Summary
Cancer stem cells (CSCs) established from surgical biopsies closely mimic the human context and can be used to investigate disease mechanisms, genetic fitness, and therapeutic evaluation. Here, we present a protocol for the derivation of primary patient-derived CSC lines from ependymal tumors. We describe the necessary steps, from surgical intervention and biopsy to the dissociation of ependymomas to derive cultures. We then detail procedures for cell line propagation and define the characteristics of these primary cancer cell lines.
For complete details on the use and execution of this protocol, please refer to Michealraj et al.1
Subject areas: Cell Biology, Cancer, Stem Cells, Cell Differentiation
Graphical abstract
Highlights
-
•
Protocol to generate patient-derived cancer stem cell lines from ependymal tumors
-
•
Steps for the expansion and maintenance of primary ependymoma cell lines
-
•
Profiling and characterization of infratentorial/supratentorial ependymoma cell lines
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Cancer stem cells (CSCs) established from surgical biopsies closely mimic the human context and can be used to investigate disease mechanisms, genetic fitness, and therapeutic evaluation. Here, we present a protocol for the derivation of primary patient-derived CSC lines from ependymal tumors. We describe the necessary steps, from surgical intervention and biopsy to the dissociation of ependymomas to derive cultures. We then detail procedures for cell line propagation and define the characteristics of these primary cancer cell lines.
Before you begin
Institutional permissions
Our protocol has been approved by the Hospital for Sick Children/SickKids Research Ethics Board, and samples were collected from consenting patients as approved by the requisite committees. To set up the collection and processing of biopsies, all relevant institutional permissions should be obtained in advance and tissues should be processed in accordance with appropriate ethical approvals.
Tumor specimens should be immediately placed in a sterile receptacle on ice for transport from the operating room to the lab for processing. Informed consent must be obtained prior to collection, and a pathologist should provide a preliminary diagnosis and release the sample prior to processing.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Collagenase/Dispase lyophilized | Roche | 10269638001 |
| PBS 1× without calcium and magnesium | WISENT | 311-010-CL |
| Poly-L-ornithine solution, 0.01% | Sigma-Aldrich | P4957 |
| Laminin from Engelbreth-Holm-Swarm murine sarcoma | Sigma-Aldrich | L2020 |
| StemPro Accutase cell dissociation reagent | Thermo Fisher Scientific | A1110501 |
| NeuroCult NS-A basal medium (human) | STEMCELL Technologies | 05750 |
| N-2 supplement (100×) | Thermo Fisher Scientific | 17502-048 |
| B-27 supplement (50×), minus vitamin A | Thermo Fisher Scientific | 12587-010 |
| Antibiotic: Antimycotic (100×) | Thermo Fisher Scientific | 15240062 |
| GlutaMAX supplement (100×) | Thermo Fisher Scientific | 35050061 |
| Bovine serum albumin solution (7.5% in DPBS) | Sigma-Aldrich | A8412 |
| Heparin sodium salt from porcine intestinal mucosa | Sigma-Aldrich | H3393 |
| Recombinant human EGF | Sigma-Aldrich | E9644 |
| Recombinant human FGF-basic | PeproTech | 100-18B |
| Dimethyl sulfoxide (DMSO) | Sigma-Aldrich | D4540 |
| Critical commercial assays | ||
| DNeasy Blood & Tissue Kit | QIAGEN | 69504 |
| EZ DNA methylation kit | Zymo Research | D5001 |
| Infinium MethylationEPIC V2.0 kit | Illumina | 20087706 |
| Experimental models: Cell lines | ||
| MDT-PFA2 | Michealraj et al.1 | Human PFA ependymoma, 0.88-year-old male, primary, balanced genome |
| MDT-PFA4 | Michealraj et al.1 | Human PFA ependymoma, 2-year-old female, primary, balanced genome |
| MDT-PFA5 | Michealraj et al.1 | Human PFA ependymoma, 1.08-year-old male, primary, balanced genome |
| MDT-PFA7 | Michealraj et al.1 | Human PFA ependymoma, 9.6-year-old male, primary, 1q gain |
| MDT-PFA9 | Michealraj et al.1 | Human PFA ependymoma, 0.92-year-old male, primary, balanced genome |
| MDT-PFA15 | Michealraj et al.1 | Human PFA ependymoma, 11.58-year-old male, primary, 1q gain |
| MDT-ST2 | Michealraj et al.1 | Human ST ependymoma, 8.3-year-old female, primary, ZFTA-RELA fusion positive |
| MDT-ST4 | Michealraj et al.1 | Human ST ependymoma, 6.6-year-old male, primary, ZFTA-RELA fusion positive |
| Software and algorithms | ||
| DKFZ methylation profiling classifier for CNS tumors | Capper et al.2 | https://www.molecularneuropathology.org/mnp/ |
| Other | ||
| Primaria 100 mm × 20 mm standard cell culture dish (Nunc EasYDish Dishes serve as an appropriate alternative) Note: Area equivalent cell culture flasks may be used in place of dishes. | Corning (Thermo Fisher Scientific) | 353803 (150466) |
| Primaria 6-well | Corning | 353846 |
| Primaria 96-well | Corning | 353872 |
| 70 μm cell strainer | Falcon | 352350 |
| 40 μm cell strainer | Falcon | 352340 |
| Cryopure tubes | Sarstedt | 72.377 |
| Mr Frosty freezing container | Thermo Fisher Scientific | 5100-0001 |
| Oxivir Tb wipes | Diversey | 5144708 |
| Multigas (CO2/O2) incubator | PHCbi | MCO-170M-PA |
Materials and equipment
-
•
Collagenase/Dispase Solution: Reconstitute 100 mg of lyophilized powder into 1 mL of sterile molecular grade H2O, and further dilute using 99 mL PBS to obtain a stock concentration of 1 mg/mL (10×). Pass the solution through a low protein binding 0.2 μm syringe filter before freezing at −20°C in 1 mL aliquots.
EPN-B (Basal) Media
| Reagent | Final concentration | Amount |
|---|---|---|
| NeuroCult NS-A basal medium (Human) | N/A | 450 mL |
| N-2 supplement (100×) | 1× | 5 mL |
| B-27 supplement, minus vitamin A (50×) | 1× | 10 mL |
| Glutamax supplement (100×) | 1× | 5 mL |
| Bovine serum albumin solution (7.5%) | 0.015% | 1 mL |
| Heparin solution (1 mg/mL) | 2 μg/mL | 1 mL |
| Antibiotic-Antimycotic (100×) - Optional | 1× | 5 mL |
Storage: Store all reagents as per manufacturer’s instructions. EPN-B media is stable for one month at 4°C.
EPN-C (Complete) Media
| Supplement EPN-B (∼500 mL) with the following: | ||
|---|---|---|
| Reagent | Final concentration | Amount |
| Recombinant human EGF (200 μg/mL) | 10 ng/mL | 25 μL |
| Recombinant human FGF-basic (4 μg/mL) | 10 ng/mL | 1.25 mL |
Storage: Growth factors should be stored at −20°C. EPN-C media is stable for ∼2 weeks at 4°C.
Note: For extended storage, only supplement EPN-B with growth factors prior to use. This can be done through the use of small volume aliquots (e.g. 50 mL).
Step-by-step method details
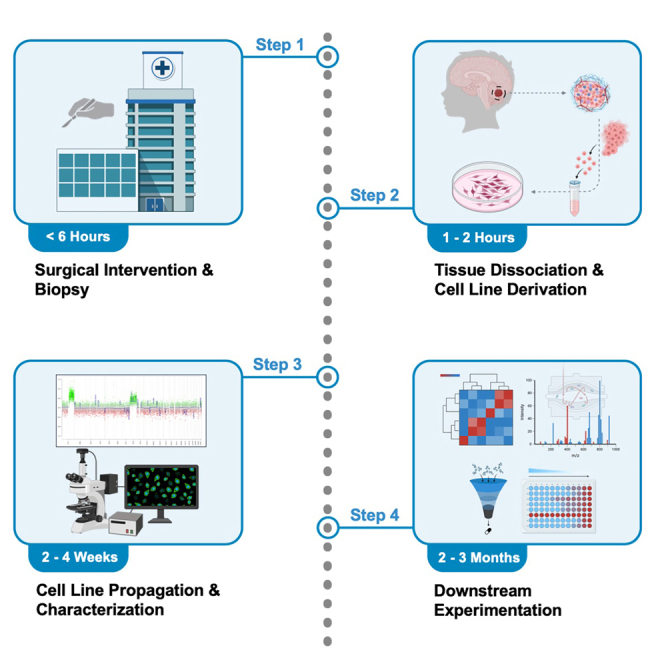
Dissociation of tumor tissue and generation of primary cultures
Timing: ∼1–2 h
This section describes the derivation of primary ependymal cancer stem cell lines and should be carried out in a biosafety cabinet under sterile conditions.
-
1.
Thoroughly clean all instruments (forceps, scalpel, and razor blade) using 70% ethanol or Oxivir Tb wipes.
-
2.
Place the fresh tissue biopsy (ideally using a minimum of 0.5 cm3, processed within 6 h of collection) into a 100 mm dish and submerge in 5 mL of PBS.
-
3.
Using a sterile blade, disaggregate the tissue through repeated mincing for several minutes until the sample appears homogeneous in solution.
-
4.
Place the homogenized suspension into a 15 mL conical tube and collect any residual fragments within the petri dish using additional PBS and a serological pipette. Add PBS to a final volume of 9 mL.
-
5.
Add one aliquot of collagenase/dispase stock solution to the conical tube to obtain a final working concentration (1:10 dilution).
Note: One stock aliquot of collagenase/dispase should be sufficient for a sample ≤1–2 cm3. The volume of dissociation solution will scale with sample size, however if the tissue is large it should be cut into individual fragments (∼1 cm3) for parallel dissociations or freezing.
-
6.
Place the conical tube at 37°C (using a water bath or by placing within an incubator) for 30 min while gently inverting every 5 min.
Note: An orbital shaker housed within an incubator (e.g., TubeRocker mini, Benchmark Scientific, M2100) or dedicated incubator with rotator (e.g., Mini Incubator with Mini Lab Roller and Rotisseries, Labnet, 15110A-MLR) can be used instead of manual inversion.
Note: Do not exceed 30 minutes as this may lower the likelihood of establishing a cell line.
-
7.
Gently break down any residual chunks of tissue using a P1000 pipette.
-
8.
Transfer the volume to a 50 mL conical tube and add 30 mL of 37°C complete media (EPN-C) to dilute the dissociation solution. Mix well using a serological pipette.
-
9.
Pass the solution through a 70 μm filter placed within a new 50 mL conical tube to remove cell aggregates and promote a homogenous single cell suspension.
Optional: This can be followed by use of a 40 μm filter.
-
10.
Take the fragments retained within the filter and wash the contents off the membrane into a poly-L-ornithine/laminin (PoL) coated 6-well plate using complete media (see plate preparation, step 17).
Note: These larger fragments have the potential to seed colonies of cells and should not be discarded.
-
11.
Centrifuge the contents of the conical tube for 5 min at 250 g. Aspirate the media and resuspend using 5 mL of complete media (scale volume to pellet size) before performing a cell count using a hemocytometer or automated cell counter and viability analyzer (>50% viable cells is desirable).
-
12.
Place a minimum of 1–2 million cells into 2 mL of complete media and dispense into a PoL coated 6-well plate. If the cell count is high, place ≥5 million cells into a 100 mm dish.
-
13.
Culture primary ependymal cancer cells under hypoxia (1% O2) using a multigas incubator. If there is an excess of cells, place an additional plate under normoxia.
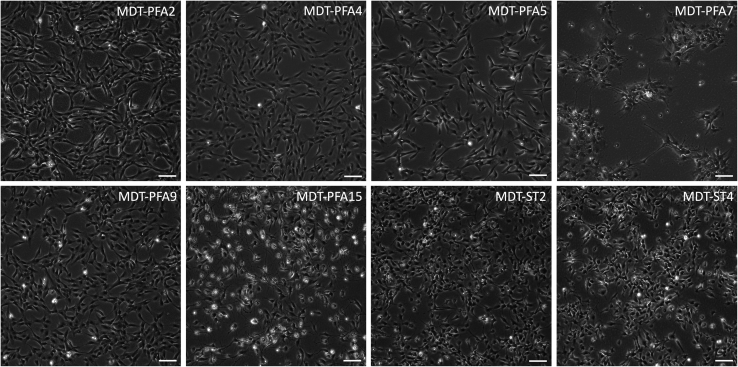
Note: Cells are expected to adhere and begin proliferating after ∼1–2 weeks in culture (Figure 1).
Note: Sustained growth of posterior fossa group A (PFA) cell lines has been demonstrated at 1% O2, however more mild hypoxic conditions (1%–5%) may warrant testing given that the partial pressure of oxygen varies within this range in accordance with vascularization and tissue depth within the brain.3 Recurrent PFAs and supratentorial (ST) ependymomas have been observed to exhibit tolerance for both hypoxic and normoxic conditions, with some lines exhibiting faster growth under normoxia.
-
14.
If the media begins to turn orange/yellow within the first week, supplement the existing media with fresh EPN-C media. There should be no media change for the first week of culture.
Note: It is best to avoid media changes when establishing cultures as the cell density is low and primary cell lines secrete growth factors that promote their proliferation and viability. Media should initially be supplemented on the basis of media acidity and cell density.
-
15.
Any additional cells should be viably frozen at a concentration ≥1 million cells/mL using EPN-C media with 10% DMSO (see cryopreservation, step 42).
-
16.
After 1–2 weeks in culture, cells can be expanded on PoL plates for downstream applications.
Note: It is recommended to cryopreserve multiple vials of early passages (P1-P5) as long-term culture will eventually lead to slowed growth and senescence.
Figure 1.
Cell morphologies of primary patient-derived ependymal cell lines
Representative images of PFA and ST primary cell lines are depicted. Scale bar = 100 μm.
Poly-L-ornithine/laminin (PoL) plate preparation
Timing: 24 h
This section describes the preparation of PoL plates for primary culture expansion.
Note: Plates should be prepared at least one day in advance of anticipated cell culture.
-
17.
Coat the entire surface of Primaria cell culture plates with poly-L-ornithine solution, scaling the volume to the area of the plate (suggested volumes: 5 mL for a 100 mm dish, 1 mL for a 6-well, 100 μL for a 96-well). Incubate for 30–40 min.
-
18.
Place a vial of laminin on ice to thaw.
Note: Laminin must be thawed slowly on ice to prevent gel formation.
-
19.
Remove the poly-L-ornithine solution and save it for repeat use (efficacy demonstrated over >5 applications).
-
20.
Add PBS to cover the surface of the plate and swirl gently to wash any remaining poly-L--ornithine solution. Remove the PBS and discard.
-
21.
Mix one full vial of laminin (1 mg) with 200 mL PBS chilled to 4°C in order to generate a working concentration of 5 μg/mL. This volume can be scaled to the number of plates required.
Note: The laminin stock can be aliquoted and frozen when less volume is required; however, aliquots should be thawed only once and not refrozen.
-
22.
Add diluted laminin solution onto poly-L-ornithine-coated plates, again scaling the volume to the surface area of the plate (suggested volumes: 10 mL for a 100 mm dish, 2 mL for a 6-well, 100 μL for a 96-well) and place into an incubator at 37°C overnight prior to use.
Note: Plates are suitable for use for up to 2 weeks.
Propagation and passaging of ependymoma stem cell lines
This section describes the expansion of primary ependymal cell lines for experimentation.
-
23.
Pre-warm EPN-C media to 37°C using a bead bath.
Thawing from a frozen stock
Timing: 1–2 weeks for ∼1–2 × 106cells freshly thawed from an early passage (<P15) vial into a 100 mm PoL-coated dish
-
24.
Obtain a vial of cells from frozen storage and thaw at 37°C using a bead bath.
-
25.
Once thawed, acclimate the cells using 1 mL of EPN-C in a drop wise manner, then top up to 10 mL in a 15 mL conical tube.
-
26.
Centrifuge the tube for 3–5 min at 250 g. Discard the supernatant and add 1 mL fresh EPN-C media onto the pellet and mix gently with a P1000 pipette.
-
27.
Assess cell count and viability using preferred methodology.
-
28.
Take the required PoL coated plates from the incubator and discard the laminin solution.
-
29.
Plate ependymal tumor stem cells at approximately 1–2 million cells per 100 mm PoL coated dish in 10 mL of EPN-C media (i.e., 1 vial per plate). Seed dropwise and swirl the dish for an even distribution. Proceed with step 39.
Note: Lower concentrations should be scaled down to smaller plates accordingly.
Passaging primary cultures
Timing: <1 h for routine passaging
-
30.Obtain plates at ∼80%–90% confluency.
-
a.Remove the conditioned media and place aside into a conical tube.
-
b.Centrifuge for 3–5 min at 250 g.
-
c.Leave aside so as not to disturb the pellet of dead cells and debris.
-
a.
Note: This supernatant will be used to supplement future passages.
-
31.Add a sufficient volume of Accutase to cover the plate surface (suggested volumes: 1–2 mL for a 100 mm dish, 500 μL for a 6-well).
-
a.Incubate and tap gently along the plate perimeter every few minutes for mechanical disruption.
-
b.Observe the cells for detachment under the microscope.
-
a.
-
32.
Add 4× the volume of sterile 37°C PBS onto the plate to neutralize the cell detachment solution and collect the cells into a conical tube.
-
33.
Centrifuge cells for 3–5 min at 250 g.
-
34.
Discard the supernatant and add 37°C EPN-C media onto the pellet. Gently resuspend the cells in 2 mL and assess cell count and viability (anticipating >95% viability).
-
35.
For routine expansion with 100 mm plates, resuspend at a concentration of 1–2 million cells per mL allowing for 1 mL to be seeded onto each plate.
Note: For ease, cells can routinely be split at a 1:3 ratio, foregoing the need for a cell count while aligning with the indicated concentration.
Note: While at low confluency (<50%) or when recovering from a recent thaw, a 1:2 split ratio is preferred for initial passages.
-
36.
Take the required PoL coated plates from the incubator and remove the laminin solution.
-
37.
For each 100 mm dish, dispense 6 mL of fresh EPN-C media and top with 3 mL of conditioned media from step 30.
Note: Conditioned media should account for approximately 20%–30% of the final volume in order to maintain growth promoting secretory factors.
-
38.
Seed 1 mL of cells dropwise into the plate, and swirl to ensure an even distribution.
-
39.
Place the plates in a multigas incubator under hypoxic conditions (1% O2).
Note: A hypoxic glove box is not required. Cells can be handled under normoxic conditions while passaging, although this time should be limited.
-
40.
Replace media every 3 days. When feeding cells in a 100 mm dish, leave 3 mL of conditioned media and replenish the remaining 7 mL with fresh 37°C EPN-C media, maintaining a total volume of 10 mL.
Note: Cells should not be passaged at >90% confluency. Cell lines can senesce if left fully confluent for greater than 24 hours.
Cryopreservation of primary ependymoma cell lines
Timing: <1 h
-
41.
Refer to steps 30–34, discarding the conditioned media from plates.
-
42.
Add cell culture grade DMSO (10% of the total volume) to the cell suspension and mix gently. The concentration should be ≥1 million cells per mL.
-
43.
Aliquot 1 mL into a cryopure tube, and place into a freezing container at −80°C. Move the cells to a liquid nitrogen tank for prolonged storage.
Note: Freezing solution may require optimization as certain lines can be sensitive to freezing. Temperamental cell lines may benefit from use of specialized freezing media (e.g., CryoStor CS10 medium, STEMCELL Technologies, 100–1061) to maximize post-thaw cell recovery and viability.
Expected outcomes
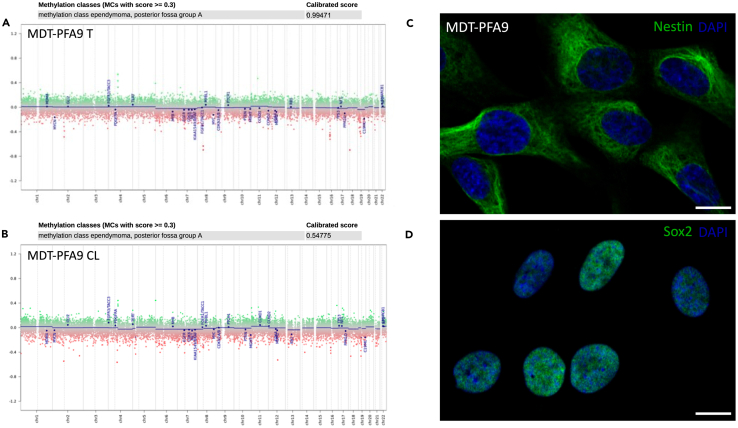
The derivation of primary cancer stem cell lines serves as a gold standard model for in vitro experimentation. Mechanistic insight and therapeutic screening require high throughput assaying for downstream translation to the preclinical setting. These cancer stem cells exhibit marker expression in line with their lineage and pathology, further recapitulating their disease entity through methylation profiling (Figure 2).2
Note: DNA extracts must undergo bisulfite conversion prior to methylation profiling using the Infinium MethylationEPIC v2.0 Kit. Approximately 10 mg of tissue should be set aside at the time of processing for DNA extraction to obtain a minimum input of 500 pg of DNA for bisulfite conversion. Methylation profiling should be carried out no earlier than passage 5 to ensure the enrichment of a homogenous population of cells.
Figure 2.
Characterization of primary cancer stem cell lines
DNA methylation array showing relevant alterations and a calibrated diagnostic score using the DKFZ CNS tumor classifier (brain classifier version v11b4, workflow version 3.2) for a PFA ependymoma tissue biopsy (A) and its corresponding cell line, MDT-PFA9 (B). Immunofluorescence of characteristic neural stem cell markers Nestin (C) and Sox2 (D) are shown for the aforementioned cancer stem cell line. Scale bar = 10 μm.
Researchers should be careful to ensure that primary cell lines are a diagnostic match to the tissues from which they are derived.
Limitations
These primary cell lines are a non-renewable resource that can senesce or terminally differentiation at late passages. As such, the period of experimentation is temporally defined and all efforts should be made to expand and cryopreserve early cultures.
Troubleshooting
Problem 1
There was no enrichment of proliferating cells subsequent to patient tissue dissociation.
Potential solution
-
•
The quality of the biopsy may be poor, whether due to necrosis or environmental exposure. Transfer time from the operating room to laboratory for processing should be minimized, with samples maintained on ice or refrigerated at 4°C (step 2).
-
•
Tissue heterogeneity may contribute to the adhesion of stromal cells or non-stem cell populations that do not proliferate. It is important to assess both the adherent cell population as well as floating cells to discriminate cell death and debris from potential neurospheres which may indicate a preference for suspension culture.
Problem 2
The cells are not expanding.
Potential solution
-
•
Slow division may be a result of cells being plated at a density that is too low; consider replating into a smaller dish to increase cell density (step 35). This may also result from a prolonged period of confluence prior to passaging (see Note, step 40).
-
•
Cells may be at a passage number that is too high; differentiated cells or cells undergoing senescence should be discarded and a fresh vial should be thawed for subsequent experiments. Experimentation should be confined below passage 20. Most primary cell lines begin to lose their proliferation capacity at high passage numbers, even with proper care and treatment (see Note, step 16).
-
•
Conditioned media should constitute 20–30% of the new volume of media upon feeding. Neglecting these secreted factors may result in a growth delay (see Note, step 37).
-
•
Cells may have been exposed to mycoplasma; if positive, MycoZap treatment can be initiated for a duration of 1–2 weeks until tests return negative, during this time contaminated incubators must be sterilized, and cells should be quarantined until they test negative.
Problem 3
There is a large fraction of floating/dead cells.
Potential solution
-
•
There may be a problem with the laminin stock. Ensure that there are no floating aggregates in thawed vial as this may reflect precipitation/gel formation (see Note, step 18). Further, the media may be acidic and require fresh EPN-C media.
-
•
Cells may have been exposed to mycoplasma; media can be tested and if positive treatment can be initiated as outlined above.
Problem 4
Cell phenotype has undergone an uncharacteristic change.
Potential solution
-
•
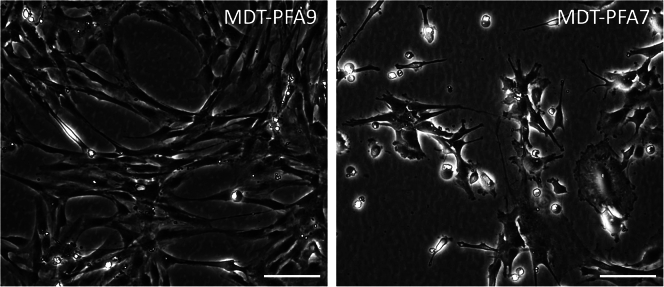
Extended periods of confluency can result in senescence, or a differentiated phenotype (Figure 3).
-
•
Aged media may have an insufficient concentration of growth factors. Regardless of cause, a fresh vial of cells should be thawed.
Figure 3.
Atypical morphologies suggesting disuse of culture
Irregular cell morphologies may include an elongated fibroblastic appearance accompanied by a stalled level of confluency (left panel) or features of differentiation (right panel). Scale bar = 100 μm.
Problem 5
Cell lines exhibit a low diagnostic score/no match after methylation profiling.
Potential solution
-
•
The methylation classifier was trained using tissues which are inherently heterogeneous, whereas the cell lines constitute a homogenized fraction of cells. If a diagnostic output is given, the top score should ideally match the expected diagnosis. If no match is obtained, ensure the certainty of diagnosis from pathology and assess characteristic markers; EZHIP expression and loss of Lysine 27 trimethylation can be determined via western blot for PFA ependymoma and C11orf95(ZFTA)-RELA fusion specific quantitative PCR primers can be used in confirming this subgroup of ST ependymomas (see expected outcomes).4,5,6,7
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Antony Michealraj (micheala@pitt.edu).
Technical contact
Technical questions on executing this protocol should be directed to and will be answered by the technical contact, Cory Richman (cory.richman@mail.utoronto.ca).
Materials availability
This study did not generate new unique reagents.
Data and code availability
This study did not generate or analyze data; original cell line derivation findings were published in Michaelraj et al.1
Acknowledgments
This work was supported by a Stand Up To Cancer (SU2C) St. Baldrick’s Pediatric Dream Team translational research grant (SU2C-AACR-DT1113) and SU2C Canada Cancer Stem Cell Dream Team research funding (SU2C-AACR-DT-19-15) provided by the Government of Canada through Genome Canada and the Canadian Institutes of Health Research, with supplementary support from the Ontario Institute for Cancer Research through funding provided by the Government of Ontario. SU2C is a program of the Entertainment Industry Foundation administered by the American Association for Cancer Research. The authors would also like to recognize the Labatt Brain Tumour Research Centre for their invaluable support. The graphical abstract was created using BioRender.
Author contributions
C.M.R. and K.A.M. contributed to the design and generation of this manuscript; P.B.D. and M.D.T. provided feedback, guidance, and funding acquisition.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Cory M. Richman, Email: cory.richman@mail.utoronto.ca.
Kulandaimanuvel Antony Michealraj, Email: micheala@pitt.edu.
References
- 1.Michealraj K.A., Kumar S.A., Kim L.J.Y., Cavalli F.M.G., Przelicki D., Wojcik J.B., Delaidelli A., Bajic A., Saulnier O., MacLeod G., et al. Metabolic Regulation of the Epigenome Drives Lethal Infantile Ependymoma. Cell. 2020;181:1329–1345.e24. doi: 10.1016/j.cell.2020.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Capper D., Jones D.T.W., Sill M., Hovestadt V., Schrimpf D., Sturm D., Koelsche C., Sahm F., Chavez L., Reuss D.E., et al. DNA methylation-based classification of central nervous system tumours. Nature. 2018;555:469–474. doi: 10.1038/nature26000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mas-Bargues C., Sanz-Ros J., Román-Domínguez A., Inglés M., Gimeno-Mallench L., El Alami M., Viña-Almunia J., Gambini J., Viña J., Borrás C. Relevance of Oxygen Concentration in Stem Cell Culture for Regenerative Medicine. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20051195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bayliss J., Mukherjee P., Lu C., Jain S.U., Chung C., Martinez D., Sabari B., Margol A.S., Panwalkar P., Parolia A., et al. Lowered H3K27me3 and DNA hypomethylation define poorly prognostic pediatric posterior fossa ependymomas. Sci. Transl. Med. 2016;8 doi: 10.1126/scitranslmed.aah6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han J., Yu M., Bai Y., Yu J., Jin F., Li C., Zeng R., Peng J., Li A., Song X., et al. Elevated CXorf67 Expression in PFA Ependymomas Suppresses DNA Repair and Sensitizes to PARP Inhibitors. Cancer Cell. 2020;38:844–856.e7. doi: 10.1016/j.ccell.2020.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jain S.U., Do T.J., Lund P.J., Rashoff A.Q., Diehl K.L., Cieslik M., Bajic A., Juretic N., Deshmukh S., Venneti S., et al. PFA ependymoma-associated protein EZHIP inhibits PRC2 activity through a H3 K27M-like mechanism. Nat. Commun. 2019;10:2146–2214. doi: 10.1038/s41467-019-09981-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parker M., Mohankumar K.M., Punchihewa C., Weinlich R., Dalton J.D., Li Y., Lee R., Tatevossian R.G., Phoenix T.N., Thiruvenkatam R., et al. C11orf95-RELA fusions drive oncogenic NF-κB signalling in ependymoma. Nature. 2014;506:451–455. doi: 10.1038/nature13109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate or analyze data; original cell line derivation findings were published in Michaelraj et al.1