Abstract
Background
Primary bilateral macronodular adrenal hyperplasia (PBMAH) is a rare cause of Cushing's syndrome. Individuals with PBMAH and glucose-dependent insulinotropic polypeptide (GIP)-dependent Cushing's syndrome due to ectopic expression of the GIP receptor (GIPR) typically harbor inactivating KDM1A sequence variants. Primary unilateral macronodular adrenal hyperplasia (PUMAH) with concomitant glucocorticoid and androgen excess has never been encountered or studied.
Methods
We investigated a woman with a large, heterogeneous adrenal mass and severe adrenocorticotropic hormone-independent glucocorticoid and androgen excess, a biochemical presentation typically suggestive of adrenocortical carcinoma. The patient presented during pregnancy (22nd week of gestation) and reported an 18-month history of oligomenorrhea, hirsutism, and weight gain. We undertook an exploratory study with detailed histopathological and genetic analysis of the resected adrenal mass and leukocyte DNA collected from the patient and her parents.
Results
Histopathology revealed benign macronodular adrenal hyperplasia. Imaging showed a persistently normal contralateral adrenal gland. Whole-exome sequencing of 4 representative nodules detected KDM1A germline variants, benign NM_001009999.3:c.136G > A:p.G46S, and likely pathogenic NM_001009999.3:exon6:c.865_866del:p.R289Dfs*7. Copy number variation analysis demonstrated an additional somatic loss of the KDM1A wild-type allele on chromosome 1p36.12 in all nodules. RNA sequencing of a representative nodule showed low/absent KDM1A expression and increased GIPR expression compared with 52 unilateral sporadic adenomas and 4 normal adrenal glands. Luteinizing hormone/chorionic gonadotropin receptor expression was normal. Sanger sequencing confirmed heterozygous KDM1A variants in both parents (father: p.R289Dfs*7 and mother: p.G46S) who showed no clinical features suggestive of glucocorticoid or androgen excess.
Conclusions
We investigated the first PUMAH associated with severe Cushing's syndrome and concomitant androgen excess, suggesting pathogenic mechanisms involving KDM1A.
Keywords: Cushing's syndrome, androgen excess, adrenocortical tumor, KDM1A, GIP receptor, MC2R, PBMAH, PUMAH
Significance.
Adrenal tumors, mostly benign and non-functioning, are detected in 5% of the general population. The detection of an adrenal mass with concomitant glucocorticoid and androgen excess is considered highly suggestive of adrenocortical carcinoma. Macronodular adrenal hyperplasia typically involves both adrenal glands with associated glucocorticoid excess. In an exploratory study, we investigated a previously unencountered case of primary unilateral macronodular adrenal hyperplasia (PUMAH) with concomitant glucocorticoid and androgen excess. We identified a germline KDM1A variant as previously reported in primary bilateral macronodular adrenal hyperplasia and glucose-dependent insulinotropic polypeptide-dependent Cushing's syndrome. This study expands the phenotypic spectrum of inactivating KDM1A variants by describing the first case of PUMAH with combined cortisol and androgen excess, associated with a KDM1A variant. These findings are essential considerations in the workup of patients with adrenal masses.
Introduction
Adrenocortical tumors can be benign or, more rarely, malignant and in either case can present with or without tumor-associated steroid excess. The likelihood of malignancy increases with the size of the tumor, and the detection of a large tumor with concomitant adrenocorticotropic hormone (ACTH)-independent glucocorticoid and androgen excess is considered highly suggestive of adrenocortical carcinoma (ACC).1
Primary bilateral macronodular adrenal hyperplasia (PBMAH) is the cause of endogenous Cushing's syndrome in <2% of cases.2 In clinical practice, PBMAH is usually diagnosed when investigating patients with clinically overt cortisol excess and bilateral adrenal incidentalomas. PBMAH with dysregulated cortisol secretion is most often sporadic, but can be associated with germline sequence variants most commonly in ARMC5.3 The frequency of the detection of germline ARMC5-inactivating mutations varies with the severity of cortisol excess, being higher with more severe hypercortisolism4-7 and, in the presence of a family history, as high as 80% if clearly familial and 15%-25% if apparently sporadic6-10.
In patients with PBMAH, cortisol excess can also be driven by an aberrant expression of G-protein-coupled receptors (GPCRs) in adrenal lesions.11,12 It includes stricto sensu ectopic expression of a variety of GPCRs, for example, glucose-dependent insulinotropic polypeptide receptor (GIPR; gastric inhibitory polypeptide receptor), or an overexpression of eutopic receptors, for example, arginine vasopressin receptor type 1a (AVPR1a) or luteinizing hormone/chorionic gonadotropin receptor (LHCGR).
Herein, we investigated a woman who presented during pregnancy with concomitant severe cortisol and androgen excess in the presence of a large unilateral adrenal mass with a normal appearance of the contralateral adrenal gland. Surprisingly, the excised specimen revealed PUMAH; the contralateral adrenal gland was radiologically normal. We undertook detailed histological, genetic, and functional analyses to study this previously unencountered presentation.
Case vignette
A previously healthy 33-year-old woman presented with a 3-year history of primary infertility. She had been diagnosed with polycystic ovary syndrome based on an 18-month history of irregular cycles, facial hirsutism, polycystic ovarian morphology on ultrasound, and mildly raised serum testosterone. After attempts of ovulation induction by metformin, ovarian drilling, and clomiphene, she eventually conceived following in vitro fertilization (IVF), which resulted in a singleton pregnancy.
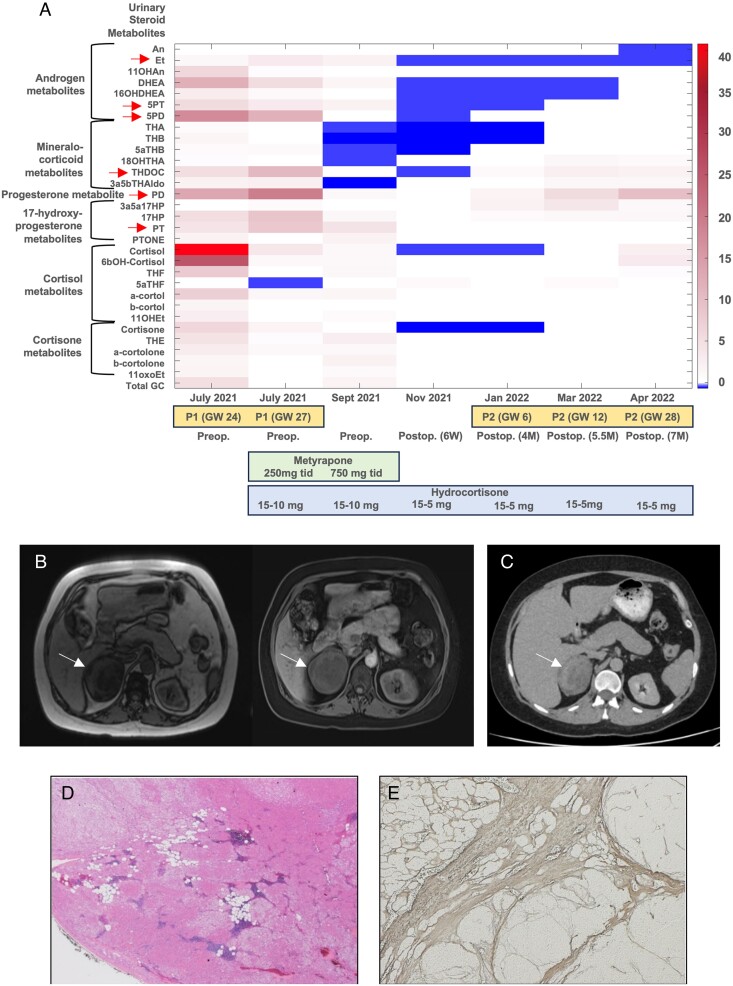
At 22-week gestation, she presented with clinically symptomatic gestational diabetes, hypertension, and hypokalemia. Florid clinical features of Cushing's syndrome, including facial plethora, thick violaceous striae, and thin bruised skin, were noticed, which retrospectively were present over the last 18 months. Random serum cortisol was 1550 nmol/L (reference 172-497), late-night salivary cortisol 18.4 nmol/L (reference <2.6), and plasma ACTH <3 ng/L (reference 0-50), consistent with ACTH-independent glucocorticoid excess. There was concomitant adrenal androgen excess with a highly increased serum DHEAS (dehydroepiandrosterone sulfate) (33.7 µmol/L; reference 2.6-13.6), while the level of serum androstenedione was in the high-to-normal range (7.0 nmol/L; reference 0.9-7.5) and that of testosterone was mildly increased (3.1 nmol/L; reference <1.9) (Table S1). Our laboratory does not provide pregnancy-specific reference ranges for serum androgens, but DHEAS is well-known to decrease significantly by mid pregnancy.13 A multi-steroid profiling analysis of 24-h urine by gas chromatography–mass spectrometry (GC–MS)14 confirmed combined glucocorticoid and androgen excess with a highly increased excretion of DHEAS, cortisol, and cortisol metabolites (Figure 1A). The urinary steroid metabolome also revealed a pattern of precursor metabolites typically observed in ACC, the so-called malignant steroid fingerprint in urine,14,15 with a highly increased excretion of the key markers pregnenediol (5PD), pregnenetriol (5PT), tetrahydro-11-deoxycorticosterone (THDOC), and pregnanetriol (PT) (Table 1, Figure 1A). Imaging showed a 7-cm right adrenal mass with signal dropout in magnetic resonance imaging out-of-phase sequences and heterogeneous appearance on unenhanced computed tomography (CT) (Figure 1B and C). Of note, no previous abdominal imaging was undertaken during the previous workup for infertility. The tumor displaced the kidney but without renal vascular involvement. No other abnormalities in the abdomen or chest were detected; the contralateral adrenal gland was of normal size and appearance.
Figure 1.
Representation of urine steroid metabolomics, imaging, and histopathological studies. A) A heatmap showing the patient's 24-h urine steroid metabolite excretion measured longitudinally by GC–MS at presentation and during further follow-up as compared to a reference range derived from 24-h urine samples collected by 73 healthy, nonpregnant women. Six urinary steroid metabolites typically found increased in patients with ACC14 are highlighted by red arrows. B) A magnetic resonance imaging (MRI) scan (T1, left; T1 with contrast, right) of the abdomen shows the right adrenal mass and normal left gland. C) A staging CT scan shows the heterogeneous right adrenal mass. No other lesions elsewhere were detected. D) and E) Coalescent hyperplastic nodules composed of foamy–clear and eosinophilic cells with foci of myelolipomatous metaplasia (D, hematoxylin–eosin 12.5×) and internodular relatively atrophic changes (E, reticulin staining 12.5×).
Table 1.
Analysis of the 24-h urine steroid metabolome in the patient at initial presentation with a large adrenal tumor at gestational week 24.
| Urinary steroid metabolites | 24-h Urinary steroid metabolite excretion in the PUMAH patient (μg/24 h) | Female Reference Range (μg/24 h) |
Range observed in 45 patients with adrenocortical carcinoma |
|---|---|---|---|
| Androsterone (An) | 767 | 859 (176-2257) | 1130 (515-2445) |
| Etiocholanolone (Et) | 4384 | 974 (255-2564) | 3671 (1171-7372) |
| 11-Hydroxyandrosterone (11OHAn) | 4357 | 328 (119-774) | 623 (346-1921) |
| Dehydroepiandrosterone (DHEA) | 24 432 | 172 (11-1972) | 612 (98-18273) |
| 16α-Hydroxy-dehydroepiandrosterone (16aDHEA) | 5126 | 289 (29-1573) | 653 (169-3168) |
| Pregnenetriol (5PT) | 2206 | 114 (30-390) | 1901 (554-7865) |
| Pregnenediol (5PD) | 8916 | 140 (44-536) | 3128 (848-14308) |
| Tetrahydro-11-dehydrocorticosterone (THA) | 213 | 76 (34-183) | 112 (56-195) |
| Tetrahydrocorticosterone (THB) | 312 | 84 (31-172) | 147 (61-380) |
| 5α-Tetrahydrocorticosterone (5aTHB) | 379 | 164 (41-335) | 155 (77-356) |
| Tetrahydro-11-deoxycorticosterone (THDOC) | 592 | 13 (5-111) | 103 (36-222) |
| 3α,5β-Tetrahydroaldosterone (3a5bTHAldo) | 129 | 30 (13-57) | 24 (11-48) |
| Pregnanediol (PD) | 16 999 | 160 (62-1286) | 839 (364-2657) |
| 3α,5α-17-Hydroxypregnanolone (3a5a17HP) | 33 | 5 (1-36) | 19 (11-44) |
| 17-Hydroxypregnanolone (17HP) | 1730 | 47 (18-331) | 511 (299-1021) |
| Pregnanetriol (PT) | 3568 | 293 (104-853) | 511 (299-1021) |
| Pregnanetriolone (PTONE) | 148 | 7 (3-63) | 32 (10-99) |
| Tetrahydro-11-deoxycortisol (THS) | 1197 | 56 (22-149) | 2151 (334-4703) |
| Cortisol (F) | 2256 | 44 (24-78) | 245 (95-701) |
| 6β-Hydroxycortisol (6bOHF) | 4633 | 103 (47-222) | 356 (187-1530) |
| Tetrahydrocortisol (THF) | 10 426 | 1035 (455-1648) | 2836 (1373-5234) |
| 5α-Tetrahydrocortisol (5aTHF) | 556 | 748 (178-1597) | 852 (450-1711) |
| α-Cortol (a-cortol) | 2286 | 201 (101-384) | 557 (279-1589) |
| β-Cortol (b-cortol) | 1393 | 301 (135-708) | 740 (316-1341) |
| 11β-Hydroxyetiocholanolone (11OHEt) | 1688 | 235 (40-606) | 366 (149-1668) |
| Cortisone (E) | 645 | 72 (37-130) | 164 (83-364) |
| Tetrahydrocortisone (THE) | 13 908 | 2100 (1035-3658) | 3701 (1909-6852) |
| α-Cortolone (a-cortolone) | 3671 | 848 (472-1459) | 1840 (868-2853) |
| β-Cortolone (b-cortolone) | 1605 | 424 (187-904) | 677 (458-1312) |
| 11-Oxoetiocholanolone (11oxoEt) | 1305 | 271 (59-626) | 484 (232-1903) |
The 7 steroids typically increased in patients with ACC14 are followed in bold and were all increased above the reference range derived from 73 healthy, nonpregnant women16 and similar to the range previously observed in 45 patients with ACC.14 Multisteroid profiling in 24-h urine was carried out by GC–MS.
The patient was urgently admitted to the antenatal ward and treated with metyrapone, labetalol, and insulin to improve fetal viability for delivery at 28-week gestation. However, fetal growth arrest and poor placental blood flow prompted an emergency cesarean section at 26 weeks. The newborn baby girl cried at birth but could not tolerate respiratory support, and neonatal death occurred 2 h after birth.
Two months after delivery, the patient underwent an uneventful laparoscopic right adrenalectomy and was discharged on oral hydrocortisone. Surprisingly, the histopathological review of the 7-cm mass revealed macronodular adrenal hyperplasia. Postoperative biochemical investigations showed low serum cortisol and androgen levels and a normalization of the urine steroid metabolome (Figure 1A, Table 1, and Table S1).
Clinical signs and symptoms of glucocorticoid excess resolved postoperatively, and regular menstrual periods resumed within 2 weeks. An unenhanced abdomen CT scan performed 2 months postoperatively showed no cause for concern and a normal left adrenal gland.
Three months post adrenalectomy, the patient spontaneously conceived. Urinary steroid excretion showed a pregnancy-typical increase in excretion of the progesterone metabolite pregnanediol (PD), as seen in the first pregnancy. However, in contrast to the first pregnancy, there was no evidence of glucocorticoid and adrenal androgen excess and no reemergence of the “malignant steroid fingerprint” in urine, with persistently normal levels of the key markers (Figure 1A). A healthy baby girl was born at 38-week gestation. An unenhanced CT scan of the abdomen undertaken 24 months post adrenalectomy continued to show a normal left adrenal gland.
Methods
We undertook detailed histopathological, genetic, and functional analysis to study this previously unencountered presentation.
Immunohistochemistry
Formalin-fixed, paraffin-embedded (FFPE) slides of lesion tissue were deparaffinized twice in xylol for 10 min, followed by rehydration in 100%, 90%, 80%, and 70% ethanol and distilled water. Nuclei were stained using Mayer's hematoxylin for 5 min and rinsed in running tap water for 5 min, and the cytoplasm was stained using eosin for 2 min, followed by a wash step in distilled water. Subsequently, the slides were washed 3 times with 100% ethanol. The slides were dried at 56 °C for 20 min and preserved in Entellan mounting medium. A microscopy analysis was performed using a Leica Aperio slide scanner (20× objective) and processed with Aperio Image Analysis software (as shown in Figure S1).
DNA and RNA isolation
Four representative adrenal nodules were carefully selected according to their macroscopic and microscopic characteristics for further investigations and named T1-T4 (as shown in Figure S2). The nodular tissue was selected and marked for DNA isolation. The high quality of the FFPE tissue was confirmed with an epithelial/stroma ratio of 9:1 and 100% vital epithelial lesion cells. For whole-exome sequencing (WES), genomic DNA was isolated from all 4 FFPE nodules using the QIAamp DNA FFPE Tissue Kit (QIAGEN) and from patient- and parents-matched leukocytes by using the Maxwell RSC Blood DNA Kit (Promega). In addition, RNA from 1 representative FFPE lesion tissue was isolated for RNA sequencing using the miRNeasy FFPE Kit (QIAGEN). DNA and RNA concentrations were determined by using a Nanodrop spectrophotometer (Thermo Fisher).
WES and RNA sequencing
For library preparation, the Twist Human Core + RefSeq + Mitochondrial Panel Kit (Twist Bioscience) was used for WES of DNA from the FFPE lesions and the matched control sample. The KAPA RNA HyperPrep Kit with RiboErase (HMR/Globin, Roche) was applied for RNA sequencing. Subsequently, sequencing analyses were performed on a NovaSeq6000 (Illumina) with a read length of 100 bp.
Data analysis for WES
An initial quality assessment of all fastq files was performed using FastQC, v0.11.9.17 Low-quality reads and adapter sequences were trimmed with TrimGalore, v0.6.4,18 powered by Cutadapt, v2.8.19
The trimmed reads were mapped to the human reference genome (hg19) using the Burrows–Wheeler Aligner, v0.7.17,20 and sorted and indexed using Picard, v2.25.0: http://broadinstitute.github.io/picard/ and SAMtools, v1.10,21 respectively. Duplicates were marked with Picard. Base recalibration was executed with GATK, v4.0.9.0. GATK, v3.8,22 was used for coverage calculations. Germline variants including substitutions and small indels were called using GATK HaplotypeCaller, v4.0.9.0. Somatic substitutions and small indels were covered by at least 20 reads, and the alternative allele was covered by at least 8 reads and comprised at least 10% of all reads. Somatic substitutions and small indels that were called by at least 2 out of 4 callers, namely GATK-Mutect2, v4.1.9.0, VarScan, v2.4.4,23 Strelka, v2.9.2,24 and Scalpel v0.5.4,25 in at least one of the nodules were visually inspected using IGV, v2.1226 in all nodules. Variants were annotated with ANNOVAR, v2019-10-24.27 Variants that alter the protein sequence or affect a splice site were considered for downstream analysis if they are rare in the population (below a frequency of 1% in the publicly available databases 1000 genomes, v2015aug_all, Exome Aggregation Consortium [ExAC, v03] without tumor samples, as well as the gnomAD exome [v2.0.1] and gnomAD_genome [v2.0.1] collection). The ACMG classification was carried out with Franklin https://franklin.genoox.com/clinical-db/home.28 Finally, we used the WES data to detect and visualize copy number variations (CNVs) with control-FREEC, v11.6,28 with a matched control to discern somatic variants from germline variants.
Data analysis for RNA sequencing (transcriptome)
Analysis of the transcriptome sequencing data was performed as we described previously.29 Additional samples from tissue of 52 adrenocortical adenomas (ACAs) and 4 normal adrenal glands (NAGs) from our previous study (deposited at EGAS00001004533) were used for comparison of the expression profile. These included 26 cortisol-producing adenomas associated with overt Cushing's syndrome, 17 cortisol-producing adenomas associated with mild autonomous cortisol secretion, and 9 endocrine-inactive adenomas.
Sanger sequencing
Primers used to amplify the regions of interest of KDM1A and MC2R genes are listed in Table S2. For the polymerase chain reaction (PCR), 50 ng of genomic DNA in a final volume of 20 µL containing 2.5 mM of MgCl2, 0.2 µM of each primer, and 200 µM of dNTPs (my-Budget 5× PCR master mix, BioBudget) was used for 30 cycles.
For KDM1A frameshift and MC2R p.M255I, denaturation at 94 °C for 20 sec, annealing at 58 °C for 30 sec, and elongation at 72 °C for 30 sec were performed. For KDM1A p.G46S, an annealing temperature of 62 °C was used. One microlitre of the PCR product was applied in an additional nested PCR using primer pair KDM1A_GS_F2 und KDM1A_GS_R.
Direct sequencing of the PCR products was performed using the QuickStart Cycle Sequencing Kit (ABSciex) on a CEQ8000 DNA Analyzer (ABSciex). PCR primers were used for sequencing. Sequencing results were analyzed with the GenomeLab Genetic Analysis System (ABSciex).
Urinary steroid metabolome profiling by GC–MS
The urinary steroid metabolome of the patient was examined at her first presentation to us mid-pregnancy and then again during follow-up prior to and following delivery and subsequent adrenalectomy and again during the subsequent complication-free second pregnancy. Steroid profiling was undertaken by GC–MS as previously described.14
Functional validation of MC2R variant
Human embryonic kidney (HEK293) cells (ATCC® CRL-1573™) were reverse-transfected using polyethyleneimine (Polysciences) with the pGLO-22f biosensor (Promega) as well as 3xHA wild-type (WT) MC2R (cDNA) or 3Xha M255I (GenScript) and MRAP-3xFlag and plated in white-walled, white-bottom 96-well plates (Corning) coated with poly-D-lysine (Merck). Forty-eight hours post-transfection, the media were aspirated and the cells were washed with assay buffer: 1× Hank's Balanced Salt Solution (HBSS), 24 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 0.1% (w/v) bovine serum albumin (BSA), 3.96 mM NaHCO3, 1 mM MgSO4, and 1.3 mM CaCl2, and replaced with 90 μL of an identical buffer supplemented with 0.45 mg/mL of firefly D-luciferin (Nanolight Technology) and equilibrated for 1 h at 28 °C. Bioluminescence was measured on the BMG CLARIOstar Plus plate reader (BMG LabTech). Six basal readings were taken before the addition of ligand; an average of these basal reads was used to calculate the fold increase before treatment for data analysis. Readings were taken for ∼30 min, with a 1-sec integration time and no lens. Data were calculated as fold change from the average basal values, the area under the curve for the basal normalized values was taken and plotted as a concentration–response curve, and the percentage was referenced to the non-treated (0%) and maximal concentration of ACTH[1-24] (1μM). Data were fit to a 4-parameter logistic fit in GraphPad prism.
Ethics
The study conducted complied with the Declaration of Helsinki. Ethical approval was obtained from the local ethics committee (PrimeAct study IRAS 261291). Written informed consent for data and sample collection, genetic analysis, and publication was obtained from the patient and her parents.
Results
Histopathology
The pathological examination showed a 7-cm right nodular adrenal mass with multiple nodules larger than 1 cm, predominantly composed of foamy–clear cells with multifocal myelolipomatous metaplasia. It revealed several coalescent non-pigmented nodules, relative internodular atrophy, and lack of necrosis, nuclear atypia, atypical mitotic figures, or vascular invasion. Without a histologically normal cortex, these findings support macronodular cortical hyperplasia and fulfill the criteria for adrenal cortical nodular hyperplasia.30,31 The combined macroscopic and microscopic appearance defined tumor-like macronodular adrenal hyperplasia30-32 (Figure 1D and E and Figure S1).
Routine genetic analysis
Targeted next-generation sequencing (Twist custom capture v8/Illumina NextSeq500) of the patient's germline DNA was carried out by the Birmingham Women's Hospital Department of Clinical Genetics. The routine, nationally predefined panel (National Genomic Test Directory unique code R160, version 6, January 2024) available for patients with nodular adrenal hyperplasia includes ARMC5, PDE11A, PDE8B, and PRKAR1A; no pathogenic variant was detected in any of those genes.
High-throughput molecular analysis
Overview of WES results
Tissue DNA was isolated from FFPE material from 4 representative adrenal nodules (T1-T4, Figure S2). CNV patterns were similar in all evaluated nodules, suggesting that they are clonally related (Figure S3). We detected 2 non-synonymous somatic variants shared in all tumors, 1 somatic mutation present in T1 only and 1 somatic mutation shared in T2 and T4. None of these variants showed a possible, probable, or likely pathogenic role. Table S3 provides the list of the detected somatic variants.
WES analysis revealed rare potentially pathogenic germline variants
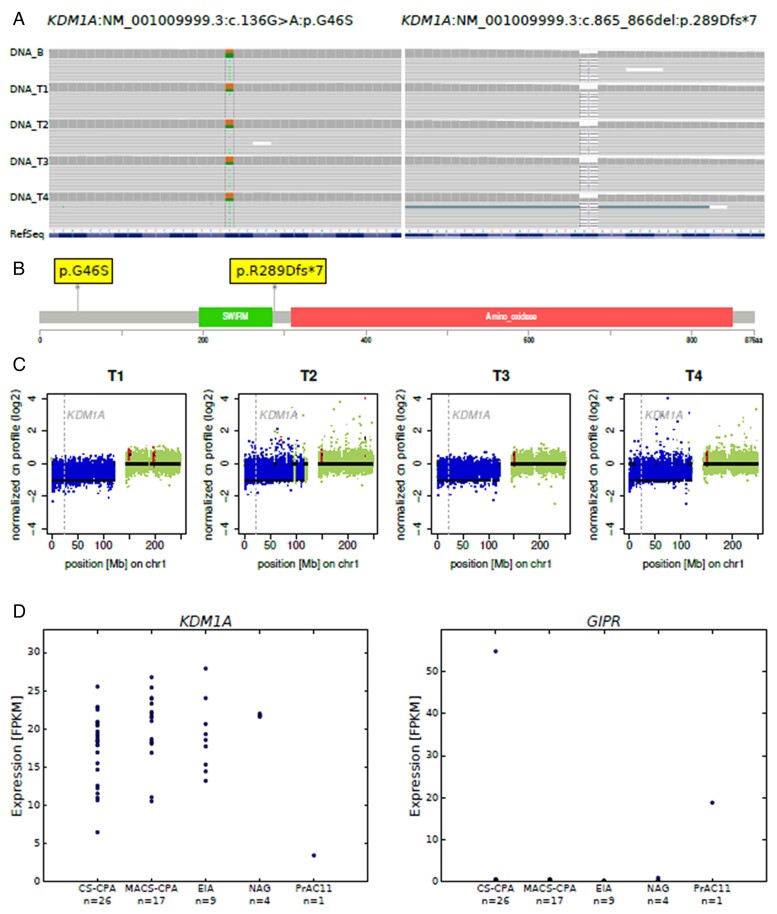
We detected several germline variants known to ClinVar, the majority with no suspected links with the patient's phenotype. However, germline variants in 2 genes were selected for further investigations due to their potential role as candidate drivers. First, we observed 2 relevant variants in the gene encoding for KDM1A (lysine-specific histone demethylase 1A). These were 1 missense variant in exon 1 (NM_001009999.3:c.136G>A:p.G46S [55.4%], conflicting pathogenicity in ClinVar, benign according to ACMG guidelines) and 1 truncating (frameshift) variant in exon 6 (KDM1A:NM_001009999.3:exon6:c.865_866del:p.R289Dfs*7 [45.5%], unknown to ClinVar, likely pathogenic according to ACMG guidelines), which causes a premature stop codon (Figure S4). They were observed in all 4 representative nodules and reference DNA. The location at the cDNA and protein level of both variants is shown in Figure 2A and B. Inactivating germline mutations in KDM1A associated with loss of heterozygosity at locus 1p have been recently described in nearly 90% of PBMAH associated with ectopic expression of the glucose-dependent insulinotropic polypeptide (GIP; gastric inhibitory polypeptide) receptor, leading to food-dependent Cushing’s syndrome.33,34 Therefore, we investigated this further and interestingly observed an additional somatic loss of the KDM1A gene locus 1p36.12 in the WT allele in all 4 nodules T1-T4 (Figure 2C).
Figure 2.
KDM1A alterations at genetic and transcriptomic analyses of representative adrenal nodules. A) An IGV visualization of the KDM1A:NM_001009999.3:c.136G>A:p.G46S and KDM1A:NM_001009999.3:exon6:c.865_866del:p.R289Dfs*7 germline variants in blood (DNA_B) and all 4 nodules (DNA_T1-T4). B) A lollipop plot showing the location of the KDM1A germline mutations on a linear KDM1A protein and its domains. C) A visualization of the log2-transformed normalized copy number (cn) profile for all nodules on chromosome1. The normal copy number status is depicted in green; copy number loss is depicted in blue. The position of the gene KDM1A is marked by the dashed gray line. D) An investigation of KDM1A and GIPR expression by transcriptome sequencing in 26 CS-cortisol-producing adenoma (CS-CPAs), 17 adenomas with mild autonomous cortisol secretion (MACS-CPAs), 9 endocrine-inactive adenomas (EIAs), 4 NAGs, and nodule 4 of the patient (PrAC11), respectively.
Moreover, we detected a germline missense variant in exon 2 of MC2R, the gene encoding for the ACTH receptor (NM_000529.2:c.765G>A:p.M255I [51.5%], uncertain in ClinVar, variant of unknown significance according to ACMG guidelines; Figure S4A and B and Table S3).
Germline variants validated by Sanger sequencing
We further confirmed the patient's germline variants in KDM1A and MC2R using Sanger sequencing. Additionally, we investigated leukocyte DNA from the patient's parents to search for germline alterations in the selected genes. We detected the germline KDM1A p.R289Dfs*7 variant in the father and the KDM1A p.G46S and MC2R p.M255I variants in the mother.
RNA-sequencing analysis implicates KDM1A as a potential driver gene
We performed RNA-sequencing (RNA-seq) analysis on 1 representative adrenal nodule. We compared the transcriptome result with a dataset available from a previous publication on adrenocortical tumors composed of 52 ACAs and 4 NAGs.29
Genetic inactivation of KDM1A in PBMAH associated with GIP-dependent Cushing's syndrome has been previously linked to loss of KDM1A and ectopic expression of GIPR, at both mRNA and protein levels.33,34 Importantly, we demonstrated deficient mRNA expression of KDM1A and high expression of GIPR in our patient compared with our dataset of ACAs and NAGs (Figure 2D). These results suggest a similar 2-hit pathogenic mechanism as described in PBMAH associated with the GIP-dependent Cushing's syndrome.
Only 1 previously collected sample from a cortisol-producing adenoma with severe Cushing's syndrome showed higher GIPR expression levels than our patient. This was from a 35-year-old woman from a published cohort,35 previously classified as showing no known driver genes at WES. We re-analyzed the genomic profile of that patient (variant calling in RNA-seq dataset) and found a frameshift variant in exon 21 of KDM1A (NM_001009999.3:c.2532_2538del:p.A846Efs*35 [85.1%]), which had not been detected at initial WES analysis.
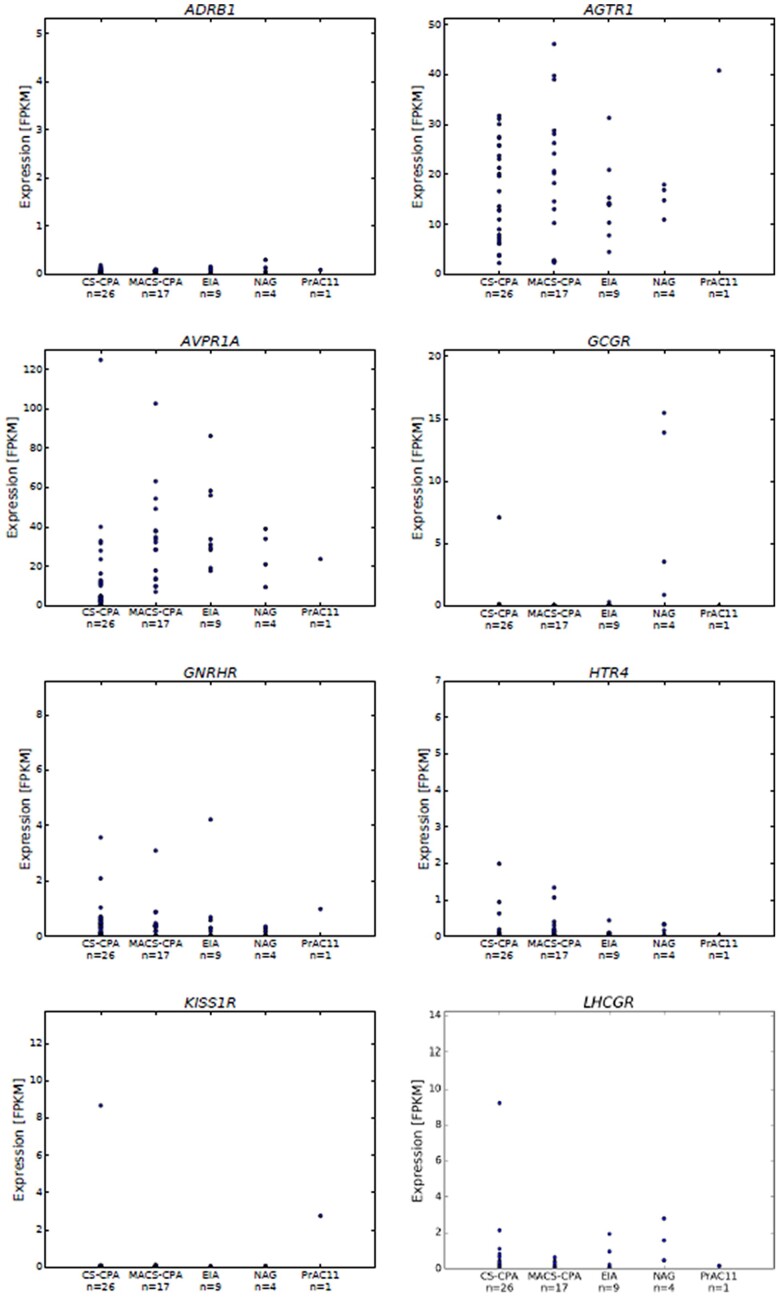
We also looked at mRNA expression of other genes potentially involved in PBMAH associated with aberrant expression of GPCRs. For most of these GPCRs, the patient's T4 sample showed expression levels similar to the ACAs, including ectopic receptors ADRB1, GNRHR or eutopic receptors AVPR1a, AGTR1, LHCGR, and HTR4, and GCGR (Figure 3). However, we found high mRNA expression of kisspeptin receptor (KISS1R) in 2 samples: the patient presented in this report and the above-described patient with a cortisol-producing adenoma associated with severe CS and high GIPR expression.35
Figure 3.
RNA-seq (transcriptome) data showing mRNA expression levels of selected relevant genes (ADRB1, AGTR1, AVPR1a, GCGR, GNRHR, HTR4, KISS1R, and LHCGR) in one of the representative nodules (T4) and an available dataset from ACAs, including 43 cortisol-producing adenomas (CPAs, 26 associated with overt Cushing’s syndrome, CS-CPAs, and 17 with mild autonomous cortisol secretion, MACS-CPAs), 9 endocrine-inactive adenomas (EIAs), and 4 NAGs.29
Functional analysis of the MC2R variant
We undertook functional experiments to test the potential biological role of the newly described germline MC2R p.M255I variant. We employed a kinetic cAMP accumulation assay using HEK293 cells transiently transfected with WT MC2R or the M255I mutation, MRAP, and the pGLO-22f cAMP biosensor. These data indicated that while the variant did not have a significantly different pEC50 in response to ACTH[1-24], it had a considerably higher maximal concentration–response (Figure S5A and B).
We also looked at the MC2R expression in our RNA-seq dataset and observed similar expression in the tumor sample from our patient compared with a subgroup of adenomas with different hormonal patterns (Figure S4C).
Evaluation of GIP-dependent Cushing's syndrome
Considering the post-adrenalectomy genetic findings, we assessed the potential presence of GIP-dependent Cushing's syndrome. Preoperatively, the patient had a profound phenotype of Cushing's syndrome with onset of symptoms 18 months prior to the IVF pregnancy. There was no suggestion that symptoms were food dependent or related to pregnancy.
The patient has 3 siblings: 40-year-old brother and 37- and 31-year-old sisters, all of whom had no symptoms or signs of glucocorticoid or androgen excess. A detailed family history assessment did not identify disorders that have been associated with KDM1A mutations, such as multiple myeloma, monoclonal gammopathy of uncertain significance, or other tumors.36-41 Clinical assessment of her parents (father 70 years old and mother 69 years old) did not reveal features of glucocorticoid or androgen excess.
The patient was not assessed for GIP-dependent Cushing's syndrome during the first pregnancy as this was not clinically suspected and the clinical course was highly suggestive of ACC. However, we were able to undertake an assessment after the surgical removal of the adrenal mass, measuring serum cortisol and plasma ACTH before and after an oral challenge with 75 g glucose; hydrocortisone was paused for 24 h prior to the test. Fasting serum cortisol was 243 nmol/L (reference 172-497) with ACTH 99.0 ng/L (reference 0-50) and glucose 4.1 mmol/L (reference 3.5-11). At 2 h after the administration of oral glucose, cortisol was 183 nmol/L (reference 172-497) with ACTH 64.0 ng/L (reference 0-50) and glucose 6.1 mmol/L (reference 3.5-11), excluding GIP-dependent cortisol secretion by the contralateral adrenal gland but does not rule out the presence of GIP-dependent cortisol excess before the removal of the adrenal mass. In patients with GIP-dependent Cushing's syndrome, fasting cortisol is normal or even low from the hypothalamic pituitary–adrenal axis suppression by the postprandial hypercortisolemia.42
Importantly, the adrenal insufficiency persists, and the patient remains dependent on glucocorticoid replacement (hydrocortisone 20 mg/day in divided doses). ACTH stimulation test undertaken 24 months post-adrenalectomy showed 0-min cortisol 228 nmol/L with ACTH 68 ng/L (reference 0-50) and 30-min post-ACTH stimulation cortisol 312 nmol/L (an adequate response is >450 nmol/L).
Discussion
We investigated the first case of PUMAH; uniquely, the patient presented with combined cortisol and androgen excess. Through comprehensive genetic and functional analysis, we showed the presence of germline alterations in KDM1A—associated with increased GIPR expression—representing a potential pathogenic driver. KDM1A variants had previously been described in patients presenting PBMAH with food-dependent Cushing's syndrome; none of them had shown evidence of androgen excess.
Androgen excess and even more so concomitant androgen and cortisol excess in the presence of an adrenal mass are considered highly suggestive for ACC.1 Only 2 previous reports describe combined cortisol and androgen excess in the context of a benign adrenal entity, 1 describing a case of PBMAH43 and 1 in a patient with a unilateral adrenal adenoma.44 In 2 further cases, isolated adrenal androgen excess was described in the context of PBMAH.45,46 These cases were studied prior to the identification of the role of KDM1A inactivation in this context, and therefore, whether they harbored KDM1A variants is not known.
In our case, we uniquely observed combined cortisol and androgen excess in the context of unilateral macronodular adrenal hyperplasia, with persistently normal appearance of the contralateral adrenal gland. The hormonal pattern, severity of the clinical presentation, and large size of the adrenal lesion led us to initially suspect underlying ACC, with a heterogeneous appearance on unenhanced CT indicative of an indeterminate adrenal mass.
Although when faced with such a presentation the clinician should always consider ACC as the underlying entity until proven otherwise, this work highlights that sizeable unilateral masses with concomitant glucocorticoid and androgen secretion are not always malignant. Furthermore, distinguishing hyperplastic and neoplastic processes can be challenging in selected cases. The critical pathological finding in our case was the tumor-like presentation of multiple macronodular cortical hyperplasia with the contralateral adrenal gland being radiologically normal. It is possible that in our patient the second somatic 1p deletion occurred in 1 adrenal, but not in the second adrenal gland, suggesting that a long-term monitoring is required to check for eventual asynchronous development of contralateral nodular hyperplasia and associated cortisol excess. Some patients with PBMAH may present with unilateral lesions,8,47 which, however, then invariably progress to clear evidence of bilateral adrenal involvement. In our case, the contralateral adrenal remained radiologically entirely normal 24 months after adrenalectomy. However, long-term follow-up is warranted.
In patients with PBMAH, clinical investigations have frequently shown abnormal cortisol secretion because of illegitimate membrane receptor expression.12 Among these receptors, ectopic adrenal GIPR expression is best described in patients with PBMAH with food-dependent Cushing's syndrome (hence now better termed GIP-dependent Cushing's syndrome). N’Diaye et al.48 reported asynchronous development of GIP-dependent macronodular adrenal hyperplasia with a cortisol increase demonstrated after an oral glucose load following resection of the adrenal mass.42 However, in contrast to our case, the contralateral adrenal harbored a small (<1 cm) unresected nodule and there was no evidence of concomitant androgen excess. In our case, we excluded GIP-dependent cortisol secretion from the contralateral adrenal gland but were not able to assess this during the first pregnancy and prior to the removal of the adrenal mass.
The likely pathogenic KDM1A frameshift germline variant (p.R289Dfs*) identified in our patient and the concomitant somatic loss of 1p36 in all nodules studied are in line with 2-hit inactivation of KDM1A, consistent with the tumor suppressor gene model of tumorigenesis. Recent studies identified the inactivation of the KDM1A gene as responsible for ectopic adrenal GIPR expression in virtually all GIP-dependent PBMAH with Cushing's syndrome.33,34 In the context of PBMAH, KDM1A acts as a tumor suppressor gene with a germline pathogenic variant associated with somatic loss of heterozygosity. The likely pathogenic KDM1A variant, p.R289Dfs*7, was also detected in the father but without features of Cushing's syndrome. Noteworthy, there was clear sexual dimorphism in the development of GIP-related Cushing's syndrome reported by Vaczlavik et al.,34 Chasseloup et al.,33 and the literature42 overall with women representing the vast majority of cases.
Undertaking transcriptome analysis, we observed an almost absent expression of KDM1A, while GIPR expression was highly increased at mRNA level but without somatic or germline variants in this gene. This profile resembles the molecular pattern reported in GIP-dependent Cushing's syndrome and corroborates a pathogenic role of these changes that have not been reported outside the context of PBMAH prior to our case.
It is recognized that in some adrenal lesions, excessive steroid production may be regulated by various aberrant receptors.11 Of note, despite the potential aggravation of the clinical phenotype during pregnancy reported in our case, we could not see any increase in LHCGR gene expression in the investigated tumor tissue. However, we observed an increased mRNA expression of KISS1R; kisspeptin has previously been shown to stimulate adrenal androgen release by the NCI-H295R ACC cell line and human fetal adrenal cells.49 Moreover, KISS1R overexpression has been observed in PBMAH associated with GIP-dependent Cushing's syndrome33 and in a patient presenting with combined adrenal cortisol and aldosterone excess.50 Interestingly, a recent paper found significantly higher circulating kisspeptin levels in pregnancies affected by fetal growth restriction;51 it is tempting to speculate whether increased kisspeptin signaling in the case of our patient aggravated the fetal growth restriction; however, it is safe to assume that cortisol excess represented the major pathogenic factor.
Finally, we also observed a novel germline M225I variant in the MC2R gene. A gain-of-function MC2R mutation (F278C) was previously identified in a single case of ACTH-independent Cushing's syndrome.52 We provide in vitro evidence that the M255I variant has an increased signal potential only at maximal (supraphysiological) concentration–response. Moreover, the mRNA expression level of MC2R in the hyperplastic adrenal tissue appears low. Therefore, considering the clinical context, that is, unilateral adrenal disease and ACTH-independent steroid excess, we are not sufficiently convinced that this variant significantly contributed to the phenotype.
A limitation of this work is the data derivation from a single patient with PUMAH. A major strength is the extensive experimental approach studying a rare case and providing novel insights as well as expanding the clinical spectrum associated with inactivating KDM1A mutations.
In conclusion, we investigated the first case of a patient with primary unilateral macronodular adrenocortical hyperplasia, PUMAH, associated with cortisol and androgen co-secretion and suggest pathogenic mechanisms involving KDM1A.
Supplementary Material
Acknowledgments
The authors thank the EU COST Action CA20122 Harmonisation for supportive networking (www.goharmonisation.com).
Contributor Information
Yasir S Elhassan, Institute of Metabolism and Systems Research, University of Birmingham, Birmingham, United Kingdom; Centre for Endocrinology, Diabetes and Metabolism, University Hospitals Birmingham NHS Foundation Trust, Birmingham Health Partners, Birmingham, United Kingdom.
Silke Appenzeller, Comprehensive Cancer Center Mainfranken, University of Würzburg, Würzburg, Germany.
Laura-Sophie Landwehr, Division of Endocrinology and Diabetes, Department of Internal Medicine I, University Hospital, University of Würzburg, Würzburg, Germany.
Juliane Lippert, Division of Endocrinology and Diabetes, Department of Internal Medicine I, University Hospital, University of Würzburg, Würzburg, Germany.
Dillon Popat, Faculty of Medicine and Dentistry, Centre for Endocrinology, William Harvey Research Institute, Queen Mary University of London, London, United Kingdom.
Lorna C Gilligan, Institute of Metabolism and Systems Research, University of Birmingham, Birmingham, United Kingdom.
Lida Abdi, MRC Laboratory of Medical Sciences, London, United Kingdom.
Edwina Goh, Birmingham Women's Hospital, Birmingham, United Kingdom.
Salvador Diaz-Cano, Department of Pathology, University Hospitals Birmingham NHS Foundation Trust, Birmingham, United Kingdom.
Stefan Kircher, Department of Pathology, University of Würzburg, Würzburg, Germany.
Susanne Gramlich, Department of Pathology, University of Würzburg, Würzburg, Germany.
Robert P Sutcliffe, The Liver Unit, Queen Elizabeth Hospital Birmingham, University Hospitals Birmingham NHS Foundation Trust, Birmingham, United Kingdom.
Shakila Thangaratinam, Institute of Metabolism and Systems Research, University of Birmingham, Birmingham, United Kingdom; Birmingham Women's Hospital, Birmingham, United Kingdom; NIHR Birmingham Biomedical Research Centre, University Hospitals Birmingham NHS Foundation Trust and University of Birmingham, Birmingham, United Kingdom.
Li F Chan, Faculty of Medicine and Dentistry, Centre for Endocrinology, William Harvey Research Institute, Queen Mary University of London, London, United Kingdom.
Martin Fassnacht, Comprehensive Cancer Center Mainfranken, University of Würzburg, Würzburg, Germany; Division of Endocrinology and Diabetes, Department of Internal Medicine I, University Hospital, University of Würzburg, Würzburg, Germany.
Wiebke Arlt, Institute of Metabolism and Systems Research, University of Birmingham, Birmingham, United Kingdom; MRC Laboratory of Medical Sciences, London, United Kingdom; Faculty of Medicine, Institute of Clinical Sciences, Imperial College London, London, United Kingdom.
Cristina L Ronchi, Institute of Metabolism and Systems Research, University of Birmingham, Birmingham, United Kingdom; Centre for Endocrinology, Diabetes and Metabolism, University Hospitals Birmingham NHS Foundation Trust, Birmingham Health Partners, Birmingham, United Kingdom.
Supplementary material
Supplementary material is available at European Journal of Endocrinology online.
Funding
Y.S.E. received support from the COST Action CA20122 Harmonisation. W.A. received funding from the Wellcome Trust (investigator award WT209492/Z/17/Z) and the Medical Research Council (MRC) (program grant MC_UP_1605/15). C.L.R. and M.F. received support from the Deutsche Forschungsgemeinschaft (grant 314061271—CRC/TRR 205). C.L.R. also received support from the AMEND ACC Research Fund 2022 and a research grant funded by HRA Pharma Rare Disease. L.F.C. received funding from the British Society of Paediatric Endocrinology and Diabetes, Barts Charity (MGU0458, G-002162), MRC/Academy of Medical Sciences Fellowship Grant G0802796, BBSRC BB/W018276/1, and MRC Collaborative Awards in Science and Engineering Studentship Grants MR/R015686/2442100.
Authors’ contributions
Yasir S. Elhassan (Conceptualization [equal], Data curation [equal], Investigation [equal], Writing—original draft [lead], Writing—review and editing [lead]), Silke Appenzeller (Data curation [equal], Formal analysis [equal]), Laura-Sophie Landwehr (Data curation [equal], Formal analysis [equal]), Juliane Lippert (Data curation [equal], Formal analysis [equal]), Dillon Popat (Data curation [equal], Formal analysis [equal]), Lorna C. Gilligan (Data curation [equal], Formal analysis [equal]), Lida Abdi (Data curation [equal], Formal analysis [equal]), Edwina Goh (Investigation [equal]), Salvador Diaz-Cano (Formal analysis [equal], Investigation [equal]), Stefan Kircher (Data curation [equal], Formal analysis [equal]), Susanne Gramlich (Data curation [equal]), Robert Sutcliffe (Investigation [equal]), Shakila Thangaratinam (Investigation [equal]), Li Chan (Formal analysis [equal]), Martin Fassnacht (Data curation [equal], Formal analysis [equal], Investigation [equal], Methodology [equal]), Wiebke Arlt (Conceptualization [equal], Data curation [equal], Formal analysis [equal], Methodology [equal], Writing—review and editing [equal]), and Cristina Ronchi (Conceptualization [equal], Data curation [equal], Formal analysis [equal], Investigation [equal], and Writing—review and editing [equal]).
Data availability
The datasets generated during the current study will be available in European Genome-phenome Archive.
References
- 1. Fassnacht M, Tsagarakis S, Terzolo M, et al. European Society of Endocrinology clinical practice guidelines on the management of adrenal incidentalomas, in collaboration with the European Network for the Study of Adrenal Tumors. Eur J Endocrinol. 2023;189(1):G1–G42. 10.1093/ejendo/lvad066 [DOI] [PubMed] [Google Scholar]
- 2. Bertherat J, Bourdeau I, Bouys L, Chasseloup F, Kamenický P, Lacroix A. Clinical, pathophysiologic, genetic, and therapeutic progress in primary bilateral macronodular adrenal hyperplasia. Endocr Rev. 2022;44(4):567–628. 10.1210/endrev/bnac034 [DOI] [PubMed] [Google Scholar]
- 3. Assié G, Libé R, Espiard S, et al. ARMC5 mutations in macronodular adrenal hyperplasia with Cushing's syndrome. N Engl J Med. 2013;369(22):2105–2114. 10.1056/NEJMoa1304603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Faucz FR, Zilbermint M, Lodish MB, et al. Macronodular adrenal hyperplasia due to mutations in an armadillo repeat containing 5 (ARMC5) gene: a clinical and genetic investigation. J Clin Endocrinol Metab. 2014;99(6):E1113–E1119. 10.1210/jc.2013-4280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Espiard S, Drougat L, Libé R, et al. ARMC5 mutations in a large cohort of primary macronodular adrenal hyperplasia: clinical and functional consequences. J Clin Endocrinol Metab. 2015;100(6):E926–E935. 10.1210/jc.2014-4204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bouys L, Vaczlavik A, Jouinot A, et al. Identification of predictive criteria for pathogenic variants of primary bilateral macronodular adrenal hyperplasia (PBMAH) gene ARMC5 in 352 unselected patients. Eur J Endocrinol. 2022;187(1):123–134. 10.1530/EJE-21-1032 [DOI] [PubMed] [Google Scholar]
- 7. Yu L, Zhang J, Guo X, Chen X, He Z, He Q. ARMC5 mutations in familial and sporadic primary bilateral macronodular adrenal hyperplasia. PLoS One. 2018;13(1):e0191602. 10.1371/journal.pone.0191602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Alencar GA, Lerario AM, Nishi MY, et al. ARMC5 mutations are a frequent cause of primary macronodular adrenal hyperplasia. J Clin Endocrinol Metab. 2014;99(8):E1501–E1509. 10.1210/jc.2013-4237 [DOI] [PubMed] [Google Scholar]
- 9. Bourdeau I, Oble S, Magne F, et al. ARMC5 mutations in a large French-Canadian family with cortisol-secreting β-adrenergic/vasopressin responsive bilateral macronodular adrenal hyperplasia. Eur J Endocrinol. 2016;174(1):85–96. 10.1530/EJE-15-0642 [DOI] [PubMed] [Google Scholar]
- 10. Gagliardi L, Schreiber AW, Hahn CN, et al. ARMC5 mutations are common in familial bilateral macronodular adrenal hyperplasia. J Clin Endocrinol Metab. 2014;99(9):E1784–E1792. 10.1210/jc.2014-1265 [DOI] [PubMed] [Google Scholar]
- 11. Kamenický P, Lacroix A. Mechanism of ectopic hormone receptors in adrenal tumors and hyperplasia. Curr Opin Endocr Metab Res. 2019;8(August):206–212. 10.1016/j.coemr.2019.08.012 [DOI] [Google Scholar]
- 12. Libé R, Coste J, Guignat L, et al. Aberrant cortisol regulations in bilateral macronodular adrenal hyperplasia: a frequent finding in a prospective study of 32 patients with overt or subclinical Cushing's syndrome. Eur J Endocrinol. 2010;163(1):129–138. 10.1530/EJE-10-0195 [DOI] [PubMed] [Google Scholar]
- 13. Makieva S, Saunders PTK, Norman JE. Androgens in pregnancy: roles in parturition. Hum Reprod Update. 2014;20(4):542–559. 10.1093/humupd/dmu008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Arlt W, Biehl M, Taylor AE, et al. Urine steroid metabolomics as a biomarker tool for detecting malignancy in adrenal tumors. J Clin Endocrinol Metab. 2011;96(12):3775–3784. 10.1210/jc.2011-1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bancos I, Taylor AE, Chortis V, et al. Urine steroid metabolomics for the differential diagnosis of adrenal incidentalomas in the EURINE-ACT study: a prospective test validation study. Lancet Diabetes Endocrinol. 2020;8(9):773–781. 10.1016/S2213-8587(20)30218-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Arlt W, Lang K, Sitch AJ, et al. Steroid metabolome analysis reveals prevalent glucocorticoid excess in primary aldosteronism. JCI Insight. 2017;2(8):e93136. 10.1172/jci.insight.93136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Andrews S. FastQC. Babraham Institute; 2010. Accessed March 15, 2022. http://www.bioinformatics.babraham.ac.uk/projects/fastqc
- 18. Krueger F. Trim Galore. Babraham Institute; 2012. Accessed March 15, 2022. http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/
- 19. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17(1):10–12. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- 20. Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li H, Handsaker B, Wysoker A, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McKenna A, Hanna M, Banks E, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Koboldt DC, Zhang Q, Larson DE, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22(3):568–576. 10.1101/gr.129684.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim S, Scheffler K, Halpern AL, et al. Strelka2: fast and accurate calling of germline and somatic variants. Nat Methods. 2018;15(8):591–594. 10.1038/s41592-018-0051-x [DOI] [PubMed] [Google Scholar]
- 25. Fang H, Bergmann EA, Arora K, et al. Indel variant analysis of short-read sequencing data with Scalpel. Nat Protoc. 2016;11(12):2529–2548. 10.1038/nprot.2016.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14(2):178–192. 10.1093/bib/bbs017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164–e164. 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Boeva V, Popova T, Bleakley K, et al. Control-FREEC: a tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics. 2012;28(3):423–425. 10.1093/bioinformatics/btr670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Di Dalmazi G, Altieri B, Scholz C, et al. RNA sequencing and somatic mutation status of adrenocortical tumors: novel pathogenetic insights. J Clin Endocrinol Metab. 2020;105(12):e4459–e4473. 10.1210/clinem/dgaa616 [DOI] [PubMed] [Google Scholar]
- 30. Blanes A, Diaz-Cano SJ. DNA and kinetic heterogeneity during the clonal evolution of adrenocortical proliferative lesions. Hum Pathol. 2006;37(10):1295–1303. 10.1016/j.humpath.2006.04.025 [DOI] [PubMed] [Google Scholar]
- 31. Blanes A, Diaz-Cano SJ. Histologic criteria for adrenocortical proliferative lesions: value of mitotic figure variability. Am J Clin Pathol. 2007;127(3):398–408. 10.1309/MCGUQ3R4A4WWN3LB [DOI] [PubMed] [Google Scholar]
- 32. Díaz-Cano SJ, de Miguel M, Blanes A, Tashjian R, Galera H, Wolfe HJ. Clonality as expression of distinctive cell kinetics patterns in nodular hyperplasias and adenomas of the adrenal cortex. Am J Pathol. 2000;156(1):311–319. 10.1016/S0002-9440(10)64732-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chasseloup F, Bourdeau I, Tabarin A, et al. Loss of KDM1A in GIP-dependent primary bilateral macronodular adrenal hyperplasia with Cushing's syndrome: a multicentre, retrospective, cohort study. Lancet Diabetes Endocrinol. 2021;9(12):813–824. 10.1016/S2213-8587(21)00236-9 [DOI] [PubMed] [Google Scholar]
- 34. Vaczlavik A, Bouys L, Violon F, et al. KDM1A inactivation causes hereditary food-dependent Cushing syndrome. Genet Med. 2022;24(2):374–383. 10.1016/j.gim.2021.09.018 [DOI] [PubMed] [Google Scholar]
- 35. Ronchi CL, Di Dalmazi G, Faillot S, et al. Genetic landscape of sporadic unilateral adrenocortical adenomas without PRKACA p.Leu206Arg mutation. J Clin Endocrinol Metab. 2016;101(9):3526–3538. 10.1210/jc.2016-1586 [DOI] [PubMed] [Google Scholar]
- 36. Wei X, Calvo-Vidal MN, Chen S, et al. Germline lysine-specific demethylase 1 (lsd1/kdm1a) mutations confer susceptibility to multiple myeloma. Cancer Res. 2018;78(10):2747–2759. 10.1158/0008-5472.CAN-17-1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Takagi S, Ishikawa Y, Mizutani A, et al. LSD1 inhibitor T-3775440 inhibits SCLC cell proliferation by disrupting LSD1 interactions with SNAG domain proteins INSM1 and GFI1B. Cancer Res. 2017;77(17):4652–4662. 10.1158/0008-5472.CAN-16-3502 [DOI] [PubMed] [Google Scholar]
- 38. Jin Y, Ma D, Gramyk T, et al. Kdm1a promotes SCLC progression by transcriptionally silencing the tumor suppressor rest. Biochem Biophys Res Commun. 2019;515(1):214–221. 10.1016/j.bbrc.2019.05.118 [DOI] [PubMed] [Google Scholar]
- 39. Mohammad HP, Smitheman KN, Kamat CD, et al. A DNA hypomethylation signature predicts antitumor activity of LSD1 inhibitors in SCLC. Cancer Cell. 2015;28(1):57–69. 10.1016/j.ccell.2015.06.002 [DOI] [PubMed] [Google Scholar]
- 40. Ramírez-Ramírez R, Gutiérrez-Angulo M, Peregrina-Sandoval J, et al. Somatic deletion of KDM1A/LSD1 gene is associated to advanced colorectal cancer stages. J Clin Pathol. 2020;73(2):107–111. 10.1136/jclinpath-2019-206128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Barlas T, Kayhan G, Yalcin MM, et al. Inactivation of KDM1A with the concurrent presence of MGUS in a patient with GIP-dependent Cushing's syndrome and the vanishing effect of octreotide therapy. JCEM Case Rep. 2024;2(Supplement_1). 10.1210/jcemcr/luad146.001 [DOI] [Google Scholar]
- 42. Lacroix A. Extensive expertise in endocrinology: glucose-dependent insulinotropic peptide–dependent Cushing's syndrome. Eur J Endocrinol. 2023;188(3):R56–R72. 10.1093/ejendo/lvad026 [DOI] [PubMed] [Google Scholar]
- 43. Albiger NM, Occhi G, Mariniello B, et al. Food-dependent Cushing's syndrome: from molecular characterization to therapeutical results. Eur J Endocrinol. 2007;157(6):771–778. 10.1530/EJE-07-0253 [DOI] [PubMed] [Google Scholar]
- 44. Tsagarakis S, Tsigos C, Vassiliou V, et al. Food-dependent androgen and cortisol secretion by a gastric inhibitory polypeptide-receptor expressive adrenocortical adenoma leading to hirsutism and subclinical Cushing's syndrome: in vivo and in vitro studies. J Clin Endocrinol Metab. 2001;86(2):583–589. 10.1210/jcem.86.2.7171 [DOI] [PubMed] [Google Scholar]
- 45. Ghayee HK, Rege J, Watumull LM, et al. Clinical, biochemical, and molecular characterization of macronodular adrenocortical hyperplasia of the zona reticularis: a new syndrome. J Clin Endocrinol Metab. 2011;96(2):E243–E250. 10.1210/jc.2010-1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Goodarzi MO, Dawson DW, Li X, et al. Virilization in bilateral macronodular adrenal hyperplasia controlled by luteinizing hormone. J Clin Endocrinol Metab. 2003;88(1):73–77. 10.1210/jc.2002-021292 [DOI] [PubMed] [Google Scholar]
- 47. Charchar HLS, Fragoso MCBV. An overview of the heterogeneous causes of cushing syndrome resulting from primary macronodular adrenal hyperplasia (PMAH). J Endocr Soc. 2022;6(5):1–10. 10.1210/jendso/bvac041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. N’Diaye N, Hamet P, Tremblay J, Boutin J-M, Gaboury L, Lacroix A. Asynchronous development of bilateral nodular adrenal hyperplasia in gastric inhibitory polypeptide-dependent Cushing's syndrome. J Clin Endocrinol Metab. 1999;84(8):2616–2622. 10.1210/jcem.84.8.5930 [DOI] [PubMed] [Google Scholar]
- 49. Katugampola H, King PJ, Chatterjee S, et al. Kisspeptin is a novel regulator of human fetal adrenocortical development and function: a finding with important implications for the human fetoplacental unit. J Clin Endocrinol Metab. 2017;102(9):3349–3359. 10.1210/jc.2017-00763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Berthon A, Settas N, Delaney A, et al. Kisspeptin deficiency leads to abnormal adrenal glands and excess steroid hormone secretion. Hum Mol Genet. 2020;29(20):3443–3450. 10.1093/hmg/ddaa215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ibanoglu MC, Oskovi-Kaplan ZA, Kara O, Ozgu-Erdinc AS, Şahin D. Relationship between kisspeptin levels in the third trimester and late-onset fetal growth restriction: a case-control study. Placenta. 2023;140:1–5. 10.1016/j.placenta.2023.07.012 [DOI] [PubMed] [Google Scholar]
- 52. Swords FM, Baig A, Malchoff DM, et al. Impaired desensitization of a mutant adrenocorticotropin receptor associated with apparent constitutive activity. Mol Endocrinol. 2002;16(12):2746–2753. 10.1210/me.2002-0099 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during the current study will be available in European Genome-phenome Archive.