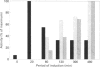Abstract
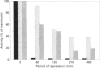
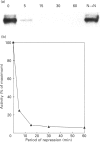
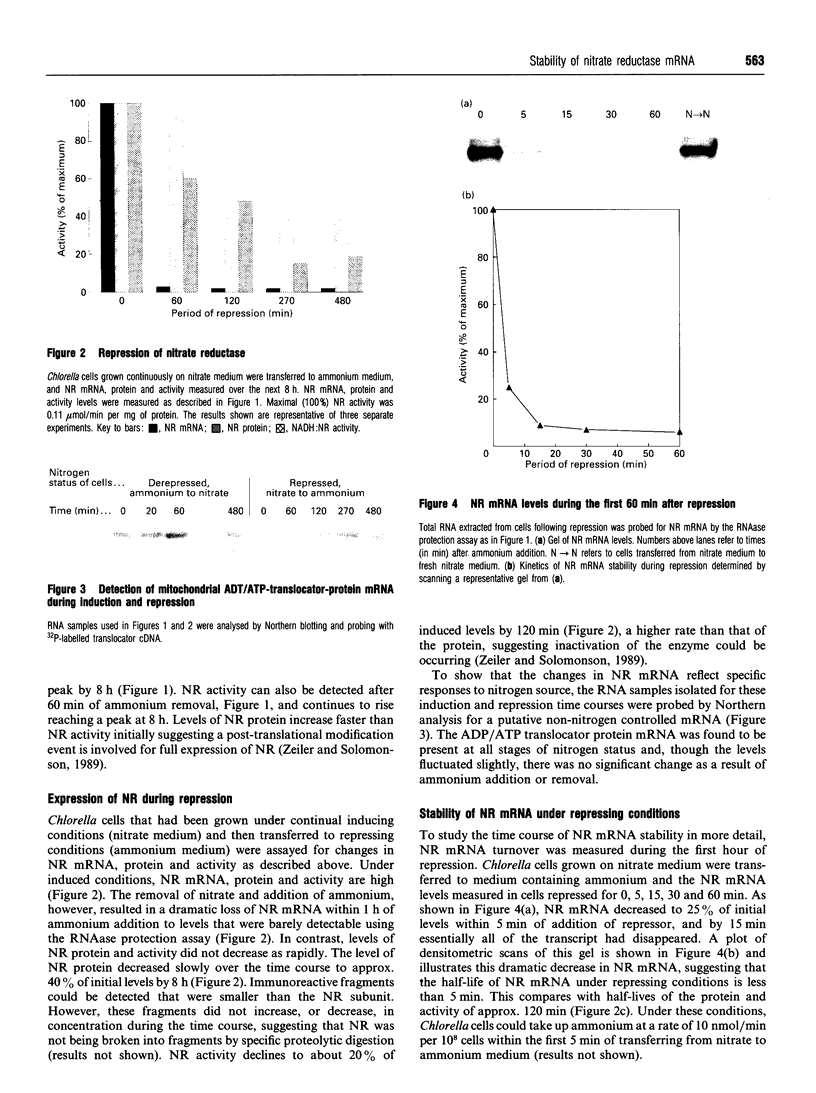
Ammonium, or a metabolite of ammonium, represses the expression of nitrate reductase (NR) in Chlorella vulgaris. The removal of ammonium and addition of nitrate (induction) resulted in a rapid (20 min) peaked synthesis of NR mRNA. Nitrate reductase protein and activity increased at a much lower rate, reaching their maxima by 8 h. Ammonium added to nitrate-grown cells resulted in a dramatic decrease in NR mRNA from a steady-state level to undetectable levels within 15 min of ammonium addition. Nitrate reductase activity and protein levels decreased to 20% and 40% of initial levels respectively over 8 h. The half-life for NR mRNA under these conditions was estimated to be less than 5 min, compared with 120 min for NR protein. Such rapid decreases in NR mRNA suggested a degradation and/or cessation in NR mRNA transcription. No apparent difference in NR mRNA-specific RNAase activity of crude cell extracts (NR-induced or repressed) was observed. However, a significant difference in the susceptibility to degradation of NR mRNA from long-term nitrate-grown cells compared with the NR mRNA isolated from short-term induced cells (20 min in nitrate) was observed. NR mRNA isolated from long-term-nitrate-grown cells was completely degraded by RNAases in cell extracts under conditions in which the NR mRNA isolated from short-term induced cells was resistant to degradation. These results suggest that mRNA stability may be an important factor in the metabolic regulation of assimilatory nitrate reductase in Chlorella.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams C. C., Stern D. B. Control of mRNA stability in chloroplasts by 3' inverted repeats: effects of stem and loop mutations on degradation of psbA mRNA in vitro. Nucleic Acids Res. 1990 Oct 25;18(20):6003–6010. doi: 10.1093/nar/18.20.6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwater J. A., Wisdom R., Verma I. M. Regulated mRNA stability. Annu Rev Genet. 1990;24:519–541. doi: 10.1146/annurev.ge.24.120190.002511. [DOI] [PubMed] [Google Scholar]
- Bohjanen P. R., Petryniak B., June C. H., Thompson C. B., Lindsten T. AU RNA-binding factors differ in their binding specificities and affinities. J Biol Chem. 1992 Mar 25;267(9):6302–6309. [PubMed] [Google Scholar]
- Calza R, Huttner E, Vincentz M, Rouzé P, Galangau F, Vaucheret H, Chérel I, Meyer C, Kronenberger J, Caboche M. Cloning of DNA fragments complementary to tobacco nitrate reductase mRNA and encoding epitopes common to the nitrate reductases from higher plants. Mol Gen Genet. 1987 Oct;209(3):552–562. doi: 10.1007/BF00331162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannons A. C., Hipkin C. R. Evidence for the transcriptional control of nitrate reductase in Candida nitratophila from in vitro translation studies. Eur J Biochem. 1987 Apr 15;164(2):383–387. doi: 10.1111/j.1432-1033.1987.tb11069.x. [DOI] [PubMed] [Google Scholar]
- Cannons A. C., Iida N., Solomonson L. P. Expression of a cDNA clone encoding the haem-binding domain of Chlorella nitrate reductase. Biochem J. 1991 Aug 15;278(Pt 1):203–209. doi: 10.1042/bj2780203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C. L., Acedo G. N., Cristinsin M., Conkling M. A. Sucrose mimics the light induction of Arabidopsis nitrate reductase gene transcription. Proc Natl Acad Sci U S A. 1992 Mar 1;89(5):1861–1864. doi: 10.1073/pnas.89.5.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C. L., Dewdney J., Kleinhofs A., Goodman H. M. Cloning and nitrate induction of nitrate reductase mRNA. Proc Natl Acad Sci U S A. 1986 Sep;83(18):6825–6828. doi: 10.1073/pnas.83.18.6825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole M. D., Mango S. E. cis-acting determinants of c-myc mRNA stability. Enzyme. 1990;44(1-4):167–180. doi: 10.1159/000468755. [DOI] [PubMed] [Google Scholar]
- Crawford N. M., Smith M., Bellissimo D., Davis R. W. Sequence and nitrate regulation of the Arabidopsis thaliana mRNA encoding nitrate reductase, a metalloflavoprotein with three functional domains. Proc Natl Acad Sci U S A. 1988 Jul;85(14):5006–5010. doi: 10.1073/pnas.85.14.5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derman E., Krauter K., Walling L., Weinberger C., Ray M., Darnell J. E., Jr Transcriptional control in the production of liver-specific mRNAs. Cell. 1981 Mar;23(3):731–739. doi: 10.1016/0092-8674(81)90436-0. [DOI] [PubMed] [Google Scholar]
- Dewan J. G. The l(+)glutamic dehydrogenase of animal tissues. Biochem J. 1938 Aug;32(8):1378–1385. doi: 10.1042/bj0321378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friemann A., Brinkmann K., Hachtel W. Sequence of a cDNA encoding the bi-specific NAD(P)H-nitrate reductase from the tree Betula pendula and identification of conserved protein regions. Mol Gen Genet. 1991 May;227(1):97–105. doi: 10.1007/BF00260713. [DOI] [PubMed] [Google Scholar]
- Gowri G., Campbell W. H. cDNA Clones for Corn Leaf NADH:Nitrate Reductase and Chloroplast NAD(P):Glyceraldehyde-3-Phosphate Dehydrogenase : Characterization of the Clones and Analysis of the Expression of the Genes in Leaves as Influenced by Nitrate in the Light and Dark. Plant Physiol. 1989 Jul;90(3):792–798. doi: 10.1104/pp.90.3.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowri G., Kenis J. D., Ingemarsson B., Redinbaugh M. G., Campbell W. H. Nitrate reductase transcript is expressed in the primary response of maize to environmental nitrate. Plant Mol Biol. 1992 Jan;18(1):55–64. doi: 10.1007/BF00018456. [DOI] [PubMed] [Google Scholar]
- Hilgarth C., Sauer N., Tanner W. Glucose increases the expression of the ATP/ADP translocator and the glyceraldehyde-3-phosphate dehydrogenase genes in Chlorella. J Biol Chem. 1991 Dec 15;266(35):24044–24047. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lorimer G. H., Gewitz H. S., Völker W., Solomonson L. P. The presence of bound cyanide in the naturally inactivated form of nitrate reductase of Chlorella vulgaris. J Biol Chem. 1974 Oct 10;249(19):6074–6079. [PubMed] [Google Scholar]
- Lu J. L., Ertl J. R., Chen C. M. Cytokinin enhancement of the light induction of nitrate reductase transcript levels in etiolated barley leaves. Plant Mol Biol. 1990 Apr;14(4):585–594. doi: 10.1007/BF00027504. [DOI] [PubMed] [Google Scholar]
- McLaren R. S., Newbury S. F., Dance G. S., Causton H. C., Higgins C. F. mRNA degradation by processive 3'-5' exoribonucleases in vitro and the implications for prokaryotic mRNA decay in vivo. J Mol Biol. 1991 Sep 5;221(1):81–95. [PubMed] [Google Scholar]
- Monod C., Goldschmidt-Clermont M., Rochaix J. D. Accumulation of chloroplast psbB RNA requires a nuclear factor in Chlamydomonas reinhardtii. Mol Gen Genet. 1992 Feb;231(3):449–459. doi: 10.1007/BF00292715. [DOI] [PubMed] [Google Scholar]
- Oaks A., Poulle M., Goodfellow V. J., Cass L. A., Deising H. The role of nitrate and ammonium ions and light on the induction of nitrate reductase in maize leaves. Plant Physiol. 1988 Dec;88(4):1067–1072. doi: 10.1104/pp.88.4.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto P. M., Fu Y. H., Marzluf G. A. Nit-3, the structural gene of nitrate reductase in Neurospora crassa: nucleotide sequence and regulation of mRNA synthesis and turnover. Mol Gen Genet. 1991 Jun;227(2):213–223. doi: 10.1007/BF00259673. [DOI] [PubMed] [Google Scholar]
- Sherman T. D., Funkhouser E. A. Induction and synthesis of nitrate reductase in Chlorella vulgaris. Arch Biochem Biophys. 1989 Nov 1;274(2):525–531. doi: 10.1016/0003-9861(89)90466-9. [DOI] [PubMed] [Google Scholar]
- Solomonson L. P., Barber M. J., Robbins A. P., Oaks A. Functional domains of assimilatory NADH:nitrate reductase from Chlorella. J Biol Chem. 1986 Aug 25;261(24):11290–11294. [PubMed] [Google Scholar]
- Solomonson L. P., Spehar A. M. Stimulation of cyanide formation by ADP and its possible role in the regulation of nitrate reductase. J Biol Chem. 1979 Apr 10;254(7):2176–2179. [PubMed] [Google Scholar]
- Takata R., Izuhara M., Akiyama K. Processing in the 5' region of the pnp transcript facilitates the site-specific endonucleolytic cleavages of mRNA. Nucleic Acids Res. 1992 Feb 25;20(4):847–850. doi: 10.1093/nar/20.4.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodorakis N. G., Cleveland D. W. Physical evidence for cotranslational regulation of beta-tubulin mRNA degradation. Mol Cell Biol. 1992 Feb;12(2):791–799. doi: 10.1128/mcb.12.2.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen Y., Green P. J. Identification and Properties of the Major Ribonucleases of Arabidopsis thaliana. Plant Physiol. 1991 Dec;97(4):1487–1493. doi: 10.1104/pp.97.4.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeiler K. G., Solomonson L. P. Regulation of Chlorella nitrate reductase: control of enzyme activity and immunoreactive protein levels by ammonia. Arch Biochem Biophys. 1989 Feb 15;269(1):46–54. doi: 10.1016/0003-9861(89)90085-4. [DOI] [PubMed] [Google Scholar]