Abstract
Immune cell infiltration and tumor-related immune molecules play key roles in tumorigenesis and tumor progression. The influence of immune interactions on the molecular characteristics and prognosis of clear cell renal cell carcinoma (ccRCC) remains unclear. A machine learning algorithm was applied to the transcriptome data from The Cancer Genome Atlas database to determine the immunophenotypic and immunological characteristics of ccRCC patients. These algorithms included single-sample gene set enrichment analyses and cell type identification. Using bioinformatics techniques, we examined the prognostic potential and regulatory networks of immune-related genes (IRGs) involved in ccRCC immune interactions. Fifteen IRGs (CCL7, CHGA, CMA1, CRABP2, IFNE, ISG15, NPR3, PDIA2, PGLYRP2, PLA2G2A, SAA1, TEK, TGFA, TNFSF14, and UCN2) were identified as prognostic IRGs associated with overall survival and were used to construct a prognostic model. The area under the receiver operating characteristic curve at 1 year was 0.927; 3 years, 0.822; and 5 years, 0.717, indicating good predictive accuracy. Molecular regulatory networks were found to govern immune interactions in ccRCC. Additionally, we developed a nomogram containing the model and clinical characteristics with high prognostic potential. By systematically examining the sophisticated regulatory mechanisms, molecular characteristics, and prognostic potential of ccRCC immune interactions, we provided an important framework for understanding the molecular mechanisms of ccRCC and identifying new prognostic markers and therapeutic targets for future research.
Keywords: Clear cell renal cell carcinoma, Immune-related gene, Prognostic model, ssGSEA, CIBERSORT
1. Introduction
Renal cell carcinoma (RCC) is one of the most common cancers of the urinary system, accounting for 2 % of all malignancies in adults, of which clear cell renal cell carcinomas (ccRCCs) constitute the majority (approximately 75 %) [1]. ccRCC is a malignant parenchymal tumor with different molecular characteristics [2]. Over the years, significant advances have been made in the treatment of ccRCC. However, these improvements often fail to yield satisfactory therapeutic results [3,4]. Metastases occur in 35 % of patients at initial diagnosis, and distant metastases occur in 30 % of patients after surgery in the early stages of the disease [[5], [6], [7]]. Therefore, further research on the mechanisms by which ccRCC occurs and develops, efforts to identify potential biomarkers, and the development of new diagnostic methods are urgently needed.
The tumor microenvironment (TME) is a complex environment that includes immune cells, stromal cells, and extracellular matrix molecules that reflect the nature of the tumor and promote its immune escape, growth, and metastasis [8,9]. Regulatory T cells and tumor-associated macrophages are associated with tumor-promoting functions in the TME [10,11]. While CD8+ T cells predominate in the early stages of tumorigenicity or development, immunosuppressive immune cells predominate as the tumor progresses [12,13]. As one of the earliest and most sensitive malignancies to immunotherapy, ccRCC has been shown to be a highly immunologically infiltrating tumor in several clinical and genomic studies [[14], [15], [16], [17]]. One study found that ccRCC has the highest cytolytic activity index compared to that of other human cancers [18]. Additionally, spontaneous degeneration found in up to 1 % of ccRCC cases is primarily regarded as immune mediated [19]. However, the molecular characteristics of tumor-immune interactions remain to be fully studied to understand the cellular and molecular composition and function of the TME in ccRCC and to further evaluate its prognostic potential in ccRCC.
Molecular characterization of tumor-immune system interactions has been systematically studied in a variety of human tumors. Based on The Cancer Genome Atlas (TCGA) database, Lin et al. found that 10 immune-related genes (IRGs) have a high prognosis for colorectal cancer and can also reflect the degree of infiltration of various immune cells [20]. Another study demonstrated that an IRG-based prognostic model is superior to two commercial prognostic markers for early non-small cell lung cancer [21]. Wang et al. developed an immune-related marker containing 15 IRGs that could better predict the prognosis of renal papillary cell carcinoma patients [22]. However, in the field of ccRCC, existing studies have only described the immune properties of ccRCC using a machine learning algorithm or from the perspective of immune cell infiltration [[23], [24], [25], [26]], and the molecular characteristics and prognostic potential of ccRCC-immune interactions have not been systematically explored. Moreover, limited IRGs models are available to predict the prognosis of ccRCC patients. Therefore, it is important to develop novel IRG markers for screening new immunotherapy targets.
Using an immunophenotypic perspective in this study, we aimed to address the molecular characterization of ccRCC-immune interactions and their prognostic potential from an immunophenotypic point of view. We systematically explored the molecular characteristics and prognostic potential associated with ccRCC immune interactions. The preliminary results revealed the complex immunological processes and biological functions of these molecules and their regulatory networks. Subsequently, we developed a predictive model and nomogram based on the IRGs, both showing high potential for diagnostic accuracy and individual risk stratification, as well as higher discriminative and predictive power than those of commonly used clinical characterization methods. Further research on immunotherapy targets and the development of novel immunotherapeutic drugs will benefit from the findings of this study, which can help individualize immunotherapy strategies for ccRCC patients.
2. Materials and methods
2.1. Data collection and collation
We downloaded transcriptome data containing 72 normal renal tissue specimens and 539 ccRCC specimens from the TCGA database (https://portal.gdc.cancer.gov/). ArrayExpress was also accessed to obtain the E-MTAB-1980 dataset (https://www.ebi.ac.uk/arrayexpress/). After this, clinical data and transcriptome data were summarized and analyzed based on the following criteria: (a) duplicate samples were deleted, (b) censored data were deleted, (c) transcriptome data expressing values less than 1 were deleted, and (d) patients whose overall survival (OS) was less than 90 days were removed.
2.2. Single‐sample gene sets enrichment analysis (ssGSEA)
The ssGSEA algorithm was used to evaluate the immune characteristics of each sample based on 29 immune gene sets associated with different immune cell types, functions, pathways, and checkpoints [27] (Table S1). Data analysis is mostly conducted using Bioconductor packages such as GSVA, limma, and GSEABase (www.bioconductor.org).
2.3. Hierarchical cluster analysis
In order to perform hierarchical cluster analysis, we used Euclidean distances and Ward's links calculated by ssGSEA for each patient's immunological characteristics. According to the clustering results, ccRCC patients with high immunity and low immunity were separated.
2.4. Estimation of stromal and immune cells in tumor tissues (ESTIMATE)
ESTIMATE is a complex algorithm based on ssGSEA that evaluates the level of infiltration of tumor cells and different normal cells by analyzing the cancer cell transcription profile [15]. Based on ccRCC expression data, we estimated the stromal cells and immune cells in each sample and computed the StromalScore, ImmuneScore, and EstimateScore. This analysis is mainly conducted through the estimate package.
2.5. Evaluating tumor-infiltrating immune cells in tumor tissues
By estimating relative subsets of RNA transcripts (CIBERSORT) and its supplied LM22 gene set, the degree of infiltration of immune cells between different groups was estimated. The CIBERSORT algorithm assesses the relative abundance of immune cells infiltrating tumors in each patient based on data from 22 tumor infiltrating lymphocyte subsets. In this case, permutations were set to 1000 in this case. P < 0.05 was used as the filtering criterion.
2.6. Differential expression IRGs analysis and evaluation of the correlation between gene modules and clinical parameters
In order to identify the differentially expressed genes, we divided the transcriptome data into the Immunity High and Immunity Low groups and using the |log2 fold change (FC)| >1.0 and the false discovery rate (FDR) < 0.05, edgeR (http://www.bioconductor.org/packages/release/bioc/html/edgeR.html) package was used to identify genes that differed. The tumor-related immune gene set was downloaded from ImmPort (www.immport.org), and the differentially expressed IRGs were extracted.
A weighted correlation network analysis (WGCNA) was performed for differentially expressed IRGs using the WGCNA package to evaluate their relationship with clinical parameters. A number of clinical parameters were analyzed, including age, gender, tumor grade, tumor stage, T stage, N stage, and M stage, as well as gene expression data. By setting the soft threshold (power), we were able to determine the optimal scale-free topological fitting model index (scale-free R2). On the basis of the topological overlap measure, we calculated gene cluster dendrograms based on the different degrees of genes. The correlation between module genes and the above clinical parameters was analyzed after clustering the modules and genes. The significance level was set at P < 0.05.
2.7. Construction and assessment of IRGs-based prognostic related model
A univariate Cox regression analysis was first used to screen for prognostic related IRGs. To further screen the data, the least absolute shrinkage and selection operator (LASSO) regression analysis was performed. Lastly, we screened IRGs with significant prognostic value using multivariate Cox regression. Significant results were determined by a P value of 0.05. We then built an IRGs-based prognostic model to predict ccRCC patients' survival times. For each patient, the following formula was used to calculate the risk score:
The formula is composed of Expression Value and Regression Coefficient. Exp stands for the expression value of each gene, and β stands for the corresponding regression coefficient. On the basis of the median risk score, TCGA patients with high immunity were divided into low-risk and high-risk subgroups. In addition, Kaplan-Meier and log-rank tests were used to compare the OS and survival status between the two groups. To study the predictive power of prognostic IRGs-based risk models, we also used the Survival ROC R package to construct a time-dependent receiver operating characteristic (ROC) curve. Furthermore, the E-MTAB-1980 cohort containing 101 samples was used as an external validation to evaluate the model's predictability and stability.
2.8. Correlation between the prognostic related model, prognostic related IRGs, and clinical parameters
We then examined whether prognostic related models could be correlated with clinical parameters to determine whether they might influence ccRCC progression. As part of our analysis, we also compared prognostic related IRGs with clinical parameters in order to determine the role of immune system in ccRCC.
2.9. A multidimensional regulatory network analysis and functional enrichment analysis of prognostic related IRGs
The Cistrome Project (www.cistrome.org) provides transcription factors (TFs) associated with tumorigenesis and progression, and the TCGA cohort provides TFs associated with ccRCC. A co-expression analysis based on |Cor| > 0.3, under P = 0.01, was conducted to examine the regulatory relationships between TFs and prognostic IRGs. Following this, the clusterProfiler package was used to perform Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes database (KEGG) enrichment analyses of these differentially expressed IRGs (http://www.bioconductor.org/packages/release/bioc/html/clusterProfiler.html). These IRGs were investigated for their biological functions and molecular mechanisms by GO annotation, and their signal regulation pathways were identified by KEGG enrichment analysis.
2.10. Construction of a nomogram
The clinical prognostic significance of each clinical parameter and risk score was assessed using Cox regression analysis and multiple regression analysis. To establish a quantitative method to predict the prognosis of patients with ccRCC, we constructed a nomogram using the rms package combining different clinical parameters and risk scores. The accuracy of the nomogram is also evaluated by drawing calibration curves at different time points. In addition, the accuracy and stability of the nomogram were further evaluated by Kaplan-Meier survival analysis and ROC analysis based on data from TCGA and E-MTAB-1980.
2.11. Comparison of IRGs-based prognostic related model with other prognostic models
We compared our immune-related model with four other immune-related models in order to determine if it was superior to other ccRCC-related models. Our ROC curve and survival curve for the entire TCGA cohort were constructed by obtaining the genes in these models from the literature.
3. Results
3.1. Immunophenotypes and tumor microenvironment in ccRCC tissues
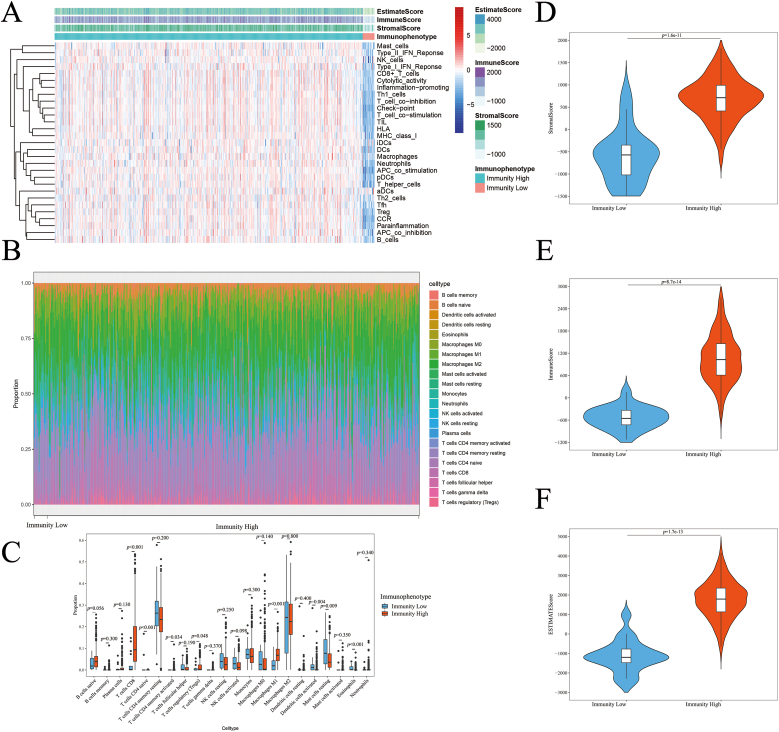
Fig. 1 illustrates the examination of IRGs in ccRCC, focusing on their molecular function and prognostic significance. A total of 72 kidney tissue specimens and 539 ccRCC specimens were downloaded from TCGA transcriptome database. Accordingly, 539 ccRCC tissues from TCGA database were analyzed for immune cell activity or enrichment, function, pathway, and checkpoints using the ssGSEA algorithm and 29 immune gene sets. Based on the hierarchical clustering results, we examined the immunity characteristics of clusters 1 (n = 519) and 2 (n = 20); the results are shown in Fig. 2. A cluster with high immunity was referred to as “Immunity High,” and a cluster with low immunity was referred to as “Immunity Low” (Fig. 2A). Each TME sample was scored, and the characteristics of the groups with Immunity High and Immunity Low were compared. The results showed that the StromalScore of the Immunity High and Immunity Low groups were 691.8 ± 21.22 and −602.9 ± 133.1, respectively. The ImmuneScore of the Immunity High and Immunity Low groups were 1066 ± 28.29 and −535.1 ± 66.03, correspondingly. The EstimateScore of the Immunity High and Immunity Low groups were 1758 ± 42.63 and −1138 ± 183.5, respectively (Fig. 2D, E, F). We used the CIBERSORT algorithm to compare the degree of immune cell infiltration between the Immunity High and Immunity Low groups (Fig. 2B). Between the two groups, the degree of infiltration of T cells CD8, CD4 naïve T cells, CD4 memory activated T cells, regulatory T cells (Tregs), macrophages M1, activated dendritic cells, resting mast cells, and eosinophils differed significantly (Fig. 2C). There was a significant difference in the infiltration of immune cells between the two groups regardless of the machine learning algorithm.
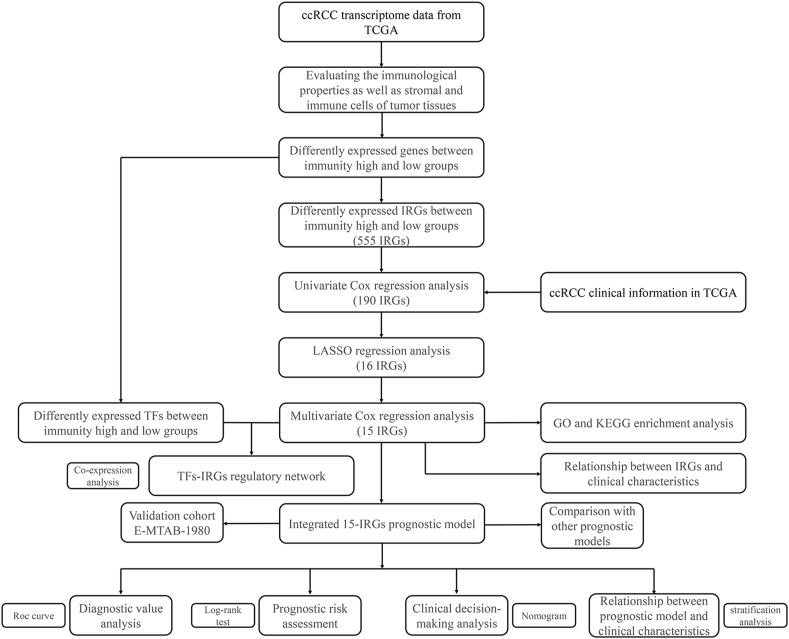
Fig. 1.
Flowchart illustrating IRG analysis in ccRCC.
Fig. 2.
Immunophenotype and tumor microenvironment of ccRCC patients.
(A) Immune characteristics and tumor microenvironment landscape in TCGA cohort determined using ssGSEA algorithm and hierarchical cluster analysis. (B) The infiltration landscape of 22 immune cells in the Immunity Low and Immunity High groups determined employing CIBERSORT algorithm. (C) Boxplot displaying the infiltration degree of 22 immune cells in the Immunity Low and Immunity High groups. (D) Violin plot depicting the StromalScore in the Immunity Low and Immunity High groups. (E) Violin plot showing the ImmuneScore in the Immunity Low and Immunity High groups. (F) Violin plot displaying the EstimateScore in the Immunity Low and Immunity High groups.
3.2. Identifying differentially expressed IRGs in ccRCC patients
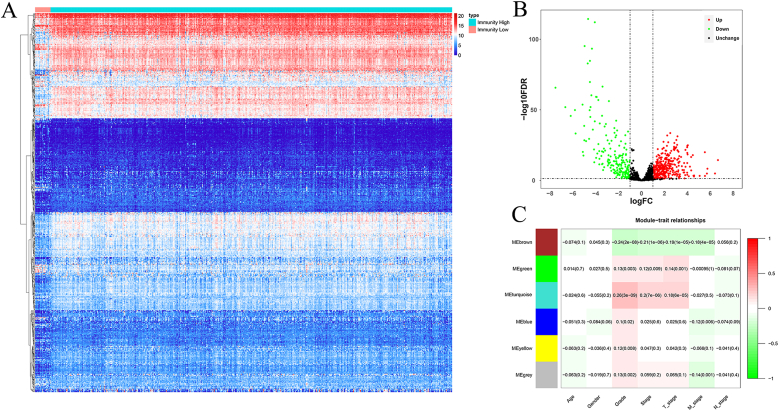
Owing to the lack of knowledge on immune-related molecular characteristics and their prognostic potential in ccRCC, we investigated the immunophenotype-specific molecular characteristics and the prognostic potential of ccRCC-immune interactions. Our first step was to obtain 2483 tumor-related immune genes from the ImmPort database. By screening for differentially expressed IRGs according to the screening criteria (|log2 FC| >1.0 and FDR<0.05), 355 upregulated IRGs and 200 downregulated IRGs were identified. As shown in Fig. 3A and B, these IRGs were expressed inconsistently.
Fig. 3.
Differential expression analysis of IRGs and evaluation of the correlation between gene modules and clinical parameters.
(A) Heatmap of the 555 differentially expressed IRGs. (B) Volcano plot of 1144 IRGs. (C) Module-trait relationships based on WGCNA analysis; red and blue indicate high expression and low expression, respectively.
3.3. Evaluation of the correlation between gene modules and clinical parameters
We utilized WGCNA to build various gene modules and evaluate the relationship between differentially expressed IRGs and clinical parameters. This analysis involved the application of WGCNA to the identified differentially expressed IRGs. Six gene modules were identified based on an optimal soft threshold. These modules were correlated with clinical parameters, including age, sex, tumor grade, tumor stage, T stage, N stage, and M stage, as shown in Fig. 3C. The results showed that all modules were associated with tumor grade (P < 0.05), with three modules showing significant correlations with tumor stage and T stage (P < 0.01); additionally, three modules displayed a notable negative correlation with M stage (P < 0.01). No significant correlations were found between age, sex, N stage, or gene modules. Consequently, further analysis of these differentially expressed IRGs is warranted because they may have implications for clinical prognosis.
3.4. Construction and evaluation of immune associated prognostic model
As a result of the univariate Cox regression analysis of these differentially expressed IRGs, we identified 190 IRGs that were associated with prognosis (Table S1). After performing LASSO regression analysis on these IRGs, 16 IRGs (CCL7, CHGA, CMA1, CRABP2, DCD, IFNE, ISG15, NPR3, PDIA2, PGLYRP2, PLA2G2A, SAA1, TEK, TGFA, TNFSF14, and UCN2) were identified as having prognostic significance (Fig. S1). These IRGs were then analyzed using multivariate Cox regression to determine the most relevant IRGs regarding prognosis, yielding 15 IRGs in total, including CCL7, CHGA, CMA1, CRABP2, IFNE, ISG15, NPR3, PDIA2, PGLYRP2, PLA2G2A, SAA1, TEK, TGFA, TNFSF14, and UCN2.
To establish a prognostic model, a multivariate Cox regression analysis was conducted on these 15 IRGs (Table 1). Each ccRCC patient was assigned a risk score based on the following formula:
| Risk score = (0.0710 × Exp CCL7) + (0.0935 × Exp CHGA) + (−0.1179 × Exp CMA1) + (0.0668 × Exp CRABP2) + (0.0999 × Exp IFNE) + (0.1679 × Exp ISG15) + (0.0095 × Exp NPR3) + (0.1025 × Exp PDIA2) + (0.0619 × Exp PGLYRP2) + (0.0078 × Exp PLA2G2A) + (0.0270 × Exp SAA1) + (−0.0207 × Exp TEK) + (−0.1503 × Exp TGFA) + (0.0949 × Exp TNFSF14) + (0.0571 × Exp UCN2) |
Table 1.
Multivariate Cox regression analysis to identify prognosis-related immune genes.
| Gene | Coef | Exp(coef) | se(coef) | z | Pr(>|z|) |
|---|---|---|---|---|---|
| CCL7 | 0.0710 | 1.0736 | 0.0588 | 1.2075 | 0.2272 |
| CHGA | 0.0935 | 1.0980 | 0.0506 | 1.8479 | 0.0646 |
| CMA1 | −0.1179 | 0.8888 | 0.0625 | −1.8863 | 0.0593 |
| CRABP2 | 0.0668 | 1.0691 | 0.0554 | 1.2062 | 0.2277 |
| IFNE | 0.0999 | 1.1051 | 0.0508 | 1.9680 | 0.0491 |
| ISG15 | 0.1679 | 1.1828 | 0.1041 | 1.6128 | 0.1068 |
| NPR3 | 0.0095 | 1.0096 | 0.0576 | 0.1651 | 0.8688 |
| PDIA2 | 0.1025 | 1.1079 | 0.0453 | 2.2615 | 0.0237 |
| PGLYRP2 | 0.0619 | 1.0639 | 0.0519 | 1.1924 | 0.2331 |
| PLA2G2A | 0.0078 | 1.0078 | 0.0371 | 0.2104 | 0.8333 |
| SAA1 | 0.0270 | 1.0273 | 0.0297 | 0.9095 | 0.3631 |
| TEK | −0.0207 | 0.9795 | 0.0848 | −0.2441 | 0.8072 |
| TGFA | −0.1503 | 0.8605 | 0.0730 | −2.0580 | 0.0396 |
| TNFSF14 | 0.0949 | 1.0996 | 0.0630 | 1.5059 | 0.1321 |
| UCN2 | 0.0571 | 1.0587 | 0.0654 | 0.8728 | 0.3828 |
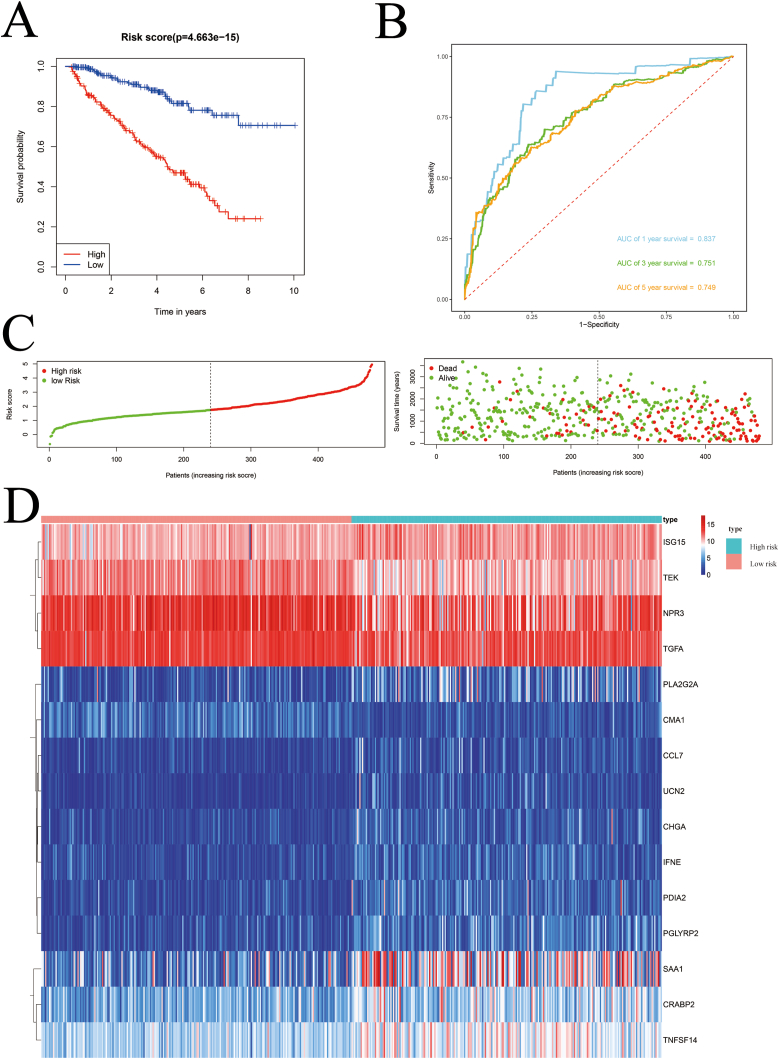
The median risk scores of 480 ccRCC patients in TCGA were used to categorize the patients into high- and low-risk subgroups.– Kaplan–Meier survival analysis showed that patients with high-risk disease had worse outcomes than those with low-risk disease (P = 4.663e-15, Fig. 4A). To further evaluate the predictive performance of the model, the areas under the ROC curves (AUCs) were calculated, which were 0.837 after 1 year, 0.751 after 3 years, and 0.749 after 5 years (Fig. 4B). Survival status and expression heat maps of TCGA cohort members are shown in Fig. 4C and D. In addition, we calculated the risk score of the E-MTAB-1980 cohort using the same formula to determine whether the model had similar predictive power in other ccRCC patient cohorts. Similarly, Kaplan–Meier survival analysis showed a worse outcome for patients with high-risk factors (P = 3.671e-04, Fig. S2A), with predicted AUCs of 0.854, 0.817, and 0.867 at 5 years (Fig. S2B). The survival status and expression heat map of each patient in the E-MTAB-1980 cohort are shown in Figs. S2C and S2D. The results indicated that the IRG-based prognostic model was stable and performed satisfactorily.
Fig. 4.
Risk score analysis on a prognostic model consisting of 15 IRGs in the TCGA cohorts.
(A) Kaplan–Meier survival curve analysis in the TCGA cohort for high-risk and low-risk groups. (B) Time-dependent ROC curves in the TCGA cohort. (C) Survival status of each patient in the TCGA cohort. (D) Expression of 15 prognostic IRGs in the TCGA cohort for high-risk and low-risk groups; red and blue indicate the high-risk and low-risk groups, respectively.
3.5. Prognostic significance of the model stratified by clinical parameters
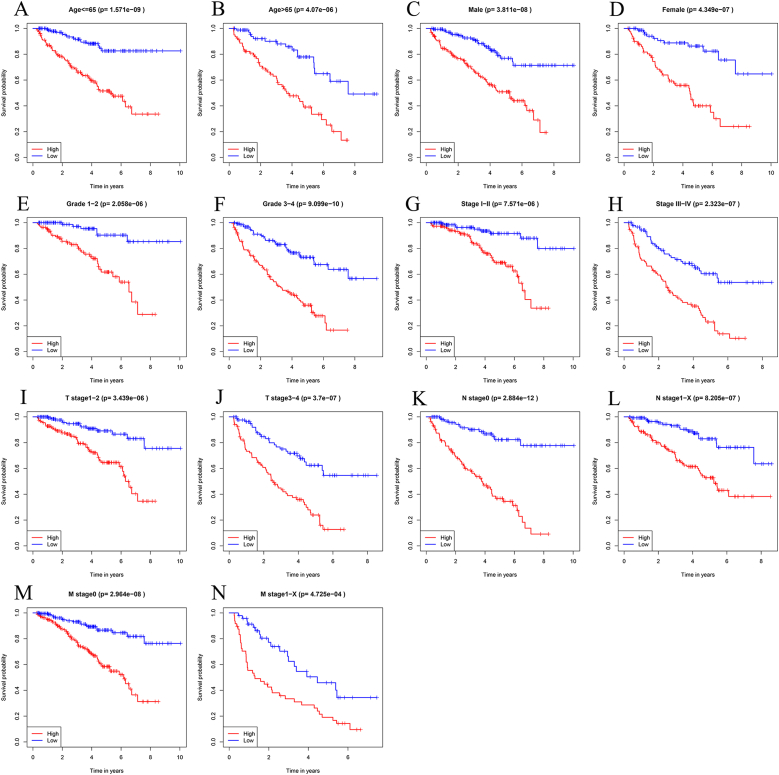
The TCGA data were stratified by age, sex, tumor grade, tumor stage, T stage, N stage, and M stage to explore the clinical significance of this model. Kaplan–Meier survival analysis showed that the OS of high-risk patients was significantly lower than that of low-risk patients (Fig. 5). Based on these results, a prediction model based on IRGs can be used to predict the prognosis of ccRCC patients regardless of clinical parameters.
Fig. 5.
Kaplan–Meier survival curve analysis stratified by various clinical parameters.
(A) Age≤65; (B) Age>65; (C) Male; (D) Female; (E) Grade1-2; (F) Grade3-4; (G) Stage Ⅰ-Ⅱ; (H) Stage Ⅲ-Ⅳ; (I) T stage1–2; (J) T stage3–4; (K) N stage0; (L) N stage1-X; (M) M stage0; and (N) M stage1-X.
3.6. Relationship between prognostic related IRGs and clinical parameters
To better understand the role of these genes in ccRCC, we examined the relationship between the 15 IRGs and clinical parameters. IFNE, NPR3, and SAA1 showed significant correlations with sex; CHGA, CMA1, IFNE, ISG15, NPR3, PDIA2, PLA2G2A, SAA1, TEK, TGFA, TNFSF14, and UCN2 were significantly associated with tumor grade; CCL7, CHGA, CMA1, CRABP2, IFNE, ISG15, NPR3, PDIA2, PGLYRP2, PLA2G2A, SAA1, TEK, TGFA, TNFSF14, and UCN2 showed significantly links to tumor stage and T stage; TEK was significantly correlated with N stage; and CCL7, CMA1, ISG15, NPR3, PDIA2, SAA1, TEK, TGFA, TNFSF14, and UCN2 were significantly associated with M stage (Table 2).
Table 2.
Relationship between prognosis-related immunity genes and clinicopathologic parameters.
| Gene | Gender (Male/Female) | Grade (G1-2/G3-4) | Stage (I-II/III-IV) | T stage (T1-T2/T3-T4) | N stage (N0/N1-X) | M stage (M0/M1-X) | |
|---|---|---|---|---|---|---|---|
| N | 318/162 | 221/257 | 292/185 | 310/170 | 213/267 | 382/96 | |
| CCL7 | t-value | 0.762 | 1.639 | 2.691 | 2.525 | 0.429 | 2.021 |
| P-value | 0.447 | 0.102 | 0.007 | 0.012 | 0.668 | 0.044 | |
| CHGA | t-value | 0.675 | 3.595 | 3.451 | 3.374 | 0.156 | 1.773 |
| P-value | 0.500 | <0.001 | <0.001 | <0.001 | 0.876 | 0.077 | |
| CMA1 | t-value | 1.409 | 4.590 | 6.814 | 5.772 | 0.602 | 5.487 |
| P-value | 0.160 | <0.001 | <0.001 | <0.001 | 0.548 | <0.001 | |
| CRABP2 | t-value | 0.868 | 1.396 | 3.088 | 3.076 | 0.785 | 0.361 |
| P-value | 0.386 | 0.163 | 0.002 | 0.002 | 0.433 | 0.718 | |
| IFNE | t-value | 2.200 | 4.068 | 3.040 | 3.204 | 1.142 | 0.483 |
| P-value | 0.028 | <0.001 | 0.003 | 0.001 | 0.254 | 0.630 | |
| ISG15 | t-value | 1.798 | 3.798 | 4.804 | 3.621 | 1.252 | 3.732 |
| P-value | 0.073 | <0.001 | <0.001 | <0.001 | 0.211 | <0.001 | |
| NPR3 | t-value | 2.240 | 4.513 | 3.996 | 3.713 | 0.369 | 3.714 |
| P-value | 0.026 | <0.001 | <0.001 | <0.001 | 0.713 | <0.001 | |
| PDIA2 | t-value | 0.549 | 3.560 | 5.851 | 4.853 | 1.238 | 4.614 |
| P-value | 0.584 | <0.001 | <0.001 | <0.001 | 0.216 | <0.001 | |
| PGLYRP2 | t-value | 0.281 | 1.465 | 4.133 | 3.278 | 0.744 | 1.501 |
| P-value | 0.779 | 0.144 | <0.001 | 0.001 | 0.457 | 0.134 | |
| PLA2G2A | t-value | 0.615 | 4.946 | 5.338 | 6.165 | 0.778 | 1.104 |
| P-value | 0.539 | <0.001 | <0.001 | <0.001 | 0.437 | 0.270 | |
| SAA1 | t-value | 3.231 | 6.930 | 7.382 | 6.598 | 0.454 | 4.251 |
| P-value | 0.001 | <0.001 | <0.001 | <0.001 | 0.650 | <0.001 | |
| TEK | t-value | 1.054 | 6.582 | 5.976 | 4.897 | 2.068 | 5.338 |
| P-value | 0.292 | <0.001 | <0.001 | <0.001 | 0.039 | <0.001 | |
| TGFA | t-value | 1.761 | 3.349 | 3.547 | 2.953 | 1.217 | 5.708 |
| P-value | 0.079 | <0.001 | <0.001 | 0.003 | 0.224 | <0.001 | |
| TNFSF14 | t-value | 1.523 | 6.451 | 4.582 | 3.850 | 0.920 | 2.008 |
| P-value | 0.128 | <0.001 | <0.001 | <0.001 | 0.358 | 0.045 | |
| UCN2 | t-value | 0.666 | 3.796 | 4.727 | 4.528 | 0.042 | 2.060 |
| P-value | 0.506 | <0.001 | <0.001 | <0.001 | 0.966 | 0.040 |
3.7. Multi-dimensional regulatory network and functional enrichment analysis of prognosis-related IRGs
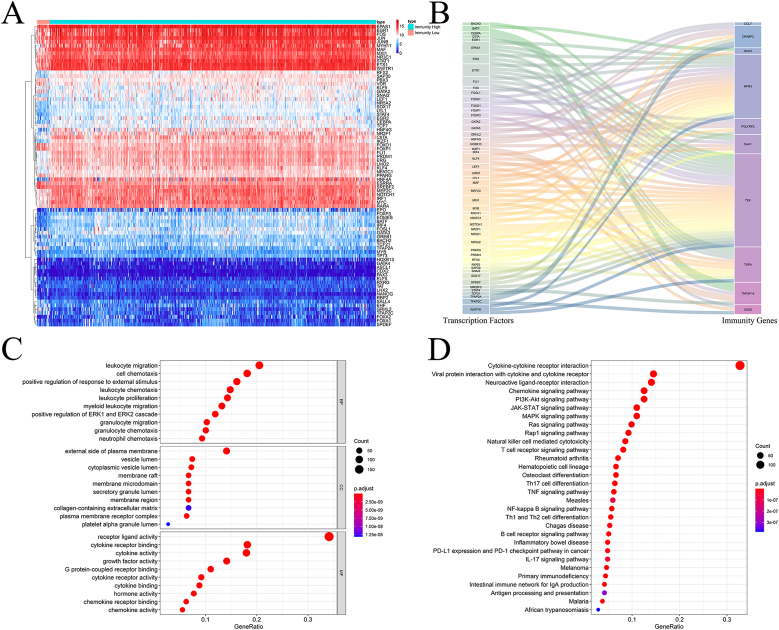
Using the prognostic model, we studied the upstream mechanism of IRGs to further explore the role of their multidimensional regulatory network in tumorigenesis and development. After differential analysis of 318 transcriptional factors (TFs) from the Cistrome Project, 79 TFs were identified, including 45 upregulated and 34 downregulated TFs (Fig. 6A). TF and IRG co-expression analyses were conducted to identify the regulatory relationships between differentially expressed TFs and prognostic indicators. A total of 50 TFs were involved in upstream regulation, as shown in Fig. 6B. The regulatory relationships between these TFs and the prognosis-related IRGs are presented in Table S2.
Fig. 6.
Multi-dimensional regulatory network and functional enrichment analysis of prognosis-related IRGs.
(A) Heatmap of the 79 differentially expressed TFs in the TCGA cohort. (B) Sankey plot of differentially expressed TFs and prognostic IRG regulatory networks. (C) GO enrichment analysis of the differentially expressed IRGs. (D) KEGG enrichment analysis of the differentially expressed IRGs.
Next, we performed GO and KEGG enrichment analyses of the differentially expressed IRGs using the ClusterProfiler package to explore the biological functions and molecular mechanisms associated with these IRGs. According to GO enrichment analysis, the molecular regulatory network can be viewed from three different perspectives. Biological process analysis revealed that these differentially expressed IRGs were significantly enriched in leukocyte migration, cell chemotaxis, positive regulation of external stimuli, leukocyte chemotaxis, leukocyte proliferation, myeloid leukocyte migration, ERK1 and ERK2 cascade activation, granulocyte migration, granulocyte chemotaxis, and neutrophil chemotaxis (Fig. 6C). These differentially expressed IRGs were greatly enriched on the external side of the plasma membrane, lumen of vesicles, membrane rafts, membrane microdomains, secretory granule lumen, membrane region, collagen-containing extracellular matrix, plasma membrane receptor complex, and alpha granule lumen of the platelets (Fig. 6C). These differentially expressed IRGs had significantly higher receptor ligand, cytokine receptor binding, cytokine, growth factor, G protein-coupled receptor binding, cytokine receptor, cytokine binding, hormone binding, chemokine receptor binding, and chemokine activities (Fig. 6C). Furthermore, KEGG analysis indicated that the differentially expressed IRGs were mainly enriched in interactions between cytokines and cytokine receptors, viral proteins and cytokine receptors, chemokine signaling pathways, JAK-STAT signaling pathways, PI3K-Akt signaling pathways, MAPK signaling pathways, Ras signaling pathways, and Rap1 signaling pathways.
We also conducted a joint analysis to evaluate the genes involved to predict the efficacy of immunotherapies (Fig. S3). Therefore, we further compared the expression of common immune checkpoint inhibitor genes (PD-1, PD-L1, and CTL4) in different patient populations by high and low risk subgroups of the IRG-based prognosis-related model to further investigate the role of our predictive model in immunotherapy. The expression of PD-1 and CTLA-4 was significantly higher in the high-risk group than in the low-risk group, however, the expression of PD-L1 showed the opposite result (Figs. S3A–C). Subsequently, we conducted a stratified survival analysis based on the risk scores from our predictive model and the expression of immune checkpoint inhibitor genes. The OS of patients in the low-risk and high-immune checkpoint genomics category was significantly superior to that of their counterparts in the high-risk and high-immune checkpoint genomics group. Furthermore, the prognosis for patients with low-risk and low-immune checkpoint genomics profiles was more favorable compared to those with high-risk and low-immune checkpoint genomics profiles (Figs. S3D–F).
3.8. Construction and validation of a nomogram
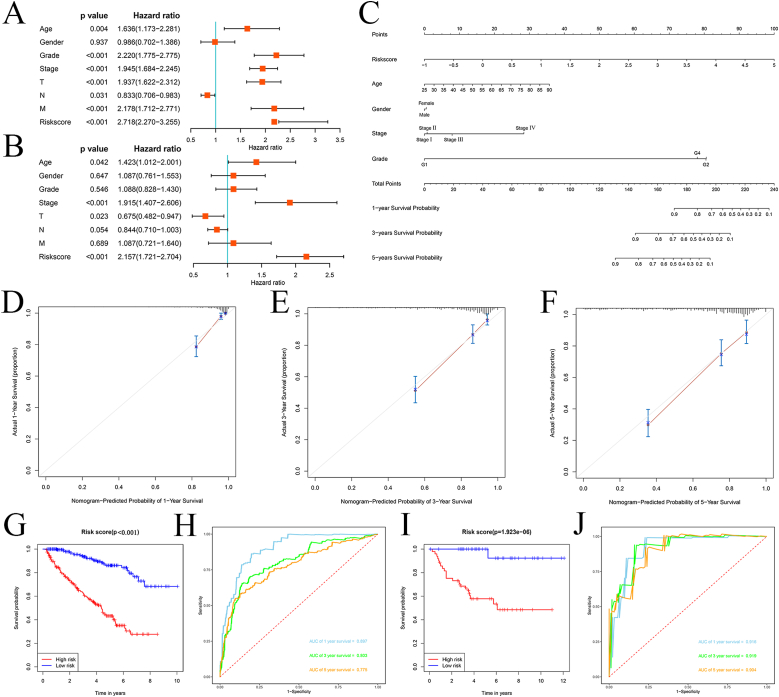
To assess the prognostic significance of different clinical parameters and risk scores in ccRCC patients, we first performed a Cox regression analysis. Age (P = 0.004), primary tumor location (P < 0.001), tumor grade (P < 0.001), tumor stage (P < 0.001), lymph node infiltration (P = 0.031), distant metastasis (P < 0.001), and risk score (P < 0.001) of ccRCC patients were significantly correlated with OS (Fig. 7A). However, multiple regression analysis indicated that age (P = 0.042), tumor stage (P < 0.001), primary tumor location (P = 0.023), and risk score (P < 0.001) were independent prognostic factors associated with OS (Fig. 7B).
Fig. 7.
Assessment of the prognostic significance of various clinical parameters, and construction of a nomogram in the TCGA cohort.
(A) Univariate Cox regression analysis of correlations between risk score and clinical parameters. (B) Multivariate Cox regression analysis of correlations between risk score and clinical parameters. (C) Nomogram for predicting the 1-, 3-, and 5-year OS of ccRCC patients. (D, E, F) Calibration curves of the nomogram predicting OS at 1-, 3-, and 5-years. (G) Kaplan–Meier survival curve analysis in the TCGA cohort based on the nomogram. (H) Time dependent ROC curve analysis in the TCGA cohort based on the nomogram. (I) Kaplan–Meier survival curve analysis in the E-MTAB-1980 cohort based on the nomogram. (J) Time dependent ROC curve analysis in the E-MTAB-1980 cohort based on the nomogram.
After combining the clinical parameters and risk scores, a nomogram was constructed using the RMS package (Fig. 7C). A 0–100 distribution was normalized by mapping the points of each variable to the corresponding horizontal line. The survival rates of ccRCC patients were calculated by drawing vertical lines between the points and total points, which served as a reference for clinical decision-making. The calibration curve also showed a good correlation between the predicted and actual values (Fig. 7D, E, and F). To extend its clinical application and availability, the nomogram was verified in TCGA and E-MTAB-1980 cohorts. In both cohorts, the nomogram was able to better distinguish between patients with poor prognosis (P < 0.001 and P = 1.923e-06, Fig. 7G–I). Based on the nomogram, the predicted AUCs for 1, 3, and 5 years were 0.897, 0.803, and 0.775, respectively, in the TCGA cohort (Fig. 7H), whereas the predicted AUCs in the E-MTAB-1980 cohort were 0.916, 0.919, and 0.904, respectively, indicating that the nomogram was very accurate and predictive.
3.9. Comparison of IRGs-based prognosis-related model with other prognostic models
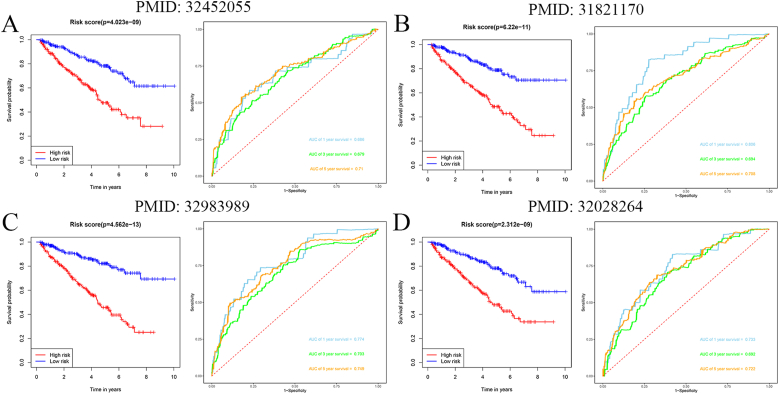
Using four different types of models, we compared our immune-related model to four-gene models [23], seven-gene models [26], six-gene models [25], and five-gene models [24]. For these models, we obtained genes from the literature and constructed the ROC and survival curves for the entire TCGA cohort. Our model was more accurate in predicting the prognosis of ccRCC than the other four models after analysis and comparison (Fig. 8).
Fig. 8.
Comparison of IRG-based prognosis-related model with other prognostic models.
(A) PMID: 32452055; (B) PMID: 31821170; (C) PMID: 32983989; and (D) PMID: 32028264.
4. Discussion
An ecosystem of adaptive and innate immune cells infiltrates the TME and regulates all aspects of tumor development [28,29]. By upregulating or downregulating IRGs at specific immune checkpoints, tumor-infiltrating immune cells can suppress cancer cells [18,30]. Numerous clinical studies have shown that ccRCC is a highly immunologically infiltrating tumor [15,31,32]. Furthermore, ccRCC exhibits early and sensitive responsiveness to immunotherapy [33,34]. Therefore, identifying objective and sensitive predictors of ccRCC is crucial for optimizing clinical diagnosis and accurately determining prognosis. Our analysis of TCGA transcriptome data for ccRCC revealed 555 differentially expressed IRGs between the Immunity High and Immunity Low groups. An immune-associated prognostic model for OS was constructed by performing Cox regression analysis and by identifying 15 prognosis-related IRGs. The roles of these prognostic IRGs in ccRCC were examined in relation to prognostic models and clinical parameters. In addition, we examined the regulatory relationships between prognostic IRGs and TFs. Bioinformatics techniques were used to systematically analyze the biological functions and molecular mechanisms of these IRGs. A nomogram combining the model and the clinical parameters was constructed and validated. Based on these results, our clinical prognostic model can predict patient prognosis and identify potential immunotherapeutic targets.
Using ssGSEA, we identified two immunophenotypic clusters: Immunity Low and Immunity High. Compared to that of patients with low immune responses, those with a high immune response had a higher percentage of stromal and immune cells infiltrating the TME. This finding is consistent with those of previous studies indicating that patients with strong immune responses have a higher number of immune and stromal cells [27]. Evidence suggests that immune cells, the extracellular matrix, growth factors, and tumor cells themselves play important roles in influencing the immune escape, immune-inflammatory interactions of tumor cells, and participating in tumor development processes [35,36]. Multivariate Cox regression, LASSO regression, and univariate Cox regression analyses were conducted on the differentially expressed IRGs. As a result, 15 prognosis-related IRGs were identified, including CCL7, CHGA, CMA1, CRABP2, IFNE, ISG15, NPR3, PDIA2, PGLYRP2, PLA2G2A, SAA1, TEK, TGFA, TNFSF14, and UCN2. Various tumors progress and metastasize via CCL7, a member of the chemokine ligand subfamily [37]. The protein CHGA is soluble in secretory vesicles of neuroendocrine, nerve, and endocrine cells and is considered a marker for neuroendocrine prostate cancer [38]. Gastric cancer is associated with immune infiltration caused by CMA1, which is expressed in mast cells [39]. CRABP2 is a protein that binds to retinoic acid and is associated with lymph node metastasis, poor overall survival, and recurrence in lung cancer patients [40]. We speculated that IFNE is a susceptibility gene for melanoma [41]. Tumorigenesis is enhanced by ubiquitin-like proteins such as ISG15, which are induced by type I interferon. It regulates the expression of oncogenes and tumor suppressors and prevents the degradation of oncogenes and tumor-related proteins by inhibiting proteasomes and lysosomes [42]. ccRCC metastasis is associated with high expression of NPR3, a natriuretic peptide receptor [43]. There is also an association between gallbladder cancer and PDIA2 expression [44]. PGLYRP2, a peptidoglycan-sensitive pattern recognition receptor, has been implicated in predicting the antitumor immune response and survival in hepatocellular carcinoma [45]. Inflammatory responses, antibacterial defense, and gastrointestinal phospholipid degradation are the main functions of PLA2G2A, an important regulator of gastric cancer invasion and metastasis [46]. A significant increase in SAA1 levels is associated with injury, inflammation, and nasopharyngeal cancer and has antiangiogenic and tumor-inhibiting effects [47]. High serum levels of soluble TEK have been identified as an independent risk factor for bladder cancer mediated by vascular endothelial cells [48]. Lung adenocarcinoma patients with high TGFA expression have a low survival rate [49]. Conversely, in patients with colorectal cancer liver metastases, overexpression of TNFSF14 leads to improved survival rates and recurrence-free survival, attributed to the immune-stimulating properties of this cytokine [50]. UCN2, a ligand of the corticotropin-releasing factor receptor, acts as an inhibitor of tumor growth by suppressing vascularization and cell proliferation [51]. These findings suggest that these 12 IRGs play a role in the occurrence and progression of ccRCC. However, many molecular mechanisms remain unknown, underscoring the need for future research to shed light on these areas. A prognostic model based on IRGs was developed, and survival analysis showed that patients in the high-risk group had worse OS than those in the low-risk group. Using this prognostic model, ccRCC patients with poor prognosis can be better identified.
Many TFs are involved in regulating the immune response, thus affecting its intensity and occurrence of the immune response. In their study [52], Zaiss et al. examined the central role of forkhead box TFs in homeostasis regulation and immune response. STAT1 expression is increased by elF4F, as demonstrated by Kingwell et al. [53]. Hence, it is noteworthy to highlight that prognosis-related IRGs and TFs form regulatory networks. Upstream regulation of prognosis-related IRGs was mediated by 50 differentially expressed TFs. Research on these TFs is warranted because they may trigger an immune response or have extensive regulatory potential in the presence of cancer.
GO and KEGG enrichment analyses indicated that these differentially expressed IRGs were associated with cell chemotaxis, receptor ligand activity, JAK-STAT signaling pathways, T cell receptor signaling pathways, PI3K-Akt signaling pathways, MAPK signaling pathways, Ras signaling pathways, and other immune response-related processes. Immune surveillance mechanisms that eliminate transformed cells are crucial to control and prevent tumor growth. Changes in the tumor microenvironment may lead to changes in the immune response, allowing transformed cells to survive undetected by immune cells and promote tumor growth [36,54,55]. Various strategies have been employed by tumors to avoid immune recognition, including downregulation of MHC class I expression on the cell surface, reduction in the ability of cytotoxic T lymphocytes to recognize tumors, and alteration of the expression of key molecules during antigen processing [56,57]. Therefore, it may be possible to limit tumor growth, invasion, and metastasis by regulating the immune response.
A nomogram comprising the model and common clinical characteristics was developed to expand the clinical applications and availability of IRG-based prognostic models. By comparing the calibration curves at different times and verifying the two cohorts, it can be concluded that this nomogram exhibited good discriminant ability in predicting survival times. Additionally, we compared our model with other relevant models and found that it was based on an immunophenotypic perspective, which matched better with the correlation between immunity and tumor growth. Furthermore, our model was more accurate in predicting ccRCC patient survival than the previous models.
In conclusion, this study provides new insights into the immunophenotypic basis of ccRCC occurrence and progression. We developed a prognostic model that can better predict survival probabilities for ccRCC patients and may serve as a biomarker for prognosis. However, this study had certain limitations. First, the results were based on a single bionomic dataset, and different platforms may yield different results. A large multicenter prospective cohort of ccRCC patients is required for further validation and evaluation of our model. Finally, we analyzed several key IRGs in ccRCC patients using bioinformatics techniques. Nonetheless, the specific molecular mechanisms by which these key IRGs are involved in ccRCC progression and development remain unclear, and further research is required to confirm these findings.
Bioinformatics techniques were used to systematically study the biological function of differentially expressed IRGs and their prognostic value in ccRCC from an immunophenotypic perspective. Moreover, we developed a model to predict the prognosis of cancer patients based on immune-associated prognostic factors. These results may provide new prognostic biomarkers and therapeutic targets for ccRCC and shed light on the mechanisms underlying this disease.
Funding
This work was supported by Science, Technology and Innovation Commission of Shenzhen Municipality (No. JCYJ20210324141404010) and Medical Youth Top Talent Program of Hubei Province (No. 2020LJRC009).
Data availability statement
The data were downloaded from TCGA database (https://portal.gdc.cancer.gov/) and ArrayExpress database (https://www.ebi.ac.uk/arrayexpress/). All the data and materials can be obtained by contacting the corresponding author.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
CRediT authorship contribution statement
Chengwei Wang: Writing – original draft, Data curation, Conceptualization. Xi Zhang: Formal analysis, Data curation. Shiqing Zhu: Formal analysis. Bintao Hu: Formal analysis. Zhiyao Deng: Formal analysis. Huan Feng: Formal analysis. Bo Liu: Formal analysis. Yang Luan: Writing – review & editing. Zhuo Liu: Writing – review & editing. Shaogang Wang: Writing – review & editing, Supervision. Jihong Liu: Writing – review & editing, Supervision. Tao Wang: Writing – review & editing, Funding acquisition. Yue Wu: Writing – review & editing, Project administration.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Not applicable.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e36156.
Contributor Information
Tao Wang, Email: twang@tjh.tjmu.edu.cn.
Yue Wu, Email: yuewutjm@hust.edu.cn.
Abbreviations
- RCC
Renal cell carcinoma
- CcRCC
Clear cell renal cell carcinoma
- TCGA
The Cancer Genome Atlas
- TME
Tumor microenvironment
- IRGs
Immune‐related genes
- SsGSEA
Single‐sample gene sets enrichment analysis
- ESTIMATE
Estimation of stromal and immune cells in tumor tissues
- CIBERSORT
Cell type identification by estimating relative subsets of RNA transcripts
- OS
Overall survival
- FC
Fold change
- FDR
False discovery rate
- WGCNA
Weighted correlation network analysis
- LASSO
Least absolute shrinkage and selection operator
- ROC:
Receiver operating characteristic
- TFs
Transcription factors
- GO
Gene ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- AUC
Area under the receiver operating characteristic curve
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Siegel R.L., Giaquinto A.N., Jemal A. Cancer statistics, 2024. CA Cancer J Clin. 2024;74(1):12–49. doi: 10.3322/caac.21820. [DOI] [PubMed] [Google Scholar]
- 2.Rouprêt M., et al. European association of urology guidelines on upper urinary tract urothelial carcinoma: 2023 update. Eur. Urol. 2023;84(1):49–64. doi: 10.1016/j.eururo.2023.03.013. [DOI] [PubMed] [Google Scholar]
- 3.Shi S.P., et al. A two-DNA methylation signature to improve prognosis prediction of clear cell renal cell carcinoma. Yonsei Med. J. 2019;60(11):1013–1020. doi: 10.3349/ymj.2019.60.11.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bi K., et al. Tumor and immune reprogramming during immunotherapy in advanced renal cell carcinoma. Cancer Cell. 2021;39(5):649–661.e5. doi: 10.1016/j.ccell.2021.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mehdi A., Riazalhosseini Y. Epigenome aberrations: emerging driving factors of the clear cell renal cell carcinoma. Int. J. Mol. Sci. 2017;18(8) doi: 10.3390/ijms18081774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen H.T., McGovern F.J. Renal-cell carcinoma. N. Engl. J. Med. 2005;353(23):2477–2490. doi: 10.1056/NEJMra043172. [DOI] [PubMed] [Google Scholar]
- 7.Jonasch E., Walker C.L., Rathmell W.K. Clear cell renal cell carcinoma ontogeny and mechanisms of lethality. Nat. Rev. Nephrol. 2021;17(4):245–261. doi: 10.1038/s41581-020-00359-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang X., et al. Role of the tumor microenvironment in PD-L1/PD-1-mediated tumor immune escape. Mol. Cancer. 2019;18(1):10. doi: 10.1186/s12943-018-0928-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denk D., Greten F.R. Inflammation: the incubator of the tumor microenvironment. Trends Cancer. 2022;8(11):901–914. doi: 10.1016/j.trecan.2022.07.002. [DOI] [PubMed] [Google Scholar]
- 10.Tay C., Tanaka A., Sakaguchi S. Tumor-infiltrating regulatory T cells as targets of cancer immunotherapy. Cancer Cell. 2023;41(3):450–465. doi: 10.1016/j.ccell.2023.02.014. [DOI] [PubMed] [Google Scholar]
- 11.Kumari N., Choi S.H. Tumor-associated macrophages in cancer: recent advancements in cancer nanoimmunotherapies. J. Exp. Clin. Cancer Res. 2022;41(1):68. doi: 10.1186/s13046-022-02272-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Z.L., et al. Single cell deciphering of progression trajectories of the tumor ecosystem in head and neck cancer. Nat. Commun. 2024;15(1):2595. doi: 10.1038/s41467-024-46912-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li K., et al. Myeloid-derived suppressor cells as immunosuppressive regulators and therapeutic targets in cancer. Signal Transduct Target Ther. 2021;6(1):362. doi: 10.1038/s41392-021-00670-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herbst R.S., et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515(7528):563–567. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshihara K., et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gebrael G., et al. Update on combined immunotherapy for the treatment of advanced renal cell carcinoma. Hum Vaccin Immunother. 2023;19(1) doi: 10.1080/21645515.2023.2193528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bukavina L., et al. Epidemiology of renal cell carcinoma: 2022 update. Eur. Urol. 2022;82(5):529–542. doi: 10.1016/j.eururo.2022.08.019. [DOI] [PubMed] [Google Scholar]
- 18.Rooney M.S., et al. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160(1–2):48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Janiszewska A.D., Poletajew S., Wasiutynski A. Spontaneous regression of renal cell carcinoma. Contemp. Oncol. 2013;17(2):123–127. doi: 10.5114/wo.2013.34613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin K., et al. Development of a prognostic index and screening of potential biomarkers based on immunogenomic landscape analysis of colorectal cancer. Aging (Albany NY) 2020;12(7):5832–5857. doi: 10.18632/aging.102979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li B., et al. Development and validation of an individualized immune prognostic signature in early-stage nonsquamous non-small cell lung cancer. JAMA Oncol. 2017;3(11):1529–1537. doi: 10.1001/jamaoncol.2017.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Z., et al. Construction of immune-related risk signature for renal papillary cell carcinoma. Cancer Med. 2019;8(1):289–304. doi: 10.1002/cam4.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao X., Yang J., Chen Y. Identification of a four immune-related genes signature based on an immunogenomic landscape analysis of clear cell renal cell carcinoma. J. Cell. Physiol. 2020;235(12):9834–9850. doi: 10.1002/jcp.29796. [DOI] [PubMed] [Google Scholar]
- 24.Hua X., et al. Identification of an immune-related risk signature for predicting prognosis in clear cell renal cell carcinoma. Aging (Albany NY) 2020;12(3):2302–2332. doi: 10.18632/aging.102746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ren S., et al. Development and validation of a clinical prognostic model based on immune-related genes expressed in clear cell renal cell carcinoma. Front. Oncol. 2020;10:1496. doi: 10.3389/fonc.2020.01496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wan B., et al. Prognostic value of immune-related genes in clear cell renal cell carcinoma. Aging (Albany NY) 2019;11(23):11474–11489. doi: 10.18632/aging.102548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He Y., et al. Classification of triple-negative breast cancers based on Immunogenomic profiling. J. Exp. Clin. Cancer Res. 2018;37(1):327. doi: 10.1186/s13046-018-1002-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grivennikov S.I., Greten F.R., Karin M. Immunity, inflammation, and cancer. Cell. 2010;140(6):883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greten F.R., Grivennikov S.I. Inflammation and cancer: triggers, mechanisms, and consequences. Immunity. 2019;51(1):27–41. doi: 10.1016/j.immuni.2019.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sayour E.J., Mitchell D.A. Manipulation of innate and adaptive immunity through cancer vaccines. Journal of Immunology Research. 2017:2017. doi: 10.1155/2017/3145742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson R.H., et al. PD-1 is expressed by tumor-infiltrating immune cells and is associated with poor outcome for patients with renal cell carcinoma. Clin. Cancer Res. 2007;13(6):1757–1761. doi: 10.1158/1078-0432.CCR-06-2599. [DOI] [PubMed] [Google Scholar]
- 32.Fiorentino V., et al. First-line ICIs in renal cell carcinoma. Hum Vaccin Immunother. 2023;19(2) doi: 10.1080/21645515.2023.2225386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Atkins M.B., Regan M., McDermott D. Update on the role of interleukin 2 and other cytokines in the treatment of patients with stage IV renal carcinoma. Clin. Cancer Res. 2004;10(18 Pt 2):6342S. doi: 10.1158/1078-0432.CCR-040029. 6S. [DOI] [PubMed] [Google Scholar]
- 34.Krishna C., et al. Single-cell sequencing links multiregional immune landscapes and tissue-resident T cells in ccRCC to tumor topology and therapy efficacy. Cancer Cell. 2021;39(5):662–677.e6. doi: 10.1016/j.ccell.2021.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turley S.J., Cremasco V., Astarita J.L. Immunological hallmarks of stromal cells in the tumour microenvironment. Nat. Rev. Immunol. 2015;15(11):669–682. doi: 10.1038/nri3902. [DOI] [PubMed] [Google Scholar]
- 36.Lei X., et al. Immune cells within the tumor microenvironment: biological functions and roles in cancer immunotherapy. Cancer Lett. 2020;470:126–133. doi: 10.1016/j.canlet.2019.11.009. [DOI] [PubMed] [Google Scholar]
- 37.Lee Y.S., Cho Y.B. CCL7 signaling in the tumor microenvironment. Adv. Exp. Med. Biol. 2020;1231:33–43. doi: 10.1007/978-3-030-36667-4_4. [DOI] [PubMed] [Google Scholar]
- 38.Li Y., et al. SRRM4 gene expression correlates with neuroendocrine prostate cancer. Prostate. 2019;79(1):96–104. doi: 10.1002/pros.23715. [DOI] [PubMed] [Google Scholar]
- 39.Shi S.P., et al. CMA1 is potent prognostic marker and associates with immune infiltration in gastric cancer. Autoimmunity. 2020;53(4):210–217. doi: 10.1080/08916934.2020.1735371. [DOI] [PubMed] [Google Scholar]
- 40.Wu J.I., et al. Crabp2 promotes metastasis of lung cancer cells via HuR and integrin beta 1/FAK/ERK signaling. Sci. Rep. 2019;9 doi: 10.1038/s41598-018-37443-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fang S., et al. Functional annotation of melanoma risk loci identifies novel susceptibility genes. Carcinogenesis. 2020;41(4):452–457. doi: 10.1093/carcin/bgz173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burks J., et al. ISG15 pathway knockdown reverses pancreatic cancer cell transformation and decreases murine pancreatic tumor growth via downregulation of PDL-1 expression. Cancer Immunol. Immunother. 2019;68(12):2029–2039. doi: 10.1007/s00262-019-02422-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li J.K., et al. Long noncoding RNA MRCCAT1 promotes metastasis of clear cell renal cell carcinoma via inhibiting NPR3 and activating p38-MAPK signaling. Mol. Cancer. 2017;16(1):111. doi: 10.1186/s12943-017-0681-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zou Q., et al. Clinicopathological features and CCT2 and PDIA2 expression in gallbladder squamous/adenosquamous carcinoma and gallbladder adenocarcinoma. World J. Surg. Oncol. 2013;11:143. doi: 10.1186/1477-7819-11-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang Z., et al. Tumor-Derived peptidoglycan recognition protein 2 predicts survival and antitumor immune responses in hepatocellular carcinoma. Hepatology. 2020;71(5):1626–1642. doi: 10.1002/hep.30924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ganesan K., et al. Inhibition of gastric cancer invasion and metastasis by PLA2G2A, a novel beta-catenin/TCF target gene. Cancer Res. 2008;68(11):4277–4286. doi: 10.1158/0008-5472.CAN-07-6517. [DOI] [PubMed] [Google Scholar]
- 47.Lung H.L., et al. SAA1 polymorphisms are associated with variation in antiangiogenic and tumor-suppressive activities in nasopharyngeal carcinoma. Oncogene. 2015;34(7):878–889. doi: 10.1038/onc.2014.12. [DOI] [PubMed] [Google Scholar]
- 48.Szarvas T., et al. Serum levels of angiogenic factors and their prognostic relevance in bladder cancer. Pathol. Oncol. Res. 2009;15(2):193–201. doi: 10.1007/s12253-008-9107-z. [DOI] [PubMed] [Google Scholar]
- 49.Wu H.J., et al. MiR-374a suppresses lung adenocarcinoma cell proliferation and invasion by targeting TGFA gene expression. Carcinogenesis. 2016;37(6):567–575. doi: 10.1093/carcin/bgw038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maker A.V., et al. Use of T-cell proliferation to predict survival and recurrence in patients with resected colorectal liver metastases. J. Clin. Oncol. 2010;28(15) [Google Scholar]
- 51.Hao Z., et al. Urocortin2 inhibits tumor growth via effects on vascularization and cell proliferation. Proc Natl Acad Sci U S A. 2008;105(10):3939–3944. doi: 10.1073/pnas.0712366105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zaiss D.M.W., Coffer P.J. Forkhead box transcription factors as context-dependent regulators of lymphocyte homeostasis. Nat. Rev. Immunol. 2018;18(11):703–715. doi: 10.1038/s41577-018-0048-9. [DOI] [PubMed] [Google Scholar]
- 53.Kingwell K. Anticancer drugs: translational target for checkpoint inhibitors. Nat. Rev. Drug Discov. 2018;17(12):863. doi: 10.1038/nrd.2018.204. [DOI] [PubMed] [Google Scholar]
- 54.Vesely M.D., Schreiber R.D. Cancer immunoediting: antigens, mechanisms, and implications to cancer immunotherapy. Ann. N. Y. Acad. Sci. 2013;1284(1):1–5. doi: 10.1111/nyas.12105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pitt J.M., et al. Targeting the tumor microenvironment: removing obstruction to anticancer immune responses and immunotherapy. Ann. Oncol. 2016;27(8):1482–1492. doi: 10.1093/annonc/mdw168. [DOI] [PubMed] [Google Scholar]
- 56.Reeves E., James E. Antigen processing and immune regulation in the response to tumours. Immunology. 2017;150(1):16–24. doi: 10.1111/imm.12675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kotsias F., Cebrian I., Alloatti A. Antigen processing and presentation. Int Rev Cell Mol Biol. 2019;348:69–121. doi: 10.1016/bs.ircmb.2019.07.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data were downloaded from TCGA database (https://portal.gdc.cancer.gov/) and ArrayExpress database (https://www.ebi.ac.uk/arrayexpress/). All the data and materials can be obtained by contacting the corresponding author.