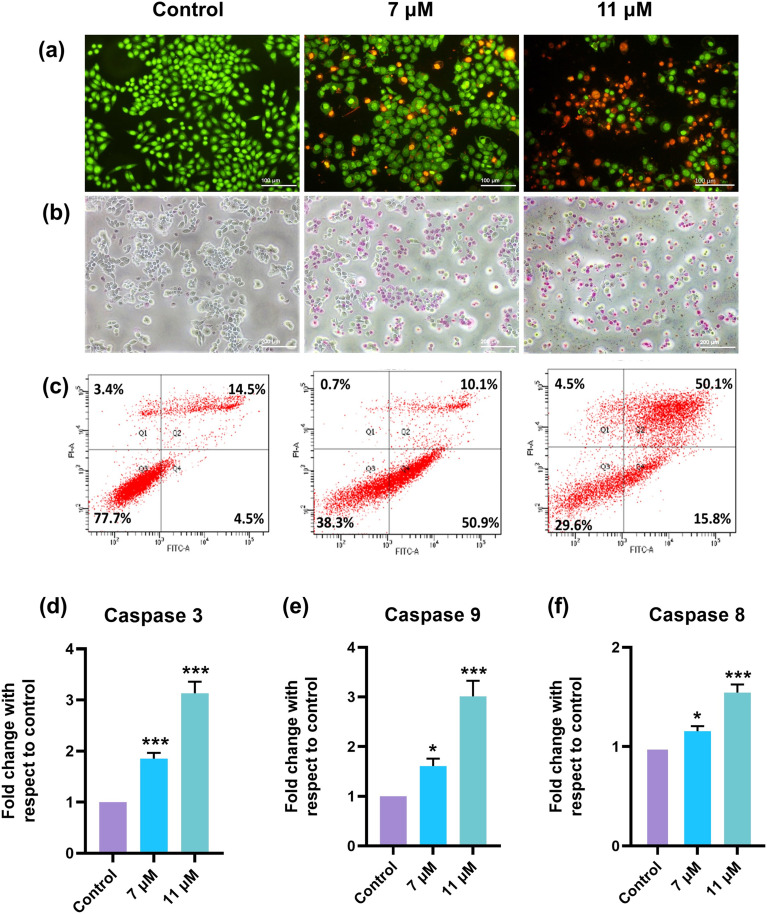
Various apoptotic assays were employed to assess the cell death mechanisms triggered by niloticin in HeLa cells. Firstly, we used acridine orange (AO) and ethidium bromide (EB) DNA-binding dyes as a dual staining method, which differentially labeled viable and dead cells, and observed the signals through fluorescence microscopy with a FITC filter. Considering the IC50 of niloticin, the apoptotic induction was conducted at two different concentrations (7 μM and 11 μM). The control cells, without niloticin treatment, were stained green as AO can penetrate the intact cell membrane of the viable cells, while the membrane-impermeant dye EB cannot. In contrast, niloticin-treated cells were stained reddish-orange because EB only enters cells with compromised membranes, binds to DNA, and exhibits a red fluorescence, indicating an early or late apoptotic stage of the cell, as shown in Fig. 3a. Furthermore, to confirm the changes in the cellular membrane due to apoptosis induction, we employed a non-fluorescent imaging method using the APOPercentage assay. In this assay, the dye enters cells with impaired membranes, staining them pink, while non-apoptotic cells remain unstained.35 The current data align with the findings of the apoptotic potential of niloticin on HeLa cells, i.e., niloticin-treated cells were stained pink, with the intensity of the color increasing at higher concentrations. Meanwhile, control cells remained unstained, as observed under the microscope (Fig. 3b). The exposure of phosphatidylserine enhances the identification of apoptotic cells, and annexin V exhibits specific binding to this phospholipid, distinguishing apoptotic cells within a cellular population. This property is leveraged to confirm the apoptotic potential of niloticin in HeLa cells using the annexin V apoptosis assay. The quantification of cells labeled with annexin V-FITC and PI found in various quadrants (Q1, Q2, Q3, Q4) in the data obtained serves as a comprehensive indicator of cells at various phases. FITC and PI negative live cells are found in Q3, with a decrease from 77.7% in control to 38.3% and 29.6% in 7 and 11 μM niloticin treatment, respectively. The lowest cell population in Q1 of the treated cells (0.7% and 4.5%) indicates that the cells have undergone necrotic cell death. In Q4, 50.9% of annexin V-labelled cells were found in the 7 μM treatment group, indicating an early apoptotic stage and a noteworthy increase in the percentage of annexin V-FITC and PI-positive cells in Q3, rising from 14.5% in the control group to 49.2% in the niloticin-treated cells, effectively indicates that the cells are in the late apoptotic stage. This shift is evident in the flow cytometry data, highlighting the impact of niloticin on apoptosis, as shown in Fig. 3c.
Fig. 3. Induction of apoptosis in HeLa cells by niloticin analysed in different concentrations of 7 and 11 μM by a) Acidine orange ethidium bromide dual staining method, b) APOP assay, c) annexin V apoptosis assay by FACS, and c) caspase expression analysis by fluorometric method. Major caspases, such as d) caspase-3, e) caspase-9 and f) caspase-8, involved in apoptotic pathways were checked and it was observed that all 3 caspases show a fold increase in its fluorescence intensity with respect to control. Results are represented as mean ± SD, *p < 0.05, **p < 0.01, ***p < 0.001 compared to the control.
Investigation of caspase-mediated apoptosis
Programmed cell death is largely orchestrated by the involvement of caspases, a distinct family of cysteine proteases. These caspases typically exist as inactive zymogens, undergoing a cascade of catalytic activation during apoptosis.36 To assess the activation profile of caspases, a fluorescence-based assay was employed, where the intensity of fluorescence indicates the expression of different caspases. The apoptotic pathway may follow the extrinsic or intrinsic pathway, and can be differentiated by examining the expression of various initiator caspases. In this study, we investigated the activation of caspase 3, an executioner caspase crucial for both the extrinsic and intrinsic modes of the apoptotic pathway. The cells were thus treated with niloticin (7 and 11 μM). Compared to the control, we observed a threefold increase in the expression of caspase 3, confirming the induction of apoptosis (Fig. 3d). We also examined the activation of the caspases associated with the initiation of apoptosis induced by niloticin. For that, we looked at the expression of caspase 8 and caspase 9, which are significant factors of the extrinsic and intrinsic pathways. Interestingly, the expression of both caspase 8 and caspase 9 was found to be increased, confirming that niloticin acts through both extrinsic and intrinsic pathways of apoptosis (Fig. 3e and f). From reviewing the literature, it is observed that certain natural compounds falling within the terpenoids category (nimbolide in MCF-7 breast cancer cells, hyperforin in glioblastoma, and carnosic acid in PC-3 prostate carcinoma cells) exhibit similar patterns of execution for both apoptotic pathways.37–39
Evaluation of apoptosis by nuclear condensation and DNA fragmentation using fluorometric and SERS analysis
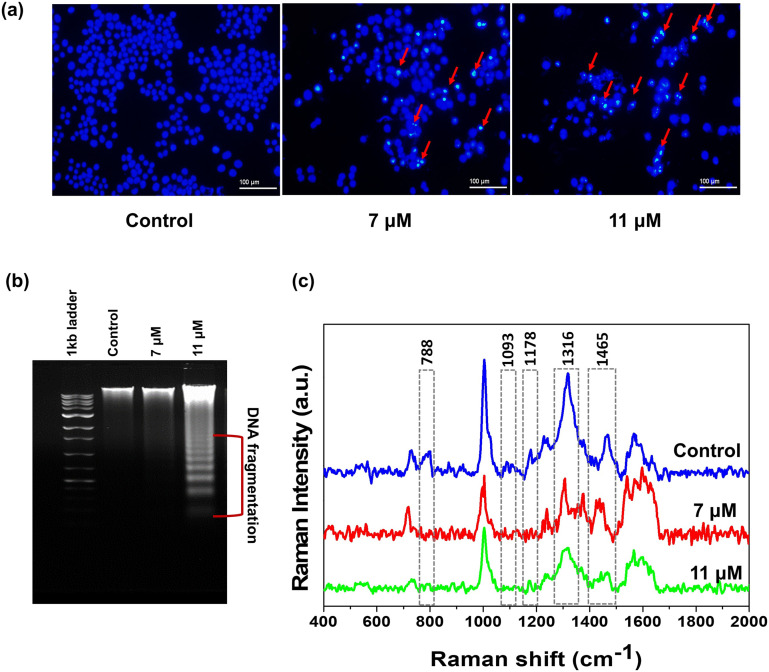
A hallmark of apoptotic events is DNA condensation, whereas the nuclei of unaffected cells retain a spherical shape with evenly distributed DNA. This feature can be utilized to illustrate the apoptotic potential of a compound using DNA-binding dyes. In this context, Hoechst 33342 staining is employed, which permeates both live and apoptotic cells, and selectively binds to adenine–thymine-rich regions of DNA in the minor groove.40 Fluorescence can be observed in both normal and condensed DNA, but with a higher intensity in the condensed DNA. Fragmentation can also be effectively detected. In our experiment, niloticin-treated (7 μM and 11 μM) HeLa cells were stained with Hoescht 33342, and analyzed using fluorescent light microscopy. The images revealed a higher number of cells with condensed nucleic acids at varying concentrations, confirming apoptosis, while the DNA in healthy cells retains its spherical shape (Fig. 4a). During the apoptotic process, the cytogenic damage is characterized by DNA fragmentation. Endonucleases cleave the DNA into internucleosomal fragments, which are indeed considered specific markers of apoptosis.41 This fragmentation pattern can be visualized through DNA laddering using agarose gel electrophoresis. Niloticin in 7 and 11 μM doses were used to treat HeLa cells. DNA isolation was performed for both control and treated cells, which was further subjected to agarose gel electrophoresis (0.8%). The fragmented DNA was visualized as stained nucleic acids using ethidium bromide (Fig. 4b). The results exhibit a distinct laddering pattern in higher concentrations of niloticin-treated cells compared to control cells, demonstrating the strong apoptotic potential of the molecule. For further confirmation of the DNA fragmentation induced by niloticin treatment, surface-enhanced Raman spectroscopic (SERS) analysis of the DNA samples from the control cells and treated cells was conducted using the 633 nm laser of a confocal Raman microscope and colloidal gold nanoparticles (AuNPs: 40–45 nm) as the SERS substrate. The enhanced Raman spectra derived from these samples showed significant differences in the pattern, especially the diminished peaks of the phosphodiester linkage and O–P–O stretching vibrations (785 cm−1 and 1093 cm−1). Moreover, the decreased intensity for the peaks of cytosine and guanine and the DNA ring breathing mode (1178 cm−1, 1316 cm−1, respectively) was also evident. As compared to the control, an enhanced peak at 1420 to 1440 cm−1 showing CH-deformation and a less intense peak for the deoxyribose vibration (1460–65 cm−1) was also noticed in the DNA isolated from the treated cells (Fig. 4c).42
Fig. 4. Nucleic acid condensation in HeLa cells upon treatment with niloticin is proved by a) Hoescht staining method, where the condensed DNA will give an intense coloration compared to the untreated DNA, b) analysing the DNA laddering pattern in both control and treated cells, in which exonucleases act on niloticin-treated cells, giving a laddering pattern analysed by agarose gel electrophoresis, and c) SERS analysis of the DNA laddering.
Apoptotic assessment based on cell cycle regulation
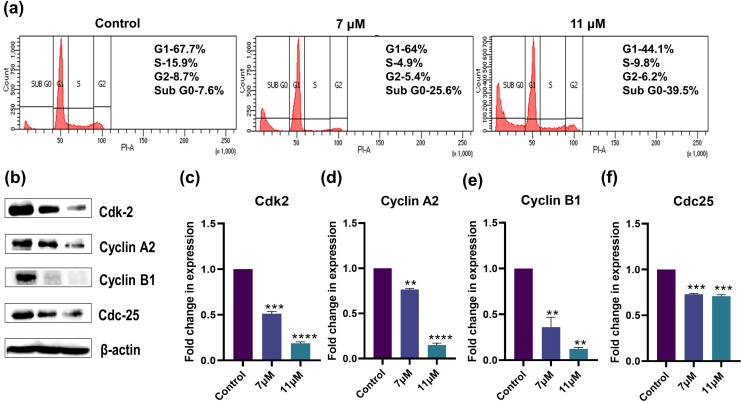
Genomic level mutations associated with cancer interfere with normal cell cycle mechanisms, compromising cell division control and resulting in the uncontrolled progression of cells.43 The effect of the drug should reestablish the cell cycle checkpoint regulation that was mutated or direct it toward apoptosis by halting the cell division. To unveil the mechanistic action of niloticin in HeLa cells, the retardation in the progression of the cell cycle is confirmed via cell cycle assay by flow cytometric analysis. The assay uses propidium iodide, a fluorescent nucleic acid dye that can enter the apoptotic cells or those in the last stages of apoptosis. This stains the DNA and gives the exact number of cell populations at different phases of the cell cycle. In the experiment, most of the cells were found in the S and G2 phases in the control. Conversely, upon treatment, the maximum cell population was restricted to the sub-G0 phase. Initially, the sub-G0 population of cells without treatment were 7.6%, which in fact increased to 25.6% at 7 μM and further to 39.5% at 11 μM concentrations of niloticin (Fig. 5a). The data confirm the cell cycle arrest at the sub-G0 phase during the advancement of the cell cycle as the percentage of cells decreased in the G0/G1, S, and G2/M phases by the effect of niloticin. Cdk-2 is a key cell cycle regulator, which has an important role in G1/S and G2/M transitions.44 Cyclin A2, essential in the activation of kinases, interacts with Cdk-2 in the G1/S checkpoint. Cyclin B1 aids cells in entering a mitotic phase, where cyclin B1–cdk2 phosphorylates many proteins for cell cycle progression.45,46 Cdc-25 promotes dephosphorylation in cdks, which is important in the progression of the cell cycle. Western blot analysis of the mentioned cell cycle proteins shows a downregulation in their expression on treatment with the compound, further affirming the cell cycle arrest by niloticin (Fig. 5b–f).
Fig. 5. Cell cycle pattern change analysis and study of the expression change in cell cycle regulatory proteins upon induction by niloticin in HeLa cells by a) Western blot of Cdk-2, cyclin A2, cyclin B1, and cdc-25, some of the major proteins in cell cycle regulation. b) Graph representing the fold change in the expression of Cdk-2, c) cyclin A2, d) cyclin B1, and e) Cdc25 in comparison with β-actin. f) Cell cycle assay using PI staining done by FACS, which confirms the sub-G0 phase arrest with a higher cell population. Results are represented as mean ± SD, **p < 0.01, ***p < 0.001 compared to the control.
Apoptotic induction through mitochondrial membrane potential
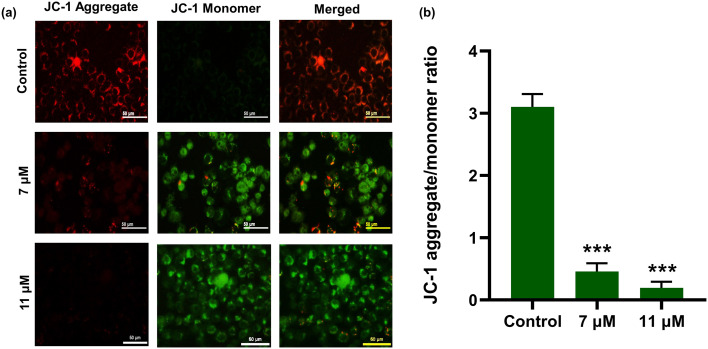
Maintenance of a stable mitochondrial membrane potential is imperative for normal cell functioning. Sustained perturbation in the transmembrane potential eventually causes severe effects on the viability of cells. The dysfunction of the mitochondrial membrane appears to induce apoptosis, where activated caspases specifically target permeabilized mitochondria. This leads to the disturbance of electron transport and the subsequent loss of the mitochondrial transmembrane potential. JC-1 dye is a cationic dye that is lipophilic in nature and can enter mitochondria. It forms J-aggregates that are reversible complexes, exhibiting red fluorescence in cells with a normal transmembrane potential. On the other hand, due to less negativity in apoptotic cells, the JC-1 dye enters at a lower concentration, where aggregates cannot be formed. Thus, the JC-1 dye retains its green fluorescence. Here, to disclose the effect of niloticin in mitochondrial functioning, the cells are induced by the compound for 24 hours. Afterwards, the JC-1 dye is added, which forms aggregates in untreated ones. In contrast, it remains as a monomer in cells where mitochondrial depolarization occurred upon treatment with niloticin (Fig. 6a). Therefore, the data clearly express the increase in green fluorescence resulting from the action of the compound as mitochondrial dysfunction occurs, which substantiates the involvement of the intrinsic (mitochondrial-mediated) mode of apoptosis.
Fig. 6. a) Analysis of the change in the mitochondrial membrane potential is done by JC-1 assay in HeLa cells after induction with Niloticin. b) Graph showing the decrease in the JC-1 aggregate-to-monomer ratio with respect to the control. Results are represented as mean ± SD, ***p < 0.001 is considered to be significant as compared to the control. Scale bar corresponds to 50 μm.
Inhibition of metastatic potential by niloticin
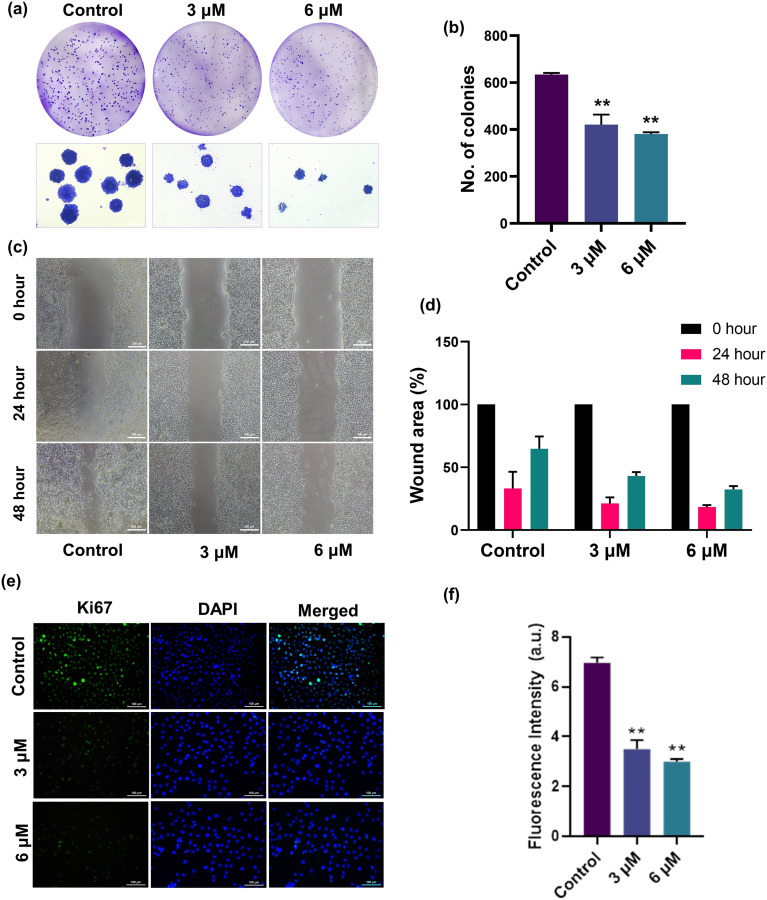
Firstly, the inhibition of clonogenic potential by niloticin in HeLa cells was assessed. The study investigates the capacity of an individual cell to develop into a colony, employing the cell survival assay to assess the reproductive potential of cells for unrestricted division and colony formation.47 The conventional colony formation assay was conducted to evaluate the suppressive effect of niloticin on the ability to form colonies. Niloticin was introduced to HeLa cells at concentrations of 3 and 6 μM, and the quantification of colonies formed was carried out using ImageJ software. The data demonstrated a noticeable decline in the colony-forming capability of Hela cells in a dose-dependent fashion from a total count of 639 in the control cells to 424 in 3 μM and 397 at 6 μM niloticin induction (Fig. 7a). The ability of the cancer cells to migrate and establish themselves in distant organs, a pivotal step in the process of metastasis, serves as the foundation for this phenomenon. This is typically assessed by examining cell movement by scratch wound assay. The assay operates on the principle that when a wound is intentionally created in a layer of cells, these cells naturally migrate to restore cell-to-cell contact. In the experiment, niloticin was treated at the concentrations of 3 and 6 μM, and then examined for 0, 24, and 48 hours of incubation. The percentage of wound closure is calculated using ImageJ software. The data clearly indicate that the percentage of wound closure is significantly high in the control compared to that in the niloticin-treated cells (Fig. 7c and d).
Fig. 7. a) Inhibition of the clonogenic potential of HeLa cells by niloticin treatment at 3 μM and 6 μM. b) Graph showing the decrease in the number of colonies with niloticin treatment compared to the control. c) Inhibition of the migratory potential of HeLa cells by niloticin treatment. d) Graph representing the wound area percentage at different time points. e) Analysis of Ki67 expression in HeLa cells at 3 μM and 6 μM niloticin treatment. f) Graph showing the fluorescence intensity of Ki67 expression of niloticin-treated cells compared to the control. Results are represented as mean ± SD, **p < 0.01 as compared to the control.
Immunofluorescence assay of Ki67
The expression of Ki67, a proliferation marker, may be utilized in cancer diagnostics.48 Here, we determined the anti-metastatic potential of niloticin by evaluating the expression of Ki67 in HeLa cells via immunofluorescence assay. Niloticin was treated at two different concentrations, and the DAPI staining was analyzed using a fluorescence microscope. The images clearly showed the downregulated expression of Ki67 in treated cells upon increasing concentration. Meanwhile, a higher expression was observed in the control cells.
Modulation of various protein expressions involved in apoptosis
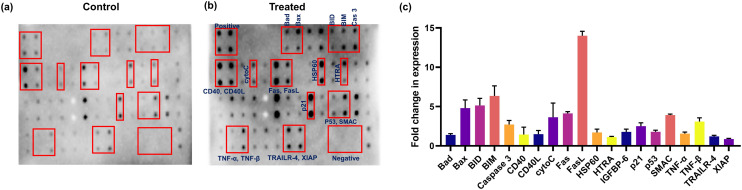
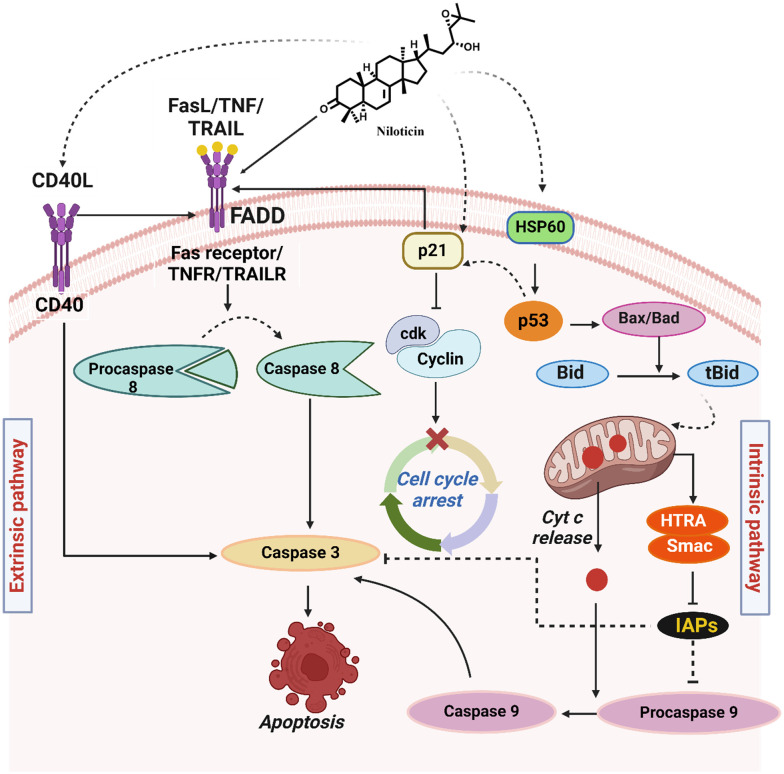
Elucidation of the underlying mechanism of action of phytomolecule emphasizes the recognition of molecular targets that could be exploited for designing efficient therapeutic entities. The mechanistic route of action of niloticin may follow any of the cell death signaling pathways, which is unraveled by identifying the upregulation and downregulation of key proteins involved in apoptosis. Herein, the expression of a broad range of proteins were analyzed using a human apoptotic array membrane, which encompasses 43 major target proteins (Fig. 8a–c). HSP60 accumulates in the cytosol to induce various signals, conferring its action to either cell death or cell survival mechanisms. In the case of apoptosis, it leads to the maturation and activation of caspase 3.49 The nuclear transcription factor, p53 is proapoptotic in function. In response to various stresses, it is induced to exert its proapoptotic properties.50 CD40 is a TNFR family member. Upon some signals, CD40 can exert a wide range of cellular responses. Here, the induction of niloticin results in the binding of CD40 to its ligand CD40L, which eventually results in the enzymatic maturation of caspase 8 and caspase 3. TNF, a pro-inflammatory cytokine, simultaneously activates cell survival and cell death mechanisms, and is the most potent inducer of apoptosis. By the action of niloticin, TNF is activated by CD40/CD40L, which is the upstream regulatory molecule.51 The binding of TNF to its TNFR induces apoptosis. Specifically, the extrinsic mode of apoptosis is executed by the death receptor TRAILR and the continued signaling to the activation of caspases for the execution of apoptosis. p53 activation enhances cell cycle arrest, either by facilitating DNA repair or routing it to apoptosis. In the present study, the induction of niloticin effectively increases the expression of p53, which could be considered as the root of a further signaling pathway, as they directly regulate a broad range of target genes. Fas induces apoptosis by the cross-linking of its own receptor, FasL. It is already known that the upregulation of the Fas receptor in stimuli to any cell death signal is p53-dependent. So, the p53 upregulation by niloticin directly induces the binding of Fas to FasL,which could be observed from the intensity of expression. Increased expression of Fas/FasL enables catalytic activation of the initiator caspase-8, which is the key factor in the extrinsic mode of apoptosis. Likewise, activation of p53 also results in Bad/Bax pro-apoptotic protein expression. This further activates Bid, which is a Bcl-2 family member protein that aids in apoptosis in its truncated form, tBid. This activates Smac, a mitochondrial protein released during cellular apoptosis, counteracts inhibitory factors (IAPs), and promotes apoptosis progression. It is already reported that the export of smac to cytosol occurs in response to the induction of cytotoxic drugs. Along with SMAC activation, HTRA (a mitochondrial factor) gets released upon an apoptotic trigger from mitochondria into the cytosol. HTRA then interacts with IAPs, which enables the caspase to be free from its inhibition.52 Cytochrome c is usually located in the inter-cristae spaces in mitochondria, which gets exported to cytosol when apoptotic signals permeabilize the mitochondrial membrane. In the cytosol, cyt c tends to activate Apaf-1, a requisite for the proteolytic activation of caspase-9, which is the critical initiator of caspase involved in the intrinsic mode of apoptosis. This is well supported by the expression analysis of cytochrome c and caspase 9. The direct p53 activation of caspase-8 and mitochondrial-mediated caspase 9 activation substantiates the concluded mode of extrinsic and intrinsic pathways of niloticin action in HeLa cells. This proteolytic maturation of caspase 8 and 9 finally results in the activation of caspase-3, the executional caspase that finally takes the cell to its demolition phase. p53 also activates p21, a key protein involved in cell cycle regulation that interacts with cyclin–cdk complexes and induces cell cycle arrest in niloticin-treated cells. The expression analysis of apoptotic proteins establishes a signaling pathway of action, and provides strong evidence for niloticin to be developed as a potent anticancer agent (Fig. 9).
Fig. 8. Apoptotic protein expression study in HeLa cells a) with and b) without treatment were analyzed using an antibody array kit. c) Comparison of the expression change in major proteins in niloticin-treated cells with respect to the control.
Fig. 9. Proposed mechanism of action of niloticin.
Conclusion
In summary, we have evaluated a detailed anticancer profiling of the tirucallane-type triterpenoid, niloticin, which exhibited the highest anticancer properties among the other four isolated triterpenoids from the stem barks of Aphanamixis polystachya against cervical cancer cells. The primary cytotoxicity assessment of niloticin was carried out in various cancer cell lines. The cervical cancer HeLa cells turned out to have an impressive IC50 value of 11 μM after 24 hours of treatment, which was adequate for apoptotic induction in HeLa cells. Molecular docking studies with major protein targets (such as TNF, Fas, p53, and caspases) exhibit reasonably high binding affinity with niloticin, as evidenced by docking scores ranging from −6.8 to −9.0 kcal mol−1. The binding stability was further evaluated through molecular dynamic simulation. To complement the in silico studies, downstream in vitro cell-based assays were employed including annexin V assay by flow cytometric analysis for validating the apoptotic potential of niloticin. In a subsequent apoptotic evaluation, DNA condensation was evident by Hoechst staining and DNA laddering supported the apoptotic potential of niloticin in HeLa cells. The SERS fingerprint analysis in the treated cells enabled the tracking of cellular DNA breakage as a complementary assessment, which was fully complemented by the DNA laddering experiment. The cell cycle analysis indicated the arrest at the sub-G0 phase. The halt in cell cycle progression was determined by the analysis of the proteins involved, which finally takes the cell into apoptosis. Another interesting fact is the involvement of both extrinsic and intrinsic modes of apoptosis induced by niloticin, analyzed by the substantial expression of caspase 3, 9, and 8. The expression of major regulatory proteins involved in the apoptosis cascade is evident in the protein dot-blot assay. Finally, the modulation in the signaling pathways involved in the cancer was studied by analyzing the different proteins involved. The activation of p53 by the induction of niloticin was the key step. p53 induced the binding of FasL to the Fas receptor, resulting in the activation of the initiator caspase 8, a prominent factor of the extrinsic pathway of apoptosis. The activation of p53 also upregulates the expression of proapoptotic proteins Bad/Bax, and further activates Bid, SMAC and HTRA, which results in the export of cytochrome c from the nucleus to cytosol. Cytochrome c then activates Apaf-1, which is a requisite for the activation of caspase 9, the key factor in the intrinsic pathway of apoptosis. Both the extrinsic and intrinsic modes finally conclude in the catalytic activation of caspase-3, taking the cell to its demolition phase. Further evaluation of niloticin in its ability to inhibit colony formation and wound healing property reflected its anti-metastatic potential. Niloticin exerting its effect in the downregulation of the proliferative marker Ki67 by immunofluorescence assay further proves its capability as an anti-metastatic potential agent. We envisaged that the naturally occurring niloticin would become a successful blueprint to generate a potential anticancer hit compound for pre-clinical studies against the efficacious management of cervical cancer.
Materials and methods
General experimental procedures and chemicals
All of the solvents were used without further purification, and were of the highest available grade. Column chromatography was performed with silica gel (100–200 mesh; Merck, Darmstadt, Germany). TLC was carried out on Merck 60 F254 silica gel plates, detecting phytomolecules under UV light or by heating after spraying samples with a p-anisaldehyde–sulfuric acid mixture. NMR spectra were obtained from a Bruker Avance 500 MHz instrument with CDCl3 and CD3OD as the solvent, and chemical shifts were expressed in δ (ppm) relative to the TMS. The HR-ESI-MS spectrum was recorded at 60 000 revolutions using a Thermo Scientific Exactive mass spectrometer, and the purity of the tirucallane-type triterpenoids was analyzed by a Waters Arc analytical HPLC instrument.
Plant material collection
The stem barks of A. polystachya were collected from Parassala, Kerala (8.34780° N and 77.1410° E), Kerala, India in April 2017. The plant material was deposited at the Department of Botany, University of Kerala, Thiruvananthapuram (voucher number 2018-06-05).
Isolation and characterization of compounds from A. polystachya
About 1.0 kg of the dried stem bark was powdered mechanically and subjected to extraction with acetone at room temperature (3 L × 3 days). Following filtration, the extract was concentrated under reduced pressure using a rotary evaporator, resulting in an approximate yield of 20 g for the acetone extract. The acetone extract was subjected to silica gel column chromatographic separation and eluted with a mixture of petroleum ether/ethyl acetate from 100 : 0 v/v to 0 : 100 v/v. Twenty-two fractions (1–22) were obtained. Fractions 5–7 were applied to a silica gel column and eluted with a mixture of petroleum ether/ethyl acetate from 9 : 1 v/v to 4 : 1 v/v, affording compounds 1 (15 mg) and 2 (32 mg). Compounds 3 (10 mg), and 4 (21 mg) were isolated from fractions 8–15 through a silica gel column with petroleum ether/ethyl acetate from 4 : 1 v/v to 7 : 3 v/v. The purification of fractions 16–19 yielded compounds 5 (18 mg) via silica gel column with petroleum ether/ethyl acetate from 7 : 3 v/v to 3 : 2 v/v. The structures of the phytochemicals were analysed through 1D and 2D NMR, HR-ESI-MS analysis, and comparison with the literature report. The purity of the compounds was checked by a Waters Arc analytical HPLC instrument, and the chromatograms are shown in the ESI† (Fig. S9, S18 and S40).
Cell culture methods
HeLa (human cervical cancer) cell line and the triple-negative human breast cancer cell line MDA-MB-231 were purchased from American Type Culture Collection (ATCC, USA). The A549 (human non-small cell lung) cancer cell line and the human pancreatic cell line PANC-1 were procured from National Centre for Cell Science (NCCS, Pune). The MCF-10A, (normal breast epithelial) cell line was obtained from Elabscience, USA. The HeLa, A549, PANC-1, and MDA-MB-231 cells were maintained in Dulbecco's modified Eagle medium (DMEM, sigma) with a supplement of 10% fetal bovine serum (FBS, himedia), 1% antibiotic antimycotic solution (Himedia). MCF-10A cells were grown in mammary epithelial cell growth medium (MEGM, Lonza) with 5% horse serum, and all the cells were maintained in 5% CO2 at 37 °C culture conditions.
Cell proliferation assay
A seeding density of 8 × 103 cells per 100 μL of DMEM media was added to 96-well plates for 24 and 48 hour proliferation studies. Niloticin at different concentrations (5 μM, 10 μM, 20 μM, 50 μM, 100 μM) were added to the plates after 24 hours of incubation. After 24 and 48 hours of incubation with the compound, 100 μL of MTT (3-[4,5-dimethylthiazole-2-yl]-2,5-diphenyltetrazolium bromide) at a concentration of 0.5 mg mL−1 in Hanks balanced salt solution (HBSS) was added to each well after removing the spent medium and washing with PBS, and then incubated at 37 °C for 2–4 hours under dark conditions. Then, the MTT solution was removed, and the sample was again washed with PBS. Finally, 100 μL of DMSO was added to each well for dissolving the formazan crystals. The conversion of the yellow-coloured MTT to a violet color was observed, and the absorbance was measured using a multimode plate reader (Synergy H1, Biotek) at 570 nm.
Molecular simulations
Initial pharmacokinetic parameters of niloticin were assessed using the SwissADME online tool.53 This tool provides valuable insights into the drug-likeness, human intestinal absorption, and lipophilicity. Molecular docking studies were conducted to elucidate the binding interactions between niloticin and key proteins involved in the apoptotic pathway. AutoDock Vina was employed for the docking simulations, utilizing the crystal structures of selected proteins from RCSB PDB – p53 (1TUP), Fas receptor (3EZQ), FasL (5L19), Bax (6EB6), CDK2 (2UZE), BCL2 (6O0K), and TNF β (1TNR). The docking score was used to screen the interactions.54 Docking simulations were visualized and analyzed using UCSF Chimera version 1.16.55 This molecular visualization software facilitated a comprehensive examination of the binding orientations and interactions between niloticin and the target proteins. The stability and dynamic behavior of the seven protein–niloticin complexes were further investigated through molecular dynamics simulations. Desmond, a component of the Schrödinger suite, was employed for these simulations, extending over 100 nanoseconds.56 The root mean square deviation (RMSD) analysis was carried out to assess the structural stability of the complexes.
Apoptotic assays
The apoptotic potential of the compound was checked by live dead assay using the ethidium bromide-acridine orange dual staining method and APO percentage assay, which distinguishes live cells from those that have undergone apoptosis. The imaging was performed by the Nikon-TS100 inverted microscope. An annexin V apoptosis assay was performed by using the FITC annexin V apoptosis detection kit (BD Pharmingen), following the assay protocol mentioned. Here, the cells were stained with annexin V and propidium iodide, and the number of apoptotic cells can be determined via flow cytometric analysis.
Caspase fluorometric assay
The activation of caspases, being a major feature of apoptosis, is studied using the caspase fluorometric assay for caspase 3, 9 and 8. A cell density of 3 × 106 was seeded on 6-well plates, and apoptosis induction was done by treatment of HeLa cells with niloticin at different concentrations. The assay was carried out following the exact protocol given in the fluorometric assay kit (Abcam). The fluorescence intensity was measured using a multimode plate reader (Synergy H1, Biotek) at an excitation wavelength of 400 nm and emission at 505 nm. This was followed for all three caspases.
Nucleic acid degradation and DNA fragmentation studies
DNA condensation was analysed using Hoescht staining. A cell density of 7 × 103 was seeded on 96-well plate. After 24 hours of treatment with the compound, Hoescht stain (1 μg mL−1) in PBS was added and imaging was performed under a DAPI filter of the Nikon-TS100 inverted microscope. Cells that had undergone apoptosis followed a laddering pattern while running in agarose gel. DNA isolation was done in HeLa cells after treatment with the compound for different concentrations using the Geneaid genomic DNA mini kit (Geneaid, cat. no. GB100), following the given protocol. The concentration of isolated DNA was analysed using the nanodrop method, and it was further normalised. The DNA was run in 0.8% agarose at 80 V and imaging was done using Chemidoc (Biorad). The DNA fragmentation pattern was then confirmed by Raman analysis. Gold nanoparticles of size 40–45 nm were used as SERS substrate that was mixed with the DNA sample in a 8 : 2 ratio. After 10 minutes of incubation, the SERS spectrum was analysed using the WITec Raman microscope (WITec, Inc., Germany) with 600 g mm−1 grating and Peltier CCD detector unit. Nanoparticle-mixed samples were excited with a 633 nm laser having 5 mW power. Spectral analysis was performed with a resolution of 3 cm−1 and 3 spectral accumulations with 5 s integration time.
Cell cycle analysis
The cell cycle arrest by the niloticin induction effect was studied using a cell cycle assay via flow cytometric analysis with the BD cycle test plus DNA kit (BD Pharmingen, cat. no. 340242). It uses propidium iodide staining, which can enter apoptotic cells and stain DNA, giving the cell population at different cell cycle phases via flow cytometric analysis. The expression of proteins involved in cell cycle regulation was studied using western blotting with the standard procedure. The protein isolation from HeLa cells in both treated and non-treated cells was done, and protein quantification was performed using the BCA assay kit (Pierce BCA Assay Kit cat no. 23225). The normalised SDS-PAGE was performed for the separation of proteins in the sample, followed by the transfer to the PVDF membrane. Blocking with 5% skim milk was performed, and membranes were incubated with respective primary antibodies (Cell Signaling Technology, USA) for 18 hours at 4 °C. After the secondary antibody (HRP conjugated) incubation, chemiluminescence was detected with (Takara Western BloT Hyper HRP Substrate) for Cdk-2, cyclin A2, cyclin B1 and Cdc-25, along with beta actin as the loading control using the Chemidoc imaging system (Biorad).
Mitochondrial membrane potential analysis
The alteration in the mitochondrial membrane potential was analysed by JC-1 assay using JC-1 (5,5,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimi-dazoylcarbocyanine iodide) dye (Sigma Aldrich), which is a cationic dye normally exhibiting green fluorescence. In cells that have undergone apoptosis, this dye will aggregate and gives a red fluorescence, which can be observed via imaging using Nikon-TS100 inverted microscope with an FITC filter.
Anti-metastatic studies
The antimetastatic potential of niloticin was checked using the clonogenic assay, where a cell density of 1 × 103 cells was seeded into 6-well plate. Once the cells attained morphology, the niloticin was treated in 3 μM and 6 μM concentrations and incubated for another 9 days at 37 °C, allowing the cells to grow into colonies. After that, the cells were fixed with 70% methanol, followed by 0.3% crystal violet staining for visualization. Subsequently, the cells were washed with PBS. Imaging was done using the Nikon-TS100 inverted microscope and further processing was done using ImageJ software. For the scratch wound assay, cells were seeded into 96-well plates. After 24 hours, when a monolayer of cells was formed, a vertical scratch was made using a 200 μL tip and washed with PBS. The compound was added in two different concentrations, and non-treated cells were taken as control. Cells were then monitored for its movement to heal the wound, and images were taken after a period of 0, 24 and 48 hours incubation using the Nikon-TS100 inverted microscope. The area of wound closure was analysed using ImageJ software.
Immunofluorescence assay of Ki67
The anti-metastatic potential of niloticin was further studied by analysis of the expression of Ki67 via immunofluorescence assay. A cell density of 7 × 103 was seeded to a 96-well plate, and niloticin was treated at two different concentrations. After 24 hours of incubation, cells were washed with PBS and fixed by 4% paraformaldehyde for 15–30 min at 37 °C. After that, the cells were treated with 0.1% Triton-X for 10 min. Cells were again washed with PBS and blocking was performed using 5% BSA in PBST for 1 hour. Again, washing with PBST was performed 3 times with 5 minutes interval. Then, the primary antibody for Ki67 was added and incubated overnight. After that, the secondary antibody was added, followed by DAPI staining, and images were visualized and captured using the Nikon-TS100 inverted fluorescent microscope.
Apoptotic protein expression
The expression of proteins that play a central role in apoptosis was determined by the human apoptotic antibody array membrane (Abcam). The experiment was conducted, strictly following the manufacturer's protocol. HeLa cells were seeded and treated with niloticin, followed by protein isolation, as outlined in the given protocol. A final volume of 1.2 ml of sample was used for analysis, as per the instruction. Imaging was done with the help of chemidoc (Biorad). Further analysis and densitometric data were obtained with the help of ImageJ software.
Abbreviations
- TNF
Tumor necrosis factor
- SERS
Surface-enhanced Raman spectroscopy
- CD40
Cluster of differentiation 40
- IAPs
Inhibitor of apoptosis proteins
- Bax
Bcl-2-associated X protein
- Cdk2
Cyclin-dependent kinase 2
- MTT
(3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide)
- FITC
Fluorescein isothiocyanate
- FACS
Fluorescence-activated cell sorting
- Cdc25
Cell division cycle 25
- JC-1
5,5,6,6′-Tetrachloro-1,1′,3,3′-tetraethylbenzimi-dazoylcarbocyanine iodide
- HSP60
Heat shock protein 60
- TRAILR
TNF-related apoptosis-inducing ligand receptor
- Bcl-2
B cell lymphoma-2
- Bax
Bcl-2-associated X protein
- Bid
Bcl-2 interacting domain
- Smac
Second mitochondria-derived activator of caspase
- Apaf-1
Apoptotic protease activating factor-1
- FBS
Fetal bovine serum
- NMR
Nuclear magnetic resonance
- HRMS
High-resolution electrospray ionization mass spectrometry
- PBS
Phosphate buffer saline
- DMSO
Dimethyl sulfoxide
- PI
Propidium iodide
Data availability
The data supporting this article have been included as part of the ESI.†
Author contributions
Anuja Gracy Joseph: experimentation, data collection, drafting, editing; Mohanan Biji: experimentation, data collection; Vishnu Priya Murali: experimentation, data collection; Daisy R. Sherin: molecular docking studies; Alisha Valsan: experimentation, drafting; Vimalkumar P. Sukumaran: experimentation; Kokkuvayil Vasu Radhakrishnan: conceptualization, editing; Kaustabh Kumar Maiti: supervision, editing.
Conflicts of interest
The authors declare no competing financial interest.
Supplementary Material
Acknowledgments
K. K. M. thanks the Council of Scientific and Industrial Research (CSIR), Govt. of India and Indian Council of Medical Research (ICMR), Govt. of India (No. 5/4-5/3/01/DHR/NEURO/2020-NCD-I) & (EM/IG/Dev.Res/2003-0001476) for research funding. AcSIR Ph.D. students A. G. J., V. K. N., M. B. and A. V. acknowledge CSIR for financial assistance through research fellowships. V. P. M. acknowledges the DHR Young Scientist Program (R.12014/22/2021) for funding. We acknowledge Dr. Rakesh K. V. and Dr. Ashalatha S. Nair for the plant material collection.
Electronic supplementary information (ESI) available: The extraction and isolation procedure, characterization, preliminary cytotoxic evaluation of extracts, and molecular docking are discussed. See DOI: https://doi.org/10.1039/d4md00318g
References
- Halim C. E. Xinjing S. L. Fan L. Vitarbo J. B. Arfuso F. Tan C. H. Narula A. S. Kumar A. P. Sethi G. Ahn K. S. Pharmacol. Res. 2019;147:104327. doi: 10.1016/j.phrs.2019.104327. [DOI] [PubMed] [Google Scholar]
- Rahmani A. H. Al Zohairy M. A. Aly S. M. Khan M. A. BioMed Res. Int. 2014;2014:1–15. doi: 10.1155/2014/761608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sailo B. L. Banik K. Girisa S. Bordoloi D. Fan L. Halim C. E. Wang H. Kumar A. P. Zheng D. Mao X. Sethi G. Kunnumakkara A. B. Cancers. 2019;11:246. doi: 10.3390/cancers11020246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banik K. Ranaware A. M. Deshpande V. Nalawade S. P. Padmavathi G. Bordoloi D. Sailo B. L. Shanmugam M. K. Fan L. Arfuso F. Sethi G. Kunnumakkara A. B. Pharmacol. Res. 2019;144:192–209. doi: 10.1016/j.phrs.2019.04.004. [DOI] [PubMed] [Google Scholar]
- Zaimy M. A. Saffarzadeh N. Mohammadi A. Pourghadamyari H. Izadi P. Sarli A. Moghaddam L. K. Paschepari S. R. Azizi H. Torkamandi S. Tavakkoly-Bazzaz J. Cancer Gene Ther. 2017;24:233–243. doi: 10.1038/cgt.2017.16. [DOI] [PubMed] [Google Scholar]
- Valsan A. Meenu M. T. Murali V. P. Malgija B. Joseph A. G. Nisha P. Radhakrishnan K. V. Maiti K. K. Exploration of Phaeanthine: A Bisbenzylisoquinoline Alkaloid Induces Anticancer Effect in Cervical Cancer Cells Involving Mitochondria-Mediated Apoptosis. ACS Omega. 2023;8(16):14799–14813. doi: 10.1021/acsomega.3c01023. doi: 10.1021/acsomega.3c01023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong S. K. L. Shanmugam M. K. Fan L. Fraser S. E. Arfuso F. Ahn K. S. Sethi G. Bishayee A. Cancers. 2019;11:611. doi: 10.3390/cancers11050611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashem S. Ali T. A. Akhtar S. Nisar S. Sageena G. Ali S. Al-Mannai S. Therachiyil L. Mir R. Elfaki I. Mir M. M. Jamal F. Masoodi T. Uddin S. Singh M. Haris M. Macha M. Bhat A. A. Biomed. Pharmacother. 2022;150:113054. doi: 10.1016/j.biopha.2022.113054. [DOI] [PubMed] [Google Scholar]
- Nuzzo G. Senese G. Gallo C. Albiani F. Romano L. D’ippolito G. Manzo E. Fontana A. Mar. Drugs. 2022;20:386. doi: 10.3390/md20060386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed S. I. A. Jantan I. Haque M. A. Naturally Occurring Immunomodulators with Antitumor Activity: An Insight on Their Mechanisms of Action. Int. Immunopharmacol. 2017:291–304. doi: 10.1016/j.intimp.2017.07.010. doi: 10.1016/j.intimp.2017.07.010. [DOI] [PubMed] [Google Scholar]
- Krishnaraju A. V. Rao C. V. Rao T. V. N. Reddy K. N. Trimurtulu G. Am. J. Infect. Dis. 2009;5:60–67. doi: 10.3844/ajidsp.2009.60.67. [DOI] [Google Scholar]
- Sadhu S. K. Phattanawasin P. Choudhuri M. S. K. Ohtsuki T. Ishibashi M. J. Nat. Med. 2006;60:258–260. doi: 10.1007/s11418-006-0047-1. [DOI] [PubMed] [Google Scholar]
- Mishra A. P. Saklani S. Chandra S. Mathur A. Milella L. Tiwari P. World J. Pharm. Pharm. Sci. 2014;3:2242–2252. [Google Scholar]
- Hossain M. S. Islam M. Jahan I. Hasan M. K. Phytomed. Plus. 2023;3:100448. doi: 10.1016/j.phyplu.2023.100448. [DOI] [Google Scholar]
- Hossain M. M. Biva J. Jahangir R. Mynol M. Vhuiyan I. Central Nervous System Depressant and Analgesic Activity of Aphanamixis Polystachya (Wall.) Parker Leaf Extract in Mice. Afr. J. Pharm. Pharmacol. 2009;3(5):282–286. [Google Scholar]
- Ganesh J. C. Venkatesha V. A. Int. J. Radiat. Biol. 2006;82:197–209. doi: 10.1080/09553000600621413. [DOI] [PubMed] [Google Scholar]
- Xu H. Jia Y. Li J. Huang X. Jiang L. Xiang T. Xie Y. Yang X. Liu T. Xiang Z. Sheng J. Biomed. Pharmacother. 2022;149:112902. doi: 10.1016/j.biopha.2022.112902. [DOI] [PubMed] [Google Scholar]
- Chen G. Liu C. Zhang M. Wang X. Xu Y. Int. J. Immunopathol. Pharmacol. 2022;36:039463202211330. doi: 10.1177/03946320221133017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reegan A. D. Stalin A. Paulraj M. G. Balakrishna K. Ignacimuthu S. Al-Dhabi N. A. Med. Chem. Res. 2016;25:1411–1419. doi: 10.1007/s00044-016-1579-x. [DOI] [Google Scholar]
- Reegan A. D. Gandhi M. R. Paulraj M. G. Balakrishna K. Ignacimuthu S. Acta Trop. 2014;139:67–76. doi: 10.1016/j.actatropica.2014.07.002. [DOI] [PubMed] [Google Scholar]
- Hong Z. L. Xiong J. Wu S. B. Zhu J. J. Hong J. L. Zhao Y. Xia G. Hu J. F. Phytochemistry. 2013;86:159–167. doi: 10.1016/j.phytochem.2012.10.008. [DOI] [PubMed] [Google Scholar]
- Yan C. Zhang Y. D. Wang X. H. Geng S. D. Wang T. Y. Sun M. Liang W. Zhang W. Q. Zhang X. D. Luo H. Fitoterapia. 2016;113:132–138. doi: 10.1016/j.fitote.2016.07.020. [DOI] [PubMed] [Google Scholar]
- Zhao W. Y. Chen J. J. Zou C. X. Zhang Y. Y. Yao G. D. Wang X. B. Huang X. X. Lin B. Song S. J. Bioorg. Chem. 2019;84:309–318. doi: 10.1016/j.bioorg.2018.11.049. [DOI] [PubMed] [Google Scholar]
- Miyake K. Tezuka Y. Awale S. Li F. Kadota S. Nat. Prod. Commun. 2010;5:1934578X1000500. [PubMed] [Google Scholar]
- Su R. Kim M. Kawaguchi H. Yamamoto T. Goto K. Taga T. Miwa Y. Kozuka M. Takahashi S. Triterpenoids from the Fruits of Phellodendron Chinese SCHNEID.: The Stereostructure of Niloticin. Chem. Pharm. Bull. 1990;38(6):1616–1619. doi: 10.1248/cpb.38.1616. [DOI] [Google Scholar]
- Wang X.-N., Fan C.-Q., Yin S., Lin L.-P., Ding J. and Yue J.-M., Cytotoxic Terpenoids from Turraea Pubescens, 2008 [Google Scholar]
- Mcchesney J. D., Dou J., Sindelar R. D., Goins D. K., Walker L. A. and Rogers R. D., Tirucallane-Type Triterpenoids: Nmr and X-Ray Diffraction Analyses of 24-Epi-Piscidinol A and Piscidinol A, 1997, vol. 27 [Google Scholar]
- Jolad S. D. Hoffmann J. J. Schram H. K. Cole J. R. Constituents of Trichilia Hispida (Meliaceae). 4. Hispidols A and B, Two New Tirucallane Triterpenoids. J. Org. Chem. 1981;46:4085–4088. doi: 10.1021/jo00333a037. [DOI] [Google Scholar]
- Mahdizadeh S. J. Thomas M. Eriksson L. A. Reconstruction of the Fas-Based Death-Inducing Signaling Complex (DISC) Using a Protein-Protein Docking Meta-Approach. J. Chem. Inf. Model. 2021;61(7):3543–3558. doi: 10.1021/acs.jcim.1c00301. doi: 10.1021/acs.jcim.1c00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatal T. K. Chaturbhuj G. U. Computational Analysis of the Binding Site(s) of TNF β-TNFR1 Complex: Implications for Designing Novel Anticancer Agents. Clin. Cancer Drugs. 2018;5(2):94–104. doi: 10.2174/2212697x06666181126111445. doi: 10.2174/2212697x06666181126111445. [DOI] [Google Scholar]
- Dengler M. A. Robin A. Y. Gibson L. Li M. X. Sandow J. J. Iyer S. Webb A. I. Westphal D. Dewson G. Adams J. M. BAX Activation: Mutations Near Its Proposed Non-Canonical BH3 Binding Site Reveal Allosteric Changes Controlling Mitochondrial Association. Cell Rep. 2019;27(2):359–373.e6. doi: 10.1016/j.celrep.2019.03.040. doi: 10.1016/j.celrep.2019.03.040. [DOI] [PubMed] [Google Scholar]
- Tondar A. Sánchez-Herrero S. Bepari A. K. Bahmani A. Calvet Liñán L. Hervás-Marín D. Biomolecules. 2024;14:544. doi: 10.3390/biom14050544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W. Ramagopal U. Cheng H. Bonanno J. B. Toro R. Bhosle R. Zhan C. Almo S. C. Crystal Structure of the Complex of Human FasL and Its Decoy Receptor DcR3. Structure. 2016;24(11):2016–2023. doi: 10.1016/j.str.2016.09.009. doi: 10.1016/j.str.2016.09.009. [DOI] [PubMed] [Google Scholar]
- Nurhayati A. P. D. Rihandoko A. Fadlan A. Ghaissani S. S. Jadid N. Setiawan E. Anti-Cancer Potency by Induced Apoptosis by Molecular Docking P53, Caspase, Cyclin D1, Cytotoxicity Analysis and Phagocytosis Activity of Trisindoline 1,3 and 4. Saudi Pharm. J. 2022;30(9):1345–1359. doi: 10.1016/j.jsps.2022.06.012. doi: 10.1016/j.jsps.2022.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guang-Yu L. Fan B. Zheng Y. C. Biomed. Environ. Sci. 2010;23:371–377. doi: 10.1016/S0895-3988(10)60078-5. [DOI] [PubMed] [Google Scholar]
- Shi Y. Protein Sci. 2004;13:1979–1987. doi: 10.1110/ps.04789804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elumalai P. Gunadharini D. N. Senthilkumar K. Banudevi S. Arunkumar R. Benson C. S. Sharmila G. Arunakaran J. Toxicol. Lett. 2012;215:131–142. doi: 10.1016/j.toxlet.2012.10.008. [DOI] [PubMed] [Google Scholar]
- Hsu F. Chen W. Wu C. Chung J. Environ. Toxicol. 2020;35:1058–1069. doi: 10.1002/tox.22942. [DOI] [PubMed] [Google Scholar]
- Kar S. Palit S. Ball W. B. Das P. K. Apoptosis. 2012;17:735–747. doi: 10.1007/s10495-012-0715-4. [DOI] [PubMed] [Google Scholar]
- Crowley L. C. Marfell B. J. Waterhouse N. J. Cold Spring Harb. Protoc. 2016;2016:prot087205. doi: 10.1101/pdb.prot087205. doi: 10.1101/pdb.prot087205. [DOI] [PubMed] [Google Scholar]
- Majtnerová P. and Roušar T., An Overview of Apoptosis Assays Detecting DNA Fragmentation, Molecular Biology Reports, Springer Netherlands, October 1, 2018, pp. 1469–1478, 10.1007/s11033-018-4258-9 [DOI] [PubMed] [Google Scholar]
- Movasaghi Z. Rehman S. Rehman I. U. Raman Spectroscopy of Biological Tissues. Appl. Spectrosc. Rev. 2007:493–541. doi: 10.1080/05704920701551530. doi: 10.1080/05704920701551530. [DOI] [Google Scholar]
- Matthews H. K. Bertoli C. de Bruin R. A. M. Cell Cycle Control in Cancer. Nat. Rev. Mol. Cell Biol. 2022:74–88. doi: 10.1038/s41580-021-00404-3. doi: 10.1038/s41580-021-00404-3. [DOI] [PubMed] [Google Scholar]
- Bačević K. Lossaint G. Achour T. N. Georget V. Fisher D. Dulić V. Sci. Rep. 2017;7:13429. doi: 10.1038/s41598-017-12868-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loukil A. Cyclin A2: At the Crossroads of Cell Cycle and Cell Invasion. World J. Biol. Chem. 2015;6(4):346. doi: 10.4331/wjbc.v6.i4.346. doi: 10.4331/wjbc.v6.i4.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss B. Harrison A. Coelho P. A. Yata K. Zernicka-Goetz M. Pines J. J. Cell Biol. 2018;217:179–193. doi: 10.1083/jcb.201612147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franken N. A. P. Rodermond H. M. Stap J. Haveman J. van Bree C. Clonogenic Assay of Cells in Vitro. Nat. Protoc. 2006;1(5):2315–2319. doi: 10.1038/nprot.2006.339. doi: 10.1038/nprot.2006.339. [DOI] [PubMed] [Google Scholar]
- Li L. T. Jiang G. Chen Q. Zheng J. N. Predic Ki67 Is a Promising Molecular Target in the Diagnosis of Cancer (Review) Mol. Med. Rep. 2015:1566–1572. doi: 10.3892/mmr.2014.2914. doi: 10.3892/mmr.2014.2914. [DOI] [PubMed] [Google Scholar]
- Chandra D. Choy G. Tang D. G. J. Biol. Chem. 2007;282:31289–31301. doi: 10.1074/jbc.M702777200. [DOI] [PubMed] [Google Scholar]
- Shen Y. and White E., P53-Dependent Apoptosis Pathways, 2001 [DOI] [PubMed] [Google Scholar]
- Arya J. S. Joseph M. M. Sherin D. R. Nair J. B. Manojkumar T. K. Maiti K. K. Exploring Mitochondria-Mediated Intrinsic Apoptosis by New Phytochemical Entities: An Explicit Observation of Cytochrome c Dynamics on Lung and Melanoma Cancer Cells. J. Med. Chem. 2019;62(17):8311–8329. doi: 10.1021/acs.jmedchem.9b01098. doi: 10.1021/acs.jmedchem.9b01098. [DOI] [PubMed] [Google Scholar]
- Van Loo G. Saelens X. Van Gurp M. MacFarlane M. Martin S. J. Vandenabeele P. Cell Death Differ. 2002;9:1031–1042. doi: 10.1038/sj.cdd.4401088. [DOI] [PubMed] [Google Scholar]
- Daina A. Michielin O. Zoete V. Sci. Rep. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O. Olson A. J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E. F. Goddard T. D. Huang C. C. Couch G. S. Greenblatt D. M. Meng E. C. Ferrin T. E. UCSF Chimera – A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Bowers K. J., Chow E., Xu H., Dror R. O., Eastwood M. P., Gregersen B. A., Klepeis J. L., Kolossvary I., Moraes M. A., Sacerdoti F. D. and Salmon J. K., Proceedings of the 2006 ACM/IEEE Conference on Supercomputing, Florida, 2006 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting this article have been included as part of the ESI.†