Abstract
Defining subcellular locations and interacting partners for proteins accelerates their functional characterization. A new in vivo tagging approach achieves both for mitochondrial matrix proteins and helps connect a key oxidoreductase to coenzyme Q biosynthesis.
Mitochondrial protein maps have guided discoveries for more than two decades, but information on tissue diversity, protein function, and sub-mitochondrial localization remains incomplete. Filling these gaps could help identify missing pathway components, reveal new mitochondrial functions, and fuel new mitochondrial disease diagnosis and treatment strategies. In this issue of Nature Chemical Biology, Park et al1. takes steps to achieving these goals through proximity labeling— a technology that helps define local proteomes and protein-protein interactions by targeting a promiscuous labeling enzyme to specific cellular locations2.
The mammalian mitochondrial proteome has proven to be complex and dynamic, with proteins whose abundances may fluctuate across different cell and tissue types and in response to changing metabolic states 3,4. Current mitochondrial compendia also reveal that approximately 20% of mitochondrial proteins lack a clear biological function, suggesting that many mitochondrial activities remain to be discovered5. Assigning functions to orphan proteins may enable the discovery of missing pathway components and ultimately provide a framework for understanding how mitochondrial function goes awry in disease6,7. One such pathway is the biosynthesis of coenzyme Q (CoQ), a redoxactive lipid required for ATP generation, antioxidant activities, and multiple other enzymatic processes in mitochondria and beyond8. Many open questions surround this pathway, including the identities of requisite enzymes and precursor transporters.
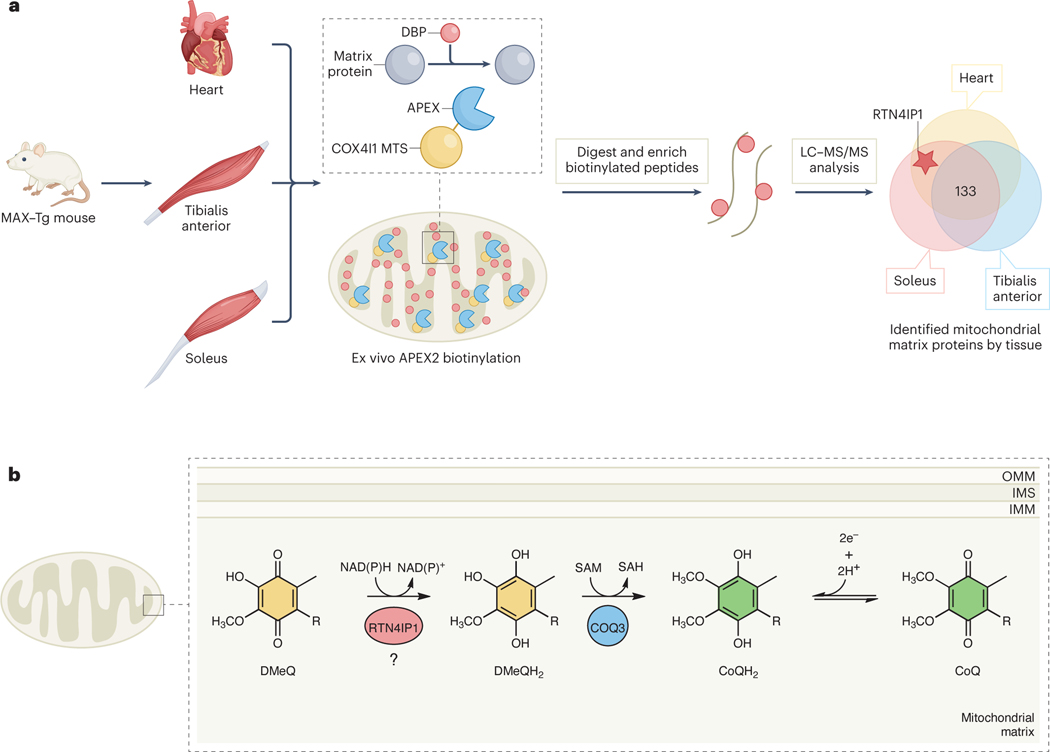
In this study, Park et al. set out to characterize the in vivo mitochondrial matrix proteome of distinct muscle tissues. To accomplish this, the authors generated whole-body and conditional muscle-specific transgenic mice, named MAX-Tg mice, in which an engineered ascorbate peroxidase (APEX2) labeling enzyme was fused to the mitochondrial-targeting sequence of the resident matrix protein COX4I1 (Fig 1a). Using the proximity labeling approach in different muscle groups — cardiac, tibialis anterior, and soleus — the authors identified 250 matrix proteins, with 133 shared between the three groups. In addition to revealing matrix proteins uniquely expressed in each tissue, their data suggest that different metabolic pathways are enriched in each tissue, such as beta oxidation in heart.
Figure 1. Mitochondrial matrix-specific proximity labeling iden fies RTN4IP1 as a novel oxidoreductase involved in CoQ biosynthesis.
A) The MAX-Tg mouse model involves localiza on of the APEX2 tag to the mitochondrial matrix via fusion to the mitochondrial targe ng sequence of COX4I1. Subsequent proximity labeling, bio n enrichment, and mass spectrometry is performed to characterize the matrix proteome across different muscle ssue types. B) The current model for RTN4IP1 func on proposes that it enables CoQ biosynthesis via the reduc on of COQ3’s DMeQ substrate.
The matrix proteome labeling patterns led the authors to the uncharacterized protein RTN4IP1/OPA10, which has strong ties to mitochondrial health and can harbor causatives mutation in human diseases including optic neuropathy9. RTN4IP1 has structural similarity to the E. coli quinone NADPH oxidoreductase, QOR10. Indeed, when purified in vitro, RTN4IP1 could use NADPH to reduce a generic quinone substrate. To explore its specific role(s) in the mitochondrial matrix, the authors once again used proximity labeling, this time to map RTN4IP1-protein interactions, and found significant enrichment of the CoQ biosynthetic enzymes COQ3 and COQ5. RTN4IP1 augmented the O-methylation activity of COQ3 in vitro, theoretically by reducing the quinone substrate prior to catalysis (Fig 1b). The authors found that deleting RTN4IP1 in cultured cells and in vivo resulted in decreased levels of CoQ, accompanied by gross defects in mitochondrial morphology and impaired OxPhos activity. Flies with muscle-specific RTN4IP1 knockdown additionally exhibited severe locomotive defects, which could be partially rescued by a small, soluble CoQ analog. Taken together, this study by Park et al. identifies a new role for RTN4IP1 as a mitochondrial matrix protein that assists CoQ biosynthesis through its oxidoreductase activity.
This work reveals the potential of in vivo proximity labeling, which could emerge as a widespread method for exploring the diversity and dynamic nature of the mitochondrial proteome at the sub-organellar level. Though this study linked an uncharacterized oxidoreductase to the CoQ biosynthesis, it also leaves some questions unanswered. COQ3 was one of many RTN4IP1 interactors identified, including COQ5, another methyltransferase in the pathway. Does RTN4IP1 support enzymatic function at multiple stages of biosynthesis? Does the lethal RTN4IP1 KO phenotype and the severe mitochondrial defects suggest a much more pervasive organellar role for RTN4IP1? More broadly, this study highlights the ability to characterize tissue-specific mitochondrial proteomes; however, understanding if and when data from these comparisons is meaningful may require further scrutiny. For CoQ biosynthesis — which occurs in every tissue in the body — one would expect CoQ-related proteins, including RTN4IP1, to be expressed at comparable levels. Are the observed enzyme abundance differences from this approach meaningful, or perhaps suggestive of experimental challenges of cross-tissue comparisons? Moving forward, the use of this proximity labeling approach to explore proteomes from new tissues/cells or to monitor how a select proteome changes in response to biological perturbations may prove even more informative. Despite these caveats, Park et al. add a unique in vivo tool to our arsenal for defining and characterizing subcellular and sub-organellar proteomes.
References
- 1.Park I, Kim K, Kim J, Kim A-K, Bae S, Jung M, Choi J, Mishra PK, Kim T-M, Kwak C, et al. (2023). In vivo mitochondrial matrix proteome profiling reveals RTN4IP1/OPA10 as an NAD(P)H oxidoreductase for Coenzyme Q biosynthesis. Nature Chemical Biology. [DOI] [PMC free article] [PubMed]
- 2.Qin W, Cho KF, Cavanagh PE, and Ting AY (2021). Deciphering molecular interactions by proximity labeling. Nat. Methods 18, 133–143. 10.1038/s41592-020-01010-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rath S, Sharma R, Gupta R, Ast T, Chan C, Durham TJ, Goodman RP, Grabarek Z, Haas ME, Hung WHW, et al. (2020). MitoCarta3.0: an updated mitochondrial proteome now with sub-organelle localization and pathway annotations. Nucleic Acids Res. 49, gkaa1011-. 10.1093/nar/gkaa1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morgenstern M, Peikert CD, Lubbert P, Suppanz I, Klemm C, Alka O, Steiert C, Naumenko N, Schendzielorz A, Melchionda L, et al. (2021). Quantitative high-confidence human mitochondrial proteome and its dynamics in cellular context. Cell Metab 33, 2464–2483 e18. 10.1016/j.cmet.2021.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sung AY, Floyd BJ, and Pagliarini DJ (2020). Systems Biochemistry Approaches to Defining Mitochondrial Protein Function. Cell Metabolism 31, 669–678. 10.1016/j.cmet.2020.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vafai SB, and Mootha VK (2012). Mitochondrial disorders as windows into an ancient organelle. Nature 491, 374–383. 10.1038/nature11707. [DOI] [PubMed] [Google Scholar]
- 7.Rensvold JW, Shishkova E, Sverchkov Y, Miller IJ, Cetinkaya A, Pyle A, Manicki M, Brademan DR, Alanay Y, Raiman J, et al. (2022). Defining mitochondrial protein functions through deep multiomic profiling. Nature 606, 382–388. 10.1038/s41586-022-04765-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guerra RM, and Pagliarini DJ (2023). Coenzyme Q biochemistry and biosynthesis. Trends Biochem Sci. 10.1016/j.tibs.2022.12.006. [DOI] [PMC free article] [PubMed]
- 9.Angebault C, Guichet P-O, Talmat-Amar Y, Charif M, Gerber S, Fares-Taie L, Gueguen N, Halloy F, Moore D, Amati-Bonneau P, et al. (2015). Recessive Mutations in RTN4IP1 Cause Isolated and Syndromic Optic Neuropathies. Am. J. Hum. Genet. 97, 754–760. 10.1016/j.ajhg.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thorn JM, Barton JD, Dixon NE, Ollis DL, and Edwards KJ (1995). Crystal Structure of Escherichia coli QOR Quinone Oxidoreductase Complexed with NADPH. J. Mol. Biol. 249, 785–799. 10.1006/jmbi.1995.0337. [DOI] [PubMed] [Google Scholar]