Abstract
Liver cancer is a global health challenge, causing a significant social-economic burden. Hepatocellular carcinoma (HCC) is the predominant type of primary liver cancer, which is highly heterogeneous in terms of molecular and cellular signatures. Early-stage or small tumors are typically treated with surgery or ablation. Currently, chemotherapies and immunotherapies are the best treatments for unresectable tumors or advanced HCC. However, drug response and acquired resistance are not predictable with the existing systematic guidelines regarding mutation patterns and molecular biomarkers, resulting in sub-optimal treatment outcomes for many patients with atypical molecular profiles. With advanced technological platforms, valuable information such as tumor genetic alterations, epigenetic data, and tumor microenvironments can be obtained from liquid biopsy. The inter- and intra-tumoral heterogeneity of HCC are illustrated, and these collective data provide solid evidence in the decision-making process of treatment regimens. This article reviews the current understanding of HCC detection methods and aims to update the development of HCC surveillance using liquid biopsy. Recent critical findings on the molecular basis, epigenetic profiles, circulating tumor cells, circulating DNAs, and omics studies are elaborated for HCC diagnosis. Besides, biomarkers related to the choice of therapeutic options are discussed. Some notable recent clinical trials working on targeted therapies are also highlighted. Insights are provided to translate the knowledge into potential biomarkers for detection and diagnosis, prognosis, treatment response, and drug resistance indicators in clinical practice.
Keywords: Hepatocellular carcinoma, Biomarkers, Cancer diagnosis, Cancer surveillance, Cancer treatment, Therapeutic option, Tissue biopsy, Liquid biopsy, Serological biomarkers, Circulating tumor cells, Cell-free DNA, Circulating tumor DNA, Non-coding RNAs, Methylation, Tumor-derived vesicles, Metabolomics, Surgical resection, TACE, HAIC, Local ablation, Liver transplantation, Targeted therapies, Immunotherapies, Artificial intelligence, Clinical trials
Introduction
Liver cancer is the second most lethal cancer worldwide, with hepatocellular carcinoma (HCC) being the most prevalent subtype, accounting for 90% of cases. Common risk factors include viral infections such as hepatitis B/C virus (HBV/HCV) infections, chronic liver diseases (CLDs) like fatty liver or cirrhosis, alcohol abuse, and metabolic diseases including diabetes. Other less common risk factors are hemochromatosis, autoimmune hepatitis, and exposure to certain environmental toxins. HCC is often challenging to diagnose due to the absence or ambiguity of symptoms. Early-stage symptoms typically encompass abdominal pain, weight loss, and fatigue, while later stages may involve jaundice, ascites, and fever. The insidious progression of chronic liver conditions further complicates the monitoring of liver diseases. Moreover, the heterogeneity of HCC at the molecular and histological levels contributes to the complexity of diagnosis and treatment. Apart from the apparent hepatitis virus, the presence of somatic alterations due to prolonged hepatic damage, aging, or inherited genetic susceptibility can result in hepatocarcinogenesis. Since patients typically do not undergo genetic testing until suspicion of liver diseases arises, these markers hold less significance in predicting HCC. Instead, they play a more substantial role in prognostic predictions and disease management. While this review does not delve into the genetic changes that lead to the development of HCC, they have been discussed extensively in other articles [1–7].
Consequently, surveillance programs and early detection tests are critical for the timely diagnosis and treatment of HCC. Screening efforts primarily target populations with multiple risk factors, such as known carriers of the hepatitis virus, individuals with cirrhosis, or those with a family history of HCC. Meta-analyses have supported the benefits of surveillance on HCC to improve patients’ survival and clinical outcomes [8, 9]. HCC can often be diagnosed using non-invasive imaging, like computer tomography (CT) / magnetic resonance imaging (MRI), due to its characteristic radiographic features, such as arterial hyperenhancement, venous washout, and capsule enhancement [10]. The Liver Imaging Reporting and Data System (LI-RADS) is an established scoring system for HCC diagnosis among HBV-infected and cirrhotic patients, with a 67% sensitivity and 93% specificity [11] for detecting tumor nodules larger than 1 cm in diameter [12]. Risk stratification is currently solely determined by LI-RADS. However, a biopsy is still necessary for patients with LR-4 or above observations without known risk factors for HCC or those with atypical imaging findings. The NCCN recommends core-needle biopsy for highly suspicious lesions for HCC that do not meet imaging criteria or specific patient categories [13]. Furthermore, the utilization of biomarkers, such as AFP, prothrombin induced by vitamin K absence or antagonist-II (PIVKA-II) (or named as des-gamma-carboxy prothrombin (DCP)), and glypican-3 (GPC3), has shown promise in improving the early detection of HCC. Only about half of the patients can be diagnosed through surveillance at a curative stage of the disease. Patients with early or intermediate-stage HCC may undergo curative therapies such as surgical resection, locoregional ablation, or liver transplantation. However, advanced-stage HCC patients are limited to chemotherapy or systemic treatments, such as chemoembolization, sorafenib, and nivolumab. Even though around half of HCC patients receive systemic therapies, the median overall survival (OS) remains limited due to the lack of effective therapeutic options.
Given the challenges in treating advanced-stage HCC, even with the advent of novel systemic therapies, it is imperative to explore improved methods for early detection, diagnosis, and prevention. To this end, significant insights into disease biology have been gleaned from genomic, transcriptomic, and epigenomic studies. These studies have led to identifying several molecular subclasses of HCC that exhibit distinct prognostic and therapeutic implications. This review aims to analyze various biomarkers that can offer valuable information on early detection, disease management, and therapeutic options. Additionally, clinical trials targeting some of these markers will be highlighted. For example, developing personalized medicine approaches, such as immunotherapy and targeted therapy, has shown potential in improving HCC treatment outcomes. Immune checkpoint inhibitors (ICIs), such as pembrolizumab and atezolizumab, have demonstrated promising results in clinical trials, while targeted therapies, such as lenvatinib and regorafenib, have exhibited significant improvements in OS. Furthermore, advancements in the understanding of the tumor microenvironment and the role of non-coding RNAs (ncRNAs), such as microRNAs (miRNAs) and long non-coding RNAs (lncRNAs), have opened new avenues for the development of novel diagnostic and therapeutic strategies. The application of liquid biopsies, mainly circulating tumor DNA (ctDNA) and extracellular vesicles, has also garnered interest as a non-invasive method for the early detection and monitoring of HCC.
In summary, developing numerous detection methods aims to gain insights through various biomarkers, enabling patients with HCC to benefit from early detection and diagnosis of the disease. This, in turn, can enhance their therapeutic regimens and increase survival probability. Consequently, this review will discuss current findings of biomarkers for early screening, detection, and diagnosis, as well as treatment options and response. Ongoing clinical trials targeting specific biomarkers will also be covered.
Current and promising diagnostic biomarkers for HCC
Current diagnostics using imaging and tissue biopsy
The American Association for the Study of Liver Diseases suggests that patients with a high risk of HCC perform ultrasound liver surveillance twice a year [14]. Although ultrasound is the most handy routine surveillance, its sensitivity is only 50%, while specificity is more than 90%. However, its detection is limited to early lesions and tumor nodules, which may reduce the therapeutic window [15]. CT and MRI are much more sensitive for diagnosing HCC, with over 90% sensitivity and specificity for tumors larger than 2 cm in diameter [16]. Despite its high sensitivity and accuracy, the cost of this cross-sectional imaging is high, and it is only feasible to be included in routine surveillance once evident signals appear.
Current stratification standards for HCC patients are based on tumor burden and cancer-related characteristics, including liver function and physical status [17]. Imaging techniques, including ultrasound, CT, and MRI, are commonly used to determine the size and number of tumor nodules in the liver. The modified Child-Pugh classification (CPC) proposed by Pugh et al. in 1973, which includes ascites, hepatic encephalopathy, prothrombin time, serum bilirubin, and albumin levels, has been used for many years to assess the remaining hepatic function in HCC-diagnosed patients [18, 19]. Still, its accuracy for use in HCC has been questioned in recent years [20, 21]. With the advancement of detection methods, improved anti-viral therapies to maintain liver function, and more prevalence of non-viral-associated HCC, more patients have been diagnosed with CPC A liver function recently, and it is no longer providing enough information for determining staging and prognosis in most cases [20, 22]. In addition to other technical limitations, it is suggested that the albumin-bilirubin (ALBI) grade is more potent in differentiating the HCC prognosis in most patients [21, 23–25]. The 2022 BCLC staging strategy recommended the ALBI score and AFP concentration for hepatic function assessment regardless of tumor burden [26]. The importance of portal hypertension in HCC is also underlined, and it often correlates with advanced conditions such as prior variceal bleeding, which should be considered, especially in patients with cirrhosis before surgical resection [27, 28].
A tissue biopsy is not usually performed unless the imaging can only provide marginal evidence of the diagnosis. For example, suppose the lesion is small and cannot be identified by imaging, or in the cases of non-cirrhotic liver disease, in that case, tissue biopsy can help make a precise diagnosis [29]. A biopsy can also help differentiate less common liver cancer subtypes from HCC and provide genetic mutation information directly from the tumor tissues [30]. It is essential in the design of precision medicine. However, tissue biopsy is an invasive method, which is potentially causing pain and bleeding in addition to liver damage. Also, there is a slight risk of inducing intrahepatic metastasis when tumor cells detach from the primary site and move to other sites along the wound [31].
After all, tissue biopsy remains a crucial tool for diagnosing HCC, as it can provide a definite diagnosis through histological evidence. Pathologists can more accurately perform tumor staging, which benefits the patient. Tissue biopsies can be used to monitor treatment response and provide prognostic information. Furthermore, it helps us know more about cancer biology, new biomarkers, and the creation of new treatments. The biopsy tissue is valuable for many pre-clinical studies to look for potential biomarkers or drug targets before the enrolment of actual patients in clinical studies. Nevertheless, due to the limitations of tissue biopsy, it is not feasible to utilize this method for surveillance. This is precisely the juncture where the potential of liquid biopsy comes to the fore, demonstrating its unique value and advantages. Details of ultrasound, imaging, and tissue biopsy are beyond the scope of this review, and the remaining sections will focus on the molecular biomarkers of HCC.
Current research in liquid biopsy diagnostics
Using liquid biopsy to screen for cancer and detect treatment response is critical as it can provide in-time information with minimal invasion to the patients [32, 33]. The most common fluid obtained in clinical settings is blood. Still, there are also other body fluids such as urine, ascites fluids, bile, saliva, etc. [34]. Current research directions aim to push forward the ability of liquid biopsy to obtain as much valuable information as a typical biopsy (Fig. 1). Large-scale HCC surveillance programs for high-risk individuals are limited to ultrasound with or without serum alpha-fetoprotein (AFP). AASLD is against the recommendation of using biomarkers to diagnose HCC, provided that the accuracy is insufficient until more solid validations in phase III and IV biomarker studies emerge [14, 35]. The following will discuss the current and potential biomarkers in various aspects of HCC to develop a more systematic and comprehensive panel for economical but sensitive HCC screening and diagnostic methods to improve patient outcomes.
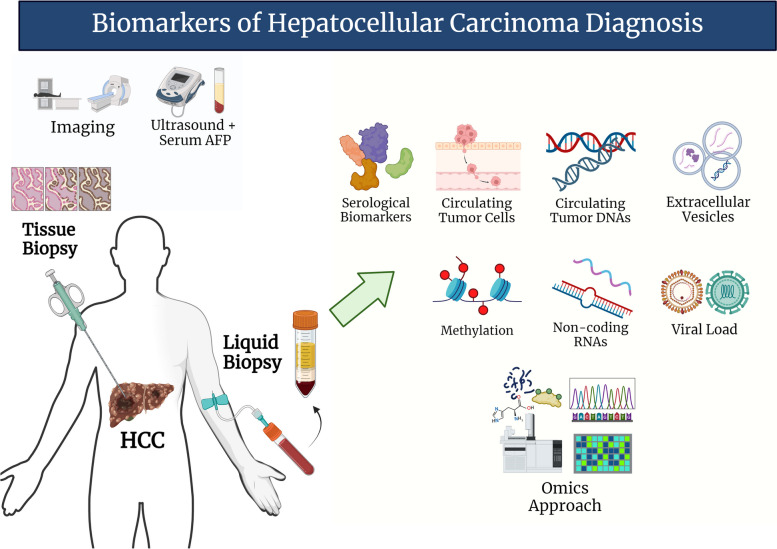
Fig. 1.
Biomarkers of HCC diagnosis. The current surveillance methodology for Hepatocellular Carcinoma (HCC) primarily involves ultrasound and serum AFP measurements, sometimes supplemented with imaging techniques. In instances where diagnosis or treatment options are ambiguous, tissue biopsy is often employed to confirm the histological structure of the tumor. Liquid biopsy offers a minimally invasive alternative for gaining insights into tumor heterogeneity. Recent research has identified potential biomarkers from diverse sources, including serological components, genetic materials, cells, and vesicles. Among these candidate biomarkers, circulating tumor DNAs and omics stand out due to their ability to reflect the complexity of HCC tumors. These technologies offer great promise in understanding the intricacies of HCC and developing more effective and personalized treatment strategies
Serological diagnostic biomarkers
Unless the doctor strongly suspects that the patients are likely to develop HCC, non-invasive measures, such as imaging and liquid biopsies, are typically performed in standard clinical practice for patients with an elevated risk of HCC. The most prevalent serum protein marker is AFP, whose expression is increased during tumorigenesis. AFP is a fetal glycoprotein, the analog of albumin in the fetus, and its expression should be reduced after birth [36]. For a long time, serum AFP levels have been considered the “gold standard” for screening tests when used in conjunction with ultrasonography [37, 38]. However, the level of AFP fluctuates a lot among patients with liver diseases. The cut-off for AFP in diagnosing HCC is generally considered to be 20 ng/mL [39]. AFP alone has a sensitivity of about 60% when the cut-off value is at 20 ng/mL, but it decreases quickly to below 50% when the value is raised to 50 ng/mL [40, 41]. It decreases to around 40 to 50% in patients with cirrhosis, while specificity remains relatively the same [15, 42]. Tumors of size smaller than 3 cm in diameter also reduce the sensitivity of AFP to about 25%, compared to more than half when > 3 cm [43]. However, this value may vary depending on the findings from the corresponding ultrasound examination. Therefore, it is generally recommended to be used with imaging techniques due to its variable sensitivity and specificity [29]. Also, the sensitivity and specificity both decrease in patients with acute and chronic liver diseases, especially hepatitis patients or HBV carriers [37, 41, 44]. Notably, elevated ALT can also impair the specificity of AFP in detecting HCC [45]. The serum ALT levels were correlated with AFP levels even in patients without HCC, and it became a confounding factor for the diagnosis. On the other hand, early-stage or small tumor HCC may not elevate AFP levels [36], so the combination with ultrasound is still likely to provide a false negative result. Consequently, in 2018, the AASLD guideline for liver diseases no longer recommended ultrasound plus AFP tests every 6 months for high-risk individuals [46]. Instead, ultrasound with or without AFP tests is suggested, as they cannot determine if AFP tests in surveillance can improve survival.
AFP-L3 is a glycosylated isoform of AFP, which reacts strongly with lens culinaris agglutinin. AFP-L3 is typically used in triple tests for pregnancy, but it also has a vital role in diagnosing HCC. AFP-L3 is usually reported as the percentage of AFP-L3 in total AFP, and 10% to 15% is the recommended cut-off [39, 47]. AFP-L3% is approved by the FDA for HCC stratification but not in surveillance programs due to its low sensitivity alone. Among patients with HCV-associated HCC, the sensitivity and specificity for AFP-L3% alone is 37% and 84.9% to 92%, but when combined with AFP, the numbers increase to 69.5% to 77% and 80.2% to 86.6% [47, 48].
In addition to AFP-L3%, PIVKA-II is introduced to the liquid biopsy test for diagnosis of HCC, especially in AFP-negative patients. PIVKA-II is detected in almost 90% of patients with HCC [49], and it is recommended that the Asia-Pacific region be included in routine surveillance programs for HCC [50]. However, similar to AFP-L3%, it is only for risk stratification but has not yet been approved by the FDA for HCC surveillance. The suggested cut-off value for PIVKA-II combined with AFP in small tumor HCC is 40 mAU/mL [43, 51]. In the Hepatitis C Antiviral Long-Term Treatment Against Cirrhosis (HALT-C) Trial, AFP combined with PIVKA-II can have a sensitivity and specificity as high as 91% and 74%, respectively. In comparison, the numbers reduce to 73% and 71% when tested 12 months before diagnosis [43]. Additionally, AFP-L3% and DCP levels are not associated with increased total AFP, making them good candidate markers for combined use with AFP for detecting HCC [47, 52].
As a result, algorithms with multiple biomarkers, such as GALAD score and HCC early detection screening (HES), are coming on stage. GALAD is calculated using gender, age, AFP, AFP-L3%, and DCP [53]. Longitudinal GALAD, which calculated the weighted average of multiple screening tests of an individual, demonstrated an area under the receiver operating characteristic (ROC) curve (AUC) of 0.85 and 0.83, even for early-stage HCC [54]. HES is a predictive model that includes results of AFP, ALT, platelets, and the age of the AFP test [55]. In the improved HES V2.0, AFP-L3% and DCP are included in the algorithm, which outperformed the sensitivity of GALAD in a cohort study in patients with hepatic cirrhosis by about 7% [55].
Additionally, several other clinically utilized biomarkers warrant mention. GPC3, a surface proteoglycan similar to AFP, usually is not detected in healthy adult liver but only in fetal liver. However, GPC3 is overexpressed in patients with HCC and is detectable in both tumor tissue and the serum [56]. It can also differentiate HCC from AFP-negative nodules, which makes it a potential biomarker on top of AFP [57, 58]. The mRNA and protein levels of GPC3 in viral-related HCC are high [59] while absent in healthy livers. However, the expression of GPC3 in non-viral HCC still needs to be clearly illustrated. The serum level of GPC3 was measured using ELISA, but the percentage of positive patients ranged from 36 to 95% in different studies [60, 61]. However, GPC3 levels may not show significant differences in early-stage HCC [62].
Besides, osteopontin (OPN) has also been reported to be a promising serological biomarker for HCC diagnosis. OPN is important in hepatic inflammation under acute or chronic liver diseases [63]. In early studies, OPN had limited or similar diagnostic value compared with AFP, but later on, it was reported to possess superior performance over AFP, with AUC ranging from 0.7 to > 0.9 [64–66]. OPN performs much better than AFP in small tumor HCC, with AUC of 0.85, sensitivity and specificity of 79.2% and 79.6% [67]. It also has an excellent diagnostic value in AFP-negative samples. The combination of OPN with AFP outperforms AFP alone.
Heat shock protein 70 (HSP70) is an oncogene that promotes tumor cell survival and proliferation and possesses anti-apoptosis effects [68]. HSP70 is located on the membrane of primary HCC cells, but it is also released into the circulation and is significantly higher than normal controls [68, 69]. The level of HSP70 can differentiate precancerous lesions with early HCC (p < 0.001) or early HCC with progressed HCC (p < 0.001), which demonstrated that it could be a sensitive biomarker for the detection and staging of HCC [70]. A recent study in 2022 suggested that when HSP70 is combined with GPC3, they have an absolute sensitivity and specificity of 100% [71]. However, the scale of this research is not large, and using HSP70 as a surveillance protein target is still not convincing.
Golgi membrane protein 1 (GOLM1, also known as Golgi protein-73, GP73) is a transmembrane protein on the Golgi complex. The level of GP73 is found to be significantly higher in patients with HCC, and it has a sensitivity and specificity of 69% and 75% for predicting HCC, which is much better than AFP (62% vs. 25%) in the same cohort to detect early HCC [72]. Its level is independent of serum AFP level [72]. Although the level of GP73 in HBV carriers and cirrhosis patients is elevated to a small extent, its sensitivity and specificity in detecting HCC remain as high as 74.6% and 97.4% when the cut-off is at 35 ng/ml [73]. The meta-analysis also reported that GP73 has the best diagnostic value for HCC among different biomarkers [74]. GP73 is also a promising therapeutic target for HBV-associated HCC patients [75].
Circulating tumor cells to predict HCC
Circulating tumor cells (CTCs) are cells isolated from tumors that travel in the circulation. Some CTCs can survive the attack by immune cells and then reach distant metastatic sites. Well-characterized CTCs are direct proof of the presence of cancer, but the isolation of CTCs is not easy. Standard techniques to isolate CTCs in related research utilize cell size, density, and immunogenicity to carry out positive and negative enrichment [76]. Since there is no universal surface marker for CTCs due to their heterogeneity, there is not yet a “golden standard” method for isolating CTCs [77]. Some CTC purification platforms allow the capturing of CTCs and then proceeding for subsequent downstream analysis, such as NGS and in situ hybridization [78].
Efforts to isolate CTCs for cancer diagnosis have been ongoing for decades. A study conducted by [79] revealed that CTCs were only present in patients with HCC but were not found in patients with CLD, cirrhosis, or healthy individuals [79]. The sensitivity is about 52%. With the advancement in the CTC capturing technology, a group from China can capture CTCs from 90.18% of patients from a cohort of 112 HCC patients with a new RNA-in situ hybridization-based platform [80]. Among the patients with HBV, two patients were detected with CTCs, which eventually developed into HCC within half a year, demonstrating its potential for diagnosing undetected tumor nodules. A study using nine candidate genes as a multi-marker panel detecting CTCs with stem-like properties in peripheral blood [81]. CD45 cells were depleted by negative enrichment, RNAs were extracted directly from the remaining sample, and qRT-PCR was performed to measure the gene expression levels. The CTC panel has an AUC of 0.88 and a sensitivity of 72.5% for detecting HCC, while the performance is also good considering early-stage HCC or AFP-negative HCC.
The current consensus on CTC isolation focuses on EpCAM+/CK+/CD45− cells [82]. EpCAM-based detection of CTCs has been employed in HCC diagnosis. Techniques like CellSearch™, an immunoaffinity-based CTC enrichment platform, have isolated CTCs in HCC patients. EpCAM+ CTCs are detected in two-thirds of HCC patients [83]. In 2014, a platform was developed by negative enrichment of CTCs, followed by quantitative PCR. It showed a three-forth consistency with the CellSearch™ system [84].
CTC profiling is an essential aspect of liquid biopsy; nevertheless, its importance in cancer diagnosis is not as high as its role in recurrence, therapeutic response, or oncology research [85]. Very few CTCs are found in the bloodstream, and their quantity is proportional to the tumor size. This makes detecting CTCs in early-stage diseases quite challenging [86]. It is almost not practical to isolate CTCs in clinical settings for the diagnosis of cancer. Instead, specific cancer-related genes in CTCs, or CSCs, are amplified and detected by reverse transcription PCR as the easiest method. The main objective of current CTC research is to enhance the sensitivity and specificity of CTC detection to achieve accurate and comprehensive molecular characterization. CTCs are not exact copies of each other but come from different tumor parts and can change under various conditions. Tumor cells in different parts of the body can also vary. Liquid biopsy using CTCs is not just a less invasive biopsy but a new way to understand tumor cells throughout the body. Since CTCs are released from the primary or distant metastatic tumors, the genetic makeup of these cells reveals information about the heterogeneity, mutations, and potential resistance to treatments. Research has shown that primary and metastatic tumors are processing high heterogeneities [87, 88]. CTC detection and isolation can help precision medicine by finding molecular characteristics and markers for targeted therapy. Furthermore, CTCs can be isolated from patients’ peripheral blood and cultured in vitro to become organoids. The tissue culture can be applied for drug response testing, checking the cellular signaling, and stimulating therapeutic strategies. It is also beneficial for new drug development.
Using cell-free DNA / circulating tumor DNA to detect HCC
Cell-free DNAs (cfDNAs) refer to nucleic acid content in the circulation, usually released from dead cells as regular cell turnover or pathologies [89, 90]. CfDNA is present in normal humans and majorly comes from blood cells [91]. Different from CTC analysis, before cfDNA analysis, all cells are removed from the peripheral blood. In patients with cancer, tissues and organs undergo uncontrolled cell deaths, which will release large amounts of nucleic acids into the circulation, contributing to the ctDNA. Since ctDNA originated from tumor cells, analysis of ctDNA content can show a collection of heterogeneous tumor cells that carry the mutations and epigenetic patterns of the tumor. Also, macrophages engulf some CTCs, releasing the nucleic acids to be cfDNA. So, ctDNA is usually regarded as fragmented DNA from tumor tissues and as part of the cfDNA content. The proportion of ctDNA in total cfDNA can differ substantially, from less than 1% to more than 90% [92]. In general, patients with HCC have higher ctDNA levels in the circulation, which is also an indicator of tumor burden. More published research has provided molecular profiling of HCC ctDNA, and its role in the screening and early detection of HCC is an emerging trend. A systematic review analyzed 15 studies with 3,686 patients, and the pooled sensitivity and specificity of using cfDNA to predict HCC is 0.83 and 0.90, while the AUC is 0.93 [93]. The following part will discuss the applications and pros and cons of ctDNA as a detection tool.
Peripheral blood is the most frequent source for extracting cfDNA, although it can also be detected in other fluids such as saliva, ascites, and urine. There is an ongoing discussion about whether to extract cfDNA from serum or plasma, with some research indicating higher concentrations in serum due to contamination from blood cell lysis. However, plasma is generally the preferred sample in most investigations [94, 95]. Storing and extracting cfDNA necessitates specialized methods because of its short half-life, with plasma separation recommended within six hours using EDTA tubes [96]. The technology of capturing cfDNA from circulation has been improved. Theoretically, ctDNA can carry the whole genome from tumor cells, providing valuable information about the mutational profiles and heterogeneity. The FDA approved a recently marketed product that analyzes patients’ blood samples for alterations in 47 genes [97]. This product is designed to detect genetic mutations that may potentially lead to certain cancers, including HCC.
Detecting cfDNA requires highly sensitive and specific techniques, which can identify various DNA-based changes that reflect the DNA status in cancer cells. The most simple tool to check the presence of ctDNA or carried mutations is quantitative PCR (Table 1). However, this is limited to relatively higher concentrations of cfDNA, and the heterogeneity of HCC may affect the significance. Bisulfite conversion is widely adapted to convert unmethylated cytosines to uracil to detect methylation on the HCC-associated genes, resulting in a difference in the DNA sequence between methylated and unmethylated regions. While digital droplet PCR offers high precision and sensitivity, it cannot cover an extensive range of sequences, making NGS more appropriate for multiple gene profiling [98, 99]. Target sequencing, whole exome sequencing [100], or ultra-deep sequencing are usually adapted in molecular biology research to study the profiling or mutation patterns within the cfDNA. Despite this, the sensitivity and specificity of NGS may be hindered by the mistakes of DNA polymerase and amplification reactions, so molecular barcoding approaches are used to identify the correct gene alterations [101] accurately. Since the cost of utilizing NGS to screen for ctDNA is high, custom-designed sequencing panels or low-depth whole-genome sequencing approaches appear to facilitate the screening [101, 102].
Table 1.
Summary of selected research output on cfDNA related to diagnosis and prognosis of HCC
| Marker | Description | Ref. | |
|---|---|---|---|
| 1 | TERT promoter |
- Modified droplet digital polymerase chain reaction G > A TERT mutation at -124 bp was found in the blood of 69% of patients. Patients with a mutation have shorter DFS (p = 0.02). Risk of disease recurrence was about three times higher in patients with the mutation (p = 0.03). |
[98] |
| 2 | With liver MRI |
- NGS of cfDNA from plasma, combined with liver MRI cfDNA was found in the blood of 36 patients with cirrhosis, and NGS detected 20 different genetic variants. Specific HCC-related gene mutations in cfDNA could help identify patients with LC at increased risk of developing HCC. |
[103] |
| 3 | 139 somatic mutations |
- Custom-designed sequencing panel with 354 target genes Number of mutations, variant allele frequency, and concentration of ctDNA linearly correlated with tumor size. Patients with mutations experienced shorter DFS, and increased the risk of recurrence (HR = 7.655, p < 0.0001). Status of cfDNA increases risk of recurrence (HR = 10.293, p < 0.0001). |
[101] |
| 4 | 5-HMC signature |
- Genome-wide 5-HMC sequencing of cfDNA Distribution pattern of 5-HMC regions can differentiate HCC patients, LC patients, and healthy volunteers. HCC score combined with protein biomarkers, achieves 92.75% accuracy in distinguishing HCC patients from LC patients. HCC score is effective in identifying high-risk patients for recurrence after surgery and serves as an independent indicator for RFS (p = 0.00865) and OS (p = 0.000739). HCC score values are positively correlated with tumor burden changes during follow-up. |
[104] |
| 5 | CNVs and SNVs |
- Target sequencing and low-coverage whole-genome sequencing Plasma samples before surgery displayed genetic variations that matched the patients’ tumor tissues, and these variations changed during follow-up, correlating with tumor burden. A strategy using comprehensive ctDNA mutation profiles was developed to accurately assess tumor burden, showing high consistency with imaging results. This approach detected tumor occurrence an average of 4.6 months earlier than imaging and outperformed serum biomarkers like AFP, AFP-L3%, and Des-Gamma-Carboxy Prothrombin (DCP). It was effective in detecting minimal residual disease (MRD) and predicting patients’ outcomes for RFS (p = 0.001) and OS (P = 0.001); combining ctDNA with DCP further improved MRD detection sensitivity. |
[105] |
| 6 | 160 SNVs |
- Target sequencing and whole exome sequencing 160 subclonal SNVs were identified in tumor tissues, peritumor tissues, and matched PBMCs, with 96.9% of these mutations also found in plasma samples. Two clusters of SNVs are related to cancer progression. Circulating levels of tumor stem somatic mutations could reflect tumor burden and predict prognosis earlier than traditional methods. HCK (p.V174M) mutation was identified as being associated with tumor recurrence and metastasis and was found to promote the migration and invasion of HCC cells. |
[100] |
| 7 | TP53, CTNNB1 |
- Customized cfDNA next-generation sequencing panel In 65% of patients, at least one pathogenic variant was found in two major HCC driver genes, TP53 and CTNNB1. There were 16 variants of TP53 and nine variants of CTNNB1 identified. The identified variants in TP53 and CTNNB1 genes are crucial in understanding the genetic basis of HCC and may help in developing targeted therapies. |
[106] |
| 8 | Mutation variant frequency change (MVFC) |
- Unique molecular identifier target sequencing on plasma and PBMCs Plasma mutations with MVFC < 0.2 were found to be enriched for tumor mutations identified in tumor tissues, and their frequency changes correlated with tumor burden. The presence of MVFC-identified tumor mutations after surgery was associated with shorter RFS, indicating MRD. Combining MVFC-identified tumor mutations and AFPimproved MRD detection (P < 0.0001). The MVFC-based identification of tumor mutations was confirmed to be applicable using a different gene panel, highlighting its potential utility in monitoring and predicting HCC outcomes. |
[107] |
| 9 | Serum DNA integrity |
- Real-time quantitative polymerase chain reaction The Triton/Heat/Phenol method was deemed effective to extract quality DNA. The integrity of serum DNA in HOC patients was significantly higher than in HBV patients or healthy individuals. The integrity of DNA was linked to factors such as tumor size, TNM stage, and the presence of lymph node and distant metastasis. |
[108] |
| 10 | TERT promoter, TP53, CTNNB1, ARID1A, TSC2, etc. |
- NGS assay with 129 validated genes Genomic alterations were found in 92.2% of advanced HCC patients, with common mutated genes including TERT promoter, TP53, CTNNB1, ARID1A, and TSC2. Paired tumor NGS was conducted on 37 patients, showing high concordance in mutations between patient-matched plasma samples. In 27% of tumor-plasma samples, alterations were detected in cfDNA analysis but not in patient-matched tumors. Higher average variant allele fraction correlated with elevated AFP, increased tumor volume, and no previous systemic therapy, but not with OS in treatment-naive patients. |
[109] |
| 11 | TERT promoter C228T, TP53, CTNNB1, PIK3CA, NRAS |
- Droplet digital PCR TERT mutation occurs in all patients with one or more ctDNA mutations. Maximal VAF is linearly correlated with larger tumor size and AFP level. ctDNA correlated with poor OS. |
[110] |
| 12 | GPBAR1 |
- Methylation-specific polymerase chain reaction Hypermethylation of the TGR5 promoter is significantly higher in HCC than CHB and HCs (p < 0.01). Increased sensitivity when combined with AFP |
[111] |
| 13 | TERT promoter, TP53, CTNNB1, AXIN1, ARID1A |
- Ultra-deep sequencing targeting HCC driver genes Patients who had more than 2 ng/mu L of cfDNA at diagnosis experienced higher mortality rates (p = 0.01). These patients had a higher number of mutated genes and more mutations in cfDNA, and associated with disease recurrence (p < 0.01). Patients with more than four mutations detected in cfDNA were at a higher risk of death (p = 0.042). |
[112] |
| 14 | UBE2Q1 |
- Methylation-specific polymerase chain reaction assay Lower methylation frequencies of UBE2Q1 from the circulating cfDNA was found in HBV-related HCC patients, compared with LC, CHB and HCs (p < 0.05). Combining with AFP increases sensitivity and specificity. |
[113] |
| 15 | cfDNA integrity |
- Quantitative PCR of ALU amplicon cfDNA integrity lower in patients with HCC or other liver cancers, but higher in benign diseases and healthy individuals |
[114] |
| 16 | TP53, CTNNB1, TERT promoter |
- Droplet digital PCR More than half of the patients were found to have at least one of the four genetic mutations. None of the mutants were detected in adjacent liver tissues among the matched tumour tissues. |
[115] |
| 17 | INK4A |
- Pyrosequencing on the CpG sites of the INK4A promoter Hypermethylation on INK4A in HCC than controls detected in the circulating cfDNA. |
[116] |
| 18 | CTNNB1, TP53, NFE2L2, ARID1A |
- Targeted sequencing of cancer associated genes Mutations are detected in majority of the HCC patients, and it does not affect the cfDNA concentrations. Higher cfDNA and mutation rates associated with shorter OS (p < 0.001). |
[117] |
| 19 | Methylation fingerprint panels |
- Genome-wide DNA methylation analysis The study identified three panels: cancer-specific biomarker panels, a pan-GI panel, and a multi-cancer prediction panel, with high AUC values ranging from 0.85 to 0.98 for detecting various gastrointestinal cancers. |
[118] |
| 20 | RNF135, LDHB |
- Methylation Sensitive High-Resolution Analysis Combined RNF135 and LDHB analysis has a sensitivity of 57% on HCC, compared with AFP at 45%. When combined with HCC, the sensitivity is 70%. |
[119] |
| 21 | cfDNA concentration and integrity |
- Real-time quantitative PCR HCC patients have higher level of cfDNA in serum than CLD and healthy controls. Increased DNA fragmentation and poor DNA integrity. |
[120] |
| 22 | HRNR, TTN |
- Whole-exome sequencing of cfDNA Single nucleotide variations were present in somatic genes in cfDNA, including in ZNF814, HRNR, ZNF492, ADAMTS12, FLG, OBSCN, TP53, and TTN. Level of cfDNA is higher in patients with HCC than chronic hepatitis patients. |
[121] |
| 23 | TERT promoter, TP53, NTRK3, JAK1 |
- Ultra-deep sequencing of all exons Mutations are detected in plasma cfDNA from plasma/serum samples of HCC patients. |
[122] |
| 24 | TERT promoter, TP53, CTNNB1, ARID2, ARID1A, NFE2L2, AXIN1, KRAS |
- Targeted sequencing with ultra-high coverage and molecular barcoding About half of the HCC patients have at least one mutations, and rate s are higher in advanced stages. TP53 has the worst survival, whereas CTNNB1 and TERT does not affect survival. Combining AFP and prothrombin improved detection rate. |
[123] |
| 25 | GNB4, Riplet |
- Methylation-specific polymerase chain reaction Significantly higher CTCs in HCC compared with HBV patients. GNB4 and Riplet methylation were different, and is able to diagnosis HCC at 0.98 when combined with CTCs. |
[124] |
| 26 | TERT promoter, CTNNB1, TP53 |
- MiSeq for targeted deep sequencing Mutations associated with vascular invasion (p = 0.04), predict shorter RFS for HCC patients (p < 0.001). |
[125] |
| 27 | Wnt signaling pathway |
- QUADAS-2 tool Hypermethylation of Wnt/Beta-catenin signaling pathway is correlated with tumor size, TNM stage, distant metastasis, and HBV infection (p < 0.05). |
[126] |
| 28 | TERT promoter, TP53, CTNNB1 |
- Ultradeep sequencing of cfDNA High levels of cfDNA showed worse PFS and OS. Mutations combined with AFP can stratify the prognosis of HCC patients. |
[127] |
| 29 | TERT promoter |
- QuantStudio 3D Digital PCR Mutation is associated with tumor diameter (p = 0.015), and tumor volume after TACE (p < 0.001). Higher cfDNA levels predict better PFS with TKI initiation (p = 0.004). |
[128] |
| 30 | TERT promoter |
- qPCR and Droplet digital PCR cfDNA increases with HCC stage, and associated with poor prognosis. Greater cfDNA is associated with better therapeutic response to treatment. |
[129] |
| 31 | Fragment length profiles |
- Next generation sequencing The fragment length profiles can differentiate ctDNA from white blood cells, non-tumor cells, and heterogeneous mutations. |
[130] |
| 32 | ctDNA and mutations |
- Whole exome sequencing ctDNA increase with disease progression. ctDNA positive predicts recurrence and extrahepatic metastasis (p = 0.01), and vascular invasion of portal vein. |
[131] |
| 33 | CTNNB1 |
- Droplet digital PCR Detection rate of HCC by CTNNB1 mutation is 9.5%, and 13.5% when tumor tissues are combined. Mutation is not related to OS. |
[132] |
| 34 | Alu247 |
- Quantitative PCR HCC cases have higher level of Alu247 and RNase P coding DNA than controls (HBeAg-negative CHB patients). No difference in cfDNA concentration, Alu115, integrity index, and mitochondrial DNA copies. |
[133] |
| 35 | TP53, CTNNB1, AXIN1, TERT promoter |
- HCC screen Identify HCC from HBsAg+ patients by combining somatic mutations detected in cfDNA with serum AFP levels. Sensitivity 100%, specificity 94%, PPV 17%. |
[134] |
| 36 | Somatic copy number aberration |
- Low-depth whole-genome sequencing Machine learning based statistical model to analyse copy number variation in ctDNA, with AUC of 0.92. |
[102] |
| 37 | TP53 R249S |
- Mass spectrometry and quantitative PCR from cfDNA Seasonal fluctuation in R249S levels, higher concentration in HBsAg+ individuals from April to July. |
[135] |
| 38 | Aberrant DNA methylation |
- DNA methylation-based digital droplet PCR Level of cfDNA is higher in patients with HCC than HC. Methylation ratio cut-off at 15.7% has an sensitivity of 78.57% for predicting HCC. |
[136] |
| 39 | GSTP1 |
- Quantitative PCR 88.5% tumour tissues, 69% non-tumor tissues, 50% ctDNA samples, were exhibiting GSTP1 promoter CpG island hypermethylation. No PBMC samples tested positive for methylation. |
[137] |
| 40 | Mutation and methylation panel |
- Mutation Capsule Plus 90% sensitivity with 94% specificity, detected four out of five HCC cases from a cohort of 311 asymptomatic HBV carrier with normal ultrasound diagnosis and AFP level. |
[138] |
| 41 | 931 mutation sites across 21 genes |
- Circulating Single-Molecule Amplification and Resequencing Technology Sensitivities of 81.25% for all stages and 66.67% for early HCC. High accuracy in differentiating AFP-negative, AFP-L3-negative, and PIVKA-II-negative HCC. |
[139] |
| 42 | Panel of HCC mutations |
- Next generation sequencing AUC 0.92, sensitivity 65%, specificity 100% regardless of AFP status. Rate of mutations is greater in recurrent HCC than non-recurrent (p < 0.05). |
[99] |
| 43 | Methylation, mutations and protein-HCC |
- M2P-HCC M2P-HCC is an independent predictor of HCC risk. AUC 0.88, combining AFP and ultrasound, yield inferior performance. |
[140] |
| 44 | TP53, CTNNB1, TERT promoter, RASSF1A |
- Urine cfDNA detection by quantitative PCR Significant quantities of ctDNA detected in urine samples. ctDNA marker detection independent of serum AFP. Combining AFP yield AUC of 0.904. |
[141] |
| 45 | NLRP7, NLRP2, NLRP3 |
- Whole-genome bisulfite sequencing Hypomethylation of programmed-cell death genes (PRGs) including NLRP7, NLRP2, and NLRP3 in HCC tissues; NLRP3 methylation levels correlated with expression level (r = 0.51). Hypomethylated PRGs can discriminate between early HCC patients and healthy controls in cfDNA analysis (AUC = 0.94), and associated with poor HCC prognosis. |
[142] |
| 46 | Intergenic and repeat reigons of HBV integration sites |
- Low-pass genome-wide bisulfite sequencing Significant enrichment of cfDNA in intergenic and repeat regions, especially in HBV integration sites. Hypomethylation only in HCC but not in hepatitis and cirrhosis patients. AUC value > 0.85 and a prediction performance of 0.954 in the validation cohort. |
[143] |
| 47 | GNB4, Riplet |
- Whole genome bisulphite sequencing Combining methylation of GNB4 and Riplet in plasma cfDNA, sensitivity, specificity, and AUC are 84%, 92%, and 0.92, superior to AFP. |
[144] |
In particular, the TERT promoter is the most common mutation in HCC. Many studies have reported that the TERT promoter mutation is associated with HCC occurrence, recurrence, and poorer survival [98, 109, 112, 125]. Apart from TERT promoter, other commonly reported mutations such as CTNNB1, TP53, AXIN1, KRAS, ARID2, etc., are also detected in cfDNA, and their presence and association with HCC development are almost the same as somatic cell mutations [101, 106, 109, 112, 123]. Besides single gene mutations, abnormal methylation on oncogenes or tumor suppressor genes is also related to oncogenesis. For example, a study uses a newly developed methylation-sensitive high-resolution analysis (MS-HRM) platform to detect the methylation levels of RNF135 and LDHB [119]. The sensitivity of the combined MS-HRM is 57%, which is better than 45% of AFP from the same cohort. When combined with AFP, the sensitivity is as high as 70%. The details of detectable methylations are discussed in the later section.
In addition, several research groups have reported single-nucleotide variants (SNVs) and copy number variants (CNVs) related to HCC in cfDNAs. Cai et al. utilized low-coverage whole-genome sequencing to perform a targeted sequencing that can check the genetic profiles of the cfDNAs from HCC patients [105]. They found that SNVs and CNVs information from ctDNA and corresponding serological biomarkers, including DCP at the cut of of 20 ng/mL, can predict clinical outcomes and detect HCC with satisfactory sensitivity (p < 0.0001).
Besides, DNA integrity has also been considered a biomarker for HCC in recent research. DNA with shorter lengths preferentially carrying the tumor-associated CNVs. Also, it is more likely to detect high levels of mitochondrial DNA in the cfDNA from HCC patients [145]. Huang et al. also reported similar findings, that cfDNA integrity is reduced in patients with HCC, and it is promising to be a biomarker for detecting HCC in surveillance with a sensitivity of 43.4% and AUC of 0.705 [114]. The cfDNA integrity is lower than those with benign nodules and healthy controls. Also, patients with HCC or other types of liver cancer have comparable cfDNA integrity (p = 0.7356) as those without tumors (p = 0.9138). CfDNA fragmentation is a nonrandom process where cfDNA fragments with specific genetic endings likely originate from HCC [146]. Detection of the tumor-associated endings, which are prevalent across the genome, may provide a simple but effective method for identifying the presence of tumors through serological surveillance.
Besides circulating isolated nucleic acids, genetic materials in plasma exosomes are also a new potential diagnostic target for HCC [147]. For instance, circular RNA Circ0006602 is one of the five circRNAs found to be up-regulated in the plasma exosomes of HCC patients compared with healthy individuals. As a diagnostic marker, the AUC of exosomal Circ0006602 to HCC patients was 0.907, with a p-value of < 0.001. The sensitivity is 77.70%, with a high specificity (93.3%) [148]. In another recent study, high-throughput sequencing was used to analyze the exosomal miRNA levels isolated from patients’ HCC tissues. After cross-validation with plasma exosomes, miR-483-5p was identified to have the power to differentiate individuals with or without HCC (AUC = 0.898), which is slightly better than the plasma miR-483-5p counterpart (AUC = 0.868) [149]. The sensitivity is, though, similar to AFP, at around 80–85%, with the specificity at 90.38%. Similar research also reported that other miRNA candidates (miR-122-5p, let-7d-5p, and miR-425-5p) had HCC early diagnostic power (AUC at 0.660 to 0.808) [150]. These studies suggest that serum exosome-derived small RNAs are potential biomarkers for HCC diagnosis in clinical settings.
Similar to CTCs, the utilization of ctDNA in clinical applications encounters obstacles. Identifying ctDNA among a large pool of cfDNA originating from other cell types presents technical difficulties. Furthermore, it remains uncertain if ctDNA can precisely reflect the genetic composition of fast-growing and highly mobile tumor cells. This is because ctDNA may overrepresent DNA from “weakened” tumor cells that are more susceptible to cell death, subsequently releasing their genetic materials into the blood. Further studies are still required in this field to optimize the full use of ctDNA as a surveillance tool.
Presence of non-coding RNAs in HCC patients
Aside from genetic modifications found in genomic DNA or cDNA, ncRNAs circulating in the bloodstream can also possess diagnostic significance. MiRNAs are short ncRNAs at about 22 bases long, and their function is to regulate gene expression in cis or trans. Given their involvement in genetic regulation, profiling circulating miRNAs holds promise as a non-invasive marker for human cancers.
In a study recruiting 513 subjects, miRNA profiles were obtained by NGS. A thirteen miRNA-based biomarker panel can differentiate clearly between HBV, HCV, and HBV-positive HCC cases [151]. Two miRNA candidates, miR-375 and miR-92a, are novel identified targets as HBV-specific miRNAs. A trio biomarker miR-25, MiR-375, and let-7f can significantly discriminate HCCs from controls, where miR-375 alone has an AUC of 0.96. Another study by Chen et al. demonstrated that the dual biomarker of miRNA-30b-5p and MINPP1 can detect HBV-positive HCC [152]. The level of MINPP1 signifies the tumorigenesis of HBV-positive patients, whereas miRNA-30b-5p can regulate the expression of MINPP1.
Sometimes, a single miRNA might not differentiate between progressive liver diseases and HCC. An analysis of miRNA expression in 168 subjects revealed intriguing miRNA expression trends in patients infected with HCV. MiR-484 demonstrated a decrease in advanced fibrosis compared to mild fibrosis and HCC, but it increased HCC relative to cirrhosis [153]. In addition, miR-524-5p showed an increase in both cirrhosis and HCC, while miR-615-5p was found to be increased in the cirrhotic group relative to the control group. Notably, miR-524 displayed considerable potential in distinguishing between cirrhosis and fibrosis. These miRNAs under study may potentially be helpful in the staging and diagnosing of HCV hepatic progression, providing a significant understanding of the disease.
In a recent scientific publication, investigators presented a process that employs DNA molecular markers to convert gene expression profiles into clinical assessments [154]. Remarkably, this approach precisely categorized HCC patients and healthy individuals using only five miRNAs obtained from blood specimens, including miR-1290, miR-4370, miR-1203, miR-221-3p, and miR-4258.
Recent findings revealed that circular RNA (circRNA) is also associated with tumor development [155, 156]. A study delved into the expression patterns of circRNAs in HCC, identifying circMTO1 as a significantly downregulated circRNA with implications for patient survival [157]. The author concluded that circMTO1’s function in suppressing HCC progression positions it as a potential drug target in HCC treatment, and its decreased presence may serve as a prognostic marker for poor patient outcomes. Besides, circSETD3 is recognized as a novel tumor suppressor of HCC and is a significant prognostic biomarker [157]. CircSETD3 can suppress the growth of HCC through the MAPK signaling pathway, and its downregulation is associated with unfavorable outcomes of HCC.
LncRNA, on the other hand, has a quite different role in tumorigenesis. They are RNA molecules with more than 200 nucleotides and have no protein-coding or potentials or only peptide-coding. They interact with oncogenes or tumor suppressor genes through epigenetic regulation and transcriptional activation or as sponges to silence miRNAs. Some lncRNAs also have a role in post-transcriptional regulation or modulating signaling pathways. It is a relatively new field to use lncRNAs as biomarkers of HCC detection, but some research articles also have significant findings. A review article 2018 discussed some findings on lncRNA as a biomarker of HCC [158]. There are indeed some reports worth mentioning in recent years. For instance, Liu et al. utilized an ultrasensitive strategy, an electrochemical nucleic acid sensor, to detect the expression of a lncRNA Highly up-regulated in liver cancer (HULC) [159]. HULC is highly expressed in patients with HCC, and its level can differentiate between HCC and control individuals [160]. Its level is also different between the HBV-positive group and the control group. Besides, research is now focusing on extracellular vesicle-derived lncRNAs as biomarkers of HCC. EV-LINC00853 displayed outstanding diagnostic ability for all-stage HCC, with higher sensitivity and specificity than AFP, particularly in early-stage and AFP-negative HCC, suggesting its potential as a novel diagnostic biomarker [161]. Another study indicated that serum small EV-derived lncRNAs, specifically EV-DLEU2, EV-SNHG1, EV-MALAT1, and EV-HOTTIP, show promising diagnostic potential for initial-stage HCC, with combined panels achieving the best results [162]. With the appearance of EV-lncRNA sequencing technology, bioinformatics approaches were introduced to analyze the potential lncRNA that can be HCC detection biomarkers. A panel of eight EV-lncRNA was identified as potential biomarkers [163]. However, further experiments are needed to validate those candidates.
Due to the lack of extensive clinical validation, ncRNAs are not being used as a detection biomarker in HCC. However, it is a valuable future research direction in diagnosing cancers because miRNA is closely related to regulating genes, especially cancer-associated genes. This can provide additional insights into the genetic makeup of the potential HCC tumors in patients with liver diseases.
Altered methylation patterns related to HCC occurrence
Apart from typical genetic mutation patterns that traditional sequencing or PCR methods can detect, epigenetic regulations on the genome also have an essential role in tumor development. Methylation is a process by which methyl groups (-CH3) are added to the DNA molecule, typically at the cytosine-guanine (CpG) dinucleotide sequences. This can change the activity of a DNA segment without changing the sequence itself. The most crucial aspect of methylation in regulating gene expression is the hypermethylation of tumor suppressor genes. And hypomethylation of oncogenes. Abnormal DNA methylation patterns can contribute to the development of cancers. Therefore, the methylation status of specific genes and their significance as a cancer biomarker has been heavily studied in recent years, with the advancement of detection technology [164] (Table 2).
Table 2.
Summary of selected research output on detection of HCC using methylation patterns
| Marker | Blood-based | Description | Ref. | |
|---|---|---|---|---|
| 1 | RASSF1A | + | Mean rate of 30% and 65% methylation in HCV-related cirrhosis and HCC tissues respectively | [165] |
| 2 | RASSF1A | Mean rate of 26.1% and 59.1% methylation in cirrhosis and HCC tissues respectively | [166] | |
| DOK1 | Mean rate of 19.6% and 56.0% methylation in cirrhosis and HCC tissues respectively | |||
| 3 | RASSF1A | + | Mean rate of 70% methylation on top of HCV-associated cirrhosis, with serum AFP combined to improve sensitivity | [167] |
| 4 | RASSF1A | + |
Hypermethylated sequences in 93% of HCC, 58% of HBV carriers, and 8% of the healthy individuals High RASSF1A at diagnosis or 1-year after resection relates to poor prognosis (p < 0.01) RASSF1A increase after cancer diagnosis (p < 0.014) |
[168] |
| 5 | CHFR, VASH2, GRID2IP, CCNJ, F12 CpG sites | + | HCC detection sensitivity was 84.5% at 95% specificity and 0.94 AUC using the count of methylated reads on combined specific genes. | [169] |
| 6 | WISP1 | + | Hypomethylation, increased plasma soluble WISP1 improved diagnostic power combining with AFP | [170] |
| 7 | Six hypermethylated CpGs sites | A combination of six hypermethylated HCC-specific CpGs sites has a 92% sensitivity predicting HCC, and 98% specificity differentiating from normal liver or other cancers | [171] | |
| 8 | Long interspersed element-1 (LINE-1) | + | Global hypomethylation measured in LINE-1 repeats in blood leukocytes DNA correlates with elevated risk of HCC (p = 0.004) | [172] |
| 9 | RASSF1A sequence | + |
Higher methylation in HCC (64.2%) than patients with LC (17.5%), CHB (5.0%) and healthy individuals (0). Associated with a worse OS (p < 0.05) |
[173] |
| 10 | CDH1, DNMT3b, ESR1 promoter | + |
Higher methylation in HBV-related HCC compared with LC, CHB and NC. Combined methylation is a better diagnostic marker than AFP. |
[174] |
| 11 | RASSF1A, E-Cadherin, RUNX3 | + |
Hypermethylation in HCC higher than cirrhosis and healthy group (p < 0.001). RASSF1A and E-Cadherin were predictors of HCC within cirrhosis cases. |
[175] |
| 12 | Methylated p16 | + |
Methylated p16 is higher in HCC than CHC, cirrhosis and healthy subjects Higher in patients with normal AFP than higher AFP |
[176] |
| 13 | TLX1, GALR1, ZNF154 | + | Multi-cancer methylation biomarkers combining three methylation markers have a sensitivity and specificity of 37.3% and 83.3% on HCC respectively. | [177] |
| 14 | 38 DNA methylation regions | + | Machine learning method for systematic analysis with a 96% of sensitivity and 98% of specificity in an independent training cohort. | [178] |
| 15 | Enzymatic methyl sequencing | + | Sequencing utilizing enzymatic conversion of unmethylated bases as a screening model to distinguish HCC patients from non-HCC individuals | [179] |
| 16 | TGR5 | + |
Hypermethylation of the TGR5 promoter is significantly higher in HCC than CHB and HCs (p < 0.01) Increased sensitivity when combined with AFP |
[111] |
| 17 | RASSF1A promoter | Hypermethylation of RASSF1A in HCC, but not NC. Reduced RASSF1A is related to TNM stage, metastasis, AFP, portal vein embolus, capsular infiltration, and multiple tumor nodes. | [180] | |
| 18 | UBE2Q1 | + | Hypomethylation of UBE2Q1 in HCC than LC (p = 0.026), CHB (p = 0.006), and HCs (p = 0.011). Negatively associated with TNM stage. Increase sensitivity with AFP combined. | [113] |
| 19 | SEPT9 | + | Increase methylation of SEPT9 in HCC than at-risk and healthy individuals (p < 0.0001) | [181] |
| 20 | INK4A | + | Hypermethylated INK4A in HCC than controls | [116] |
| 21 | ELF, RASSF1A, p16, GSTP1 | Hypermethylation in tumor than non-tumor tissues (p < 0.05), and also combined markers (p < 0.001). Increase sensitivity with AFP combined | [182] | |
| 22 | APC, GSTP1, FASSF1A, SFRP1 | + |
Hypermethylation in all four genes in HCC than normal or benign controls. Methylated RASSF1A is a prognostic marker of overall survival. |
[183] |
| 23 | APC | Methylation status of APC complement with AFP to predict HCC | [184] | |
| 24 | MT1M and MT1G | + |
Hypermethylated MT1M and MT1G in HCC than CHB and NC group (p < 0.001), with a specificity of 94.6%. MT1M promoter methylation positively correlates with tumor size (p < 0.001), and metastasis when combined with MT1G. |
[185] |
| 25 | P16 | + | P16 methylation increases from benign liver disease to HCC progression. | [186] |
| 26 | HOXA1, TSPYL5, B3GALT6 | + | The multi-target HCC blood test utilized three methylation markers and demonstrated high concordance (> = 97%) to predict HCC without significant interference observed. | [187] |
| 27 | Methylation Fingerprint Panel | + | The study identified three panels: cancer-specific biomarker panels, a pan-GI panel, and a multi-cancer prediction panel, with high AUC values ranging from 0.85 to 0.98 for detecting various gastrointestinal cancers. | [118] |
| 28 | RNF135, LDHB | + | Combined RNF135 and LDHB methylation level analysis has a sensitivity of 57% on HCC, compared with AFP at 45%. When combined with HCC, the sensitivity is 70%. | [119] |
| 29 | SEPT9 | + | A significantly higher copy number of methylated SEPT9 was observed in the HCC group than in the control group (p < 0.001) | [188] |
| 30 | SEPT9 | + | SEPT9 methylation pattern is a better predictor than serum AFP for diagnostic performance. | [189] |
| 31 | IGFBP7 | + | Frequency of serum IGFBP7 promoter methylation is higher in HCC than in CHB and HC controls (p < 0.001). | [190] |
| 32 | GNB4, Riplet | + | Circulating tumor cells combined with methylation patterns have a sensitivity of 88.2% and an AUC value of 0.98. | [124] |
| 33 | CCND1 | + | The methylation status of the CCND1 promoter outperforms serum AFP in both AUC and specificity to predict HBV-HCC versus CHB. | [191] |
| 34 | SOX1, VIM | + | Higher frequency of SOX and VIM promoter methylation than LC, CHB, and HC subjects (p < 0.001). | [192] |
| 35 | LINE-1, RASSF1A | + |
LINE-1 was hypomethylated in 66.7% and RASSFIA promoter was hypermethylated in 73.3% of HCC serum. Associated with HBsAg positivity, tumor size, AFP, and lymph node metastasis. |
[193] |
| 36 | BARD1, MAGEB3, BRUNOL5, FXYD6, TET1, TSPAN5, DPPA5, KIAA1210, and LSP1 | + |
Neighboring CpG sites on 9 genes are predictable for prospective HCC development from HBV-negative cirrhotic patients. DPPA5, KIAA1210, LSP1 are hypermethylated, while BARD1, MAGEB3, BRUNOL5, FXYD6, TET1, TSPAN5 are hypomethylated, compared with controls. |
[194] |
| 37 | EXO1 | DNA methylation status in five CpG islands of the EXO1 gene was associated with the prognosis of HCC | [195] | |
| 38 | Wnt/beta-catenin signaling pathway | + | Hypermethylation of Wnt/Beta-catenin signaling pathway is correlated with tumor size, TNM stage, distant metastasis, and HBV infection (p < 0.05). | [126] |
| 39 | SEPT9 | + | Serum methylated SEPT9 test has a high diagnostic accuracy for HCC on cirrhotic patients (AUROC 0.944, p < 0.0001), and is the only variable associated with HCC diagnosis in this cohort. | [196] |
| 40 | RASSF1A, SOCS1 | + | RASSF1A and SOCS-1 methylation were detected in 40% and 38% of HCC patients. RASSF1A/SOCS-1/AFP panel detects HCC at sensitivity of 86% and specificity of 75%. | [197] |
| 41 | CCND2 | + | CCND2 methylation is significantly higher in HCC patients (p < 0.001). Advanced HCC is associated with higher CCND2 methylation and lower CCND2 mRNA levels than early-stage disease. | [198] |
| 42 | GSTP1 | + | Higher GSTP1 promoter region methylation frequency in pre-ACHBLF patients compared to CHB and HCs. Lower GSTP1 mRNA levels in pre-ACHBLF patients. Increased ACHBLF incidence in pre-ACHBLF patients with methylated GSTP1. | [199] |
| 43 | MAGE-A1, MAGE-A3 | Different methylation patterns in several CpG sites among the MAGE-A1 and MAGE-A3 promoters in HCC cells. Clinical sensitivity and specificity were 91.2% and 100%. | [200] | |
| 44 | KLHL35, PAX5, PENK, and SPDYA | These genes are significantly more highly methylated in HCC than in non-cancerous liver tissue, irrespective of the hepatitis virus status. LINE-1 hypomethylation was prevalent in HCC. | [201] | |
| 45 | A1AT, SERPINA1 | + | More fully-methylated SERPINA1 promoters in control than HCC samples, and higher hemimethylation in stage I compared to stage II and III HCC. Higher AFP and A1AT levels in hemimethylated patients (p < 0.001). | [202] |
| 46 | TFPI2 | Methylation of TFPI2 gene was detected in 44.9% of primary HCC samples, 10.7% of the corresponding non-tumorous liver samples, and 5.0% of the normal liver samples. Lower expression of TFPI2 is correlated with TNM stage, and methylation is associated with poorer prognosis (p < 0.001). | [203] | |
| 47 | RUNX3, p16, RASSF1A, CDH1 | + | 88%, 100%, 50%, and 13% of HCC patients were detected with hypermethylation of RUNX3, p16, RASSF1A and CDH1. The inclusion of RUNX3 in the gene panel can potentiate the detection of advanced cancer. | [204] |
| 48 | MAPK10 | Methylation of MAPK10 detected in 58% of HCC cell lines, and 666% of primary HCC tissues, results in downregulated expression of MAPK10 proteins. | [205] | |
| 49 | SOX11 | + | Higher methylation of SOX11 promoter in HCC patients (69.4%) compared to CHB patients (13.6%) and healthy controls (10.7%). Significant difference in SOX11 promoter methylation between HCC patients with vascular invasion and those without. SOX11 methylation demonstrates a 69% sensitivity in distinguishing HCC from CHB, higher than the 57% sensitivity of serum AFP. | [206] |
| 50 | HCCS1 | + | HCCS1 promoter methylation frequency higher in HCC patients compared to CHB patients and healthy controls (P < 0.001). HCCS1 promoter methylation associated with TNM stage (P = 0.01). 62.5% sensitivity for serum HCCS1 promoter methylation in discriminating HCC from CHB, compared to 55% for AFP alone, and sensitivity combined with AFP level is 81.7%. | [207] |
| 51 | Sat2 | Correlation between hypomethylation Sat2 with a breakpoint in chromosome 1(p < 0.001). Sat2 demethylation play a role in early stage of liver carcinogenesis. | [208] | |
| 52 | Sat2, LINE-1 | Sat2 hypomethylation associated with HCC risk. LINE-1 is not associated with HCC risk by age. Decrease in Sat2 methylation and LINE-1 hypomethylation associated with increased risk of HCC for HBsAg carriers. | [209] | |
| 53 | Sat2, LINE-1 | + | Negative relationship between urinary aflatoxin B albumin levels and LINE-1 and Sat2 methylation. It is associated with the risk of HCC development. | [210] |
| 54 | SFRP2 | + | SFRP2 methylation levels significantly higher in patients with HBV-HCC than in those with CHB and healthy controls (p < 0.001). SFRP2 mRNA level significantly lower in HCC group compared to the others (p < 0.05). SFRP2 methylation level showed better diagnostic value than AFP. | [211] |
| 55 | SGIP1 | + | Elevated levels of SGIP1 methylation in HCC patients associated with poorer OS, PFS, and MFS compared to those with low levels (p < 0.05). | [212] |
| 56 | SCAND3, Myo1g | + | SCAND3 and Myo1g methylation were high in HCC cell lines and tissues, and serum SCAND3, Myo1g, and SCAND3 + Myo1g methylation values showed better detection and early detection of HCC than AFP alone. In the AFP-negative HCC group, SCAND3 and Myo1g methylation also can predict diagnosis. | [213] |
| 57 | LZP | Methylation of LZP promoter decreases mRNA expression, and is negatively related to the HCC status. | [214] | |
| 58 | ATM | Higher ATM promoter methylation in HCC tissues, and associated with ATM expression (p < 0.001). Methylation of ATM promoter associated with better outcomes in locally advanced HCC patients who received radiotherapy | [215] | |
| 59 | IL-6 | + | IL-6 promoter methylation levels lower in HCC patients than in CHB patients (p < 0.001). IL-6 promoter methylation level is an independent factor in HCC development, and its diagnostic value is superior to AFP. Combined use of AFP and IL-6 methylation level improves AUC (0.773). | [216] |
| 60 | CDO1 | + | CDO1 promoter methylation frequency higher in HBV-related HCC than in LC, CHB, and healthy controls (p < 0.001). Higher frequency of CDO1 promoter methylation in advanced stages HCC compared to early stages. Improved diagnostic value combined with serum AFP. | [217] |
| 61 | CDH13 | + | Higher methylation frequency of CDH13 promoter in HCC patients compared to NCs and CHB groups (p < 0.05). Methylation level of CDH13 promoter influenced by TBil, ALB, and AFP. Combined methylated CDH13 level and AFP level show better diagnostic score (AUC = 0.796). CDH13 methylation is an independent predictor for HCC prognosis (p < 0.05). | [218] |
| 62 | FES | FES hypermethylation correlated with tumor size, serum AFP, and tumor differentiation (p < 0.005). Both FES hypermethylation and protein downregulation associated with PFS and OS of HCC patients. | [219] | |
| 63 | NLRP7, NLRP2, NLRP3 | + | Hypomethylation of programmed-cell death genes (PRGs) including NLRP7, NLRP2, and NLRP3 in HCC tissues; NLRP3 methylation levels correlated with expression level (r = 0.51). Hypomethylated PRGs can discriminate between early HCC patients and healthy controls in cfDNA analysis (AUC = 0.94), and associated with poor HCC prognosis. | [142] |
| 64 | DBX2, THY1 | + | DBX2 and THY1 are hypomethylated in HCC. Diagnostic sensitivity and specificity of DBX2 for differentiating healthy controls and early stage HCC were 88.89% and 87.10% respectively. Diagnostic sensitivity and specificity of THY1 were 85.19% and 80.65% respectively. | [220] |
| 65 | F-box protein 43 promoter | + | F-box protein 43 promoter methylation levels were significantly lower in HCC PBMCs than in chronic hepatitis B and healthy control PBMCs (P < 0.001). This was superior to those of AFP levels in the diagnosis of HCC. Combination of F-box protein 43 promoter methylation and AFP levels improved the AUC to 0.888 with sensitivity of 76.42% and specificity of 86.08%. | [221] |
| 66 | RASSF1A, p16, p15 | + | Abnormal methylation was found in blood samples 1 to 9 years before HCC was diagnosed. RASSF1A had the highest rate of increased methylation (70%), followed by p16 (44%) and p15 (22%). Combined risk factors and methylation markers, the accuracy of predicting HCC was 89%, with 84% sensitivity and 94% specificity | [222] |
| GNB4, Riplet | + | In tissue validation, GNB4 and Riplet had an AUC of 100%, with 100% sensitivity and specificity for detecting any-stage HCC. In blood tests, this combination showed a high sensitivity of 84.39% and specificity of 91.92%, with an AUC of 92.51% for detecting any-stage HCC. The dual-marker panel was more sensitive for detecting stage I HCC than AFP. It had a high sensitivity (70.27%) for detecting a single tumour of size 3 cm or less. | [144] |
Hypermethylation of Ras Association Domain Family Member 1A (RASSF1A) is one of the most commonly identified methylation markers for HCC. RASSF1A is a tumor suppressor gene that regulates cell progression by apoptosis, microtubule stability, and cell proliferation. The CpG island of the RASSF1A promoter is usually highly methylated in HCC patients, ranging from 40 to 93%, varying from different studies [168, 197]. This hypermethylation of RASSF1A in HCC patients can be detected in both tumor tissues [166] and liquid biopsy using circulating cfDNA [168]. When combined with serum AFP, methylation of RASSF1A is a better diagnostic biomarker for HCC, especially in patients with hepatitis infection, with a sensitivity and specificity of 65.0% to 85.0% and 70.0% to 100%, respectively [167, 182]. Notably, methylation is also detected on the RASSF1A gene of cirrhotic liver tissues, but not as high as the tumor tissues [175]. Among the cirrhosis cases, RASASF1A could be considered a prediction biomarker for developing HCC [175]. A study has demonstrated that the methylation extent of RASSF1A increases during CLD development, from chronic hepatitis B to cirrhosis and finally into HCC [173]. Consistently, a cohort study with patients recruited in a cancer screening study from Zhang et al. revealed that abnormal methylation of RASSF1A can be detected as early as 1 to 9 years before HCC is diagnosed [222]. Therefore, screening RASSF1A methylation status is considered an effective measure for early detection of HCC in patients with high cancer risk, such as CLD or HBV carriers [180, 222, 223]. However, there are also challenges to validating RASSF1A hypermethylation as a cancer biomarker due to contradictory data on methylation frequency in various malignant diseases for cfDNA and gDNA. Differences in the CpG sites analyzed could cause these discrepancies, the cancer stages of the patients, and the timing of RASSF1A methylation in tumorigenesis. Furthermore, inconsistencies in the association between RASSF1A methylation and clinicopathological parameters suggest that other genes’ aberrant methylation may significantly influence tumorigenesis in some cancers [224].
Additionally, methylation of the CDKN2A, coding for the tumor suppressor protein p16, is a well-studied HCC detection biomarker. P16, a tumor suppressor protein, is vital for controlling the cell cycle by blocking the actions of CDKs. By inhibiting these enzymes, p16 helps avert uncontrolled cell growth and division, which could lead to tumor formation. Hypermethylation of CDKN2A results in a lower level of p16, which is more prevalent in patients with HCC than in chronic hepatic cirrhosis and healthy control people [176]. The levels of p16 increase with HCC progression from benign liver disease [186]. It is also a potent marker as it can help differentiate HCC among patients with normal AFP than higher AFP levels [176]. Like RASSF1A, p16 methylation was found in 44% of patients before HCC diagnosis [222].
The septin 9 (SEPT9) gene, a crucial regulator of cell division, functions as a tumor suppressor. Its excessive methylation is associated with the progression of liver cancer [225, 226]. The SEPT9 gene is expressed in all healthy tissues, but its expression is reduced or silenced due to abnormal promoter hypermethylation in liver cancer [227]. An extensive epigenome analysis of 304 liver cancer tissue specimens identified SEPT9 as a crucial epi-driver gene in liver cancer development, primarily due to the hypermethylation of the SEPT9-promoter [225]. Increased methylation of SEPT9 is found in HCC patients than in healthy individuals [181], and it’s a better predictor than serum AFP for diagnosis of HCC. It is also reported that a higher copy number of SEPT9 methylation was presented in HCC than in control individuals [188].
Glutathione S-transferase pi 1 (GSTP1) is a protein detoxifying harmful substances, and its hypermethylation is often observed in HCC. This hypermethylation reduces the GSTP1 enzyme’s production, impairing detoxification processes and potentially contributing to HCC development and progression [228]. GSTP1 hypermethylation and AFP level can be a high-sensitivity biomarker for detecting HCC at 93.55% sensitivity [182]. It is also consistent with RASSF1A and is hypermethylated in tumors compared to benign control groups [183]. In patients with acute-on-chronic hepatitis B liver failure (ACHBLF), which is a condition of acute regression of liver function due to CLDs such as HBV infection, GSTP1 methylation frequency was significantly higher than in chronic hepatitis B patients and healthy individuals [199]. The higher methylation and lower expression of GSTP1 are associated with ACHBLF incidence.
A repetitive DNA sequence known as a retrotransposon, long interspersed nuclear element-1 (LINE-1), is found in the genome. In the case of HCC, LINE-1 exhibits overexpression and hypomethylation. This hypomethylation in LINE-1 elements can result in genomic instability, higher mutation rates, and changes in gene expression, which can all contribute to the development and progression of cancer. Research has demonstrated a connection between LINE-1 hypomethylation and unfavorable outcomes in individuals with liver cancer. More than 50% of HCC patients demonstrate an overall hypomethylation in LINC-1 [201]. This pattern is also observed in cfDNA, ctDNA, and blood leukocyte DNA [172, 193]. Additionally, satellite 2 DNA (Sat2), located in the 1q12 pericentromeric region, is a sequence with reduced methylation in cancer cells and contributes to chromosomal instability [229]. Recent research has revealed that Sat2 and similar heterochromatic regions can generate ncRNAs [230], which might have a role in responding to stress and facilitating heterochromatin reformation. Nevertheless, the influence of DNA hypomethylation on Sat2 expression remains uncertain, as different studies have produced inconsistent results. Nonetheless, several studies have reported that Sat2 hypomethylation is associated with HCC risk. A significant relationship has been identified between Sat2 sequences with reduced methylation and increased 1q copies having a 1q12 breakpoint. This decrease in methylation is thought to change how CpG-rich satellite DNA interacts with chromatin proteins, causing heterochromatin to decondense, break, and form 1q abnormally. This implies that the demethylation of Sat2 could be involved in the initial phases of the development of liver cancer [208]. Meanwhile, aflatoxin is a well-described carcinogen leading to HCC. Aflatoxin B1 albumin is a metabolite under aflatoxin exposure and can be excreted through urine. A cohort study with 1140 cancer-free participants revealed a reverse association between the aflatoxin concentration and global LINE-1 and Sat2 methylation [210]. This demonstrates that there may be an epigenetic regulation effect of aflatoxin, which results in HCC development. Urinary aflatoxin metabolite level and DNA methylation can be a biomarker of potential HCC occurrence. On the other hand, a prospective case-control study measuring methylation levels in leukocyte DNA suggested that Sat2 hypomethylation is associated with the risk of HCC, but LINE-1 is not associated with HCC risk by age [209]. The association is stronger within HBsAg carrier between Sat2 and LINE-1 hypomethylation and the risk of HCC.
Other reported methylated genes that can be potentially regarded as biomarkers of HCC are listed in Table 2.
Presence of tumor-derived vesicles in the circulation
Extracellular vesicles (EVs) are tiny, membrane-bound capsules containing various organ components [231]. There are three main types of EVs: microvesicles, exosomes, and apoptotic bodies, each varying in size and composition and released from cells in different ways [232]. EVs contain various cellular components such as proteins, DNA, RNA, and lipids [233]. It plays a crucial role in cell-to-cell communication and is involved in essential biological processes like inflammation and tissue remodeling [234]. Various liver cells, such as hepatocytes, hepatic stellate cells, and endothelial cells, are released into the bloodstream. The EVs released by the tumor cells are tumor-derived vesicles [235]. LncRNAs, which regulate biological and genetic functions, have been found in EVs and linked to numerous diseases. The composition of EVs can change in response to different stimuli, making them potential biomarkers for HCC. Liquid biopsy methods are now widely used to detect the presence of EVs, providing valuable information about the occurrence and progression of HCC [235].
Change in viral load in viral hepatitis-related HCC patients
Pre-treatment serological data were analyzed in a study of 125 HBV-positive HCC patients with chemotherapy. Upon analyzing the virological data alongside conventional clinical variables, factors such as high total bilirubin, HCV infection, and elevated HBV DNA levels significantly influenced survival outcomes [236]. Notably, an exploratory analysis revealed a higher incidence of severe hepatitis during chemotherapy in patients with high pretreatment HBV DNA levels. On the other hand, Choi and the others reported that the association of viral load on patients with HBV but not cirrhosis, with or without prior anti-viral treatment, is non-linear [237, 238]. The risk of developing into HCC is the highest at around 6 to 7 logIU/mL. The risk decreases with increasing baseline viral load. It is worth mentioning that CLDs with viral infection are alone the dangerous risk factor leading to HCC. Therefore, the clinical importance of checking the viral load as a potential prognostic biomarker is crucial. Several viral factors contributing to HCC, including HBsAg, HBeAg, viral DNA levels, viral genotypes, and viral mutations, may be considered integratively and provide essential information for surveillance program development [239].
Potential metabolomics clues for HCC diagnosis
On the other hand, metabolic profiling is also an emerging research direction in the biomarkers of HCC diagnosis. With the advancement of Omics technology, metabolic profiles from hepatitis patients and HCC patients were characterized and compared. High classification accuracy models could be derived from these multi-omics data [76]. “Omics” technologies, which include genomics, transcriptomics, proteomics, and metabolomics, provide an in-depth molecular and biochemical analysis of a biological entity, whether it’s an organism, tissue, or specific cell type. Metabolomics, which pertains to the exhaustive study of lightweight metabolites within a biological sample, holds several unique advantages. It is more closely aligned with the phenotype, is not restricted to specific species, and involves fewer molecules than other omics technologies. The metabolite profile serves as a comprehensive summary of the preceding omics profiles, which makes it particularly effective in identifying minor alterations in metabolic pathways and shifts from homeostasis [240]. Techniques such as MS and nuclear magnetic resonance (NMR) spectroscopy have been effectively used in metabolomic analysis.
Gas chromatography (GC)-MS-based metabolomics is commonly used to analyze patients’ serum. In a study involving 20 HCC patients and 20 normal subjects, serum metabolome was analyzed. Metabolites, including butanoic acid, L-valine, glycerol, etc., significantly differentiate the two groups. The specificity is 100%, and the total accuracy is 75% [241]. The same group also investigated using urine metabolomics via GC/MS, yielding an AUC of 0.9275 for detecting HCC from control groups combined with AFP [242]. Another metabolic profiling study unveiled 43 serum metabolites and 31 urinary metabolites in HCC patients, which impact crucial metabolic pathways [243]. Specific metabolites, including bile acids, histidine, and inosine, exhibited notable statistical variances and considerable fold changes, hinting at their prospective utility as HCC biomarkers. Nonetheless, it was observed that irregular levels of specific bile acids were linked with conditions such as liver cirrhosis and hepatitis. The research also distinguished HCC patients with low AFP values from healthy subjects, utilizing a set of metabolite markers. Moreover, membrane phospholipids, measured by LC-MS/MS, were shown to discriminate HCC from adjacent liver tissues with an AUC of 0.963 [243].
Although the technology has matured for more than a decade already, the detailed metabolic profiles of HCC patients have not yet been well characterized due to the heterogeneity of this disease. The training and validation cohort have not been large enough to validate the power of specific metabolic panels for HCC diagnosis. With the advancement of analytical tools, the omics approach is inevitably essential to precision medicine.
Omics approach to identify novel biomarkers
The general approach of using omics technologies to diagnose HCC involves analyzing an array of biological molecules, such as proteins, metabolites, and modified RNA sites, to identify specific biomarkers and molecular profiles associated with the disease [244]. This comprehensive and integrative method allows for early detection and improved understanding of HCC pathogenesis, ultimately leading to better patient outcomes and targeted treatment options [245]. By overcoming challenges in data analysis, identification, and accessibility, omics technologies can revolutionize the field of HCC diagnosis and management. Mass spectrometry (MS)-based omics technologies have the potential to diagnose HCC by analyzing biological molecules related to disease development, such as proteomics techniques that identify specific serum protein panels [246]. However, there are challenges to overcome, such as the rapid identification of metabolites, proteins, and modified RNA sites and the development of feasible in-house databases and powerful bioinformatics tools [247, 248]. Addressing these challenges and making multi-omic strategies accessible to non-specialist groups could lead to a better understanding of HCC pathogenesis and improved human health outcomes.
Additionally, employing bioinformatics analysis on the current dataset is a valuable method for the secondary discovery of potential targets. Establishing prediction models based on cfDNA, methylation profiling, and somatic copy number aberrations data is a viable strategy [102, 249–251]. In recent years, the integration of machine learning algorithms has substantially advanced the field of biomarker discovery [178, 252]. Besides the bioinformatics approach to the studies of epigenetics and ncRNAs [253, 254], spatial proteomics, or transcriptomics [253, 254] can also contribute to the disease prognosis and management. However, it is crucial to recognize that bioinformatics should be utilized as a supplementary tool in biomarker research rather than a primary source due to the absence of experimental evidence. Consequently, further experimental and clinical validations must be conducted to ensure the efficacy of the identified biomarkers.
Taken together, many promising targets can be considered as new liquid biopsy biomarkers for detecting HCC. However, there is still a long way to look for a new universal serological HCC detection biomarker using liquid biopsy to replace AFP completely.
Biomarkers for treatment options and therapeutic response
Some recent review articles have discussed the representative findings of molecular biomarkers to predict HCC prognosis and the current staging system to predict HCC prognosis [32, 35, 255, 256]. The following part highlights the current understanding of biomarkers that can provide insights into treatment options and therapeutic responses (Fig. 2).
Fig. 2.
Biomarkers for therapeutic options. To facilitate the decision on the best treatment for HCC patients, biomarkers predicting therapeutic response to different treatments, including surgical resection, TACE, HAIC, liver transplantation, targeted therapies, and immunotherapies, are being investigated. Some of the significant aspects of the studies are highlighted in the illustration, and the predicted response and correlated clinical outcomes are drawn on the diagram
Biomarkers related to surgical resection
While the surgical resection guidelines are beyond this review’s scope, we will discuss biomarkers for postoperative monitoring and prediction of recurrence. A recent study revealed that serum AFP level greater than 400 ng/mL is a risk factor for OS and recurrence-free survival (RFS) (HR: 1.512) [257], which is consistent with previous studies that AFP decrease after surgery is an excellent prognostic indicator [258, 259]. Another study has combined the level of sulfite oxidase with AFP, which can predict postoperative outcomes and recurrence risk at sensitivity and specificity of 93.8% and 95.2%, respectively [260].
On the other hand, Yamamoto et al. compared the predictive values of AFP, AFP-L3%, DCP, and GP73 in postoperative patients [261]. It was found that DCP is the best prognostic marker for HCC development, but AFP-L3% may be more significant for early recognition of tumor recurrence. Another study investigating the significance of combining these three tumor markers found that the number of elevated markers post-surgery predicts survival and recurrence [262]. A multivariable analysis using large-scale clinical data has identified that males, large tumors, multi-nodules, high albumin-bilirubin, and high serum AFP are the predictors of early recurrence after hepatectomy [263].
It was shown that GPC3-positive patients with HCC have worse survival than GPC3-negative patients [264]. In general, higher GPC3 expression is associated with poorer prognosis in HCC patients, no matter who is HBV-related, post-operative, or not [265–267]. Similar results were observed for serum GPC3, in which a higher level signifies a shorter OS and disease-free survival (DFS) after surgical treatment [268, 269], although there are also controversial results [270]. Consequently, GPC3 is being investigated for its potential when combined with other biomarkers for the prognostic indication. The expression of GPC3 and OPN in HCC tissue was a negative predictor for patients with HBV-related small HCC tumors who have undergone surgical resection [271]. On the other hand, Feng et al. discovered a link between the expression of GPC3 and CK-19 in HCC tissue and local recurrence in patients with post-surgical resection for HCC [272].
Apart from the conventional serological markers, there are also reports of other potential biomarkers. A study found that platelet count (PLT) and five platelet-related factors can predict HCC recurrence, especially late recurrence, in non-cirrhotic patients [263]. Ascites, PLT levels, alkaline phosphatase levels, and tumor sizes were independently associated with the risk of HCC recurrence. However, larger cohorts are needed to validate these findings. NcRNAs are also within the research scope. The HCCseek-23 panel with eight miRNA showed 81% sensitivity and 83% specificity for early-stage HCC diagnosis and 93% sensitivity for identifying AFP-negative HCC. Combined with serum biomarkers, they were significantly associated with DFS. Furthermore, lncRNA MVIH has also been reported to be a predictor. Overexpression of MVIH is correlated with poor RFS (p < 0.001) and OS (p = 0.007) of postoperative HCC patients [273]. One study found that circFOXK2 is highly expressed in HCC tissues and correlates with poor outcomes in patients undergoing radical hepatectomy (p = 0.0005) [274]. The research revealed that circFOXK2 promotes HCC progression and activates the Warburg effect by upregulating specific proteins and miRNA sponges, making it a potential prognostic marker and drug target for HCC.
Identifying CTCs with specific genes can also help healthcare providers predict patients’ responses to treatment. For instance, MAGE3, surviving, and CEA could be used to indicate the effectiveness of cryosurgery on unresectable HCC [275]. CTCs are associated with tumor diffusion and thrombosis, and the presence and number of CTCs are associated with worse survival [79]. A study focusing on the EMT properties of CTCs found that CTC count and mesenchymal characteristics of CTCs are associated with early recurrence or postoperative DFS [80]. During the process of EMT, the expression of EpCAM is usually down-regulated, and it becomes no longer the only reliable marker for the enrichment of CTCs [276]. Another study has shown consistent results that CTCs with high EMT phenotypes are more likely to be metastatic. More epithelial markers, such as CD133, vimentin, HER2, and PSMA, are provided for the positive enrichment of these stem-like CTCs [277]. Additionally, HCC patients with CTCs detected are associated with vascular invasion, elevated AFP levels, disease progression, recurrence, and poorer prognosis [278–280].
On the other hand, Chen and others performed the unique molecular identifier target sequencing on plasma and PBMCs from HCC patients [107]. They found that mutation variant frequency changes (MVFCs) correlate with shorter RFS, thus suggesting minimum residual disease (MRD). MRD refers to the small number of tumor cells that remain in the primary site after treatment, usually surgical resection. That tiny tumor volume is not generally detectable by conventional detection strategies, such as ultrasound, imaging, or serological tests. MRD is a condition in which patients are susceptible to recurrence before average time and is usually accompanied by shorter RFS and OS. Chen defined the MRD-positive patients as MVFC mutations favorable in ctDNA. The prognostic evaluation is greatly improved when combined with serum AFP and DCP (p < 0.0001). The design of their platform is also compatible with different gene panels, so it is theoretically applicable to other samples or diseases.
Biomarkers related to transarterial chemoembolization and hepatic arterial infusion chemotherapy
Transarterial Chemoembolization (TACE) is a medical process where a catheter is used to directly administer chemotherapy, typically doxorubicin, into the tumor’s feeding arteries. This is then followed by blocking these arteries [281]. This procedure has a relatively low mortality rate related to treatment, which is less than 5%, and it has been found to extend the median survival rate to somewhere between 11 and 20 months [282]. Notably, this treatment does not yield practical results for over 40% of patients who are left to depend on less potent systemic therapies [283]. Hence, discovering predictive biomarkers for TACE treatment response could greatly benefit patients. This would enable them to transition to alternative therapies promptly without unnecessary delay. Pyruvate kinase muscle isozyme M2 (PKM2) is reported to be associated with resistance (p < 0.01) and survival to TACE (27.5 m vs. undefined, p < 0.0001) [284]. Elevated PKM2 predicts a poor therapeutic outcome. Furthermore, cfDNA and ctDNA levels are also suggested to be associated with the failure of the TACE procedure, with a sensitivity of 80% and a specificity of 95.6% [285]. A recent study on 132 patients showed that DNA-dependent protein kinase (DNA-PKcs) is associated with poorer survival (4.5 vs. 16.9 m, p = 0.011) [286]. The expression of DNA-PKcs is related to doxorubicin resistance.
Like TACE, Hepatic arterial infusion chemotherapy (HAIC) delivers the chemotherapies into the liver, but instead of the arteries, the vein leading to nearby organs will be blocked. A study found that serum VEGF levels in advanced HCC patients can predict the therapeutic response and survival for receiving cisplatin and 5-FU via HAIC [287]. In a recent Phase III clinical trial testing the efficacy of HAIC-oxaliplatin plus fluorouracil (HAIC-FO), a 15-gene mutation panel was discovered that can predict the responsiveness to the treatment. 83% of patients were identified to have a longer OS (19.3 vs. 10.6 months, p = 0.002) [288].
Biomarkers related to local ablation
Local ablation involves using electrical equipment to heat and destroy tumor tissues at the local site, typically through radiofrequency. At temperatures exceeding 60 °C, the probe burns the tumor tissues and adjacent inoperable or metastatic tissues. This procedure is sometimes performed via endoscopy, or in the case of larger tumors, a laparotomy may be required. The decision to utilize local ablation for HCC treatment primarily depends on tumor size, ascertained through CT or MRI. The Child–Pugh score is also occasionally considered, with only patients classified as grade A or B deemed suitable for ablation therapies [289]. AFP level is perceived as an indicator of whether patients will experience recurrence following ablation [290]. A comprehensive retrospective study involving nearly 3,000 patients who underwent tumor ablation revealed that additional biomarkers, including tumor size and numbers, serum DCP, platelet counts, Child-Pugh score, and HCV viral load, are associated with distant recurrence [291]. An Italian survey corroborated these findings, reporting that advanced age is an independent predictor of poor OS apart from AFP levels, while ALT can prognosticate recurrence [292]. More studies suggest AFP-L3%, DCP, neutrophil to lymphocyte ratio, ferritin, and specific miRNAs correlate with the OS and DFS after ablation, but further research is awaited to confirm their clinical significance [293].
Biomarkers related to liver transplantation
Liver Transplantation is considered when patients have unresectable HCC tumors by conventional surgery, but the tumors are still not widespread, usually at T1. Patients have to have good health conditions and normal liver function. As liver transplantation involves the donation of liver tissues from another individual and is a complicated procedure, it is precious. It requires special attention before the clinician and surgeon decide to operate on the patient. Therefore, reliable biomarkers are needed to help determine if a patient is suitable for transplantation and the predicted prognosis after the surgery. AFP levels are usually considered only in current clinical guidelines for therapeutic options. For example, patients with AFP levels above 1000 ng/mL are not considered for liver transplantation due to poor predicted prognosis after operation [46, 294, 295]. Although AFP is one of the most widely used biomarkers for HCC diagnosis, its stability as an indicator of disease progression and the reference range has been controversial. Studies report a positive correlation between AFP mRNA level and tumor size, circulating malignant hepatocytes, and metastases. However, controversial studies have also been reported [296–299], limiting its significance as a prognostic marker for HCC.
Parameters, including tumor sizes, numbers, serum AFP levels, and tumor uptake on FDG PET/CT, are recognized post-transplantation predictors of tumor recurrence in HCC patients [300–303]. A combined set of criteria, including tumor sizes, numbers, serum AFP levels, and maximum tumor-to-background ratio, is the most effective way to predict post-transplant recurrence and select candidates for living donor liver transplantation compared to conventional criteria sets [304]. A study of 103 patients with HCC found that poorly differentiated tumors and high AFP levels were linked with poorer survival (p < 0.05) and higher recurrence (p < 0.05) after liver transplantation [305]. The research suggests that these tumor biological characteristics should be taken into account before performing the procedure for patients with HBV-associated HCC. This information can help assess HCC patient survival time after liver transplantation. A 30-year comprehensive study calculated the recurrence rate of 865 HCC patients receiving liver transplantation. Multivariate factors predicting recurrence were developed, including tumor grade, macrovascular and microvascular invasion, tumors outside Milan criteria, radiographic maximum diameter, and specific pre-transplant neutrophil-to-lymphocyte ratios [281]. The situation is highly individual and requires more clinical information to help make decisions.
Biomarkers related to targeted therapies
For patients who have unresectable HCC tumors and who are also not suitable for liver transplantation, systemic therapies are the first-line treatment. Sorafenib is the most commonly used first-line drug, but the development of sorafenib resistance is usually observed. The study of sorafenib resistance is a hot topic, and many molecular mechanisms leading to sorafenib resistance have been discovered [306]. Therefore, it is crucial to predict the drug response to systemic therapies in HCC patients [307]. In a study using data from the SORAMIC study, researchers utilized EV-based proteomics to predict treatment response [308]. Low GPX3 and high ARHGAP1 in plasma predicted better treatment response to sorafenib with AUC 0.97. From another phase III SHARP trial, the plasma Tregs and CD56Dim NK cells are correlated with shorter OS for patients receiving sorafenib (p < 0.05) [309]. When sorafenib is combined with mFOLFOX, plasma sVEGFR21 is associated with shorter time-to-progression in HCC patients. Consistent results were observed that VEGF and HIF in HCC have poor predicted treatment outcomes (p < 0.001) and OS (p < 0.001) [310]. Apart from typical serological markers, ncRNAs are also at the hot spot. For instance, the lncRNA Linc01056 is found to be downregulated in HCC tumors, where its expression level is related to sorafenib sensitivity [311]. Besides, from a genomic study using patient-derived tissues, a sub-type of β‐Catenin with high Male predominance has low miRNA expression and is sensitive to sorafenib [312]. Similarly, the circulating level of another miRNA, miR-518d-5p, is reported to be a potential sorafenib resistance biomarker in BCLC-C HCC patients [313]. On the other hand, the immune microenvironment also affects systemic drugs’ therapeutic effect. A study showed that high CD4+ T effector/Treg ratios and IFN-γ production by CD8+Ki67+ T cells are potential biomarkers for improved survival and response to sorafenib therapy [310]. Similarly, pERK+/pAkt- CTCs were discovered to be correlated with better prognosis in HCC patients receiving sorafenib treatment [314].
Lenvatinib is sometimes regarded as the second-line therapy after sorafenib resistance has arisen. High expression of FAT1 and LRP1B genes in ctDNA is associated with poor PFS and a higher recurrence rate [315]. Also, serum ST6GAL1, a tumor-secreted protein, is positively correlated with better survival of Lenvatinib [316]. Likewise, a higher level of circMED27 in the serum of HCC patients also correlates with worse prognosis and clinical outcomes when treated with Lenvatinib [317]. Some other biomarkers related to treating HCC with targeted therapies are reviewed in some recent articles [7, 318, 319].
Biomarkers related to immunotherapies
The immune-suppressive environment is one of the reasons for the poor treatment response of HCC. A recent clinical study measured the serum AFP and CRP levels from patients with HCC treated with anti-PD-(L)-1 therapies. The two parameters were identified as independent predictive markers and developed as the CRAFITY score. The CRAFITY score is associated with survival and therapeutic response in those HCC patients [320]. In 2024, a different study examined the significance of AFP trends in forecasting responses to systemic treatment in HCC patients undergoing bevacizumab combined with immunotherapy [321]. The retrospective evaluation of 536 HCC individuals revealed three unique AFP patterns in both AFP-low and AFP-high groups, with a quick decrease in AFP following treatment associated with improved patient outcomes (HR: 0.2 in AFP-low, and 0.04 in AFP-high class). These results highlight the possibility of using AFP trends as an independent biomarker for predicting clinical results, offering a dynamic prediction tool for systemic therapy prognosis in HCC patients and assisting in clinical decision-making. Weng et al. discovered that SLAMF7 deficiency-caused CCL2 signaling can modulate macrophage repolarization and result in an immunosuppressive response (p = 0.00034) [322]. SLAMF7 could be used as a biomarker and drug target to predict patient immunotherapy response. Winograd et al. also found that PD-L1+ CTCs could help identify HCC patients likely to respond to immunotherapy [323].
A detailed molecular analysis was carried out on tumor biopsy samples from 60 HCC patients insensitive to sorafenib treatment to understand better the genomic characteristics of HCC patients treating anti-PD-1 [324]. The research found a 10% overall response rate to the therapy, with key contributing factors including female gender, PD-L1 positivity, and a low neutrophil-to-lymphocyte ratio. Notably, CTNNB1 mutations and up-regulation of MET were only detected in those who did not respond to treatment, underscoring their potential as predictive biomarkers. Therefore, these biomarkers merit further exploration to improve our knowledge of their potential clinical application in managing HCC patients receiving immunotherapy. Non-coding RNAs such as RNA methylation and circRNA can also serve as biomarkers to predict the effectiveness of immunotherapies in HCC [325]. For instance, the circuit can inhibit the immune response to HCC tumors and stimulate tumor growth via the Snail/DPP4/CXCL10 axis [326].
Notably, atezolizumab (Anti-PD-L1) combined with bevacizumab (Anti-VEGF) (AtezoBev) is one of the standard therapeutic options for unresectable HCC. However, an integrated molecular analysis from samples of patients enrolled in a clinical trial demonstrated that the pre-existing immunity population, with CD274 expression (p = 0.0011), T effector signature (p = 0.0035), and density of intratumoral CD8+ T cells (p = 0.053) as the predictors of better therapeutic response to this combination therapy [327]. Conversely, a high Treg to effector T cells ratio and expression of GPC3 and AFP worsen the clinical benefit. The serum AFP and albumin-bilirubin (ALBI) levels can be biomarkers to predict the response to AtezoBev [328]. The multivariate analysis established an independent correlation between OS and the ALBI-AFP combination, where patients in the ALBI2-AFP non-responders category exhibited the poorest prognosis.
A stage-specific immune classifier was developed by Shi et al. based on patients’ samples [329]. This panel of markers can detect early-stage HCC with a performance superior to serum AFP. It also provides information about whether the patients are resistant to anti-PD-1 monotherapies. Similarly, another study reported that β-catenin activation can result in immune escape of the HCC tumor, and the patients will be more resistant to anti-PD-1 immunotherapies [330]. Bioinformatics analysis also provides potential prognostic biomarkers and therapeutic targets for HCC [331]. Research on novel biomarkers for predicting immunotherapy responses is emerging, and it is anticipated that a more precise understanding will be attained through advanced technologies that analyze the tumor immune landscape, such as single-cell RNA sequencing (scRNA-seq), spatial transcriptomics and multi-omics approaches.
With the help of scRNA-seq, the immunosuppressive microenvironment can be well characterized, and more insight into the interaction between the immune cells can be illustrated. For instance, He et al. utilized the multi-dimensional scRNA-seq to analyze samples from AFP-positive and AFP-negative HCC and adjacent normal tissues [332]. The sequencing results showed that APC-positive tumors are associated with multifaceted immune distortion, including T cell exhaustion and tumor-associated macrophages (TAMs) elevation. When looking further into details, SPP1+ TAMs are responsible for the T cell suppression. On the other hand, the CTCs are also regulating the immune microenvironment through intercellular signaling. CCL5 overexpression in CTCs can recruit regulatory T cells, which help the HCC tumor escape from immune response and metastasis [332]. Furthermore, scRNA-seq has offered insights demonstrating that patients showing PR or SD experienced immune shifts toward cytotoxic T cells. In contrast, patients with PD showed increased numbers of CD14+ and CD16+ monocytes and the upregulation of neutrophil-associated pathways[324]. These observations imply that cytotoxic T cell infiltration, increased activated CD8+ T cells during anti-PD-1 treatment, and the suppression of neutrophil-associated markers could be critical markers for predicting and enhancing response to anti-PD-1 therapy in HCC patients.
The microbiome, defined as the collective genomes of the microbiota within a host, is increasingly acknowledged for its role in immune regulation and responses to various cancer treatments [333, 334]. Significantly, the gut microbiome and the liver interact bidirectionally through the gut-liver axis, enabling gut microbiota and their byproducts to both directly and indirectly impact gene expression in hepatocytes, tumor cells, and non-tumor cells, including immune cells [335–337]. A metagenomic sequencing research study revealed that fecal samples from immunotherapy responders exhibited higher taxa richness and gene counts [338]. It concludes that gut microbiome variations could significantly influence the responses of HCC patients to anti-PD-1 immunotherapy. Interestingly, a meta-analysis involving 1,056 HCC patients revealed that antibiotic-induced reduction in microbiota diversity does not affect OS in HCC patients on ICIs. However, more extensive and better-controlled studies are warranted [339]. A prospective clinical study explores the relationship between the gut microbiome and combined immunotherapy in patients with HCC [340]. Initial findings showed higher gut microbiome diversity in immunotherapy responders, but this difference faded by week 6 of treatment, while certain gut bacteria showed correlations with specific serum metabolites. The study suggests that gut microbiome biomarkers and serum metabolites could help identify HCC patients who would benefit from immunotherapy. Another study reported that antibiotic exposure around the time of treatment initiation has been associated with adverse prognostic implications in patients receiving AtezoBev [341].
In addition to the microbiome, gut metabolites have also been associated with immunotherapeutic responses. In a recent study investigating the connection between gut microbiota and metabolites in predicting outcomes for unresectable HCC patients undergoing treatment with ICIs, researchers discovered significant differences in fecal bacteria between patients with objective response (OR) and those with progressive disease (PD) before starting immunotherapy [342]. The study emphasizes the relationship between fecal microbiota, bile acids, and immunotherapy outcomes for HCC. It showcases the potential of gut microbiota and metabolites as biomarkers for predicting treatment outcomes in ICI-treated HCC patients. Research exploring the impact of the microbiome on immunotherapy responses in HCC patients is still in its early stages. The current consensus suggests that the microbiome modulates the immune environment and response. However, the complex mechanisms that drive this interaction and the potential association between diversity and sensitivity remain unexplored.
Candidate drug targets in HCC and recent clinical trials
Despite ongoing improvements in disease diagnostic platforms, it is crucial that treatment options concurrently evolve to enhance patient prognosis. Notably, conventional targeted drugs, such as sorafenib, Lenvatinib, and regorafenib, have been associated with the rapid development of drug resistance. To combat this, current research in systemic therapies for HCC primarily focuses on combination therapies supplemented with additional targeted inhibitors [343]. Research is exploring the inhibition of multiple molecular targets to boost therapeutic efficacy and longevity, potentially broadening the applicable patient demographic [344]. For example, CD147 is a glycoprotein reported to be highly expressed, and its presence enhances the malignancy of HCC [345, 346]. The signaling machinery involving CD147 is related to the tumor cell properties [346, 347], but efforts have been made to target CD147 using antibodies [348]. On the other hand, GC33 demonstrated anti-tumor activity in both animal models and phase I clinical trials, believed to regulate GPC3 expression in HCC, thus potentially benefiting HCC patients [349]. It was thought to control the GPC3 expression in HCC and has the potential clinical benefit to HCC patients. However, subsequent phase Ib and II studies revealed no clinical benefits within the HCC population, even with concurrent sorafenib treatment [350, 351]. In response to these findings, recent clinical trials have commenced investigations into combinatorial treatments involving other molecular inhibitors alongside targeted drugs. In one phase II trial (NCT01246986), TGF-βRI inhibitor LY2157299 was administered with sorafenib or ramucirumab, a VEGFA inhibitor [352]. The results indicated a correlation between lower TGF-β1 levels and extended survival in HCC patients. Another avenue of exploration is the ICI AK104 (PD-1/CTLA-4 bispecific antibody), which, when combined with Lenvatinib as a first-line treatment for unresectable HCC patients in a Phase II study (NCT04444167), exhibited promising anti-tumor activity. No toxicities or unforeseen safety issues were reported, suggesting a promising direction for future HCC treatment strategies [353]. Additionally, apart from being the conventional HCC biomarkers, T-cell receptors specific for AFP peptide are also engineered to be tested in clinical trials [354–356]. However, the studies are still in the early stages, and there is yet to validate the safety and efficacy of this TCR-gene transfer therapy.
Indeed, AtezoBev has become a significant focus in HCC research. Numerous recent clinical trials across various phases are investigating the potential of AtezoBev for treating different sub-categories of HCC. In contrast to anti-PD-1 monotherapy, the combination of atezolizumab and bevacizumab has shown promising responses in intrahepatic lesions, similar to those observed in extrahepatic lesions. This combination may potentially overcome the immune-tolerant hepatic microenvironment in patients with advanced HCC, offering a new avenue for treatment and improved outcomes. Ongoing clinical trials are also examining the application of AtezoBev in different contexts. For example, the Kirros trial (NCT06096779) investigates the impact of Bevacizumab within the AtezoBev combination on patients with advanced or metastatic HCC accompanied by cirrhosis. Moreover, the CCGLC-001 trial (NCT05713994) evaluates the efficacy of AtezoBev in HAIC. In contrast, the CCGLC-008 trial (NCT05717738) investigates the combined therapy of TACE, anti-VEGF antibodies or pan-target anti-angiogenic drugs, and anti-PD-1/PD-L1 antibodies for advanced HCC patients who are not suitable for radical treatment, resection, transplantation, or ablation. Examining AtezoBev in these clinical settings underscores its potential as a promising treatment option for HCC patients, expanding the range of therapeutic approaches and possibly improving patient outcomes. More recent completed and ongoing clinical trials testing on targeted therapies are listed in Table 3.
Table 3.
Recent clinical trials on targeted drugs from 2020
| Interventions | Trial Name/Identifier | Patient no. | Study Type | Primary Endpoint | Study Status | Ref. | |
|---|---|---|---|---|---|---|---|
| 1 | SHR-1210 (Anti-PD-1), Apatinib | RESCUE / NCT03463876 | 190 | Phase II | ORR: 34.3%, median PFS: 5.7 m | Completed | [357] |
| 2 | Pembrolizumab, MTL-CEBPA (C/EBPα small activating RNA) | TIMEPOINT / NCT04105335 | 108 | Phase I | No AE reported, Anti-tumor activity in 26.7% | Completed | [358] |
| 3 | FGF401 (FGFR4 inhibitor), PDR001 (anti-PD1 IgG4 antibody) | NCT02325739 | 172 | Phase I/II | DLT: 8.6%, TTP: 2.63 m (Phase I), ORR: 11.4% | Completed | [359] |
| 4 | TACE (Sorafenib) | NCT02504983 | 200 | Phase IV | Time to CR: 14.4 m (TACE + Sora) vs. not reach (TACE only) | Completed | [360] |
| 5 | BLU-554 (FGFR4 inhibitor), CS1001 (Anti-PD-1) | NCT04194801 | 26 | Phase Ib/II | ORR: 20% | Completed | [361] |
| 6 | Dovitinib | EU-CTR 2011-002445-36 | 25 | Phase II | ORR: 48% | Completed | [362] |
| 7 | Tivozanib | NCT01835223 | 33 | Phase Ib/II |
PFS: 24 wk, OS: 9 m |
Completed | [363] |
| 8 | LY2157299 (TGF-βRI inhibitor), Sorafenib, Ramucirumab | NCT01246986 | 204 | Phase II |
OS: 7.3–16.8 m TTP: 2.7–4.2 m |
Completed | [364] |
| 9 | Cabozantinib | CELESTIAL / NCT01908426 | 707 | Phase III | OS: 10.2 m vs. 8 m (placebo) | Completed | [365] |
| 10 | Nivolumab, BMS-813160 (CCR2/5-inhibitor), BMS-986253 (anti-IL-8) | NCT04123379 | 48 | Phase II | STN: 8.3% | Completed | [366] |
| 11 | Donafenib, Tislelizumab | NCT06232759 | 56 | Phase II |
ORR: 80.4%, DCR: 90.2% |
Completed | [367] |
| 12 | AK104 (Anti-PD-1/CTLA-4), Lenvatinib | NCT04444167 | 59 | Phase II |
ORR: 35.5–35.7%, DOR 13.6–13.67 m, PFS: 8.6–9.8 m, OS: 27.1 m |
Completed | [353] |
| 13 | AtezoBev | NCT04862949 | 124 | Observational | ORR: 29.8% | Completed | [368] |
| 14 | TACE: PD-1/PD-L1 inhibitors + VEGF-TKI/bevacizumab | CHANCE2201 / NCT05332821 | 474 | Observational |
OS: 15.9–22.6 m, PFS: 7.4–9.9 m, ORR: 22.9–41.2% |
Completed | [352] |
| 15 | Atezolizumab, Bevacizumab, UCPVax (anti-telomerase vaccine) | TERTIO / NCT05528952 | 105 | Phase II | ORR | On-going | [369] |
| 16 | Chidamide (Histone deacetylase inhibitor), Regorafenib | NCT05770882 | 46 | Phase Ib/II | AE, ORR, PFS | Ongoing | [370] |
| 17 | Cabozantinib | CaPture / NCT04767906 | 40 | Phase II | TT | Ongoing | [371] |
| 18 | Anti-PD-1 antibody, Lenvatinib | NCT06311929 | 300 | Phase IV | DFS | Ongoing | [372] |
| 19 | PTX-9908 (CXCR4 inhibitor) | NCT03812874 | 50 | Phase I/II | AE, DLT | Ongoing | [373] |
| 20 | Budigalimab, Livmoniplimab, Lenvatinib, Sorafenib | LIVIGNO-1 / NCT05822752 | 120 | Phase II | BOR | Ongoing | [374] |
| 21 | Serplulimab, Bevacizumab | NCT06370065 | 35 | Phase II | ORR | Ongoing | [375] |
| 22 | TACE: PD-1/PD-L1 inhibitors + VEGF-TKI/bevacizumab | CHANCE2202 / NCT05332496 | 220 | Observational | PFS, OS | Ongoing | [376] |
| 23 | Zabadinostat (class I HDAC inhibitor), Geptanolimab, Lenvatinib, Sorafenib | NCT05873244 | 44 | Phase II | PFS | Ongoing | [377] |
| 24 | ETN101 (mTKI) | NCT06326502 | 50 | Phase I | DLT | Ongoing | [378] |
| 25 | Anlotinib hydrochloride, Penpulimab | NCT05862337 | 480 | Phase III | RFS | Ongoing | [379] |
| 26 | Camrelizumab, Lenvatinib | NCT04443309 | 53 | Phase I/II | ORR | Ongoing | [380] |
| 27 | Ablation: Lenvatinib + anti-PD-1 | NCT05803928 | 70 | Phase I | RFS | Ongoing | [381] |
| 28 | Cryoablation: Tislelizumab, Lenvatinib | CASTLE-10 / NCT05897268 | 25 | Phase II | ORR | Ongoing | [382] |
| 29 | TACE: Lenvatinib, iodion-125 | NCT04967495 | 171 | Phase I | OS | Ongoing | [383] |
| 30 | SBRT: AtezoBev | NCT05488522 | 18 | Phase I | DLT | Ongoing | [384] |
| 31 | Liver transplantation: Anti-PD-1 inhibitors | NCT05475613 | 59 | Phase II | PFS | Ongoing | [385] |
| 32 | Tislelizumab, Sitravatinib | NCT05407519 | 40 | Phase II | RFS | Ongoing | [386] |
| 33 | Penpulimab, TQB2618 (TIM-3 inhibitor), Anlotinib Hydrochloride | NCT05975645 | 40 | Phase I | ORR | Ongoing | [387] |
| 34 | Tislelizumab, regorafenib | NCT04183088 | 125 | Phase II | AE, ORR, PFS | Ongoing | [388] |
| 35 | Adebrelimab, Lenvatinib | HSBRT2401 / NCT06261125 | 60 | Phase II | PFS | Ongoing | [389] |
| 36 | Lenvatinib, VIC1911 (Aurora kinase A inhibitor) | NCT05718882 | 30 | Phase II | PFS | Ongoing | [390] |
| 37 | Pembrolizumab, Lenvatinib | PLENTY202001 / NCT04425226 | 192 | Phase II | RFS | Ongoing | [391] |
| 38 | iNKT Cells, Anti-PD-1, Regorafenib | NCT05962450 | 84 | Phase II | PFS | Ongoing | [392] |
| 39 | Oncorine (H101), Tislelizumab, Lenvatinib | NCT05675462 | 25 | Phase I | DLT, MTD, AE | Ongoing | [393] |
| 40 | Durvalumab, Tremelimumab | NCT05809869 | 25 | Phase II | BOR, RAE | Ongoing | [394] |
| 41 | Regorafenib, ICIs | NCT05573282 | 50 | Observational | OS, PFS | Ongoing | [395] |
| 42 | Regorafenib, PD-1 inhibitor | NCT05048017 | 20 | Phase II | PFS | Ongoing | [396] |
| 43 | TACE: TKIs | CHANCE-CHESS 2302 / NCT05704192 | 373 | Observational | OS | Ongoing | [397] |
| 44 | SBRT: Durvalumab | NCT04913480 | 37 | Phase II | PFS | Ongoing | [398] |
| 45 | AtezoBev | Kirros / NCT06096779 | 120 | Phase II | AE | Ongoing | [399] |
| 46 | Ipilimumab, Pembrolizumab, Durvalumab, Idarubicin, Bevacizumab | NCT06482801 | 90 | Phase II | AE, PFS, ORR, DOR | Ongoing | [400] |
| 47 | Palbociclib | NCT06478927 | 22 | / | PFS | Ongoing | [401] |
| 48 | OTX-2002 (mRNA MYC inhibitor), TKIs, ICI | MYCHELANGELO I / NCT05497453 | 190 | Phase I/II | DLT, MTD, AE, ORR, DOR | Ongoing | [402] |
| 49 | Atezolizumab, Cabozantinib, Lenvatinib | NCT05168163 | 122 | Phase II | OS, PFS | Ongoing | [403] |
| 50 | Tegavivint (TBL1 inhibitor), Pembrolizumab | NCT05797805 | 108 | Phase I/II | AE, DLT, | Ongoing | [404] |
| 51 | Toripalimab plus Lenvatinib | NCT04368078 | 76 | Phase II | ORR | Ongoing | [405] |
| 52 | Donafenib + Envafolimab | NCT06498622 | 45 | Phase II | RFS | Ongoing | [406] |
| 53 | TACE: Sintilimab (IBI308) | NCT04297280 | 25 | Phase II | ORR | Ongoing | [407] |
| 54 | HAIC: Tremelimumab + Durvalumab, Lenvatinib | NCT06364007 | 20 | Phase II | ORR | Ongoing | [408] |
| 55 | HAIC: Camrelizumab, TKIs | NCT05135364 | 48 | Phase II | PFS | Ongoing | [409] |
| 56 | HAIC: AtezoBev, Bevacizumab Biosimilar IBI305 + Sintilimab | CCGLC-001 / NCT05713994 | 300 | Observational | Number of Patients Amendable to Curative Surgical Interventions | Ongoing | [410] |
| 57 | Futibatinib, Pembrolizumab | NCT04828486 | 25 | Phase II | PFS | Ongoing | [411] |
| 58 | Lenvatinib | NCT05103904 | 19 | Phase II | ORR | Ongoing | [412] |
| 59 | Atezolizumab, Bevacizumab, Durvalumab, Tremelimumab | REFINE-IO / NCT06117891 | 300 | Observational | OS | Ongoing | [413] |
| 60 | TACE: Lenvatinib, Anti-PD-1 antibody, Bevacizumab Biosimilar IBI305 plus sintilimab, AtezoBev, apatinib plus camrelizumab, Sorafenib, Donafenib, Regorafenib | CCGLC-008 / NCT05717738 | 300 | Observational | Number of Patients Amendable to Curative Surgical Interventions | Ongoing | [414] |
| 61 | Radiotherapy: TKIs, Anti-PD-1 | NCT06061445 | 22 | / | OS, ORR | Ongoing | [415] |
| 62 | DNAJB1-PRKACA peptide vaccine, Nivolumab, Ipilimumab | NCT04248569 | 56 | Phase I | AE, CD4/CD8 interferon production | Ongoing | [416] |
| 63 | HAIC: Lenvatinib, Tislelizumab, Toripalimab, Sintilimab, Camrelizumab | NCT06333561 | 300 | Observational | OS | Ongoing | [417] |
| 64 | Durvalumab, Lenvatinib | Dulect2020-1 / NCT04443322 | 20 | / | PFS, RFS | Ongoing | [418] |
| 65 | Trametinib (MEK inhibitor), Everolimus (mTOR inhibitor), Lenvatinib | NCT04803318 | 100 | Phase II | AE | Ongoing | [419] |
| 66 | Bevacizumab, Atezolizumab, Tislelizumab, Toripalimab, Sintilimab, Camrelizumab | NCT06323382 | 240 | Observational | PFS | Ongoing | [420] |
| 67 | Radiation: Durvalumab, Tremelimumab | NCT04430452 | 21 | Phase II | ORR | Ongoing | [421] |
| 68 | Microspheres: Chemodrug, IL2, PD1, CTLA4, VEGF antibodies | NCT04770207 | 100 | Phase II | AE | Ongoing | [422] |
| 69 | Tislelizumab | HESTIA / NCT05622071 | 50 | Phase II | ORR | Ongoing | [423] |
| 70 | Q702 (TKI), Pembrolizumab | NCT05438420 | 120 | Phase I/II | AE, Tumor response | Ongoing | [424] |
| 71 | Cabozantinib | NCT04204850 | 20 | Phase II | DCR | Ongoing | [425] |
| 73 | Anlotinib hydrochloride, TQB2450 (anti-PD-L1) | NCT04888546 | 20 | Phase I/II | pCR, ORR | Ongoing | [426] |
Estimated numbers
AE Adverse Events, BOR Best Overall Response, DCR Disease Control Rate, DFS Disease-Free Survival, DLT Dose Limiting Toxicity, DOR Duration of remission, m month, MPR Major Pathologic Response, MTD Maximum Tolerated Dose, mTKI multiple Tyrosine Kinase Inhibitor, OSRR Organ-Specific Response Rate, pCR pathological Complete Response Rate, RAE Rate of Abscopal Effect, RFS Recurrence-Free Survival, STN Significant Tumor Necrosis, TT Time on Treatment, TTP Time to Progression
In conclusion, exploring combination therapies represents a promising future in advancing HCC treatment strategies. As research in this field continues to evolve, it holds the potential to substantially improve patient prognosis and redefine the therapeutic landscape for HCC.
Discussions and perspectives
As the prevalence of non-heritable risk factors for HCC continues to increase, it is anticipated that HCC will become more widespread in the coming decades. Despite advancements in targeted drug therapies, the predicted survival rate for HCC patients has not improved significantly. This presents a substantial burden for both the medical and social communities. It is not feasible to perform hepatic CT or MRI for every individual with CLDs or poor lifestyle habits. Consequently, there is an urgent need to identify easily quantifiable biomarkers for early detection or therapeutic indicators through liquid biopsy. This review highlights and discusses significant and recent research findings in studying HCC biomarkers.
As medical knowledge advances, the heterogeneity of HCC has become increasingly apparent to healthcare professionals. It is now widely understood that there is no universal cure for all cancers; instead, personalized medicine is the key to successful treatment. To design the most effective therapeutic regimen for each patient, it is essential to accurately identify their genetic alterations, tumor phenotypes, and immune microenvironment makeup. Ideally, comprehensive diagnostic and monitoring information could be obtained from a single blood sample, significantly reducing the need for invasive procedures such as tissue biopsy. This non-invasive approach would greatly benefit patients, eliminating unnecessary discomfort and potential complications associated with more invasive diagnostic methods.
This review does not intend to offer specific clinical recommendations to clinicians; instead, it aims to provide insights into the current research directions of biomarkers that may contribute to patients’ well-being. By showcasing the latest findings in biomarker detection using liquid biopsy, we aim to promote a deeper understanding of these innovative techniques and their implications for the future of HCC diagnosis and treatment. Furthermore, continued research and collaboration in this field will be instrumental in advancing personalized medicine and ultimately improving patient outcomes. As our understanding of HCC biomarkers evolves, we anticipate that these advancements will significantly impact the way healthcare professionals approach the management of this complex disease.
Artificial intelligence (AI) is the most promising technological advance in the 21st century. AI and machine learning have been widely applied to nearly every aspect of R&D. In terms of the research in HCC, with improved computing power, the deep learning models can integrate vast amounts of data, including patients’ healthcare records, biochemical test results, imaging, biomarkers, and corresponding clinical outcomes, that are impossible to be analyzed as a whole before. Combining AI-predicted models and histochemical morphological analysis can accurately detect the disease and predict the prognosis and treatment response [427, 428]. Future directions will include validating the stability and interpretability of the AI-generated results and extending the database to a more extensive collection to generate general diagnostic standards.
In conclusion, identifying and validating reliable and easily quantifiable biomarkers for HCC are crucial in addressing the increasing burden of this disease. Using liquid biopsy techniques for detecting these biomarkers offers a non-invasive, cost-effective, and efficient alternative to traditional diagnostic methods. Moreover, understanding the complex interplay between genetic alterations, tumor phenotypes, and immune microenvironments will be essential for developing personalized treatment strategies to improve patient outcomes significantly. As research into HCC biomarkers continues to progress, it is imperative that we also investigate novel technologies and methodologies to enhance the sensitivity and specificity of these diagnostic tools. This includes exploring the potential of combining multiple biomarkers to improve diagnostic accuracy or using advanced computational methods to analyze complex datasets to identify new biomarkers. By working together, we can overcome the challenges posed by HCC and significantly improve the lives of patients affected by this devastating disease.
Authors’ contributions
YT.C. conceptualized the study, collected literature, prepared the figures and drafted the manuscript; C.Z. edited the manuscript and figures; J.W., P.L., L.X., H.Y., Y.F. edited the manuscript; ZC.C. and N.W. conceived the idea, design the study, drafted the manuscript.
Funding
This work is financially supported by the University Research Committee of The University of Hong Kong (Project Code: 109000349), the Research Grant Committee of Hong Kong (Project Code: 17119621 and 17111424), The Health and Medical Research Fund (Project Code: 21222151, 19201591 and 15162961) and Innovation and Technology Fund (Project Code: PRP/028/22FX).
Availability of data and materials
This published article and its supplementary information files include all data generated or analyzed during this study.
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Zhe-Sheng Chen, Email: chenz@stjohns.edu.
Ning Wang, Email: ckwang@hku.hk.
References
- 1.Bianco C, Jamialahmadi O, Pelusi S, Baselli G, Dongiovanni P, Zanoni I, et al. Non-invasive stratification of hepatocellular carcinoma risk in non-alcoholic fatty liver using polygenic risk scores. J Hepatol. 2021;74(4):775–82. 10.1016/j.jhep.2020.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Toh MR, Wong EYT, Wong SH, Ng AWT, Loo L-H, Chow PK-H, et al. Global epidemiology and genetics of hepatocellular carcinoma. Gastroenterology. 2023;164(5):766–82. 10.1053/j.gastro.2023.01.033 [DOI] [PubMed] [Google Scholar]
- 3.Fujiwara N, Kubota N, Crouchet E, Koneru B, Marquez CA, Jajoriya AK, et al. Molecular signatures of long-term hepatocellular carcinoma risk in nonalcoholic fatty liver disease. Sci Transl Med. 2022;14(650):eabo4474. 10.1126/scitranslmed.abo4474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li L, Wang H. Heterogeneity of liver cancer and personalized therapy. Cancer Lett. 2016;379(2):191–7. 10.1016/j.canlet.2015.07.018 [DOI] [PubMed] [Google Scholar]
- 5.Whitfield JB, Schwantes-An TH, Darlay R, Aithal GP, Atkinson SR, Bataller R, et al. A genetic risk score and diabetes predict development of alcohol-related cirrhosis in drinkers. J Hepatol. 2022;76(2):275–82. 10.1016/j.jhep.2021.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nahon P, Bamba-Funck J, Layese R, Trépo E, Zucman-Rossi J, Cagnot C, et al. Integrating genetic variants into clinical models for hepatocellular carcinoma risk stratification in cirrhosis. J Hepatol. 2023;78(3):584–95. 10.1016/j.jhep.2022.11.003 [DOI] [PubMed] [Google Scholar]
- 7.Llovet JM, Pinyol R, Kelley RK, El-Khoueiry A, Reeves HL, Wang XW, et al. Molecular pathogenesis and systemic therapies for hepatocellular carcinoma. Nat Cancer. 2022;3(4):386–401. 10.1038/s43018-022-00357-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singal AG, Zhang E, Narasimman M, Rich NE, Waljee AK, Hoshida Y, et al. HCC surveillance improves early detection, curative treatment receipt, and survival in patients with cirrhosis: a meta-analysis. J Hepatol. 2022;77(1):128–39. 10.1016/j.jhep.2022.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kansagara D, Papak J, Pasha AS, O’Neil M, Freeman M, Relevo R, et al. Screening for hepatocellular carcinoma in chronic liver disease: a systematic review. Ann Intern Med. 2014;161(4):261–9. 10.7326/M14-0558 [DOI] [PubMed] [Google Scholar]
- 10.Jiang HY, Chen J, Xia CC, Cao LK, Duan T, Song B. Noninvasive imaging of hepatocellular carcinoma: from diagnosis to prognosis. World J Gastroenterol. 2018;24(22):2348–62. 10.3748/wjg.v24.i22.2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li L, Hu Y, Han J, Li Q, Peng C, Zhou J. Clinical application of liver imaging reporting and data system for characterizing liver neoplasms: a meta-analysis. Diagnostics (Basel). 2021;11(2):323. 10.3390/diagnostics11020323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Villanueva A. Hepatocellular carcinoma. N Engl J Med. 2019;380(15):1450–62. 10.1056/NEJMra1713263 [DOI] [PubMed] [Google Scholar]
- 13.Brown ZJ, Tsilimigras DI, Ruff SM, Mohseni A, Kamel IR, Cloyd JM, et al. Management of hepatocellular carcinoma: a review. JAMA Surg. 2023;158(4):410–20. 10.1001/jamasurg.2022.7989 [DOI] [PubMed] [Google Scholar]
- 14.Marrero JA, Kulik LM, Sirlin CB, Zhu AX, Finn RS, Abecassis MM, et al. Diagnosis, staging, and management of hepatocellular carcinoma: 2018 practice guidance by the American Association for the Study of Liver Diseases. Hepatology. 2018;68(2):723–50. 10.1002/hep.29913 [DOI] [PubMed] [Google Scholar]
- 15.Tzartzeva K, Obi J, Rich NE, Parikh ND, Marrero JA, Yopp A, et al. Surveillance imaging and alpha fetoprotein for early detection of hepatocellular carcinoma in patients with cirrhosis: a meta-analysis. Gastroenterology. 2018;154(6):1706-18.e1. 10.1053/j.gastro.2018.01.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Terzi E, Ayuso C, Piscaglia F, Bruix J. Liver imaging reporting and data system: review of pros and cons. Semin Liver Dis. 2022;42(1):104–11. 10.1055/s-0041-1732356 [DOI] [PubMed] [Google Scholar]
- 17.Sørensen JB, Klee M, Palshof T, Hansen HH. Performance status assessment in cancer patients. An inter-observer variability study. Br J Cancer. 1993;67(4):773–5. 10.1038/bjc.1993.140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Child CG, Turcotte JG. Surgery and portal hypertension. Major Probl Clin Surg. 1964;1:1–85. [PubMed] [Google Scholar]
- 19.Pugh RNH, Murray-Lyon IM, Dawson JL, Pietroni MC, Williams R. Transection of the oesophagus for bleeding oesophageal varices. Br J Surg. 2005;60(8):646–9. 10.1002/bjs.1800600817 [DOI] [PubMed] [Google Scholar]
- 20.Kumada T, Toyoda H, Tada T, Yasuda S, Tanaka J. Changes in background liver function in patients with hepatocellular carcinoma over 30 years: comparison of Child-Pugh classification and albumin bilirubin grade. Liver Cancer. 2020;9(5):518–28. 10.1159/000507933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dominguez DA, Wong P, Melstrom LG. Existing and emerging biomarkers in hepatocellular carcinoma: relevance in staging, determination of minimal residual disease, and monitoring treatment response: a narrative review. Hepatobil Surg Nutr. 2023;13(1):39–55. 10.21037/hbsn-22-526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tateishi R, Uchino K, Fujiwara N, Takehara T, Okanoue T, Seike M, et al. A nationwide survey on non-B, non-C hepatocellular carcinoma in Japan: 2011–2015 update. J Gastroenterol. 2019;54(4):367–76. 10.1007/s00535-018-1532-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson PJ, Berhane S, Kagebayashi C, Satomura S, Teng M, Reeves HL, et al. Assessment of liver function in patients with hepatocellular carcinoma: a new evidence-based approach-the ALBI grade. J Clin Oncol. 2015;33(6):550–8. 10.1200/JCO.2014.57.9151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuo YH, Wang JH, Hung CH, Rau KM, Wu IP, Chen CH, et al. Albumin-Bilirubin grade predicts prognosis of HCC patients with sorafenib use. J Gastroenterol Hepatol. 2017;32(12):1975–81. 10.1111/jgh.13783 [DOI] [PubMed] [Google Scholar]
- 25.Pinato DJ, Sharma R, Allara E, Yen C, Arizumi T, Kubota K, et al. The ALBI grade provides objective hepatic reserve estimation across each BCLC stage of hepatocellular carcinoma. J Hepatol. 2017;66(2):338–46. 10.1016/j.jhep.2016.09.008 [DOI] [PubMed] [Google Scholar]
- 26.Reig M, Forner A, Rimola J, Ferrer-Fàbrega J, Burrel M, Garcia-Criado Á, et al. BCLC strategy for prognosis prediction and treatment recommendation: the 2022 update. J Hepatol. 2022;76(3):681–93. 10.1016/j.jhep.2021.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hackl C, Schlitt HJ, Renner P, Lang SA. Liver surgery in cirrhosis and portal hypertension. World J Gastroenterol. 2016;22(9):2725–35. 10.3748/wjg.v22.i9.2725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berardi G, Morise Z, Sposito C, Igarashi K, Panetta V, Simonelli I, et al. Development of a nomogram to predict outcome after liver resection for hepatocellular carcinoma in Child-Pugh B cirrhosis. J Hepatol. 2020;72(1):75–84. 10.1016/j.jhep.2019.08.032 [DOI] [PubMed] [Google Scholar]
- 29.European Association for the Study of the Liver. EASL Clinical Practice Guidelines: Management of hepatocellular carcinoma. J Hepatol. 2018;69(1):182–236. 10.1016/j.jhep.2018.03.019. 10.1016/j.jhep.2018.03.019 [DOI] [PubMed] [Google Scholar]
- 30.Vogel A, Cervantes A, Chau I, Daniele B, Llovet JM, Meyer T, et al. Hepatocellular carcinoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2018;29(Suppl 4):iv238–55. 10.1093/annonc/mdy308 [DOI] [PubMed] [Google Scholar]
- 31.Szpakowski JL, Drasin TE, Lyon LL. Rate of seeding with biopsies and ablations of hepatocellular carcinoma: a retrospective cohort study. Hepatol Commun. 2017;1(9):841–51. 10.1002/hep4.1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee YT, Fujiwara N, Yang JD, Hoshida Y. Risk stratification and early detection biomarkers for precision HCC screening. Hepatology. 2023;78(1):319–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson P, Zhou Q, Dao DY, Lo YMD. Circulating biomarkers in the diagnosis and management of hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2022;19(10):670–81. 10.1038/s41575-022-00620-y [DOI] [PubMed] [Google Scholar]
- 34.Ye Q, Ling S, Zheng S, Xu X. Liquid biopsy in hepatocellular carcinoma: circulating tumor cells and circulating tumor DNA. Mol Cancer. 2019;18(1):114. 10.1186/s12943-019-1043-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li J, Han X, Yu XN, Xu ZZ, Yang GS, Liu BQ, et al. Clinical applications of liquid biopsy as prognostic and predictive biomarkers in hepatocellular carcinoma: circulating tumor cells and circulating tumor DNA. J Exp Clin Cancer Res. 2018;37(1):213. 10.1186/s13046-018-0893-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Galle PR, Foerster F, Kudo M, Chan SL, Llovet JM, Qin S, et al. Biology and significance of alpha-fetoprotein in hepatocellular carcinoma. Liver Int. 2019;39(12):2214–29. 10.1111/liv.14223 [DOI] [PubMed] [Google Scholar]
- 37.Lok AS, Lai CL. alpha-Fetoprotein monitoring in Chinese patients with chronic hepatitis B virus infection: role in the early detection of hepatocellular carcinoma. Hepatology. 1989;9(1):110–5. 10.1002/hep.1840090119 [DOI] [PubMed] [Google Scholar]
- 38.Collier J, Sherman M. Screening for hepatocellular carcinoma. Hepatology. 1998;27(1):273–8. 10.1002/hep.510270140 [DOI] [PubMed] [Google Scholar]
- 39.Hanif H, Ali MJ, Susheela AT, Khan IW, Luna-Cuadros MA, Khan MM, et al. Update on the applications and limitations of alpha-fetoprotein for hepatocellular carcinoma. World J Gastroenterol. 2022;28(2):216–29. 10.3748/wjg.v28.i2.216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gambarin-Gelwan M, Wolf DC, Shapiro R, Schwartz ME, Min AD. Sensitivity of commonly available screening tests in detecting hepatocellular carcinoma in cirrhotic patients undergoing liver transplantation. Am J Gastroenterol. 2000;95(6):1535–8. 10.1111/j.1572-0241.2000.02091.x [DOI] [PubMed] [Google Scholar]
- 41.Trevisani F, D’Intino PE, Morselli-Labate AM, Mazzella G, Accogli E, Caraceni P, et al. Serum alpha-fetoprotein for diagnosis of hepatocellular carcinoma in patients with chronic liver disease: influence of HBsAg and anti-HCV status. J Hepatol. 2001;34(4):570–5. 10.1016/S0168-8278(00)00053-2 [DOI] [PubMed] [Google Scholar]
- 42.Singal A, Volk ML, Waljee A, Salgia R, Higgins P, Rogers MA, et al. Meta-analysis: surveillance with ultrasound for early-stage hepatocellular carcinoma in patients with cirrhosis. Aliment Pharmacol Ther. 2009;30(1):37–47. 10.1111/j.1365-2036.2009.04014.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lok AS, Sterling RK, Everhart JE, Wright EC, Hoefs JC, Di Bisceglie AM, et al. Des-gamma-carboxy prothrombin and alpha-fetoprotein as biomarkers for the early detection of hepatocellular carcinoma. Gastroenterology. 2010;138(2):493–502. 10.1053/j.gastro.2009.10.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Di Bisceglie AM, Hoofnagle JH. Elevations in serum alpha-fetoprotein levels in patients with chronic hepatitis B. Cancer. 1989;64(10):2117–20. [DOI] [PubMed] [Google Scholar]
- 45.Chen J, Wu M, Gong J, Liu Z, He G, Zhu H, et al. Influence of alanine transaminase levels on alpha-fetoprotein for predicting hepatocellular carcinoma in patients with hepatitis B infection. Biomed Res Int. 2020;2020:2043715. 10.1155/2020/2043715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heimbach JK, Kulik LM, Finn RS, Sirlin CB, Abecassis MM, Roberts LR, et al. AASLD guidelines for the treatment of hepatocellular carcinoma. Hepatology. 2018;67(1):358–80. 10.1002/hep.29086 [DOI] [PubMed] [Google Scholar]
- 47.Sterling RK, Jeffers L, Gordon F, Venook AP, Reddy KR, Satomura S, et al. Utility of Lens culinaris agglutinin-reactive fraction of alpha-fetoprotein and des-gamma-carboxy prothrombin, alone or in combination, as biomarkers for hepatocellular carcinoma. Clin Gastroenterol Hepatol. 2009;7(1):104–13. 10.1016/j.cgh.2008.08.041 [DOI] [PubMed] [Google Scholar]
- 48.Liu S, Sun L, Yao L, Zhu H, Diao Y, Wang M, et al. Diagnostic performance of AFP, AFP-L3, or PIVKA-II for hepatitis C virus-associated hepatocellular carcinoma: a multicenter analysis. J Clin Med. 2022;11(17):5075. 10.3390/jcm11175075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weitz IC, Liebman HA. Des-gamma-carboxy (abnormal) prothrombin and hepatocellular carcinoma: a critical review. Hepatology. 1993;18(4):990–7. 10.1002/hep.1840180434 [DOI] [PubMed] [Google Scholar]
- 50.Kim DY, Toan BN, Tan CK, Hasan I, Setiawan L, Yu ML, et al. Utility of combining PIVKA-II and AFP in the surveillance and monitoring of hepatocellular carcinoma in the Asia-Pacific region. Clin Mol Hepatol. 2023;29(2):277–92. 10.3350/cmh.2022.0212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seo SI, Kim HS, Kim WJ, Shin WG, Kim DJ, Kim KH, et al. Diagnostic value of PIVKA-II and alpha-fetoprotein in hepatitis B virus-associated hepatocellular carcinoma. World J Gastroenterol. 2015;21(13):3928–35. 10.3748/wjg.v21.i13.3928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang AE, Leven EA, Grinspan LT, Villanueva A. Novel biomarkers and strategies for HCC diagnosis and care. Clin Liver Dis (Hoboken). 2024;23(1):e0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johnson PJ, Pirrie SJ, Cox TF, Berhane S, Teng M, Palmer D, et al. The detection of hepatocellular carcinoma using a prospectively developed and validated model based on serological biomarkers. Cancer Epidemiol Biomark Prev. 2014;23(1):144–53. 10.1158/1055-9965.EPI-13-0870 [DOI] [PubMed] [Google Scholar]
- 54.Singal AG, Tayob N, Mehta A, Marrero JA, El-Serag H, Jin Q, et al. GALAD demonstrates high sensitivity for HCC surveillance in a cohort of patients with cirrhosis. Hepatology. 2022;75(3):541–9. 10.1002/hep.32185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.El-Serag HB, Kanwal F, Davila JA, Kramer J, Richardson P. A new laboratory-based algorithm to predict development of hepatocellular carcinoma in patients with hepatitis C and cirrhosis. Gastroenterology. 2014;146(5):1249-55.e1. 10.1053/j.gastro.2014.01.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Capurro M, Wanless IR, Sherman M, Deboer G, Shi W, Miyoshi E, et al. Glypican-3: a novel serum and histochemical marker for hepatocellular carcinoma. Gastroenterology. 2003;125(1):89–97. 10.1016/S0016-5085(03)00689-9 [DOI] [PubMed] [Google Scholar]
- 57.Llovet JM, Chen Y, Wurmbach E, Roayaie S, Fiel MI, Schwartz M, et al. A molecular signature to discriminate dysplastic nodules from early hepatocellular carcinoma in HCV cirrhosis. Gastroenterology. 2006;131(6):1758–67. 10.1053/j.gastro.2006.09.014 [DOI] [PubMed] [Google Scholar]
- 58.Wang XY, Degos F, Dubois S, Tessiore S, Allegretta M, Guttmann RD, et al. Glypican-3 expression in hepatocellular tumors: diagnostic value for preneoplastic lesions and hepatocellular carcinomas. Hum Pathol. 2006;37(11):1435–41. 10.1016/j.humpath.2006.05.016 [DOI] [PubMed] [Google Scholar]
- 59.Saber MA, AbdelHafiz SM, Khorshed FE, Aboushousha TS, Hamdy HE, Seleem MI, et al. Differential expression of Glypican-3 and Insulin-Like growth factor-II mRNAs and Alpha-Fetoprotein and Ki-67 markers in HCV related hepatocellular carcinomas in Egyptian patients. Asian Pac J Cancer Prev. 2017;18(1):121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang W, Zhao L-J, Wang Y, Tao Q-Y, Feitelson MA, Zhao P, et al. Application of HBx-induced anti-URGs as early warning biomarker of cirrhosis and HCC. Cancer Biomark. 2012;11(1):29–39. 10.3233/CBM-2012-0261 [DOI] [PubMed] [Google Scholar]
- 61.Abdelgawad IA, Mossallam GI, Radwan NH, Elzawahry HM, Elhifnawy NM. Can Glypican3 be diagnostic for early hepatocellular carcinoma among Egyptian patients? Asian Pac J Cancer Prev. 2013;14(12):7345–9. 10.7314/APJCP.2013.14.12.7345 [DOI] [PubMed] [Google Scholar]
- 62.Nault JC, Guyot E, Laguillier C, Chevret S, Ganne-Carrie N, N’Kontchou G, et al. Serum proteoglycans as prognostic biomarkers of hepatocellular carcinoma in patients with alcoholic cirrhosis. Cancer Epidemiol Biomarkers Prev. 2013;22(8):1343–52. 10.1158/1055-9965.EPI-13-0179 [DOI] [PubMed] [Google Scholar]
- 63.Wen Y, Jeong S, Xia Q, Kong X. Role of osteopontin in liver diseases. Int J Biol Sci. 2016;12(9):1121–8. 10.7150/ijbs.16445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Morales-Ibanez O, Domínguez M, Ki SH, Marcos M, Chaves JF, Nguyen-Khac E, et al. Human and experimental evidence supporting a role for osteopontin in alcoholic hepatitis. Hepatology. 2013;58(5):1742–56. 10.1002/hep.26521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Abu El Makarem MA, Abdel-Aleem A, Ali A, Saber R, Shatat M, Rahem DA, et al. Diagnostic significance of plasma osteopontin in hepatitis C virus-related hepatocellular carcinoma. Ann Hepatol. 2011;10(3):296–305. 10.1016/S1665-2681(19)31541-8 [DOI] [PubMed] [Google Scholar]
- 66.Shen Q, Fan J, Yang XR, Tan Y, Zhao W, Xu Y, et al. Serum DKK1 as a protein biomarker for the diagnosis of hepatocellular carcinoma: a large-scale, multicentre study. Lancet Oncol. 2012;13(8):817–26. 10.1016/S1470-2045(12)70233-4 [DOI] [PubMed] [Google Scholar]
- 67.Zhu M, Zheng J, Wu F, Kang B, Liang J, Heskia F, et al. OPN is a promising serological biomarker for hepatocellular carcinoma diagnosis. J Med Virol. 2020;92(12):3596–603. 10.1002/jmv.25704 [DOI] [PubMed] [Google Scholar]
- 68.Gehrmann M, Cervello M, Montalto G, Cappello F, Gulino A, Knape C, et al. Heat shock protein 70 serum levels differ significantly in patients with chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma. Front Immunol. 2014;5:307. 10.3389/fimmu.2014.00307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang B, Lan T, Xiao H, Chen Z-H, Wei C, Chen L-F, et al. The expression profiles and prognostic values of HSP70s in hepatocellular carcinoma. Cancer Cell Int. 2021;21(1):286. 10.1186/s12935-021-01987-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chuma M, Sakamoto M, Yamazaki K, Ohta T, Ohki M, Asaka M, et al. Expression profiling in multistage hepatocarcinogenesis: identification of HSP70 as a molecular marker of early hepatocellular carcinoma. Hepatology. 2003;37(1):198–207. 10.1053/jhep.2003.50022 [DOI] [PubMed] [Google Scholar]
- 71.Mohamed SA, Tealeb A-SMI. The role of heat shock protein 70 and glypican 3 expression in early diagnosis of hepatocellular carcinoma. Egypt J Pathol. 2022;42(2):112–6. 10.4103/egjp.egjp_21_22 [DOI] [Google Scholar]
- 72.Marrero JA, Romano PR, Nikolaeva O, Steel L, Mehta A, Fimmel CJ, et al. GP73, a resident Golgi glycoprotein, is a novel serum marker for hepatocellular carcinoma. J Hepatol. 2005;43(6):1007–12. 10.1016/j.jhep.2005.05.028 [DOI] [PubMed] [Google Scholar]
- 73.Mao Y, Yang H, Xu H, Lu X, Sang X, Du S, et al. Golgi protein 73 (GOLPH2) is a valuable serum marker for hepatocellular carcinoma. Gut. 2010;59(12):1687–93. 10.1136/gut.2010.214916 [DOI] [PubMed] [Google Scholar]
- 74.Pang BY, Leng Y, Wang X, Wang YQ, Jiang LH. A meta-analysis and of clinical values of 11 blood biomarkers, such as AFP, DCP, and GP73 for diagnosis of hepatocellular carcinoma. Ann Med. 2023;55(1):42–61. 10.1080/07853890.2022.2153163 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 75.Huang F, Guo J, Zhao N, Hou M, Gai X, Yang S, et al. PTEN deficiency potentiates HBV-associated liver cancer development through augmented GP73/GOLM1. J Transl Med. 2024;22(1):254. 10.1186/s12967-024-04976-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bankó P, Lee SY, Nagygyörgy V, Zrínyi M, Chae CH, Cho DH, et al. Technologies for circulating tumor cell separation from whole blood. J Hematol Oncol. 2019;12(1):48. 10.1186/s13045-019-0735-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ferreira MM, Ramani VC, Jeffrey SS. Circulating tumor cell technologies. Mol Oncol. 2016;10(3):374–94. 10.1016/j.molonc.2016.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wu Y, Park KJ, Deighan C, Amaya P, Miller B, Pan Q, et al. Multiparameter evaluation of the heterogeneity of circulating tumor cells using integrated RNA in situ hybridization and immunocytochemical analysis. Front Oncol. 2016;6:234. 10.3389/fonc.2016.00234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vona G, Estepa L, Béroud C, Damotte D, Capron F, Nalpas B, et al. Impact of cytomorphological detection of circulating tumor cells in patients with liver cancer. Hepatology. 2004;39(3):792–7. 10.1002/hep.20091 [DOI] [PubMed] [Google Scholar]
- 80.Qi L-N, Xiang B-D, Wu F-X, Ye J-Z, Zhong J-H, Wang Y-Y, et al. Circulating tumor cells undergoing EMT provide a metric for diagnosis and prognosis of patients with hepatocellular carcinoma. Cancer Res. 2018;78(16):4731–44. 10.1158/0008-5472.CAN-17-2459 [DOI] [PubMed] [Google Scholar]
- 81.Guo W, Sun Y-F, Shen M-N, Ma X-L, Wu J, Zhang C-Y, et al. Circulating tumor cells with stem-like phenotypes for diagnosis, prognosis, and therapeutic response evaluation in hepatocellular carcinoma. Clin Cancer Res. 2018;24(9):2203–13. 10.1158/1078-0432.CCR-17-1753 [DOI] [PubMed] [Google Scholar]
- 82.Coumans FAW, Doggen CJM, Attard G, de Bono JS, Terstappen L. All circulating EpCAM+CK+CD45- objects predict overall survival in castration-resistant prostate cancer. Ann Oncol. 2010;21(9):1851–7. 10.1093/annonc/mdq030 [DOI] [PubMed] [Google Scholar]
- 83.Sun YF, Xu Y, Yang XR, Guo W, Zhang X, Qiu SJ, et al. Circulating stem cell-like epithelial cell adhesion molecule-positive tumor cells indicate poor prognosis of hepatocellular carcinoma after curative resection. Hepatology. 2013;57(4):1458–68. 10.1002/hep.26151 [DOI] [PubMed] [Google Scholar]
- 84.Guo W, Yang XR, Sun YF, Shen MN, Ma XL, Wu J, et al. Clinical significance of EpCAM mRNA-positive circulating tumor cells in hepatocellular carcinoma by an optimized negative enrichment and qRT-PCR-based platform. Clin Cancer Res. 2014;20(18):4794–805. 10.1158/1078-0432.CCR-14-0251 [DOI] [PubMed] [Google Scholar]
- 85.Zhao H, Ling Y, He J, Dong J, Mo Q, Wang Y, et al. Potential targets and therapeutics for cancer stem cell-based therapy against drug resistance in hepatocellular carcinoma. Drug Resist Updates. 2024;74:101084. 10.1016/j.drup.2024.101084 [DOI] [PubMed] [Google Scholar]
- 86.Okajima W, Komatsu S, Ichikawa D, Miyamae M, Ohashi T, Imamura T, et al. Liquid biopsy in patients with hepatocellular carcinoma: circulating tumor cells and cell-free nucleic acids. World J Gastroenterol. 2017;23(31):5650–68. 10.3748/wjg.v23.i31.5650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lu Y, Yang A, Quan C, Pan Y, Zhang H, Li Y, et al. A single-cell atlas of the multicellular ecosystem of primary and metastatic hepatocellular carcinoma. Nat Commun. 2022;13(1):4594. 10.1038/s41467-022-32283-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sun Y, Wu P, Zhang Z, Wang Z, Zhou K, Song M, et al. Integrated multi-omics profiling to dissect the spatiotemporal evolution of metastatic hepatocellular carcinoma. Cancer Cell. 2024;42(1):135-56.e17. 10.1016/j.ccell.2023.11.010 [DOI] [PubMed] [Google Scholar]
- 89.Rostami A, Lambie M, Yu CW, Stambolic V, Waldron JN, Bratman SV. Senescence, necrosis, and apoptosis govern circulating cell-free DNA release kinetics. Cell Rep. 2020;31(13):107830. 10.1016/j.celrep.2020.107830 [DOI] [PubMed] [Google Scholar]
- 90.Lo YMD, Han DSC, Jiang P, Chiu RWK. Epigenetics, fragmentomics, and topology of cell-free DNA in liquid biopsies. Science. 2021;372(6538):eaaw3616. 10.1126/science.aaw3616 [DOI] [PubMed] [Google Scholar]
- 91.Sun K, Jiang P, Chan KC, Wong J, Cheng YK, Liang RH, et al. Plasma DNA tissue mapping by genome-wide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proc Natl Acad Sci U S A. 2015;112(40):E5503–12. 10.1073/pnas.1508736112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Corcoran RB, Chabner BA. Application of cell-free DNA analysis to cancer treatment. N Engl J Med. 2018;379(18):1754–65. 10.1056/NEJMra1706174 [DOI] [PubMed] [Google Scholar]
- 93.Yinzhong W, Miaomiao W, Xiaoxue T, Qian W, Meng Q, Junqiang L. Diagnostic accuracy of circulating-free DNA for the determination of hepatocellular carcinoma: a systematic review and meta-analysis. Expert Rev Mol Diagn. 2023;23(1):63–9. 10.1080/14737159.2023.2167555 [DOI] [PubMed] [Google Scholar]
- 94.Zinkova A, Brynychova I, Svacina A, Jirkovska M, Korabecna M. Cell-free DNA from human plasma and serum differs in content of telomeric sequences and its ability to promote immune response. Sci Rep. 2017;7(1):2591. 10.1038/s41598-017-02905-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Martignano F. Cell-free DNA: an overview of sample types and isolation procedures. Methods Mol Biol. 2019;1909:13–27. 10.1007/978-1-4939-8973-7_2 [DOI] [PubMed] [Google Scholar]
- 96.van Dessel LF, Beije N, Helmijr JC, Vitale SR, Kraan J, Look MP, et al. Application of circulating tumor DNA in prospective clinical oncology trials - standardization of preanalytical conditions. Mol Oncol. 2017;11(3):295–304. 10.1002/1878-0261.12037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Winstead E. FDA authorizes blood test for assessing risk of hereditary cancers. NCI; 2023. [updated November 2, 2023]. Available from: https://www.cancer.gov/news-events/cancer-currents-blog/2023/fda-blood-test-hereditary-cancer-risk.
- 98.Ako S, Nouso K, Kinugasa H, Matsushita H, Terasawa H, Adachi T, et al. Human telomerase reverse transcriptase gene promoter mutation in serum of patients with hepatocellular carcinoma. Oncology. 2020;98(5):311–7. 10.1159/000506135 [DOI] [PubMed] [Google Scholar]
- 99.Xiong Y, Xie CR, Zhang S, Chen J, Yin ZY. Detection of a novel panel of somatic mutations in plasma cell-free DNA and its diagnostic value in hepatocellular carcinoma. Cancer Manag Res. 2019;11:5745–56. 10.2147/CMAR.S197455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cai ZX, Chen G, Zeng YY, Dong XQ, Lin MJ, Huang XH, et al. Circulating tumor DNA profiling reveals clonal evolution and real-time disease progression in advanced hepatocellular carcinoma. Int J Cancer. 2017;141(5):977–85. 10.1002/ijc.30798 [DOI] [PubMed] [Google Scholar]
- 101.An Y, Guan YF, Xu YP, Han YX, Wu C, Bao CH, et al. The diagnostic and prognostic usage of circulating tumor DNA in operable hepatocellular carcinoma. Am J Transl Res. 2019;11(10):6462–74. [PMC free article] [PubMed] [Google Scholar]
- 102.Tao KS, Bian ZY, Zhang Q, Guo X, Yin C, Wang Y, et al. Machine learning-based genome-wide interrogation of somatic copy number aberrations in circulating tumor DNA for early detection of hepatocellular carcinoma. Ebiomedicine. 2020;56:102811. 10.1016/j.ebiom.2020.102811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Alunni-Fabbroni M, Weber S, Öcal O, Seidensticker M, Mayerle J, Malfertheiner P, et al. Circulating cell-free DNA combined to magnetic resonance imaging for early detection of HCC in patients with liver cirrhosis. Cancers. 2021;13(3):521. 10.3390/cancers13030521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cai Z, Zhang J, He Y, Xia L, Dong X, Chen G, et al. Liquid biopsy by combining 5-hydroxymethylcytosine signatures of plasma cell-free DNA and protein biomarkers for diagnosis and prognosis of hepatocellular carcinoma. Esmo Open. 2021;6(1):100021. 10.1016/j.esmoop.2020.100021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cai ZX, Chen G, Zeng YY, Dong XQ, Li ZL, Huang YB, et al. Comprehensive liquid profiling of circulating tumor DNA and protein biomarkers in long-term follow-up patients with hepatocellular carcinoma. Clin Cancer Res. 2019;25(17):5284–94. 10.1158/1078-0432.CCR-18-3477 [DOI] [PubMed] [Google Scholar]
- 106.Chae H, Sung PS, Choi H, Kwon A, Kang D, Kim Y, et al. Targeted next-generation sequencing of plasma cell-free DNA in Korean patients with hepatocellular carcinoma. Ann Lab Med. 2021;41(2):198–206. 10.3343/alm.2021.41.2.198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chen G, Peng F, Dong XQ, Cai ZX, Li ZL, He L, et al. Identification of tumor mutations in plasma based on mutation variant frequency change (MVFC). Mol Oncol. 2023;17(9):1871–83. 10.1002/1878-0261.13498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chen H, Sun LY, Zheng HQ, Zhang QF, Jin XM. Total serum DNA and DNA integrity: diagnostic value in patients with hepatitis B virus-related hepatocellular carcinoma. Pathology. 2012;44(4):318–24. 10.1097/PAT.0b013e328353a24c [DOI] [PubMed] [Google Scholar]
- 109.Cowzer D, White JB, Chou JF, Chen PJ, Kim TH, Khalil DN, et al. Targeted molecular profiling of circulating cell-free DNA in patients with advanced hepatocellular carcinoma. JCO Precis Oncol. 2023;7:e2300272. 10.1200/PO.23.00272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ge ZH, Helmijr JCA, Jansen M, Boor PPC, Noordam L, Peppelenbosch M, et al. Detection of oncogenic mutations in paired circulating tumor DNA and circulating tumor cells in patients with hepatocellular carcinoma. Transl Oncol. 2021;14(7):101073. 10.1016/j.tranon.2021.101073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Han LY, Fan YC, Mu NN, Gao S, Li F, Ji XF, et al. Aberrant DNA methylation of G-protein-coupled bile acid receptor Gpbar1 (TGR5) is a potential biomarker for hepatitis B virus associated hepatocellular carcinoma. Int J Med Sci. 2014;11(2):164–71. 10.7150/ijms.6745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Higuera M, Vargas-Accarino E, Torrens M, Gregori J, Salcedo MT, Martínez-Campreciós J, et al. Ultra deep sequencing of circulating cell-free DNA as a potential tool for hepatocellular carcinoma management. Cancers. 2022;14(16):3875. 10.3390/cancers14163875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hu N, Fan XP, Fan YC, Chen LY, Qiao CY, Han LY, et al. Hypomethylated ubiquitin-conjugating enzyme2 Q1 (UBE2Q1) gene promoter in the serum is a promising biomarker for hepatitis B virus-associated hepatocellular carcinoma. Tohoku J Exp Med. 2017;242(2):93–100. 10.1620/tjem.242.93 [DOI] [PubMed] [Google Scholar]
- 114.Huang A, Zhang X, Zhou SL, Cao Y, Huang XW, Fan J, et al. Plasma circulating cell-free DNA integrity as a promising biomarker for diagnosis and surveillance in patients with hepatocellular carcinoma. J Cancer. 2016;7(13):1798–803. 10.7150/jca.15618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Huang A, Zhang X, Zhou SL, Cao Y, Huang XW, Fan J, et al. Detecting circulating tumor DNA in hepatocellular carcinoma patients using droplet digital PCR is feasible and reflects intratumoral heterogeneity. J Cancer. 2016;7(13):1907–14. 10.7150/jca.15823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Huang GM, Krocker JD, Kirk JL, Merwat SN, Ju H, Soloway RD, et al. Evaluation of INK4A promoter methylation using pyrosequencing and circulating cell-free DNA from patients with hepatocellular carcinoma. Clin Chem Lab Med. 2014;52(6):899–909. 10.1515/cclm-2013-0885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jiao JJ, Sanchez J, Thompson EJ, Mao XZ, McCormick JB, Fisher-Hoch SP, et al. Somatic mutations in circulating cell-free DNA and risk for hepatocellular carcinoma in Hispanics. Int J Mol Sci. 2021;22(14):7411. 10.3390/ijms22147411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kandimalla R, Xu JF, Link A, Matsuyama T, Yamamura K, Parker MI, et al. EpiPanGI Dx: a cell-free DNA methylation fingerprint for the early detection of gastrointestinal cancers. Clin Cancer Res. 2021;27(22):6135–44. 10.1158/1078-0432.CCR-21-1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kim SC, Kim DW, Cho EJ, Lee JY, Kim J, Kwon C, et al. A circulating cell-free DNA methylation signature for the detection of hepatocellular carcinoma. Mol Cancer. 2023;22(1):164. 10.1186/s12943-023-01872-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kumar S, Nadda N, Paul S, Gamanagatti S, Dash NR, Vanamail P, et al. Evaluation of the cell-free DNA integrity index as a liquid biopsy marker to differentiate hepatocellular carcinoma from chronic liver disease. Front Mol Biosci. 2022;9:1024193. 10.3389/fmolb.2022.1024193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kunadirek P, Chuaypen N, Jenjaroenpun P, Wongsurawat T, Pinjaroen N, Sirichindakul P, et al. Cell-free DNA analysis by whole-exome sequencing for hepatocellular carcinoma: a pilot study in Thailand. Cancers. 2021;13(9):2229. 10.3390/cancers13092229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Labgaa I, Villacorta-Martin C, D’Avola D, Craig AJ, von Felden J, Martins SN, et al. A pilot study of ultra-deep targeted sequencing of plasma DNA identifies driver mutations in hepatocellular carcinoma. Oncogene. 2018;37(27):3740–52. 10.1038/s41388-018-0206-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lee HW, Kim E, Cho KJ, Park HJ, Seo J, Lee H, et al. Applications of molecular barcode sequencing for the detection of low-frequency variants in circulating tumour DNA from hepatocellular carcinoma. Liver Int. 2022;42(10):2317–26. 10.1111/liv.15356 [DOI] [PubMed] [Google Scholar]
- 124.Liang WJ, Xu ZG, Kong FY, Huang X, Xiao YX, Zhou W, et al. Circulating tumour cell combined with DNA methylation for early detection of hepatocellular carcinoma. Front Genet. 2022;13:1065693. 10.3389/fgene.2022.1065693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Liao WJ, Yang HY, Xu HF, Wang YN, Ge PL, Ren JJ, et al. Noninvasive detection of tumor-associated mutations from circulating cell-free DNA in hepatocellular carcinoma patients by targeted deep sequencing. Oncotarget. 2016;7(26):40481–90. 10.18632/oncotarget.9629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ma XY, Wang Z, Wang SY, Tian Y, Xie B, Li J, et al. The assessment of circulating tumor DNA associated with Wnt/β-catenin signaling pathway as a diagnostic tool for liver cancer: a systematic review and meta-analysis. Expert Rev Anticancer Ther. 2024;24(3–4):155–67. 10.1080/14737140.2024.2312246 [DOI] [PubMed] [Google Scholar]
- 127.Matsumae T, Kodama T, Myojin Y, Maesaka K, Sakamori R, Takuwa A, et al. Circulating cell-free DNA profiling predicts the therapeutic outcome in advanced hepatocellular carcinoma patients treated with combination immunotherapy. Cancers. 2022;14(14):3367. 10.3390/cancers14143367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Muraoka M, Maekawa S, Katoh R, Komiyama Y, Nakakuki N, Takada H, et al. Usefulness of cell-free human telomerase reverse transcriptase mutant DNA quantification in blood for predicting hepatocellular carcinoma treatment efficacy. Hepatol Commun. 2021;5(11):1927–38. 10.1002/hep4.1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Nakatsuka T, Nakagawa H, Hayata Y, Wake T, Yamada T, Kinoshita MN, et al. Post-treatment cell-free DNA as a predictive biomarker in molecular-targeted therapy of hepatocellular carcinoma. J Gastroenterol. 2021;56(5):456–69. 10.1007/s00535-021-01773-4 [DOI] [PubMed] [Google Scholar]
- 130.Nguyen VC, Nguyen TH, Phan TH, Tran THT, Pham TTT, Ho TD, et al. Fragment length profiles of cancer mutations enhance detection of circulating tumor DNA in patients with early-stage hepatocellular carcinoma. BMC Cancer. 2023;23(1):233. 10.1186/s12885-023-10681-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ono A, Fujimoto A, Yamamoto Y, Akamatsu S, Hiraga N, Imamura M, et al. Circulating tumor DNA analysis for liver cancers and its usefulness as a liquid biopsy. Cell Mol Gastroenterol Hepatol. 2015;1(5):516–34. 10.1016/j.jcmgh.2015.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Oversoe SK, Clement MS, Weber B, Gronbæk H, Hamilton-Dutoit SJ, Sorensen BS, et al. Combining tissue and circulating tumor DNA increases the detection rate of a CTNNB1 mutation in hepatocellular carcinoma. BMC Cancer. 2021;21(1):376. 10.1186/s12885-021-08103-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Papatheodoridi A, Chatzigeorgiou A, Chrysavgis L, Lembessis P, Loglio A, Facchetti F, et al. Circulating cell-free DNA species affect the risk of hepatocellular carcinoma in treated chronic hepatitis B patients. J Viral Hepatitis. 2021;28(3):464–74. 10.1111/jvh.13446 [DOI] [PubMed] [Google Scholar]
- 134.Qu CF, Wang YT, Wang P, Chen K, Wang MJ, Zeng HM, et al. Detection of early-stage hepatocellular carcinoma in asymptomatic HBsAg-seropositive individuals by liquid biopsy. Proc Natl Acad Sci U S A. 2019;116(13):6308–12. 10.1073/pnas.1819799116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Villar S, Le Roux-Goglin E, Gouas DA, Plymoth A, Ferro G, Boniol M, et al. Seasonal variation in TP53R249S-mutated serum DNA with aflatoxin exposure and hepatitis B virus infection. Environ Health Perspect. 2011;119(11):1635–40. 10.1289/ehp.1103539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wang J, Yang L, Diao YJ, Liu JY, Li JJ, Li R, et al. Circulating tumour DNA methylation in hepatocellular carcinoma diagnosis using digital droplet PCR. J Int Med Res. 2021;49(3):300060521992962. 10.1177/0300060521992962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wang JH, Qin Y, Li B, Sun ZL, Yang B. Detection of aberrant promoter methylation of GSTP1 in the tumor and serum of Chinese human primary hepatocellular carcinoma patients. Clin Biochem. 2006;39(4):344–8. 10.1016/j.clinbiochem.2006.01.008 [DOI] [PubMed] [Google Scholar]
- 138.Wang P, Song QQ, Ren J, Zhang WL, Wang YT, Zhou L, et al. Simultaneous analysis of mutations and methylations in circulating cell-free DNA for hepatocellular carcinoma detection. Sci Transl Med. 2022;14(672):eabp8704. 10.1126/scitranslmed.abp8704 [DOI] [PubMed] [Google Scholar]
- 139.Wu T, Fan R, Bai J, Yang Z, Qian YS, Du LT, et al. The development of a cSMART-based integrated model for hepatocellular carcinoma diagnosis. J Hematol Oncol. 2023;16(1):1. 10.1186/s13045-022-01396-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yu X, Lei XZ. Application of the multi-omics liquid biopsy method M2P-HCC in early liver cancer screening for high-risk individuals with hepatitis B-related liver cancer. Diagnostics. 2023;13(15):2484. 10.3390/diagnostics13152484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zhang A, Lee TJ, Jain S, Su YH, editors. Urine as an alternative to blood for cancer liquid biopsy and precision medicine. In: IEEE International Conference on Bioinformatics and Biomedicine (BIBM) - Human Genomics; 2018 Dec 03–06. Madrid: IEEE (Institute of Electrical and Electronics Engineers); 2018. 10.1109/BIBM.2018.8621453.
- 142.Zhang HK, Dong PL, Fan HL, Liang H, Zhang K, Zhao YQ, et al. Gene body hypomethylation of pyroptosis-related genes NLRP7, NLRP2, and NLRP3 facilitate non-invasive surveillance of hepatocellular carcinoma. Funct Integr Genomics. 2023;23(2):198. 10.1007/s10142-023-01114-z [DOI] [PubMed] [Google Scholar]
- 143.Zhang HK, Dong PL, Guo SC, Tao CC, Chen W, Zhao WM, et al. Hypomethylation in HBV integration regions aids non-invasive surveillance to hepatocellular carcinoma by low-pass genome-wide bisulfite sequencing. BMC Med. 2020;18(1):200. 10.1186/s12916-020-01667-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhao YT, Zhao L, Jin HF, Xie Y, Chen LYH, Zhang W, et al. Plasma methylated GNB4 and Riplet as a novel dual-marker panel for the detection of hepatocellular carcinoma. Epigenetics. 2024;19(1):2299044. 10.1080/15592294.2023.2299044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Jiang P, Chan CWM, Chan KCA, Cheng SH, Wong J, Wong VW-S, et al. Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc Natl Acad Sci. 2015;112(11):E1317–25. 10.1073/pnas.1500076112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Jiang P, Sun K, Tong YK, Cheng SH, Cheng THT, Heung MMS, et al. Preferred end coordinates and somatic variants as signatures of circulating tumor DNA associated with hepatocellular carcinoma. Proc Natl Acad Sci. 2018;115(46):E10925–33. 10.1073/pnas.1814616115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zeng Y, Hu S, Luo Y, He K. Exosome cargos as biomarkers for diagnosis and prognosis of hepatocellular carcinoma. Pharmaceutics. 2023;15(9):2365. 10.3390/pharmaceutics15092365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Guo S, Hu CX, Zhai XY, Sun D. Circular RNA 0006602 in plasma exosomes: a new potential diagnostic biomarker for hepatocellular carcinoma. Am J Transl Res. 2021;13(6):6001–15. [PMC free article] [PubMed] [Google Scholar]
- 149.Lin J, Lin WS, Bai YN, Liao YL, Lin QY, Chen LF, et al. Identification of exosomal hsa-miR-483-5p as a potential biomarker for hepatocellular carcinoma via microRNA expression profiling of tumor-derived exosomes. Exp Cell Res. 2022;417(2):113232. 10.1016/j.yexcr.2022.113232 [DOI] [PubMed] [Google Scholar]
- 150.Rui T, Zhang XB, Guo JF, Xiang AZ, Tang N, Liu J, et al. Serum-exosome-derived miRNAs serve as promising biomarkers for HCC diagnosis. Cancers. 2023;15(1):205. 10.3390/cancers15010205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Li L-M, Hu Z-B, Zhou Z-X, Chen X, Liu F-Y, Zhang J-F, et al. Serum microRNA profiles serve as novel biomarkers for HBV infection and diagnosis of HBV-positive hepatocarcinoma. Cancer Res. 2010;70(23):9798–807. 10.1158/0008-5472.CAN-10-1001 [DOI] [PubMed] [Google Scholar]
- 152.Chen WB, Jiang JJ, Gong L, Shu ZY, Xiang DR, Zhang XJ, et al. Hepatitis B virus P protein initiates glycolytic bypass in HBV-related hepatocellular carcinoma via a FOXO3/miRNA-30b-5p/MINPP1 axis. J Exp Clin Cancer Res. 2021;40(1):1–8. 10.1186/s13046-020-01803-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.El-Maraghy SA, Adel O, Zayed N, Yosry A, El-Nahaas SM, Gibriel AA. Circulatory miRNA-484, 524, 615 and 628 expression profiling in HCV mediated HCC among Egyptian patients; implications for diagnosis and staging of hepatic cirrhosis and fibrosis. J Adv Res. 2020;22:57–66. 10.1016/j.jare.2019.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zhang L, Liu Q, Guo Y, Tian L, Chen K, Bai D, et al. DNA-based molecular classifiers for the profiling of gene expression signatures. J Nanobiotechnol. 2024;22(1):189. 10.1186/s12951-024-02445-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kristensen LS, Jakobsen T, Hager H, Kjems J. The emerging roles of circRNAs in cancer and oncology. Nat Rev Clin Oncol. 2022;19(3):188–206. 10.1038/s41571-021-00585-y [DOI] [PubMed] [Google Scholar]
- 156.Zhang F, Jiang J, Qian H, Yan Y, Xu W. Exosomal circRNA: emerging insights into cancer progression and clinical application potential. J Hematol Oncol. 2023;16(1):67. 10.1186/s13045-023-01452-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Han D, Li J, Wang H, Su X, Hou J, Gu Y, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66(4):1151–64. 10.1002/hep.29270 [DOI] [PubMed] [Google Scholar]
- 158.Klingenberg M, Matsuda A, Diederichs S, Patel T. Non-coding RNA in hepatocellular carcinoma: mechanisms, biomarkers and therapeutic targets. J Hepatol. 2017;67(3):603–19. 10.1016/j.jhep.2017.04.009 [DOI] [PubMed] [Google Scholar]
- 159.Liu F, Xiang G, Jiang D, Zhang L, Chen X, Liu L, et al. Ultrasensitive strategy based on PtPd nanodendrite/nano-flower-like@GO signal amplification for the detection of long non-coding RNA. Biosens Bioelectron. 2015;74:214–21. 10.1016/j.bios.2015.06.021 [DOI] [PubMed] [Google Scholar]
- 160.Xie H, Ma H, Zhou D. Plasma HULC as a promising novel biomarker for the detection of hepatocellular carcinoma. Biomed Res Int. 2013;2013:136106. 10.1155/2013/136106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kim SS, Baek GO, Ahn HR, Sung S, Seo CW, Cho HJ, et al. Serum small extracellular vesicle-derived LINC00853 as a novel diagnostic marker for early hepatocellular carcinoma. Mol Oncol. 2020;14(10):2646–59. 10.1002/1878-0261.12745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kim SS, Baek GO, Son JA, Ahn HR, Yoon MK, Cho HJ, et al. Early detection of hepatocellular carcinoma via liquid biopsy: panel of small extracellular vesicle-derived long noncoding RNAs identified as markers. Mol Oncol. 2021;15(10):2715–31. 10.1002/1878-0261.13049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Li Y, Zhao J, Yu S, Wang Z, He X, Su Y, et al. Extracellular vesicles long RNA sequencing reveals abundant mRNA, circRNA, and lncRNA in human blood as potential biomarkers for cancer diagnosis. Clin Chem. 2019;65(6):798–808. 10.1373/clinchem.2018.301291 [DOI] [PubMed] [Google Scholar]
- 164.Zhu Q, Xie J, Mei W, Zeng C. Methylated circulating tumor DNA in hepatocellular carcinoma: a comprehensive analysis of biomarker potential and clinical implications. Cancer Treat Rev. 2024;128:102763. 10.1016/j.ctrv.2024.102763 [DOI] [PubMed] [Google Scholar]
- 165.Abou Zeid AA, El-Sayed ET, Ahdy JK, Tawfik MR. Ras association domain family 1A gene promoter methylation as a biomarker for chronic viral hepatitis C-related hepatocellular carcinoma. Cureus J Med Sci. 2023;15(9):e45687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Araújo OC, Rosa AS, Fernandes A, Niel C, Villela-Nogueira CA, Pannain V, et al. RASSF1A and DOK1 promoter methylation levels in hepatocellular carcinoma, cirrhotic and non-cirrhotic liver, and correlation with liver cancer in Brazilian patients. PLoS One. 2016;11(4):e0153796. 10.1371/journal.pone.0153796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Azab NI, Abd El Kariem HM, Mowafi T, Fouad HF, El Abd AM. Blood Ras-association domain family 1 A gene methylation status in some liver diseases. Life Sci J Acta Zhengzhou Univ Overseas Ed. 2011;8(2):531–9. [Google Scholar]
- 168.Chan KCA, Lai PBS, Mok TSK, Chan HLY, Ding CM, Yeung SW, et al. Quantitative analysis of circulating methylated DNA as a biomarker for hepatocellular carcinoma. Clin Chem. 2008;54(9):1528–36. 10.1373/clinchem.2008.104653 [DOI] [PubMed] [Google Scholar]
- 169.Cheishvili D, Wong C, Karim MM, Kibria MG, Jahan N, Das PC, et al. A high-throughput test enables specific detection of hepatocellular carcinoma. Nat Commun. 2023;14(1):3306. 10.1038/s41467-023-39055-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Chen LM, Xiang L, Sun WJ, Zhai YJ, Gao S, Fan YC, et al. Diagnostic value of the hypomethylation of the WISP1 promoter in patients with hepatocellular carcinoma associated with hepatitis B virus. Tohoku J Exp Med. 2020;252(4):297–307. 10.1620/tjem.252.297 [DOI] [PubMed] [Google Scholar]
- 171.Cheng JM, Wei DK, Ji Y, Chen LL, Yang LG, Li G, et al. Integrative analysis of DNA methylation and gene expression reveals hepatocellular carcinoma-specific diagnostic biomarkers. Genome Med. 2018;10(1):42. 10.1186/s13073-018-0548-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Di JZ, Han XD, Gu WY, Wang Y, Zheng Q, Zhang P, et al. Association of hypomethylation of LINE-1 repetitive element in blood leukocyte DNA with an increased risk of hepatocellular carcinoma. J Zhejiang Univ Sci B. 2011;12(10):805–11. 10.1631/jzus.B1000422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Dong XY, He H, Zhang WY, Yu DJ, Wang XJ, Chen YM. Combination of serum RASSF1A methylation and AFP is a promising non-invasive biomarker for HCC patient with chronic HBV infection. Diagn Pathol. 2015;10:133. 10.1186/s13000-015-0317-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Dou CY, Fan YC, Cao CJ, Yang Y, Wang K. Sera DNA Methylation of CDH1, DNMT3b and ESR1 promoters as biomarker for the early diagnosis of hepatitis B virus-related hepatocellular carcinoma. Dig Dis Sci. 2016;61(4):1130–8. 10.1007/s10620-015-3975-3 [DOI] [PubMed] [Google Scholar]
- 175.El-Bendary M, Nour D, Arafa M, Neamatallah M. Methylation of tumour suppressor genes RUNX3, RASSF1A and E-Cadherin in HCV-related liver cirrhosis and hepatocellular carcinoma. Br J Biomed Sci. 2020;77(1):35–40. 10.1080/09674845.2019.1694123 [DOI] [PubMed] [Google Scholar]
- 176.Elsewify WA, Hassan EA, Mekky MA, Abd El-Rehim A, Sayed Z, Malek MOA, et al. Usefulness of circulating methylated p16 as a noninvasive molecular biomarker for hepatitis C-related hepatocellular carcinoma with normal serum alpha-fetoprotein levels. Int J Gen Med. 2020;13:147–55. 10.2147/IJGM.S249272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Funderburk K, Bang-Christensen SR, Miller BF, Tan H, Margolin G, Petrykowska HM, et al. Evaluating stacked methylation markers for blood-based multicancer detection. Cancers. 2023;15(19):4826. 10.3390/cancers15194826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Gonçalves E, Gonçalves-Reis M, Pereira-Leal JB, Cardoso J. DNA methylation fingerprint of hepatocellular carcinoma from tissue and liquid biopsies. Sci Rep. 2022;12(1):11512. 10.1038/s41598-022-15058-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Guo P, Zheng HL, Li YH, Li YT, Xiao Y, Zheng J, et al. Hepatocellular carcinoma detection via targeted enzymatic methyl sequencing of plasma cell-free DNA. Clin Epigenetics. 2023;15(1):2. 10.1186/s13148-022-01420-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Hu L, Chen G, Yu HP, Qiu XQ. Clinicopathological significance of RASSF1A reduced expression and hypermethylation in hepatocellular carcinoma. Hepatol Int. 2010;4(1):423–32. 10.1007/s12072-010-9164-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Huang F, Yang GW, Jiang HQ, Chen XN, Yang YH, Yu Q, et al. Role of plasma methylated SEPT9 for predicting microvascular invasion and tumor proliferation in hepatocellular carcinoma. Technol Cancer Res Treat. 2022;21:15330338221144510. 10.1177/15330338221144510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Huang WQ, Li T, Yang WL, Chai XJ, Chen KF, Wei L, et al. Analysis of DNA methylation in plasma for monitoring hepatocarcinogenesis. Genet Test Mol Biomarkers. 2015;19(6):295–302. 10.1089/gtmb.2014.0292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Huang ZH, Hu Y, Hua D, Wu YY, Song MX, Cheng ZH. Quantitative analysis of multiple methylated genes in plasma for the diagnosis and prognosis of hepatocellular carcinoma. Exp Mol Pathol. 2011;91(3):702–7. 10.1016/j.yexmp.2011.08.004 [DOI] [PubMed] [Google Scholar]
- 184.Jain S, Chang TT, Hamilton JP, Lin SY, Lin YJ, Evans AA, et al. Methylation of the CpG sites only on the sense strand of the APC gene is specific for hepatocellular carcinoma. PLoS One. 2011;6(11):e26799. 10.1371/journal.pone.0026799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Ji XF, Fan YC, Gao S, Yang Y, Zhang JJ, Wang K. MT1M and MT1G promoter methylation as biomarkers for hepatocellular carcinoma. World J Gastroenterol. 2014;20(16):4723–9. 10.3748/wjg.v20.i16.4723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Jiang CG, Chen Q, Wu LN, Wang G, Ma JZ. The innovative regularity and role of p16 methylation in blood during HCC development. J Cancer. 2018;9(11):1925–31. 10.7150/jca.23968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Johnson AM, Dudek JM, Edwards DK, Myers TA, Joseph P, Laffin JJ, et al. Analytical validation of a novel multi-target blood-based test to detect hepatocellular carcinoma. Expert Rev Mol Diagn. 2021;21(11):1245–52. 10.1080/14737159.2021.1981290 [DOI] [PubMed] [Google Scholar]
- 188.Kotoh Y, Suehiro Y, Saeki I, Hoshida T, Maeda M, Iwamoto T, et al. Novel liquid biopsy test based on a sensitive methylated SEPT9 assay for diagnosing hepatocellular carcinoma. Hepatol Commun. 2020;4(3):461–70. 10.1002/hep4.1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Li BL, Huang H, Huang RH, Zhang W, Zhou GP, Wu Z, et al. SEPT9 gene methylation as a noninvasive marker for hepatocellular carcinoma. Dis Markers. 2020;2020:6289063. 10.1155/2020/6289063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Li F, Fan YC, Gao S, Sun FK, Yang Y, Wang K. Methylation of serum insulin-like growth factor-binding protein 7 promoter in hepatitis B virus-associated hepatocellular carcinoma. Genes Chromosom Cancer. 2014;53(1):90–7. 10.1002/gcc.22120 [DOI] [PubMed] [Google Scholar]
- 191.Liu HH, Fang Y, Wang JW, Yuan XD, Fan YC, Gao S, et al. Hypomethylation of the cyclin D1 promoter in hepatitis B virus-associated hepatocellular carcinoma. Medicine (Baltimore). 2020;99(20):e20326. 10.1097/MD.0000000000020326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Liu XY, Fan YC, Gao S, Zhao J, Chen LY, Li F, et al. Methylation of SOX1 and VIM promoters in serum as potential biomarkers for hepatocellular carcinoma. Neoplasma. 2017;64(5):745–53. 10.4149/neo_2017_513 [DOI] [PubMed] [Google Scholar]
- 193.Liu ZJ, Huang Y, Wei L, He JY, Liu QY, Yu XQ, et al. Combination of LINE-1 hypomethylation and RASSF1A promoter hypermethylation in serum DNA is a non-invasion prognostic biomarker for early recurrence of hepatocellular carcinoma after curative resection. Neoplasma. 2017;64(5):795–802. 10.4149/neo_2017_519 [DOI] [PubMed] [Google Scholar]
- 194.Lubecka K, Flower K, Beetch M, Qiu J, Kurzava L, Buvala H, et al. Loci-specific differences in blood DNA methylation in HBV-negative populations at risk for hepatocellular carcinoma development. Epigenetics. 2018;13(6):605–26. 10.1080/15592294.2018.1481706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Ma JX, Jin JP, Lu HS, Zhang J, Li YL, Cai XF. Exonuclease 1 is a potential diagnostic and prognostic biomarker in hepatocellular carcinoma. Front Mol Biosci. 2022;9:889414. 10.3389/fmolb.2022.889414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Oussalah A, Rischer S, Bensenane M, Conroy G, Filhine-Tresarrieu P, Debard R, et al. Plasma mSEPT9: a novel circulating cell-free DNA-based epigenetic biomarker to diagnose hepatocellular carcinoma. EBioMedicine. 2018;30:138–47. 10.1016/j.ebiom.2018.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Pasha HF, Mohamed RH, Radwan MI. RASSF1A and SOCS1 genes methylation status as a noninvasive marker for hepatocellular carcinoma. Cancer Biomark. 2019;24(2):241–7. 10.3233/CBM-181638 [DOI] [PubMed] [Google Scholar]
- 198.Qian Y, Wang JW, Fang Y, Yuan XD, Fan YC, Gao S, et al. Measurement of cyclin D2 (CCND2) gene promoter methylation in plasma and peripheral blood mononuclear cells and alpha-fetoprotein levels in patients with hepatitis B virus-associated hepatocellular carcinoma. Med Sci Monit. 2020;26:e927444. 10.12659/MSM.927444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Qiao CY, Li F, Teng Y, Zhao J, Hu N, Fan YC, et al. Aberrant GSTP1 promoter methylation predicts poor prognosis of acute-on-chronic hepatitis B pre-liver failure. Clin Exp Med. 2018;18(1):51–62. 10.1007/s10238-017-0466-1 [DOI] [PubMed] [Google Scholar]
- 200.Qiu G, Fan JC, He YS. 5′ CpG island methylation analysis identifies the MAGE-A1 and MAGE-A3 genes as potential markers of HCC. Clin Biochem. 2006;39(3):259–66. 10.1016/j.clinbiochem.2006.01.014 [DOI] [PubMed] [Google Scholar]
- 201.Shitani M, Sasaki S, Akutsu N, Takagi H, Suzuki H, Nojima M, et al. Genome-wide analysis of DNA methylation identifies novel cancer-related genes in hepatocellular carcinoma. Tumor Biology. 2012;33(5):1307–17. 10.1007/s13277-012-0378-3 [DOI] [PubMed] [Google Scholar]
- 202.Sultan MQ, Charfeddine B, Al-Salih ARH. Evaluation of the diagnostic performance of Alpha-1-Antitrypsin in early detection of hepatocellular carcinoma. Cell Mol Biol (Noisy-le-grand). 2023;69(14):177–85. 10.14715/cmb/2023.69.14.29 [DOI] [PubMed] [Google Scholar]
- 203.Sun FK, Sun Q, Fan YC, Gao S, Zhao J, Li F, et al. Methylation of tissue factor pathway inhibitor 2 as a prognostic biomarker for hepatocellular carcinoma after hepatectomy. J Gastroenterol Hepatol. 2016;31(2):484–92. 10.1111/jgh.13154 [DOI] [PubMed] [Google Scholar]
- 204.Tan SH, Ida H, Lau QC, Goh BC, Chieng WS, Loh M, et al. Detection of promoter hypermethylation in serum samples of cancer patients by methylation-specific polymerase chain reaction for tumour suppressor genes including RUNX3. Oncol Rep. 2007;18(5):1225–30. [PubMed] [Google Scholar]
- 205.Tang LP, Zhu SS, Peng WY, Yin XD, Tan C, Yang YY. Epigenetic identification of mitogen-activated protein kinase 10 as a functional tumor suppressor and clinical significance for hepatocellular carcinoma. PeerJ. 2021;9:e10810. 10.7717/peerj.10810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Teng Y, Fan YC, Mu NN, Zhao J, Sun FK, Wang K. Serum SOX11 promoter methylation is a novel biomarker for the diagnosis of Hepatitis B virus-related hepatocellular carcinoma. Neoplasma. 2016;63(3):419–26. 10.4149/311_151029N552 [DOI] [PubMed] [Google Scholar]
- 207.Tian MM, Fan YC, Zhao J, Gao S, Zhao ZH, Chen LY, et al. Hepatocellular carcinoma suppressor 1 promoter hypermethylation in serum. A diagnostic and prognostic study in hepatitis B. Clin Res Hepatol Gastroenterol. 2017;41(2):171–80. 10.1016/j.clinre.2016.10.003 [DOI] [PubMed] [Google Scholar]
- 208.Wong N, Lam WC, Lai PB, Pang E, Lau WY, Johnson PJ. Hypomethylation of chromosome 1 heterochromatin DNA correlates with q-arm copy gain in human hepatocellular carcinoma. Am J Pathol. 2001;159(2):465–71. 10.1016/S0002-9440(10)61718-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Wu HC, Wang Q, Yang HI, Tsai WY, Chen CJ, Santella RM. Global DNA methylation levels in white blood cells as a biomarker for hepatocellular carcinoma risk: a nested casecontrol study. Carcinogenesis. 2012;33(7):1340–5. 10.1093/carcin/bgs160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Wu HC, Wang Q, Yang HI, Tsai WY, Chen CJ, Santella RM. Global DNA methylation in a population with aflatoxin B1 exposure. Epigenetics. 2013;8(9):962–9. 10.4161/epi.25696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Xiang L, Chen LM, Zhai YJ, Sun WJ, Yang JR, Fan YC, et al. Hypermethylation of secreted frizzled related protein 2 gene promoter serves as a noninvasive biomarker for HBV-associated hepatocellular carcinoma. Life Sci. 2021;270:119061. 10.1016/j.lfs.2021.119061 [DOI] [PubMed] [Google Scholar]
- 212.Xie GF, Xu YX, Xu F, Sun LY, Ye ZL, Ma JJ, et al. Plasma SGIP1 methylation in diagnosis and prognosis prediction in hepatocellular carcinoma. Neoplasma. 2021;68(1):62–70. 10.4149/neo_2020_200623N657 [DOI] [PubMed] [Google Scholar]
- 213.Xu F, Zhang LL, Xu YX, Song D, He WT, Ji XM, et al. Hypermethylation of SCAND3 and Myo1g gene are potential diagnostic biomarkers for hepatocellular carcinoma. Cancers. 2020;12(8):2332. 10.3390/cancers12082332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Xu ZG, Du JJ, Zhang X, Cheng ZH, Ma ZZ, Xiao HS, et al. A novel liver-specific zona pellucida domain containing protein that is expressed rarely in hepatocellular carcinoma. Hepatology. 2003;38(3):735–44. 10.1053/jhep.2003.50340 [DOI] [PubMed] [Google Scholar]
- 215.Yan XJ, Wu TY, Tang M, Chen DL, Huang MY, Zhou SC, et al. Methylation of the ataxia telangiectasia mutated gene (ATM) promoter as a radiotherapy outcome biomarker in patients with hepatocellular carcinoma. Medicine (Baltimore). 2020;99(4):e18823. 10.1097/MD.0000000000018823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Yang JR, Wang J, Li HM, Gao S, Fan YC, Wang K. IL-6 promoter hypomethylation acts as a diagnostic biomarker in hepatitis B virus-associated hepatocellular carcinoma. Front Oncol. 2022;12:746643. 10.3389/fonc.2022.746643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Yang Y, Fan YC, Gao S, Dou CY, Zhang JJ, Sun FK, et al. Methylated cysteine dioxygenase-1 gene promoter in the serum is a potential biomarker for hepatitis B virus-related hepatocellular carcinoma. Tohoku J Exp Med. 2014;232(3):187–94. 10.1620/tjem.232.187 [DOI] [PubMed] [Google Scholar]
- 218.Yuan XD, Wang JW, Fang Y, Qian Y, Gao S, Fan YC, et al. Methylation status of the T-cadherin gene promotor in peripheral blood mononuclear cells is associated with HBV-related hepatocellular carcinoma progression. Pathol Res Pract. 2020;216(5):152914. 10.1016/j.prp.2020.152914 [DOI] [PubMed] [Google Scholar]
- 219.Zhang C, Ge S, Wang J, Jing XT, Li HL, Mei SY, et al. Epigenomic profiling of DNA methylation for hepatocellular carcinoma diagnosis and prognosis prediction. J Gastroenterol Hepatol. 2019;34(10):1869–77. 10.1111/jgh.14694 [DOI] [PubMed] [Google Scholar]
- 220.Zhang PJ, Wen XY, Gu F, Deng XX, Li J, Dong J, et al. Methylation profiling of serum DNA from hepatocellular carcinoma patients using an Infinium Human Methylation 450 BeadChip. Hepatol Int. 2013;7(3):893–900. 10.1007/s12072-013-9437-0 [DOI] [PubMed] [Google Scholar]
- 221.Zhang Y, Wang JW, Su X, Li JE, Wei XF, Yang JR, et al. F-box protein 43 promoter methylation as a novel biomarker for hepatitis B virus-associated hepatocellular carcinoma. Front Microbiol. 2023;14:1267844. 10.3389/fmicb.2023.1267844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Zhang YJ, Wu HC, Shen J, Ahsan H, Tsai WY, Yang HL, et al. Predicting hepatocellular carcinoma by detection of aberrant promoter methylation in serum DNA. Clin Cancer Res. 2007;13(8):2378–84. 10.1158/1078-0432.CCR-06-1900 [DOI] [PubMed] [Google Scholar]
- 223.Zhang ZY, Chen P, Xie H, Cao PG. Using circulating tumor DNA as a novel biomarker to screen and diagnose hepatocellular carcinoma: a systematic review and meta-analysis. Cancer Med. 2020;9(4):1349–64. 10.1002/cam4.2799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Raos D, Ulamec M, Bojanac AK, Bulic-Jakus F, Jezek D, Sincic N. Epigenetically inactivated RASSF1A as a tumor biomarker. Bosn J Basic Med Sci. 2021;21(4):386–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Villanueva A, Portela A, Sayols S, Battiston C, Hoshida Y, Méndez-González J, et al. DNA methylation-based prognosis and epidrivers in hepatocellular carcinoma. Hepatology. 2015;61(6):1945–56. 10.1002/hep.27732 [DOI] [PubMed] [Google Scholar]
- 226.Kakehashi A, Ishii N, Shibata T, Wei M, Okazaki E, Tachibana T, et al. Mitochondrial prohibitins and septin 9 are implicated in the onset of rat hepatocarcinogenesis. Toxicol Sci. 2011;119(1):61–72. 10.1093/toxsci/kfq307 [DOI] [PubMed] [Google Scholar]
- 227.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. 10.1126/science.1260419 [DOI] [PubMed] [Google Scholar]
- 228.Zhang W, Dai J, Hou G, Liu H, Zheng S, Wang X, et al. SMURF2 predisposes cancer cell toward ferroptosis in GPX4-independent manners by promoting GSTP1 degradation. Mol Cell. 2023;83(23):4352-69.e8. 10.1016/j.molcel.2023.10.042 [DOI] [PubMed] [Google Scholar]
- 229.Tilman G, Arnoult N, Lenglez S, Van Beneden A, Loriot A, De Smet C, et al. Cancer-linked satellite 2 DNA hypomethylation does not regulate Sat2 non-coding RNA expression and is initiated by heat shock pathway activation. Epigenetics. 2012;7(8):903–13. 10.4161/epi.21107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Probst AV, Okamoto I, Casanova M, El Marjou F, Le Baccon P, Almouzni G. A strand-specific burst in transcription of pericentric satellites is required for chromocenter formation and early mouse development. Dev Cell. 2010;19(4):625–38. 10.1016/j.devcel.2010.09.002 [DOI] [PubMed] [Google Scholar]
- 231.van Niel G, D’Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018;19(4):213–28. 10.1038/nrm.2017.125 [DOI] [PubMed] [Google Scholar]
- 232.Jeppesen DK, Fenix AM, Franklin JL, Higginbotham JN, Zhang Q, Zimmerman LJ, et al. Reassessment of exosome composition. Cell. 2019;177(2):428-45.e18. 10.1016/j.cell.2019.02.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Guo X, Gao C, Yang D-H, Li S. Exosomal circular RNAs: a chief culprit in cancer chemotherapy resistance. Drug Resist Updates. 2023;67:100937. 10.1016/j.drup.2023.100937 [DOI] [PubMed] [Google Scholar]
- 234.Adamczyk AM, Leicaj ML, Fabiano MP, Cabrerizo G, Bannoud N, Croci DO, et al. Extracellular vesicles from human plasma dampen inflammation and promote tissue repair functions in macrophages. J Extracell Vesicles. 2023;12(6):e12331. 10.1002/jev2.12331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Bebelman MP, Smit MJ, Pegtel DM, Baglio SR. Biogenesis and function of extracellular vesicles in cancer. Pharmacol Ther. 2018;188:1–11. 10.1016/j.pharmthera.2018.02.013 [DOI] [PubMed] [Google Scholar]
- 236.Yeo W, Mo FK, Chan SL, Leung NW, Hui P, Lam WY, et al. Hepatitis B viral load predicts survival of HCC patients undergoing systemic chemotherapy. Hepatology. 2007;45(6):1382–9. 10.1002/hep.21572 [DOI] [PubMed] [Google Scholar]
- 237.Choi WM, Kim GA, Choi J, Choi GH, Lee YB, Sinn DH, et al. Non-linear association of baseline viral load with on-treatment hepatocellular carcinoma risk in chronic hepatitis B. Gut. 2024;73(4):649–58. [DOI] [PubMed] [Google Scholar]
- 238.Choi WM, Yip TC, Kim WR, Yee LJ, Brooks-Rooney C, Curteis T, et al. Chronic hepatitis B baseline viral load and on-treatment liver cancer risk: a multinational cohort study of HBeAg-positive patients. Hepatology. 2024;80(2):428–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Riveiro-Barciela M, Pericàs JM, Buti M. How to interpret viral markers in the management of chronic hepatitis B infection. Clin Microbiol Infect. 2022;28(3):355–61. 10.1016/j.cmi.2021.10.020 [DOI] [PubMed] [Google Scholar]
- 240.Yoshida M, Hatano N, Nishiumi S, Irino Y, Izumi Y, Takenawa T, et al. Diagnosis of gastroenterological diseases by metabolome analysis using gas chromatography–mass spectrometry. J Gastroenterol. 2012;47(1):9–20. 10.1007/s00535-011-0493-8 [DOI] [PubMed] [Google Scholar]
- 241.Xue R, Lin Z, Deng C, Dong L, Liu T, Wang J, et al. A serum metabolomic investigation on hepatocellular carcinoma patients by chemical derivatization followed by gas chromatography/mass spectrometry. Rapid Commun Mass Spectrom. 2008;22(19):3061–8. 10.1002/rcm.3708 [DOI] [PubMed] [Google Scholar]
- 242.Wu H, Xue R, Dong L, Liu T, Deng C, Zeng H, et al. Metabolomic profiling of human urine in hepatocellular carcinoma patients using gas chromatography/mass spectrometry. Anal Chim Acta. 2009;648(1):98–104. 10.1016/j.aca.2009.06.033 [DOI] [PubMed] [Google Scholar]
- 243.Chen T, Xie G, Wang X, Fan J, Qiu Y, Zheng X, et al. Serum and urine metabolite profiling reveals potential biomarkers of human hepatocellular carcinoma. Mol Cell Proteomics. 2011;10(7):M110.004945. 10.1074/mcp.M110.004945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Chew V, Chuang CH, Hsu C. Translational research on drug development and biomarker discovery for hepatocellular carcinoma. J Biomed Sci. 2024;31(1):22. 10.1186/s12929-024-01011-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Niu F, Wang DC, Lu J, Wu W, Wang X. Potentials of single-cell biology in identification and validation of disease biomarkers. J Cell Mol Med. 2016;20(9):1789–95. 10.1111/jcmm.12868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Xing X, Cai L, Ouyang J, Wang F, Li Z, Liu M, et al. Proteomics-driven noninvasive screening of circulating serum protein panels for the early diagnosis of hepatocellular carcinoma. Nat Commun. 2023;14(1):8392. 10.1038/s41467-023-44255-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Nie W, Yan L, Lee YH, Guha C, Kurland IJ, Lu H. Advanced mass spectrometry-based multi-omics technologies for exploring the pathogenesis of hepatocellular carcinoma. Mass Spectrom Rev. 2016;35(3):331–49. 10.1002/mas.21439 [DOI] [PubMed] [Google Scholar]
- 248.Wang Y, Xie Y, Qian L, Ding R, Pang R, Chen P, et al. RAB42 overexpression correlates with poor prognosis, immune cell infiltration and chemoresistance. Front Pharmacol. 2024;15:1445170. 10.3389/fphar.2024.1445170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Xu RH, Wei W, Krawczyk M, Wang WQ, Luo HY, Flagg K, et al. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat Mater. 2017;16(11):1155-#x0002B; 10.1038/nmat4997 [DOI] [PubMed] [Google Scholar]
- 250.Huang H, Ren Z, Gao X, Hu X, Zhou Y, Jiang J, et al. Integrated analysis of microbiome and host transcriptome reveals correlations between gut microbiota and clinical outcomes in HBV-related hepatocellular carcinoma. Genome Med. 2020;12(1):102. 10.1186/s13073-020-00796-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Wang X, Zhang A, Sun H. Power of metabolomics in diagnosis and biomarker discovery of hepatocellular carcinoma. Hepatology. 2013;57(5):2072–7. 10.1002/hep.26130 [DOI] [PubMed] [Google Scholar]
- 252.Lee T, Rawding PA, Bu J, Hyun S, Rou W, Jeon H, et al. Machine-learning-based clinical biomarker using cell-free DNA for hepatocellular carcinoma (HCC). Cancers. 2022;14(9):2061. 10.3390/cancers14092061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Li M, Wang L, Cong L, Wong CC, Zhang X, Chen H, et al. Spatial proteomics of immune microenvironment in nonalcoholic steatohepatitis-associated hepatocellular carcinoma. Hepatology. 2024;79(3):560–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Zhang S, Deshpande A, Verma BK, Wang H, Mi H, Yuan L, et al. Integration of clinical trial spatial multiomics analysis and virtual clinical trials enables immunotherapy response prediction and biomarker discovery. Cancer Res. 2024;84(16):2734–48. 10.1158/0008-5472.CAN-24-0943 [DOI] [PubMed] [Google Scholar]
- 255.Rebouissou S, Nault JC. Advances in molecular classification and precision oncology in hepatocellular carcinoma. J Hepatol. 2020;72(2):215–29. 10.1016/j.jhep.2019.08.017 [DOI] [PubMed] [Google Scholar]
- 256.Trevisani F, Vitale A, Kudo M, Kulik L, Park JW, Pinato DJ, et al. Merits and boundaries of the BCLC staging and treatment algorithm: learning from the past to improve the future with a novel proposal. J Hepatol. 2024;80(4):661–9. 10.1016/j.jhep.2024.01.010 [DOI] [PubMed] [Google Scholar]
- 257.Zhang XP, Jiang N, Zhu L, Lin ZY, Guo WX, Chen X, et al. Short-term and long-term outcomes after robotic versus open hepatectomy in patients with large hepatocellular carcinoma: a multicenter study. Int J Surg. 2024;110(2):660–7. 10.1097/JS9.0000000000000873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Liu G, Wang K, Li J, Xia Y, Lu L, Wan X, et al. Changes in serum alpha fetoprotein in patients with recurrent hepatocellular carcinoma following hepatectomy. J Gastroenterol Hepatol. 2015;30(9):1405–11. 10.1111/jgh.12953 [DOI] [PubMed] [Google Scholar]
- 259.Wei W-X, Yang Z-S, Lu L-H, Li J, Lei Z-Q, Wang K, et al. Long-term survival after partial hepatectomy for sub-stage patients with intermediate stage hepatocellular carcinoma. Int J Surg. 2018;56:256–63. 10.1016/j.ijsu.2018.06.020 [DOI] [PubMed] [Google Scholar]
- 260.Jin GZ, Yu WL, Dong H, Zhou WP, Gu YJ, Yu H, et al. SUOX is a promising diagnostic and prognostic biomarker for hepatocellular carcinoma. J Hepatol. 2013;59(3):510–7. 10.1016/j.jhep.2013.04.028 [DOI] [PubMed] [Google Scholar]
- 261.Yamamoto K, Imamura H, Matsuyama Y, Kume Y, Ikeda H, Norman GL, et al. AFP, AFP-L3, DCP, and GP73 as markers for monitoring treatment response and recurrence and as surrogate markers of clinicopathological variables of HCC. J Gastroenterol. 2010;45(12):1272–82. 10.1007/s00535-010-0278-5 [DOI] [PubMed] [Google Scholar]
- 262.Toyoda H, Kumada T, Tada T, Niinomi T, Ito T, Kaneoka Y, et al. Prognostic significance of a combination of pre- and post-treatment tumor markers for hepatocellular carcinoma curatively treated with hepatectomy. J Hepatol. 2012;57(6):1251–7. 10.1016/j.jhep.2012.07.018 [DOI] [PubMed] [Google Scholar]
- 263.Chan AWH, Zhong J, Berhane S, Toyoda H, Cucchetti A, Shi K, et al. Development of pre and post-operative models to predict early recurrence of hepatocellular carcinoma after surgical resection. J Hepatol. 2018;69(6):1284–93. 10.1016/j.jhep.2018.08.027 [DOI] [PubMed] [Google Scholar]
- 264.Shirakawa H, Suzuki H, Shimomura M, Kojima M, Gotohda N, Takahashi S, et al. Glypican-3 expression is correlated with poor prognosis in hepatocellular carcinoma. Cancer Sci. 2009;100(8):1403–7. 10.1111/j.1349-7006.2009.01206.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Wang L, Pan L, Yao M, Cai Y, Dong Z, Yao D. Expression of oncofetal antigen glypican-3 associates significantly with poor prognosis in HBV-related hepatocellular carcinoma. Oncotarget. 2016;7(27):42150. 10.18632/oncotarget.9892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Ning S, Bin C, Na H, Peng S, Yi D, Xiang-hua Y, et al. Glypican-3, a novel prognostic marker of hepatocellular cancer, is related with postoperative metastasis and recurrence in hepatocellular cancer patients. Mol Biol Rep. 2012;39(1):351–7. 10.1007/s11033-011-0745-y [DOI] [PubMed] [Google Scholar]
- 267.Yorita K, Takahashi N, Takai H, Kato A, Suzuki M, Ishiguro T, et al. Prognostic significance of circumferential cell surface immunoreactivity of glypican-3 in hepatocellular carcinoma. Liver Int. 2011;31(1):120–31. 10.1111/j.1478-3231.2010.02359.x [DOI] [PubMed] [Google Scholar]
- 268.Haruyama Y, Yorita K, Yamaguchi T, Kitajima S, Amano J, Ohtomo T, et al. High preoperative levels of serum glypican-3 containing N-terminal subunit are associated with poor prognosis in patients with hepatocellular carcinoma after partial hepatectomy. Int J Cancer. 2015;137(7):1643–51. 10.1002/ijc.29518 [DOI] [PubMed] [Google Scholar]
- 269.Wang YL, Zhu ZJ, Teng DH, Yao Z, Gao W, Shen ZY. Glypican-3 expression and its relationship with recurrence of HCC after liver transplantation. World J Gastroenterol. 2012;18(19):2408–14. 10.3748/wjg.v18.i19.2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Wang Y, Shen Z, Zhu Z, Han R, Huai M. Clinical values of AFP, GPC3 mRNA in peripheral blood for prediction of hepatocellular carcinoma recurrence following OLT: AFP, GPC3 mRNA for prediction of HCC. Hepat Mon. 2011;11(3):195. [PMC free article] [PubMed] [Google Scholar]
- 271.Yu M-C, Lee Y-S, Lin S-E, Wu H-Y, Chen T-C, Lee W-C, et al. Recurrence and poor prognosis following resection of small hepatitis B-related hepatocellular carcinoma lesions are associated with aberrant tumor expression profiles of glypican 3 and osteopontin. Ann Surg Oncol. 2012;19:455–63. 10.1245/s10434-011-1946-2 [DOI] [PubMed] [Google Scholar]
- 272.Feng J, Zhu R, Chang C, Yu L, Cao F, Zhu G, et al. CK19 and glypican 3 expression profiling in the prognostic indication for patients with HCC after surgical resection. PLoS One. 2016;11(3):e0151501. 10.1371/journal.pone.0151501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Yuan SX, Yang F, Yang Y, Tao QF, Zhang J, Huang G, et al. Long noncoding RNA associated with microvascular invasion in hepatocellular carcinoma promotes angiogenesis and serves as a predictor for hepatocellular carcinoma patients’ poor recurrence-free survival after hepatectomy. Hepatology. 2012;56(6):2231–41. 10.1002/hep.25895 [DOI] [PubMed] [Google Scholar]
- 274.Zheng J, Yan X, Lu T, Song W, Li Y, Liang J, et al. CircFOXK2 promotes hepatocellular carcinoma progression and leads to a poor clinical prognosis via regulating the Warburg effect. J Exp Clin Cancer Res. 2023;42(1):63. 10.1186/s13046-023-02624-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Shi J, Li Y, Liang S, Zeng J, Liu G, Mu F, et al. Circulating tumour cells as biomarkers for evaluating cryosurgery on unresectable hepatocellular carcinoma. Oncol Rep. 2016;36(4):1845–51. 10.3892/or.2016.5050 [DOI] [PubMed] [Google Scholar]
- 276.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9(4):265–73. 10.1038/nrc2620 [DOI] [PubMed] [Google Scholar]
- 277.Satelli A, Brownlee Z, Mitra A, Meng QH, Li S. Circulating tumor cell enumeration with a combination of epithelial cell adhesion molecule- and cell-surface vimentin-based methods for monitoring breast cancer therapeutic response. Clin Chem. 2015;61(1):259–66. 10.1373/clinchem.2014.228122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Kelley RK, Magbanua MJM, Butler TM, Collisson EA, Hwang J, Sidiropoulos N, et al. Circulating tumor cells in hepatocellular carcinoma: a pilot study of detection, enumeration, and next-generation sequencing in cases and controls. BMC Cancer. 2015;15:206. 10.1186/s12885-015-1195-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Yu JJ, Xiao W, Dong SL, Liang HF, Zhang ZW, Zhang BX, et al. Effect of surgical liver resection on circulating tumor cells in patients with hepatocellular carcinoma. BMC Cancer. 2018;18(1):835. 10.1186/s12885-018-4744-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Shen J, Wang WS, Zhu XL, Ni CF. High epithelial cell adhesion molecule-positive circulating tumor cell count predicts poor survival of patients with unresectable hepatocellular carcinoma treated with transcatheter arterial chemoembolization. J Vasc Interv Radiol. 2018;29(12):1678–84. 10.1016/j.jvir.2018.07.030 [DOI] [PubMed] [Google Scholar]
- 281.Agopian VG, Harlander-Locke M, Zarrinpar A, Kaldas FM, Farmer DG, Yersiz H, et al. A novel prognostic nomogram accurately predicts hepatocellular carcinoma recurrence after liver transplantation: analysis of 865 consecutive liver transplant recipients. J Am Coll Surg. 2015;220(4):416–27. 10.1016/j.jamcollsurg.2014.12.025 [DOI] [PubMed] [Google Scholar]
- 282.Marelli L, Stigliano R, Triantos C, Senzolo M, Cholongitas E, Davies N, et al. Transarterial therapy for hepatocellular carcinoma: which technique is more effective? A systematic review of cohort and randomized studies. Cardiovasc Intervent Radiol. 2007;30:6–25. 10.1007/s00270-006-0062-3 [DOI] [PubMed] [Google Scholar]
- 283.Gillmore R, Stuart S, Kirkwood A, Hameeduddin A, Woodward N, Burroughs AK, et al. EASL and mRECIST responses are independent prognostic factors for survival in hepatocellular cancer patients treated with transarterial embolization. J Hepatol. 2011;55(6):1309–16. 10.1016/j.jhep.2011.03.007 [DOI] [PubMed] [Google Scholar]
- 284.Martin SP, Fako V, Dang H, Dominguez DA, Khatib S, Ma L, et al. PKM2 inhibition may reverse therapeutic resistance to transarterial chemoembolization in hepatocellular carcinoma. J Exp Clin Cancer Res. 2020;39(1):99. 10.1186/s13046-020-01605-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Sefrioui D, Verdier V, Savoye-Collet C, Beaussire L, Ghomadi S, Gangloff A, et al. Circulating DNA changes are predictive of disease progression after transarterial chemoembolization. Int J Cancer. 2022;150(3):532–41. 10.1002/ijc.33829 [DOI] [PubMed] [Google Scholar]
- 286.Cornell L, Munck JM, Alsinet C, Villanueva A, Ogle L, Willoughby CE, et al. DNA-PK—a candidate driver of hepatocarcinogenesis and tissue biomarker that predicts response to treatment and survival. Clin Cancer Res. 2015;21(4):925–33. 10.1158/1078-0432.CCR-14-0842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Niizeki T, Sumie S, Torimura T, Kurogi J, Kuromatsu R, Iwamoto H, et al. Serum vascular endothelial growth factor as a predictor of response and survival in patients with advanced hepatocellular carcinoma undergoing hepatic arterial infusion chemotherapy. J Gastroenterol. 2012;47(6):686–95. 10.1007/s00535-012-0555-6 [DOI] [PubMed] [Google Scholar]
- 288.Lyu N, Wang X, Li JB, Lai JF, Chen QF, Li SL, et al. Arterial chemotherapy of oxaliplatin plus fluorouracil versus sorafenib in advanced hepatocellular carcinoma: a biomolecular exploratory, randomized, phase III trial (FOHAIC-1). J Clin Oncol. 2022;40(5):468–80. 10.1200/JCO.21.01963 [DOI] [PubMed] [Google Scholar]
- 289.Llovet JM, Pinyol R, Yarchoan M, Singal AG, Marron TU, Schwartz M, et al. Adjuvant and neoadjuvant immunotherapies in hepatocellular carcinoma. Nat Rev Clin Oncol. 2024;21(4):294–311. 10.1038/s41571-024-00868-0 [DOI] [PubMed] [Google Scholar]
- 290.Doyle A, Gorgen A, Muaddi H, Aravinthan AD, Issachar A, Mironov O, et al. Outcomes of radiofrequency ablation as first-line therapy for hepatocellular carcinoma less than 3 cm in potentially transplantable patients. J Hepatol. 2019;70(5):866–73. 10.1016/j.jhep.2018.12.027 [DOI] [PubMed] [Google Scholar]
- 291.Shiina S, Tateishi R, Arano T, Uchino K, Enooku K, Nakagawa H, et al. Radiofrequency ablation for hepatocellular carcinoma: 10-year outcome and prognostic factors. Am J Gastroenterol. 2012;107(4):569–77; quiz 78. 10.1038/ajg.2011.425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Pompili M, Saviano A, de Matthaeis N, Cucchetti A, Ardito F, Federico B, et al. Long-term effectiveness of resection and radiofrequency ablation for single hepatocellular carcinoma ≤3 cm. Results of a multicenter Italian survey. J Hepatol. 2013;59(1):89–97. 10.1016/j.jhep.2013.03.009 [DOI] [PubMed] [Google Scholar]
- 293.Canale M, Ulivi P, Foschi FG, Scarpi E, De Matteis S, Donati G, et al. Clinical and circulating biomarkers of survival and recurrence after radiofrequency ablation in patients with hepatocellular carcinoma. Crit Rev Oncol Hematol. 2018;129:44–53. 10.1016/j.critrevonc.2018.06.017 [DOI] [PubMed] [Google Scholar]
- 294.Hameed B, Mehta N, Sapisochin G, Roberts JP, Yao FY. Alpha-fetoprotein level> 1000 ng/mL as an exclusion criterion for liver transplantation in patients with hepatocellular carcinoma meeting the Milan criteria. Liver Transpl. 2014;20(8):945–51. 10.1002/lt.23904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Berry K, Ioannou GN. Serum alpha-fetoprotein level independently predicts posttransplant survival in patients with hepatocellular carcinoma. Liver Transpl. 2013;19(6):634–45. 10.1002/lt.23652 [DOI] [PubMed] [Google Scholar]
- 296.Matsumura M, Shiratori Y, Niwa Y, Tanaka T, Ogura K, Okudaira T, et al. Presence of alpha-fetoprotein mRNA in blood correlates with outcome in patients with hepatocellular carcinoma. J Hepatol. 1999;31(2):332–9. 10.1016/S0168-8278(99)80232-3 [DOI] [PubMed] [Google Scholar]
- 297.Lemoine A, Le Bricon T, Salvucci M, Azoulay D, Pham P, Raccuia J, et al. Prospective evaluation of circulating hepatocytes by alpha-fetoprotein mRNA in humans during liver surgery. Ann Surg. 1997;226(1):43–50. 10.1097/00000658-199707000-00006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Jin J, Niu X, Zou L, Li L, Li S, Han J, et al. AFP mRNA level in enriched circulating tumor cells from hepatocellular carcinoma patient blood samples is a pivotal predictive marker for metastasis. Cancer Lett. 2016;378(1):33–7. 10.1016/j.canlet.2016.04.033 [DOI] [PubMed] [Google Scholar]
- 299.Kong SY, Park JW, Kim JO, Lee NO, Lee JA, Park KW, et al. Alpha-fetoprotein and human telomerase reverse transcriptase mRNA levels in peripheral blood of patients with hepatocellular carcinoma. J Cancer Res Clin Oncol. 2009;135(8):1091–8. 10.1007/s00432-009-0549-9 [DOI] [PubMed] [Google Scholar]
- 300.Mazzaferro V, Regalia E, Doci R, Andreola S, Pulvirenti A, Bozzetti F, et al. Liver transplantation for the treatment of small hepatocellular carcinomas in patients with cirrhosis. N Engl J Med. 1996;334(11):693–700. 10.1056/NEJM199603143341104 [DOI] [PubMed] [Google Scholar]
- 301.Onaca N, Davis GL, Goldstein RM, Jennings LW, Klintmalm GB. Expanded criteria for liver transplantation in patients with hepatocellular carcinoma: a report from the International Registry of Hepatic Tumors in Liver Transplantation. Liver Transpl. 2007;13(3):391–9. 10.1002/lt.21095. 10.1002/lt.21095 [DOI] [PubMed] [Google Scholar]
- 302.Hong G, Suh K-S, Suh S-W, Yoo T, Kim H, Park M-S, et al. Alpha-fetoprotein and 18F-FDG positron emission tomography predict tumor recurrence better than Milan criteria in living donor liver transplantation. J Hepatol. 2016;64(4):852–9. 10.1016/j.jhep.2015.11.033 [DOI] [PubMed] [Google Scholar]
- 303.Kim Y-I, Paeng JC, Cheon GJ, Suh K-S, Lee DS, Chung J-K, et al. Prediction of posttransplantation recurrence of hepatocellular carcinoma using metabolic and volumetric indices of 18F-FDG PET/CT. J Nucl Med. 2016;57(7):1045–51. 10.2967/jnumed.115.170076 [DOI] [PubMed] [Google Scholar]
- 304.Kang YK, Choi JY, Paeng JC, Kim YI, Kwon HW, Cheon GJ, et al. Composite criteria using clinical and FDG PET/CT factors for predicting recurrence of hepatocellular carcinoma after living donor liver transplantation. Eur Radiol. 2019;29(11):6009–17. 10.1007/s00330-019-06239-z [DOI] [PubMed] [Google Scholar]
- 305.Li WX, Li Z, Gao PJ, Gao J, Zhu JY. Histological differentiation predicts post-liver transplantation survival time. Clin Res Hepatol Gastroenterol. 2014;38(2):201–8. 10.1016/j.clinre.2013.11.002 [DOI] [PubMed] [Google Scholar]
- 306.Lu Y, Chan Y-T, Wu J, Feng Z, Yuan H, Li Q, et al. CRISPR/Cas9 screens unravel miR-3689a-3p regulating sorafenib resistance in hepatocellular carcinoma via suppressing CCS/SOD1-dependent mitochondrial oxidative stress. Drug Resist Updates. 2023;71:101015. 10.1016/j.drup.2023.101015 [DOI] [PubMed] [Google Scholar]
- 307.Yu J, Park R, Kim R. Promising novel biomarkers for hepatocellular carcinoma: diagnostic and prognostic insights. J Hepatocell Carcinoma. 2023;10:1105–27. 10.2147/JHC.S341195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Shuen TWH, Alunni-Fabbroni M, Öcal E, Malfertheiner P, Wildgruber M, Schinner R, et al. Extracellular vesicles may predict response to radioembolization and sorafenib treatment in advanced hepatocellular carcinoma: an exploratory analysis from the SORAMIC trial. Clin Cancer Res. 2022;28(17):3890–901. 10.1158/1078-0432.CCR-22-0569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Goyal L, Zheng H, Abrams TA, Miksad R, Bullock AJ, Allen JN, et al. A phase II and biomarker study of sorafenib combined with modified FOLFOX in patients with advanced hepatocellular carcinoma. Clin Cancer Res. 2019;25(1):80–9. 10.1158/1078-0432.CCR-18-0847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.El Shorbagy S, abuTaleb F, Labib HA, Ebian H, Harb OA, Mohammed MS, et al. Prognostic significance of VEGF and HIF-1 α in hepatocellular carcinoma patients receiving sorafenib versus metformin sorafenib combination. J Gastrointest Cancer. 2021;52(1):269–79. 10.1007/s12029-020-00389-w [DOI] [PubMed] [Google Scholar]
- 311.Chan YT, Wu J, Lu Y, Li Q, Feng Z, Xu L, et al. Loss of lncRNA LINC01056 leads to sorafenib resistance in HCC. Mol Cancer. 2024;23(1):74. 10.1186/s12943-024-01988-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Lee SH, Yim SY, Jeong YS, Li QX, Kang SH, Sohn BH, et al. Consensus subtypes of hepatocellular carcinoma associated with clinical outcomes and genomic phenotypes. Hepatology. 2022;76(6):1634–48. 10.1002/hep.32490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Fernández-Tussy P, Rodríguez-Agudo R, Fernández-Ramos D, Barbier-Torres L, Zubiete-Franco I, Davalillo SL, et al. Anti-miR-518d-5p overcomes liver tumor cell death resistance through mitochondrial activity. Cell Death Dis. 2021;12(6):555. 10.1038/s41419-021-03827-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Li J, Shi L, Zhang X, Sun B, Yang Y, Ge N, et al. pERK/pAkt phenotyping in circulating tumor cells as a biomarker for sorafenib efficacy in patients with advanced hepatocellular carcinoma. Oncotarget. 2016;7(3):2646–59. 10.18632/oncotarget.6104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Fu Y, Yang Z, Hu Z, Yang Z, Pan Y, Chen J, et al. Preoperative serum ctDNA predicts early hepatocellular carcinoma recurrence and response to systemic therapies. Hepatol Int. 2022;16(4):868–78. 10.1007/s12072-022-10348-1 [DOI] [PubMed] [Google Scholar]
- 316.Myojin Y, Kodama T, Maesaka K, Motooka D, Sato Y, Tanaka S, et al. ST6GAL1 is a novel serum biomarker for lenvatinib-susceptible FGF19-driven hepatocellular carcinoma. Clin Cancer Res. 2021;27(4):1150–61. 10.1158/1078-0432.CCR-20-3382 [DOI] [PubMed] [Google Scholar]
- 317.Zhang P, Sun H, Wen P, Wang Y, Cui Y, Wu J. circRNA circMED27 acts as a prognostic factor and mediator to promote lenvatinib resistance of hepatocellular carcinoma. Mol Ther Nucleic Acids. 2022;27:293–303. 10.1016/j.omtn.2021.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Wang Y, Deng B. Hepatocellular carcinoma: molecular mechanism, targeted therapy, and biomarkers. Cancer Metastasis Rev. 2023;42(3):629–52. 10.1007/s10555-023-10084-4 [DOI] [PubMed] [Google Scholar]
- 319.Yan T, Yu L, Zhang N, Peng C, Su G, Jing Y, et al. The advanced development of molecular targeted therapy for hepatocellular carcinoma. Cancer Biol Med. 2022;19(6):802–17. 10.20892/j.issn.2095-3941.2021.0661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Scheiner B, Pomej K, Kirstein MM, Hucke F, Finkelmeier F, Waidmann O, et al. Prognosis of patients with hepatocellular carcinoma treated with immunotherapy - development and validation of the CRAFITY score. J Hepatol. 2022;76(2):353–63. 10.1016/j.jhep.2021.09.035 [DOI] [PubMed] [Google Scholar]
- 321.Yang Z, Fu Y, Wang Q, Pan Y, Wang J, Chen J, et al. Dynamic changes of serum α-fetoprotein predict the prognosis of bevacizumab plus immunotherapy in hepatocellular carcinoma. Int J Surg. 2024. 10.1097/JS9.0000000000001860. [DOI] [PubMed]
- 322.Weng J, Wang Z, Hu Z, Xu W, Sun JL, Wang F, et al. Repolarization of immunosuppressive macrophages by targeting SLAMF7-regulated CCL2 signaling sensitizes hepatocellular carcinoma to immunotherapy. Cancer Res. 2024;84(11):1817–33. 10.1158/0008-5472.CAN-23-3106 [DOI] [PubMed] [Google Scholar]
- 323.Winograd P, Hou S, Sadeghi S, Finn R, DiPardo B, Li Q, et al. Evaluation of hepatocellular carcinoma circulating tumor cells expressing programmed death-ligand 1. HPB. 2018;20:S2–3. 10.1016/j.hpb.2018.02.004 [DOI] [Google Scholar]
- 324.Hong JY, Cho HJ, Sa JK, Liu X, Ha SY, Lee T, et al. Hepatocellular carcinoma patients with high circulating cytotoxic T cells and intra-tumoral immune signature benefit from pembrolizumab: results from a single-arm phase 2 trial. Genome Med. 2022;14(1):1. 10.1186/s13073-021-00995-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Li Y, Jin H, Li Q, Shi L, Mao Y, Zhao L. The role of RNA methylation in tumor immunity and its potential in immunotherapy. Mol Cancer. 2024;23(1):130. 10.1186/s12943-024-02041-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Huang XY, Zhang PF, Wei CY, Peng R, Lu JC, Gao C, et al. Circular RNA circMET drives immunosuppression and anti-PD1 therapy resistance in hepatocellular carcinoma via the miR-30-5p/snail/DPP4 axis. Mol Cancer. 2020;19(1):92. 10.1186/s12943-020-01213-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Zhu AX, Abbas AR, de Galarreta MR, Guan Y, Lu S, Koeppen H, et al. Molecular correlates of clinical response and resistance to atezolizumab in combination with bevacizumab in advanced hepatocellular carcinoma. Nat Med. 2022;28(8):1599–611. 10.1038/s41591-022-01868-2 [DOI] [PubMed] [Google Scholar]
- 328.Campani C, Bamba-Funck J, Campion B, Sidali S, Blaise L, Ganne-Carrié N, et al. Baseline ALBI score and early variation of serum AFP predicts outcomes in patients with HCC treated by atezolizumab-bevacizumab. Liver Int. 2023;43(3):708–17. 10.1111/liv.15487 [DOI] [PubMed] [Google Scholar]
- 329.Shi J, Liu J, Tu X, Li B, Tong Z, Wang T, et al. Single-cell immune signature for detecting early-stage HCC and early assessing anti-PD-1 immunotherapy efficacy. J Immunother Cancer. 2022;10(1):e003133. 10.1136/jitc-2021-003133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Ruiz M, Bresnahan E, Molina-Sánchez P, Lindblad KE, Maier B, Sia D, et al. β-catenin activation promotes immune escape and resistance to anti-pd-1 therapy in hepatocellular carcinoma. Cancer Discov. 2019;9(8):1124–41. 10.1158/2159-8290.CD-19-0074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Ye Z, Wang Y, Yuan R, Ding R, Hou Y, Qian L, et al. Vesicle-mediated transport-related genes predict the prognosis and immune microenvironment in hepatocellular carcinoma. J Cancer. 2024;15(12):3645–62. 10.7150/jca.94902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.He H, Chen S, Fan Z, Dong Y, Wang Y, Li S, et al. Multi-dimensional single-cell characterization revealed suppressive immune microenvironment in AFP-positive hepatocellular carcinoma. Cell Discov. 2023;9(1):60. 10.1038/s41421-023-00563-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Sepich-Poore GD, Zitvogel L, Straussman R, Hasty J, Wargo JA, Knight R. The microbiome and human cancer. Science. 2021;371(6536):eabc4552. 10.1126/science.abc4552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.McQuade JL, Daniel CR, Helmink BA, Wargo JA. Modulating the microbiome to improve therapeutic response in cancer. Lancet Oncol. 2019;20(2):e77–91. 10.1016/S1470-2045(18)30952-5 [DOI] [PubMed] [Google Scholar]
- 335.Schwabe RF, Greten TF. Gut microbiome in HCC - mechanisms, diagnosis and therapy. J Hepatol. 2020;72(2):230–8. 10.1016/j.jhep.2019.08.016 [DOI] [PubMed] [Google Scholar]
- 336.Silveira MAD, Bilodeau S, Greten TF, Wang XW, Trinchieri G. The gut-liver axis: host microbiota interactions shape hepatocarcinogenesis. Trends Cancer. 2022;8(7):583–97. 10.1016/j.trecan.2022.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Kang Y, Cai Y, Yang Y. The gut microbiome and hepatocellular carcinoma: implications for early diagnostic biomarkers and novel therapies. Liver Cancer. 2022;11(2):113–25. 10.1159/000521358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Zheng Y, Wang T, Tu X, Huang Y, Zhang H, Tan D, et al. Gut microbiome affects the response to anti-PD-1 immunotherapy in patients with hepatocellular carcinoma. J Immunother Cancer. 2019;7(1):193. 10.1186/s40425-019-0650-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Zhang L, Chen C, Chai D, Li C, Guan Y, Liu L, et al. The association between antibiotic use and outcomes of HCC patients treated with immune checkpoint inhibitors. Front Immunol. 2022;13:956533. 10.3389/fimmu.2022.956533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Wu H, Zheng X, Pan T, Yang X, Chen X, Zhang B, et al. Dynamic microbiome and metabolome analyses reveal the interaction between gut microbiota and anti-PD-1 based immunotherapy in hepatocellular carcinoma. Int J Cancer. 2022;151(8):1321–34. 10.1002/ijc.34118 [DOI] [PubMed] [Google Scholar]
- 341.Fulgenzi CAM, et al. Effect of early antibiotic exposure on survival of patients receiving atezolizumab plus bevacizumab but not sorafenib for unresectable HCC: A sub-analysis of the phase III IMbrave150 study. JCO. 2023;41:597–597. 10.1200/JCO.2023.41.4_suppl.597. 10.1200/JCO.2023.41.4_suppl.597 [DOI] [Google Scholar]
- 342.Lee PC, Wu CJ, Hung YW, Lee CJ, Chi CT, Lee IC, et al. Gut microbiota and metabolites associate with outcomes of immune checkpoint inhibitor-treated unresectable hepatocellular carcinoma. J Immunother Cancer. 2022;10(6):e004779. 10.1136/jitc-2022-004779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Nault JC, Villanueva A. Biomarkers for hepatobiliary cancers. Hepatology. 2021;73(Suppl 1):115–27. 10.1002/hep.31175 [DOI] [PubMed] [Google Scholar]
- 344.Wang B, Qi FZ, Chen P, Qian LM, Su HS, Wang Y, et al. Hypoxia-activated selectivity-improved anti-PKM2 antibody combined with prodrug TH-302 for potentiated targeting therapy in hepatocellular carcinoma. Int J Biol Sci. 2024;20(5):1634–51. 10.7150/ijbs.92211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Qi S, Su L, Li J, Zhao P, Zhang Q, Niu X, et al. YIPF2 is a novel Rab-GDF that enhances HCC malignant phenotypes by facilitating CD147 endocytic recycle. Cell Death Dis. 2019;10(6):462. 10.1038/s41419-019-1709-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Zhao P, Zhang W, Wang SJ, Yu XL, Tang J, Huang W, et al. HAb18G/CD147 promotes cell motility by regulating annexin II-activated RhoA and Rac1 signaling pathways in hepatocellular carcinoma cells. Hepatology. 2011;54(6):2012–24. 10.1002/hep.24592 [DOI] [PubMed] [Google Scholar]
- 347.Qi S, Su L, Li J, Zhang C, Ma Z, Liu G, et al. Arf6-driven endocytic recycling of CD147 determines HCC malignant phenotypes. J Exp Clin Cancer Res. 2019;38(1):471. 10.1186/s13046-019-1464-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Qi FZ, Su HS, Wang B, Qian LM, Wang Y, Wang CH, et al. Hypoxia-activated ADCC-enhanced humanized anti-CD147 antibody for liver cancer imaging and targeted therapy with improved selectivity. MedComm (2020). 2024;5(3):e512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Zhu AX, Gold PJ, El-Khoueiry AB, Abrams TA, Morikawa H, Ohishi N, et al. First-in-man phase I study of GC33, a novel recombinant humanized antibody against glypican-3, in patients with advanced hepatocellular carcinoma. Clin Cancer Res. 2013;19(4):920–8. 10.1158/1078-0432.CCR-12-2616 [DOI] [PubMed] [Google Scholar]
- 350.Abou-Alfa GK, Yen CJ, Hsu CH, O’Donoghue J, Beylergil V, Ruan S, et al. Phase Ib study of codrituzumab in combination with sorafenib in patients with non-curable advanced hepatocellular carcinoma (HCC). Cancer Chemother Pharmacol. 2017;79(2):421–9. 10.1007/s00280-017-3241-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Abou-Alfa GK, Puig O, Daniele B, Kudo M, Merle P, Park JW, et al. Randomized phase II placebo controlled study of codrituzumab in previously treated patients with advanced hepatocellular carcinoma. J Hepatol. 2016;65(2):289–95. 10.1016/j.jhep.2016.04.004 [DOI] [PubMed] [Google Scholar]
- 352.Jin ZC, Chen JJ, Zhu XL, Duan XH, Xin YJ, Zhong BY, et al. Immune checkpoint inhibitors and anti-vascular endothelial growth factor antibody/tyrosine kinase inhibitors with or without transarterial chemoembolization as first-line treatment for advanced hepatocellular carcinoma (CHANCE2201): a target trial emulation study. EClinicalMedicine. 2024;72:102622. 10.1016/j.eclinm.2024.102622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Bai L, Sun M, Xu A, Bai Y, Wu J, Shao G, et al. Phase 2 study of AK104 (PD-1/CTLA-4 bispecific antibody) plus lenvatinib as first-line treatment of unresectable hepatocellular carcinoma. J Clin Oncol. 2021;39(15_suppl):4101. 10.1200/JCO.2021.39.15_suppl.4101 [DOI] [Google Scholar]
- 354.Hussein MS, Li Q, Mao R, Peng Y, He Y. TCR T cells overexpressing c-Jun have better functionality with improved tumor infiltration and persistence in hepatocellular carcinoma. Front Immunol. 2023;14:1114770. 10.3389/fimmu.2023.1114770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.A phase 1, single-arm, open-label, dose-escalation study of AFP specific T cell receptor transduced T cells injection (HRYZ-T102) in patients with AFP positive advanced hepatocellular carcinoma and other solid tumors. 2024. Available from: https://clinicaltrials.gov/study/NCT06515314.
- 356.Sun L, Guo H, Jiang R, Lu L, Liu T, He X. Engineered cytotoxic T lymphocytes with AFP-specific TCR gene for adoptive immunotherapy in hepatocellular carcinoma. Tumour Biol. 2016;37(1):799–806. 10.1007/s13277-015-3845-9 [DOI] [PubMed] [Google Scholar]
- 357.Xu J, Shen J, Gu S, Zhang Y, Wu L, Wu J, et al. Camrelizumab in combination with apatinib in patients with advanced hepatocellular carcinoma (RESCUE): a nonrandomized, open-label, phase II trial. Clin Cancer Res. 2021;27(4):1003–11. 10.1158/1078-0432.CCR-20-2571 [DOI] [PubMed] [Google Scholar]
- 358.Hashimoto A, Sarker D, Reebye V, Jarvis S, Sodergren MH, Kossenkov A, et al. Upregulation of C/EBPα inhibits suppressive activity of myeloid cells and potentiates antitumor response in mice and patients with cancer. Clin Cancer Res. 2021;27(21):5961–78. 10.1158/1078-0432.CCR-21-0986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Chan SL, Schuler M, Kang YK, Yen CJ, Edeline J, Choo SP, et al. A first-in-human phase 1/2 study of FGF401 and combination of FGF401 with spartalizumab in patients with hepatocellular carcinoma or biomarker-selected solid tumors. J Exp Clin Cancer Res. 2022;41(1):189. 10.1186/s13046-022-02383-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Chen WT, Lin SM, Lee WC, Wu TJ, Lin CC, Shen CH, et al. GALNT14 genotype-guided chemoembolization plus sorafenib therapy in hepatocellular carcinoma: a randomized trial. Hepatol Int. 2022;16(1):148–58. 10.1007/s12072-021-10283-7 [DOI] [PubMed] [Google Scholar]
- 361.Zhou M, Zhu S, Xu C, Liu B, Shen J. A phase Ib/II study of BLU-554, a fibroblast growth factor receptor 4 inhibitor in combination with CS1001, an anti-PD-L1, in patients with locally advanced or metastatic hepatocellular carcinoma. Invest New Drugs. 2023;41(1):162–7. 10.1007/s10637-023-01335-w [DOI] [PubMed] [Google Scholar]
- 362.Woei AJF, Weijl NI, Burgmans MC, Fariña Sarasqueta A, van Wezel JT, Wasser M, et al. Neoadjuvant treatment with angiogenesis-inhibitor dovitinib prior to local therapy in hepatocellular carcinoma: a phase II study. Oncologist. 2021;26(10):854–64. 10.1002/onco.13901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Fountzilas C, Gupta M, Lee S, Krishnamurthi S, Estfan B, Wang K, et al. A multicentre phase 1b/2 study of tivozanib in patients with advanced inoperable hepatocellular carcinoma. Br J Cancer. 2020;122(7):963–70. 10.1038/s41416-020-0737-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Giannelli G, Santoro A, Kelley RK, Gane E, Paradis V, Cleverly A, et al. Biomarkers and overall survival in patients with advanced hepatocellular carcinoma treated with TGF-βRI inhibitor galunisertib. PLoS One. 2020;15(3):e0222259. 10.1371/journal.pone.0222259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Kelley RK, Ryoo BY, Merle P, Park JW, Bolondi L, Chan SL, et al. Second-line cabozantinib after sorafenib treatment for advanced hepatocellular carcinoma: a subgroup analysis of the phase 3 CELESTIAL trial. ESMO Open. 2020;5(4):e000714. 10.1136/esmoopen-2020-000714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Venturini N, Marron T, Casanova-Acebes M, Mandeli J, Doroshow D, Lucas N, et al. 629 neoadjuvant nivolumab combined with CCR2/5 inhibitor or anti-IL-8 antibody in non-small cell lung cancer and hepatocellular carcinoma. J Immunother Cancer. 2022;10(Suppl 2):A661. [Google Scholar]
- 367.Cai M, Hong X, Guo Y, Shi W, Huang W, Liang L, et al. 173P A phase II trial of donafenib plus sintilimab for advanced stage hepatocellular carcinoma. Ann Oncol. 2024;35:S79. 10.1016/j.annonc.2024.05.182 [DOI] [Google Scholar]
- 368.Cheon J, Jung S, Kang B, Kim H, Kim C, Chon H. Organ-specific responses to atezolizumab plus bevacizumab in patients with advanced hepatocellular carcinoma. J Clin Oncol. 2022;40(16_suppl):4076. 10.1200/JCO.2022.40.16_suppl.4076 [DOI] [Google Scholar]
- 369.Vienot A, Jacquin M, Rebucci-Peixoto M, Pureur D, Ghiringhelli F, Assenat E, et al. Evaluation of the interest to combine a CD4 Th1-inducer cancer vaccine derived from telomerase and atezolizumab plus bevacizumab in unresectable hepatocellular carcinoma: a randomized non-comparative phase II study (TERTIO - PRODIGE 82). BMC Cancer. 2023;23(1):710. 10.1186/s12885-023-11065-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.A phase Ib/II, two-part, non-randomized, open-label study to evaluate the safety, tolerability, efficacy, and pharmacokinetics of chidamide in combination with regorafenib in patients with hepatocellular carcinoma. 2023. Available from: https://clinicaltrials.gov/study/NCT05770882.
- 371.Cabozantinib treatment in a phase II study for patients with hepatocellular carcinoma (HCC) refractory to PD-1 inhibitors. 2021. Available from: https://clinicaltrials.gov/study/NCT04767906.
- 372.VETC-based precision adjuvant therapy for postoperative hepatocellular carcinoma: a prospective multicenter cohort study. 2024. Available from: https://clinicaltrials.gov/study/NCT06311929.
- 373.Phase I/II study of PTX-9908 injection as an inhibitor of cancer progression in patients with non-resectable hepatocellular carcinoma following transarterial chemoembolization treatment. 2019. Available from: https://clinicaltrials.gov/study/NCT03812874.
- 374.A phase 2, randomized study to evaluate the optimized dose, safety, and efficacy of livmoniplimab in combination with budigalimab for locally advanced or metastatic hepatocellular carcinoma (HCC) patients who have progressed after an immune checkpoint inhibitor containing regimen in first-line HCC. 2023. Available from: https://clinicaltrials.gov/study/NCT05822752.
- 375.Efficacy and safety of serplulimab combined with bevacizumab biosimilar and HAIC in advanced hepatocellular carcinoma (HCC) patients. 2024. Available from: https://clinicaltrials.gov/study/NCT06370065.
- 376.Efficacy and safety of transarterial chemoembolization in combination with PD-1/PD-L1 inhibitors and molecular target therapies (VEGF-TKI/Bevacizumab) for intermediate stage HCC. 2022. Available from: https://clinicaltrials.gov/study/NCT05332496.
- 377.Epigenetic therapeutics to overcome resistance against immune checkpoint inhibitors in hepatocellular carcinoma: a proof-of-concept clinical trial. 2023. Available from: https://clinicaltrials.gov/study/NCT05873244.
- 378.An open-label, multi-center, dose-escalation phase 1 study to evaluate the safety, tolerability, and pharmacokinetics of ETN101 in patients with advanced hepatocellular carcinoma. 2024. Available from: https://clinicaltrials.gov/study/NCT06326502.
- 379.A randomized, double-blind, placebo-controlled, multicenter phase 3 study of anlotinib hydrochloride capsules combined with penpulimab injection in patients with high risk of relapse after radical surgery or ablation of hepatocellular carcinoma (HCC). 2023. Available from: https://clinicaltrials.gov/study/NCT05862337.
- 380.A single-arm, non-randomized, single-center study to evaluate lenvatinib in combination with camrelizumab as first-line therapy in patients with advanced hepatocellular carcinoma. 2020. Available from: https://clinicaltrials.gov/study/NCT04443309.
- 381.Ablation in combination with lenvatinib and anti-PD-1 antibodies in patients with early recurrence after radical resection/ablation of HCC: a prospective, randomized, controlled clinical study. 2023. Available from: https://clinicaltrials.gov/study/NCT05803928.
- 382.A phase II study to evaluate the efficacy and safety of cryoablation combined with tislelizumab plus lenvatinib as first-line treatment in patients with advanced hepatocellular carcinoma. 2023. Available from: https://clinicaltrials.gov/study/NCT05897268.
- 383.Transarterial chemoembolization combined with lenvatinib and Iodion-125 seeds brachytherapy for hepatocellular carcinoma with portal vein branch tumor thrombus: a single center, prospective, randomized control trail. 2021. Available from: https://clinicaltrials.gov/study/NCT04967495.
- 384.A phase I study of SBRT vaccination with atezolizumab and bevacizumab for patients with advanced hepatocellular carcinoma. 2022. Available from: https://clinicaltrials.gov/study/NCT05488522.
- 385.A prospective, single-arm study of downstaging protocol containing immunotherapy for HCC beyond the Milan criteria before liver transplantation. 2022. Available from: https://clinicaltrials.gov/study/NCT05475613.
- 386.A open-label, multi-center, single arm study to evaluate the efficacy and safety of tislelizumab combined with sitravatinib as adjuvant therapy in participants with hepatocellular carcinoma who are at high risk of recurrence after curative hepatic resection. 2022. Available from: https://clinicaltrials.gov/study/NCT05407519.
- 387.A phase Ib clinical study to evaluate the efficacy and safety of TQB2618 injection combined with penpulimab injection and anlotinib hydrochloride capsules as first-line treatment for advanced hepatocellular carcinoma. 2023. Available from: https://clinicaltrials.gov/study/NCT05975645.
- 388.Regorafenib plus tislelizumab as first-line systemic therapy for patients with advanced hepatocellular carcinoma (HCC). 2019. Available from: https://clinicaltrials.gov/study/NCT04183088.
- 389.Efficacy and safety of stereotactic body radiotherapy followed by adebrelimab and lenvatinib for hepatocellular carcinoma with abdominal lymph node metastases: a two-arm, phase II trial. 2024. Available from: https://clinicaltrials.gov/study/NCT06261125.
- 390.Safety and efficacy of lenvatinib combined with VIC-1911 in the treatment of lenvatinib-unresponsive or lenvatinib-resistant hepatocellular carcinoma. 2023. Available from: https://clinicaltrials.gov/study/NCT05718882.
- 391.Safety and efficacy study of pembrolizumab in combination with lenvatinib in participants with hepatocellular carcinoma (HCC) before liver transplant as neoadjuvant therapY--PLENTY randomized clinical trial. 2020. Available from: https://clinicaltrials.gov/study/NCT04425226. [DOI] [PMC free article] [PubMed]
- 392.An open label, randomized, controlled, clinical trial of adoptive autologous invariant natural killer T cells for the treatment of progressed hepatocellular carcinoma continuing on PD-1 inhibitor therapy. 2023. Available from: https://clinicaltrials.gov/study/NCT05962450.
- 393.A phase 1b dose-escalation and cohort-expansion study of the safety/tolerability, and efficacy of oncolytic virotherapy plus PD-1 inhibitor and lenvatinib for patients with advanced hepatocellular carcinoma. 2022. Available from: https://clinicaltrials.gov/study/NCT05675462.
- 394.A phase 2 study on immune checkpoint inhibitors and radioembolisation for previously untreated metastatic hepatocellular carcinoma. 2023. Available from: https://clinicaltrials.gov/study/NCT05809869.
- 395.Observation study of sequential regorafenib combined with immunocheckpoint inhibitors after hepatic artery infusion chemotherapy for advanced hepatocellular carcinoma. 2022. Available from: https://clinicaltrials.gov/study/NCT05573282.
- 396.Regorafenib combined with PD-1 inhibitor therapy for second-line treatment of hepatocellular carcinoma: a single arm, nonrandomized, single center clinical study. 2021. Available from: https://clinicaltrials.gov/study/NCT05048017.
- 397.Non-invasive CT-based hepatic venous pressure gradient assessment for predicting the prognosis of hepatocellular carcinoma with transarterial chemoembolization (CHANCE-CHESS 2302): a multicenter retrospective study. 2023. Available from: https://clinicaltrials.gov/study/NCT05704192.
- 398.A multi-centre single-arm phase II study on durvalumab (MEDI 4736) with stereotactic body radiation therapy (SBRT) in patients with inoperable/unresectable hepatocellular carcinoma. 2021. Available from: https://clinicaltrials.gov/study/NCT04913480.
- 399.A phase II, open-label, multi-cohort, multicenter study in patients with unresectable hepatocellular carcinoma and Child-Pugh B7 and B8 cirrhosis. 2023. Available from: https://clinicaltrials.gov/study/NCT06096779.
- 400.Intra-tumor delivery of double checkpoint inhibitors, chemodrug, and/or bevacizumab therapy as first line for hepatocellular carcinoma. 2024. Available from: https://clinicaltrials.gov/study/NCT06482801.
- 401.A single-arm, prospective, open clinical study of palbociclib for backline treatment of advanced hepatocellular carcinoma. 2024. Available from: https://clinicaltrials.gov/study/NCT06478927.
- 402.Phase 1/2 open-label study to evaluate the safety, tolerability, pharmacokinetics, pharmacodynamics, and preliminary antitumor activity of OTX-2002 as a single agent and in combination with standard of care in patients with hepatocellular carcinoma and other solid tumor types known for association with the MYC oncogene. 2022. Available from: https://clinicaltrials.gov/study/NCT05497453.
- 403.A phase II randomized study of atezolizumab plus multi-kinase inhibitor versus multi-kinase inhibitor alone in subjects with unresectable, advanced hepatocellular carcinoma who previously received atezolizumab plus bevacizumab. 2021. Available from: https://clinicaltrials.gov/study/NCT05168163.
- 404.A phase 1/2 exploratory study of the TBL1 inhibitor, tegavivint (BC2059), in patients with advanced hepatocellular carcinoma. 2023. Available from: https://clinicaltrials.gov/study/NCT05797805.
- 405.Lenvatinib combined toripalimab in advanced hepatocellular carcinoma: a single-center, single-arm, non-randomized clinical study. 2020. Available from: https://clinicaltrials.gov/study/NCT04368078.
- 406.Adjuvant treatment of patients with high risk of recurrent hepatocellular carcinoma after radical surgery with donafenib in combination with envafolimab: a prospective, multicentre, single-arm phase II clinical study. 2024. Available from: https://clinicaltrials.gov/study/NCT06498622.
- 407.A phase II study of transcatheter arterial chemoembolization (TACE) combined with anti-PD-1 antibody (IBI308) in patients with advanced hepatocellular carcinoma. 2020. Available from: https://clinicaltrials.gov/study/NCT04297280.
- 408.Phase II study evaluating the efficacy of tremelimumab (T) plus durvalumab (D) with lenvatinib combined with concurrent hepatic arterial infusion chemotherapy (HAIC) in patients (Pts) with unresectable hepatocellular carcinoma (uHCC). 2024. Available from: https://clinicaltrials.gov/study/NCT06364007.
- 409.Hepatic artery infusion chemotherapy (HAIC) combined with camrelizumab and tyrosine kinase inhibitor for unresectable hepatocellular carcinoma after transcatheter arterial chemoembolization (TACE) failure: a single-arm and open-label prospective study. 2021. Available from: https://clinicaltrials.gov/study/NCT05135364.
- 410.Combined hepatic arterial infusion chemotherapy, tyrosine kinase inhibitor/ anti-VEGF antibody, and anti-PD-1/ PD-L1 antibody as conversion therapy for unresectable hepatocellular carcinoma. 2023. Available from: https://clinicaltrials.gov/study/NCT05713994. [DOI] [PMC free article] [PubMed]
- 411.Phase II study of FGFR inhibitor futibatinib in combination with anti-PD-1 antibody pembrolizumab in patients with advanced or metastatic hepatocellular carcinoma with FGF19 expression after first line therapy. 2021. Available from: https://clinicaltrials.gov/study/NCT04828486.
- 412.Lenvatinib in recurrent hepatocellular carcinoma after liver transplantation. 2021. Available from: https://clinicaltrials.gov/study/NCT05103904.
- 413.An observational study in patients with unresectable hepatocellular carcinoma (uHCC) following treatment with atezolizumab plus bevacizumab (AB) or with another approved immuno-oncology immune checkpoint inhibitor combination in first-line. 2023. Available from: https://clinicaltrials.gov/study/NCT06117891.
- 414.Combined transarterial chemoembolization, tyrosine kinase inhibitor/ anti-VEGF antibody, and anti-PD-1/ PD-L1 antibody as conversion therapy for advanced hepatocellular carcinoma: a multicenters, real-world, ambispective cohort study. 2023. Available from: https://clinicaltrials.gov/study/NCT05717738.
- 415.A study to evaluate the effects of radiotherapy combined with tyrosine kinase inhibitor (TKI) and anti-PD-1 antibody for stage IIIA hepatocellular carcinoma with portal vein tumor thrombus (PVTT). 2023. Available from: https://clinicaltrials.gov/study/NCT06061445.
- 416.A pilot study of a DNAJB1-PRKACA fusion kinase peptide vaccine combined with nivolumab and ipilimumab for patients with fibrolamellar hepatocellular carcinoma. 2020. Available from: https://clinicaltrials.gov/study/NCT04248569.
- 417.Efficacy of HAIC combined with lenvatinib and PD-1 inhibitor in infiltrative hepatocellular carcinoma: an observational, real-world study. 2024. Available from: https://clinicaltrials.gov/study/NCT06333561.
- 418.Safety and efficacy study of durvalumab in combination with lenvatinib in participants with locally advanced and metastatic hepatocellular carcinoma-- DULECT2020-1 trial. 2020. Available from: https://clinicaltrials.gov/study/NCT04443322.
- 419.Trametinib combined with everolimus and lenvatinib in the treatment of recurrent/refractory advanced solid tumors: a phase II clinical trial. 2021. Available from: https://clinicaltrials.gov/study/NCT04803318.
- 420.Efficacy of locoregional therapy combined with bevacizumab and PD1/L1 inhibitor in advanced hepatocellular carcinoma: a multicenter, observational, real-world study. 2024. Available from: https://clinicaltrials.gov/study/NCT06323382.
- 421.Phase II trial of palliative hypofractionated radiotherapy followed by durvalumab (MEDI4736) with/without tremelimumab for advanced hepatocellular carcinoma after progression on prior PD-1 inhibition. 2020. Available from: https://clinicaltrials.gov/study/NCT04430452.
- 422.Intra-tumor injection of drug-eluting microspheres loading with chemodrug plus checkpoint inhibitors and IL2 for treatment of advanced solid tumors. 2020. Available from: https://clinicaltrials.gov/study/NCT04770207.
- 423.A multicentric national phase II trial assessing TIslelizumab in monotherapy for patients with hepatocellular carcinoma Child-Pugh B and ALBI grade 1 or 2 liver function score. 2022. Available from: https://clinicaltrials.gov/study/NCT05622071.
- 424.A phase 1B/2, open-label study of Q702 in combination with intravenous pembrolizumab in patients with selected advanced solid tumors. 2022. Available from: https://clinicaltrials.gov/study/NCT05438420.
- 425.A phase II trial of cabozantinib in the treatment of recurrent hepatocellular carcinoma post liver transplant. 2019. Available from: https://clinicaltrials.gov/study/NCT04204850.
- 426.A single-arm, multicenter phase Ib clinical trial of the efficacy and safety of TQB2450 injection combined with anlotinib hydrochloride capsule neoadjuvant in the treatment of resectable hepatocellular carcinoma with a high risk of recurrence or metastasis. 2021. Available from: https://clinicaltrials.gov/study/NCT04888546.
- 427.Zeng Q, Klein C, Caruso S, Maille P, Laleh NG, Sommacale D, et al. Artificial intelligence predicts immune and inflammatory gene signatures directly from hepatocellular carcinoma histology. J Hepatol. 2022;77(1):116–27. 10.1016/j.jhep.2022.01.018 [DOI] [PubMed] [Google Scholar]
- 428.Calderaro J, Seraphin TP, Luedde T, Simon TG. Artificial intelligence for the prevention and clinical management of hepatocellular carcinoma. J Hepatol. 2022;76(6):1348–61. 10.1016/j.jhep.2022.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This published article and its supplementary information files include all data generated or analyzed during this study.
No datasets were generated or analysed during the current study.