Abstract
Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL), also known as APO2L, has emerged as a highly potential anticancer agent because of its capacity to effectively trigger apoptosis in tumor cells by specifically binding to either of its death receptors (DR4 or DR5) while having no adverse effects on normal cells. Nevertheless, its practical use has been hindered by its inefficient pharmacokinetics characteristics, the challenges involved in its administration and delivery to targeted cells, and the resistance exhibited by most cancer cells towards TRAIL. Gene therapy, as a promising approach would be able to potentially circumvent TRAIL-based cancer therapy challenges mainly through localized TRAIL expression and generating a bystander impact. Among different strategies, using nanoparticles in TRAIL gene delivery allows for precise targeting, and overcoming TRAIL resistance by combination therapy. In this review, we go over potential mechanisms by which cancer cells achieve resistance to TRAIL and provide an overview of different carriers for delivering of the TRAIL gene to resistant cancer cells, focusing on different types of nanoparticles utilized in this context. We will also explore the challenges, and investigate future perspectives of this nanomedicine approach for cancer therapy.
Keywords: TRAIL, Gene delivery, Nanoparticles, Cancer cell resistance
1. Introduction: TRAIL signaling and therapeutic challenges
Cancer is a major cause of mortality worldwide; undoubtedly, it is desperately in need of more efficient and targeted treatments. In this regard, causing tumor cells to undergo apoptosis, or programmed cell death while protecting healthy cells, is one of the more promising approaches for treating cancer [1,2]. Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) therapy has demonstrated efficacy in several cancers, including colorectal cancer [3], triple-negative breast cancer [4], prostate cancer [5], and non-small cell lung cancer (NSCLC) [6]. Among all ligands triggering apoptosis, TRAIL, a member of the tumor necrosis factor (TNF) superfamily, has attracted considerable attention due to its ability to selectively induce apoptosis in the majority of cancer cells while showing a minor impact on normal cells [7,8]. It is also used in combination with other types of chemotherapy agents to enhance the treatment efficacy. Bortezomib, a proteasome inhibitor, has been shown to enhance TRAIL-induced apoptosis in various cancer types. It sensitizes cancer cells by targeting molecules such as DR4, DR5, c-FLIP, NF-κB, p21, and p27. Additionally, Bortezomib influences Bcl-2 family members, including Bcl-2, Bax, Bak, Bcl-xL, Bik, and Bim, contributing to its synergistic effect with TRAIL. This combination effectively induces apoptosis in resistant cancer cells, highlighting Bortezomib as a promising co-therapeutic agent with TRAIL gene therapy [[9], [10], [11]]. In addition, TRAIL gene therapy in combination with conventional sorafenib treatment attenuated HCC progression as well as liver fibrosis [12].
TRAIL interacts with five receptors on the cell surface, including two main receptors, TRAIL-R1 (DR4) and TRAIL-R2 (DR5), which possess intracellular death domains required for initiating apoptotic signaling. Additionally, it has three antagonist receptors, TRAIL-R3 (DcR1), TRAIL-R4 (DcR2), and osteoprotegerin (OPG), which lack functional death domains and consequently inhibit TRAIL-induced apoptosis [13,14].
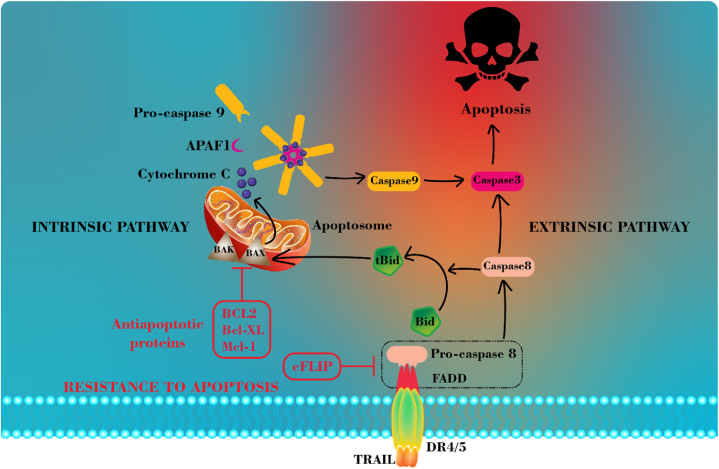
Binding TRAIL to its main death receptors (DR4 and DR5) triggers the initiation of the death signaling pathway. As demonstrated in Fig. 2, this process involves the recruitment of the adaptor protein Fas-associated death domain (FADD) to the death domain of DR4 and DR5, forming a death-inducing signaling complex (DISC). FADD subsequently recruits pro-caspase-8/10 to the DISC by interacting with their death effector domains (DEDs), as both FADD and caspases possess DEDs that can interact with one another [15,16]. Subsequently, caspase 8 is activated at the DISC and undergoes homodimerization. Following the release of active caspase-8 homodimers from the DISC, they cleave and activate the downstream substrates in the apoptotic pathway [17,18].
Fig. 2.
Schematic representation of intrinsic and extrinsic TRAIL-mediated apoptotic signaling pathways and the molecular mechanism of TRAIL resistance in cancer cells.
Considering the ability of TRAIL to selectively induce apoptosis in cancer cells, primarily due to the high expression of its main receptors (DR4 and DR5) in various types of cancer [19,20], using TRAIL as a potential cancer therapeutic agent has attracted significant attention. TR AIL selectively induces apoptosis in cancer cells while sparing normal cells. Studies show that Smac peptides sensitize various tumor cells, including glioblastoma, to TRAIL-induced apoptosis. In an intracranial glioma model, combined treatment with Smac peptides and TRAIL completely eradicated tumors and improved mouse survival without harming normal brain tissue [21]. However, its clinical applications have faced diverse obstacles including extremely poor blood circulation, short serum half-life, and rapid renal clearance. The challenges related to TRAIL delivery to the target cells and developing resistance in some cancer cells are another factors hindering its practical use in clinics [13,22]. A great deal of research has been done thus far to improve TRAIL stability and activity and put these major obstacles under control [23,24]. Developing death receptor-specific monoclonal antibodies [25,26], covalently linking TRAIL to molecules with favorable features [27], or employing diverse nano-based gene and protein delivery systems [23,28,29] were some of the strategies employed.
One of the promising approaches is leveraging nanoparticles to deliver TRAIL gene to cancer cells [30]. In this approach, which involves the delivery of nucleic acids into tumor cells, the limitations of recombinant TRAIL protein can be overcome. One of the primary advantages of delivering TRAIL-encoding DNA into tumor cells compared to the direct applying of protein is its potential to release TRAIL protein locally [31,32]. Moreover, the co-administration of drugs that sensitize or regulate apoptotic pathways could potentially be employed through gene delivery to address the resistance of cancer cells to TRAIL [30].
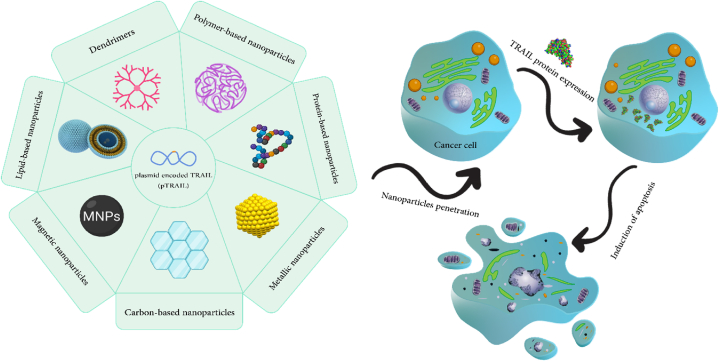
To date, numerous articles have reviewed the delivery of TRAIL gene by nanoparticles; based on our knowledge none of them has specifically addressed the crucial aspects of targeting TRAIL-resistant cancer cells. To fill this gap in the literature, we concentrate on elucidating gene delivery strategies to resistant cancer cells to TRAIL-induced apoptosis. By narrow focusing on this type of cancer cells, a novel prospective has been considered. This review, initially discusses possible processes via which cancer cells develop resistance to TRAIL. The mechanisms explored in this section encompass dysfunction in TRAIL receptors and endocytosis, DISC assembly, FADD defects, overexpression of cFLIP, loss of Bax/Bak functionality, overexpression of Bcl-2/Bcl-xL, overexpression of inhibitor of apoptosis proteins (IAPs), and activation of mitogen-activated protein kinases (MAPKs) or nuclear factor-kappa B (NF-κB) subunits. Next, we will highlight the various types of nanoparticles utilized to deliver the TRAIL gene to cancer cells exhibiting varying degrees of resistance to this type of therapy. We will also look at the potential applications of this nanomedicine strategy for cancer treatment in the future. Discussed nanoparticles in the subsequent sections are demonstrated in Fig. 1.
Fig. 1.
Schematic illustration of nanoparticle-based gene delivery systems utilized in TRAIL-based cancer therapy.
1.1. Mechanisms of cancer cells resistance to TRAIL
TRAIL is considered as a promising anticancer agent, TRAIL-resistance, nevertheless, has been found in many malignant cells. Several mechanisms have been proposed to explain the occurrence of TRAIL resistance in cancer cells, which will be explored in the subsequent discussion.
DR4 and DR5, two agonistic death receptors, located on chromosome 8p21-22, a common location for allelic deletions in several human malignancies, transmit apoptotic signals when TRAIL binds to them. Consequently, resistance may result from mutation or dysfunction in these two receptors [33,34]. Homozygous death receptor DR4 gene deletion has been reported in the FaDu nasopharyngeal cancer cell line [35]. The Homozygous deletion in DR4 attributed to its cytoplasmic death domain, which is crucial for transmitting apoptotic signals. Consequently, FaDu cells with this mutation are unable to initiate apoptosis and exhibit resistance to the cytotoxic effects of TRAIL. The paper states that by re-introducing the wild-type DR4 to the cells, scientists were able to restore their sensitivity to TRAIL [35]. Another research investigation demonstrated that a dominant-negative mutation in DR4 is responsible for the resistance of MG-63 cells to TRAIL, inhibiting the signaling pathway [36].
Several studies have demonstrated that death receptors 4 and 5 endocytosis could serve as a further mechanism for TRAIL resistance in specific cancer cells. Zhang et al. conducted a comparative analysis of the apoptotic response in six human breast cancer cell lines employing recombinant human TRAIL and antibodies targeting DR4 or DR5 [37]. The cell lines BT474 and T47D, which lacked cell surface expression of both DR4 and DR5, were discovered to demonstrate total resistance to Recombinant human TRAIL (rhTRAIL), expressing homotrimeric TRAIL. They suggested that DR4 and DR5 may undergo constitutive endocytosis through clathrin-dependent pathways, leading to a lack of expression on the cell surface and resistance to apoptosis triggered by TRAIL [37]. Furthermore, it was observed that the expression of DR4 and DR5 on the cell surface did not always indicate the cell's susceptibility to TRAIL-induced apoptosis. MCF7 cells exhibit the presence of both DR4 and DR5 on their cell surface, although they display resistance to rhTRAIL. The author ascribed this phenomenon to the defects occurring downstream of TRAIL death receptors [37]. Another study by the same investigators, demonstrated that TRAIL induced internalization and cleavage of DR4, in MDA-MB-231 human breast cancer cells. This process hampered TRAIL's capacity to induce apoptosis in this cell line [38].
Another possible mechanism for TRAIL resistance could be related to DISC assembly. DISC formation is an essential step in the apoptosis signaling pathway triggered by TRAIL. It involves the participation of many molecules such as TRAIL death receptors, FADD, and caspase-8. Any dysfunction or downregulation of the DISC components might result in resistance to TRAIL [33,39]. On the other hand, some anti-apoptotic proteins can regulate or interfere with this process. By binding to FADD through its DED, the cellular-FLICE inhibitory protein (c-FLIP) possesses this capability to inhibit transducing apoptotic signal, As an example. The higher affinity of FADD to the c-FLIP/caspase 8 heterodimer compared to the caspase 8 homodimer enables this binding process [33,40]. IAP family proteins are another member of the anti-apoptotic proteins, regulating the apoptotic pathway through binding to caspases 3,7, and 9, resulting in their inactivation [40,41].
Another resistance strategy to TRAIL, in type II cells, may occur through the intrinsic pathway, in which the proapoptotic protein Bid (BH3-interacting domain death agonist), is cleaved by caspase-8 and then truncated Bid (tBid) transmits apoptotic signals in mitochondria. The Bax and Bak proteins in the mitochondrial pathway can induce cell death by activating Apaf-1 and pro-caspase-9. Moreover, the regulation of intrinsic apoptosis is mediated by anti-apoptotic proteins belonging to the Bcl-2 family [42,43]. Thus, the resistance of cancer cells to TRAIL can be achieved through Overexpression of Bcl-xL and Bcl-2 or any mutation in the proapoptotic Bax or Bak genes resulting in their deactivation. NF-κB and mitogen-activated protein (MAP) kinases also play essential role in TRAIL resistance [33]. According to one research, by binding to DR5 on the cell surface, TRAIL can activate the NFκB pathway in B16F10 mouse melanoma cells, which are entirely resistant to TRAIL. The outcomes demonstrated that TRAIL may cause MMP-9 synthesis and B16F10 cell proliferation in addition to inducing B16F10 cell lung metastasis in vivo [44]. Intrinsic and extrinsic TRAIL signaling apoptotic pathways and the molecular mechanism of cancer cell resistance is summarized in Fig. 2.
It is important to note that the discussion around the various mechanisms of cancer cell resistance to TRAIL is very controversial. For instance, there are different results about the dependency of TRAIL-activated apoptosis on FADD [33]. Moreover, a single cell line might demonstrate diverse responses to TRAIL therapy under different conditions. It was shown that if TRAIL-susceptible cancer cells frequently undergo the administration of TRAIL protein, it might lead to the generation of TRAIL-resistant cells [45]. Furthermore, tumor microenvironment (TME) can significantly affect TRAIL signaling and turn the TRAIL-sensitive cells into resistant ones. For instance, interaction of cancer cells with their stromal TME can develop resistance in these cancer cells [39,46]. In this regard, the cellular response to TRAIL depends on the environmental conditions, and the cell lines discussed in this study can potentially be or become resistant to TRAIL-based therapy.
1.2. Overcoming TRAIL resistance
Over the past few decades, multiple approaches have been developed and put into practice to tackle the issue of TRAIL resistance. The utilization of agents that enhance cellular sensitivity to TRAIL in combination therapy has demonstrated remarkable potential in overcoming TRAIL resistance.
In one study, the researchers, to overcom the cancer cell resistance to TRAIL, used a triple combination of TRAIL, AT406 and rocaglamide (ART) therapy [47]. Given that the AT406 functions as an inhibitor of IAPs and rocaglamide inhibits the expression of c-FLIP, the researchers anticipate that their triple combination treatment (ART) would successfully reduce the levels of c-FLIP and IAPs, and promote the activation of caspase-8 and caspase-3 in a majority of cancer cell lines. ART was evaluated on 18 distinct TRAIL-resistant cell lines, and it successfully overcame TRAIL resistance in 15 of the 18 solid tumor cell lines examined, including HT29, HeLa, Hep G2, A549, and U87. Out of the 18 cell lines, only three cell lines (MCF7, U251, and U373) maintained a high resistance level even after being exposed to ART for 72 h. The authors propose that the resistance of these cell lines may be attributed to the low level expression of procaspase 8 and 3 [47]. The resistance in cervical cancer (HeLa cells) was successfully overcome by employing a combined therapy of TRAIL and YM155, a potent survivin inhibitor. This therapeutic approach effectively downregulated the mRNA and protein expression levels of cFLIP and survivin [48]. According to another study, researchers assert that the utilization of combination therapy including, TRAIL and Artonin E, can potentially overcome resistance in TRAIL-refractory colorectal cancer (LoVo cells) by DR5 upregulation and cFLIP downregulation [49]. Another research also has demonstrated that Goniothalamin can sensitize the target cells to TRAIL through the similar approach [50].
1.3. TRAIL-based gene delivery systems
Over the last years, gene therapy has emerged as a potential therapeutic option for treating cancer. Several strategies for cancer gene therapy have been developed, including suicide genes, gene silencing, microRNA-mediated therapy, anti-angiogenesis therapy, and immunotherapy [29,51,52]. However, gene therapy still needs promotion to overcome obstacles, primarily targeting delivery. Of all the available solutions to deal with these obstacles, developing advanced gene delivery systems is of great importance. Therefore, different types of gene delivery systems have been explored to enhance the tumor targeting of gene delivery [[53], [54], [55]]. They can be classified into two main categories: viral and non-viral vectors.
1.4. Viral-based vectors
Viral-based vectors are systems for delivering genes through viruses using the virus's capacity to insert its DNA into host cells. High transfection efficiency, sustained expression, and the potential of some of them to infect both dividing and non-dividing cells are benefits of viral vectors [55,56]. Viral vectors are often genetically engineered to avoid toxicity while maintaining the ability to encode the inserted gene and produce desired protein, making them useful as therapeutic agents [57]. Research has been conducted extensively on delivering the TRAIL gene to cancer cells via viral vectors. Among all, adenoviruses are at the forefront of viral-based delivery studies [[58], [59], [60], [61], [62]]. The Main drawback in the clinical application of TRAIL is its short half-life and rapid elimination from plasma, requiring high concentrations for effective treatment [24]. To overcome this issue, researchers have developed adenoviral vectors that can deliver TRAIL directly to the target cells, ensuring a more consistent and effective therapy. The potential of TRAIL in inducing apoptosis in prostate cancer cells has been demonstrated in a comprehensive study, involving eight cell lines and primary cultures of normal prostate epithelial cells. Although the combination of soluble TRAIL protein with doxorubicin resulted in cytotoxicity in prostate originated resistant cancer cells, delivery of TRAIL gene by Adenovirus vector resulted in significant cytotoxicity of TRAIL, remarkable lower dosage, in studied cells without co-treatment with other chemotherapeutic agents such as doxorubicin, highlighting the importance of viral vectors in optimizing TRAIL gene delivery. Beside, tissue-specific promoters, such as probasin or PSA, can further enhance the specificity and safety of TRAIL gene therapy. Ref: Resistance of prostate cancer cells to soluble TNF-related apoptosis-inducing ligand.
(TRAIL/Apo2L) can be overcome by doxorubicin or adenoviral delivery of full-length TRAIL [63].
1.5. Non-viral vectors
Despite all the advantages of viral delivery, they suffer from some limitations regarding payload capacity, insertional mutagenesis, toxicity, and immunogenicity. Their application is further restricted by the challenge of large-scale manufacture in clinical practice and costly production techniques [[64], [65], [66]]. In comparison with viral vectors, non-viral vectors do not suffer such limitations. In the last decades, plenty of approaches, such as utilization of exosomes [67,68], cell-based delivery systems [69,70] and microbubbles [71] have been suggested for efficiently deliver nucleic acids into target cells.
Nanoparticle-mediated gene delivery of TRAIL has also been extensively employed in TRAIL research, which is comprehensively reviewed in the subsequent sections and summarized in Table 1, focusing on their potential application in gene delivery to TRAIL-resistant malignancies. Additionally, the physicochemical and biological properties of each nanoparticle utilized is summarized in Table 2.
Table 1.
Nanoparticle-based platforms utilized in TRAIL gene delivery for the treatment of various cancers.
| Gene Delivery Platform | Form of NPs/Polymer/Material | Loaded Molecules | Cancer Type (cell line) |
In Vitro/In Vivo Applications | Mode of Delivery | Ref |
|---|---|---|---|---|---|---|
| Polymeric-based a | TPGS-b- (PCL-ran-PGA Copolymer/PEI | TRAIL/Endostatin genes | Cervical Cancer (HeLa) | In-vitro and In-vivo | Combination therapy | [85] |
| PEI-PLGA | pTRAIL/GA | Breast Cancer (MCF-7 and MDA-MB-231) | In-vitro | Combination therapy | [78] | |
| 4T1 | In-vivo | |||||
| LMW-PEI | pTRAIL/Monensin | Colon Cancer (HCT8/ADR) | In-vitro and In-vivo | Combination therapy | [86] | |
| PEGylated PEI | pTRAIL | Breast Cancer (MDA-MB-231) | In-vitro and In-vivo | Combination therapy with Wtmn | [87] | |
| PEI/PEG | pTRAIL | Melanoma Cancers (B16) | In-vitro | Combination with PDT | [88] | |
| PEI | pTRAIL | Cervical Cancer (HeLa) | In-vitro and In-vivo | Monotherapy | [89] | |
| PBAE-PEI | pTRAIL/EMB | Breast Cancer (MDA-MB-231) | In-vitro | Combination therapy | [79] | |
| Poly (β-amino ester) | sTRAIL/HDAC inhibitors | Liver Cancer (HepG2) | In-vitro and In-vivo | Combination therapy | [90] | |
| Poly (β-amino ester) | TRAIL/vorinostat | Liver Cancer (HepG2) | In-vitro and In-vivo | Combination therapy | [91] | |
| B-PDEAEA | TRAIL/SAHA | Lung Cancer (A549) | in-vitro | Combination therapy | [93] | |
| PADDAC/PEGylated Lipid | pTRAIL | Cervical/Lung/Liver Cancers (HeLa/A549/HepG2) | In-vitro and In-vivo | Monotherapy | [94] | |
| Dendrimers | PAMAM | TRAIL plasmid | Colon Cancer (C26) | In-vitro and In-vivo | Monotherapy | [97] |
| PAMAM | pTRAIL | Osteosarcoma (MG-63) | In-vitro and In-vivo | Monotherapy | [98] | |
| Lipid-based Nps | Lipid/Calcium/Phosphate/Protamine NPs | pTRAIL/sorafenib | Hepatocellular Carcinoma (Hep3B, JHH-7, HCA-1) | In-vitro and In-vivo | Combination therapy | [12] |
| Arginine-Conjugated Tocopherol Lipid Vesicle | pTRAIL | Glioblastoma (U87) | In-vitro | Monotherapy | [104] | |
| Cationic Lipid | pTRAIL/NO | Liver Cancer (HepG2) | in-vitro | Combination therapy | [105] | |
| Liposome | pEGFP-hTRAIL/PTX | Glioblastoma (U87 MG) | In-vitro and In-vivo | Combination therapy | [106] | |
| Liposome | FL/TRAIL genes | Colorectal Cancer (Lovo) | In-vitro and In-vivo | Combination therapy | [107] | |
| Carbon-based NPs | Carbon Dots/mPEG-PEI-DMMA | pTRAIL | Breast Cancer (MCF-7) | In-vitro and In-vivo | Monotherapy | [114] |
| branched PEI carbon dots | pTRAIL–GFP | Lung Cancer (A549) | In-vitro | Monotherapy | [115] | |
| Magnetic NPs | MNP/Chitosan | pCEM-TRAIL | Melanoma (B16F10) | In-vitro and In-vivo | Monotherapy | [119] |
| HA-SPIONs | TRAIL Gene | Glioblastoma (U87MG) | In-vitro and In-vivo | Monotherapy | [120] | |
| Protein-based NPs | Zein NPs | PTEN/TRAIL Genes | Liver Cancer (HepG2) | In-vitro and In-vivo | Combination therapy | [124] |
| Metallic NPs | Gold NPs | pTRAIL | Hepatocellular Carcinoma (Hep3B) | In-vivo | Monotherapy | [127] |
Nanoparticles.
Table 2.
Comparative analysis of physicochemical and biological properties of nanoparticle-based platforms in TRAIL gene delivery for cancer treatment.
| Gene Delivery Platform | Form of NPs/Polymer/Material | Loaded Molecules | Nanoparticle zeta potential (mv) | Complex zeta potential (mv) | Complex diameter (nm) | Nanoparticle diameter (nm) | Ref |
|---|---|---|---|---|---|---|---|
| Polymeric-based NPs |
TPGS-b- (PCL-ran-PGA Copolymer/PEI | TRAIL/Endostatin genes | 23.65 | 16.54 | 236.31 | 272.97 | [67] |
| PEI-PLGA | pTRAIL/GA | 50.3 | −0.6 | 107 | 121 | [68] | |
| LMW-PEI | pTRAIL/Monensin | NR | 35 | NRa | 170 | [69] | |
| PEGylated PEI | pTRAIL | NR | NR | NR | 130 | [70] | |
| PEI/PEG | pTRAIL | NR | 3.4 | 119 | 200 | [71] | |
| PEI | pTRAIL | NR | 3.4 | NR | 112.5 | [72] | |
| PBAE-PEI | pTRAIL/EMB | 34 | NR | 85 | NR | [73] | |
| Poly (β-amino ester) | sTRAIL/HDAC inhibitors | NR | 16 | NR | 200 | [74] | |
| Poly (β-amino ester) | TRAIL/vorinostat | NR | 10.5 to 25 | NR | 250 | [75] | |
| B-PDEAEA | TRAIL/SAHA | NR | NR | NR | NR | [77] | |
| PADDAC/PEGylated Lipid | pTRAIL | 40 | −4.2 | NR | 110 | [78] | |
| Dendrimers | PAMAM | TRAIL plasmid | 9.6 to 48.8 | NA | 74 to 177 | 142.9 | [81] |
| PAMAM | pTRAIL | NR | NR | NR | 200 | [82] | |
| Lipid-based Nps | Lipid/Calcium/Phosphate/Protamine NPs | pTRAIL/sorafenib | NR | 6 | NR | 102.9 | [88] |
| Arginine-Conjugated Tocopherol Lipid Vesicle | pTRAIL | 25 | 20 | 169 | <200 | [89] | |
| Cationic Lipid | pTRAIL/NO | 45.8 | 25 to 33 | 116.3 | 235–250 | [90] | |
| Liposome | pEGFP-hTRAIL/PTX | NR | 8.2 to 38.7 | NR | 131.4 to 257.7 | [91] | |
| Liposome | FL/TRAIL genes | 30.45 | 31 | 361 | 334 to 345 | [92] | |
| Carbon-based NPs | Carbon Dots/mPEG-PEI-DMMA | pTRAIL | 20.5 | −8.77 | 34.3 | 171 | [97] |
| branched PEI carbon dots | pTRAIL–GFP | 15 | NR | <10 | NR | [98] | |
| Magnetic NPs | MNP/Chitosan | pCEM-TRAIL | NR | −25 | NR | 200 to 250 | [102] |
| HA-SPIONs | TRAIL Gene | −32 | 26 | 130 | 243 | [103] | |
| Protein-based NPs | Zein NPs | PTEN/TRAIL Genes | NR | −37.5 to −24.3 | NR | 132.5 to 238.5 | [107] |
| Metallic NPs | Gold NPs | pTRAIL | 18.3 | −17 | 5.7 | 10.4 | [110] |
Not-reported.
1.6. Polymer-based nanoparticles
Polymers are widely utilized nanoparticles in gene delivery, possibly owing to their diverse characteristics in terms of structure, molecular weight, and composition [72]. Possessing linear, branching, or dendritic features, Cationic polymers are frequently used in gene delivery-related research. The reason is behind the positive charge on the outer layer of these polymers, facilitating electrostatic interaction as well as nucleic acid condensation. This binding reduces the size of DNA or RNA molecules, offering more protection and helping cellular uptake [[72], [73], [74], [75]].
Most synthetic and natural polymers benefit from their exceptional properties such as biocompatibility and biodegradability which are of significant importance for biological research. Moreover, “proton sponge effect” a mechanism by which polyplex would be released from the endosome into the cytoplasm has been demonstrated by the majority of polymers. Additionally, the surface functionality of the polymer is crucial in the process of conjugating biomolecules to target cancer cells for therapeutic purposes [72,73,76].
In addition, green biomaterials, such as chitosan, hyaluronic acid, and liposomes, have shown great promise in gene delivery applications due to their biocompatibility, biodegradability, and minimal environmental impact. These materials facilitate efficient delivery of therapeutic genes like TRAIL to cancer cells, enhancing targeted gene expression and inducing apoptosis with reduced toxicity. Their natural origins and sustainable properties make them ideal candidates for advancing gene therapy in oncology. The challenge posed by TRAIL-resistant human cancers has spurred efforts to devise more effective TRAIL-based combination therapies. Various chemotherapeutic agents and natural compounds have demonstrated promise in sensitizing tumor cells, thus overcoming TRAIL resistance and enhancing therapeutic outcomes [77]. Simultaneously delivery of plasmid coded TRAIL gene and gambogic acid, a natural compound, by using hyaluronic acid-PEI-PLGA system promoted the cellular uptake of cargoes into triple-negative breast cancer (TNBC) cells through CD44-dependenet endocytosis pathway evoking apoptosis both invitro and in mice model [78]. The specific binding between Hyaluronic acid and CD44 leads to a significant increase internalization of TRAIL plasmid and Embelin, an anticancer drug, to TNBC cells compared to non-TNBC cells [79].
Due to the promising capabilities of polymer-based nanoparticles in gene delivery, they have been extensively utilized to transfer the TRAIL gene to cancer cells. The most commonly used polymers in gene delivery are Poly(L-lysine) (PLL), polyethyleneimine (PEI), poly [(2-dimethylamino) ethyl methacrylate] (pDMAEMA), poly(β-amino ester)s, Polyethylene glycol (PEG), Dendrimers, and Chitosan [72].
Compared to other cationic polymers, polyethyleneimine (PEI) is considered the gold standard for non-viral gene delivery vectors due to its unique structural characteristics [80]. PEI is traditionally synthesized via ring-opening polymerization, resulting in a branched structure with primary amines at the termini, secondary amines along the polymer chain, and tertiary amines at the branch points [81]. While all cationic polymers contain amine groups, the specific positioning of these groups on the backbone of the PEI chain, rather than on the side chains, enhances its effectiveness for gene delivery purposes. Additionally, the presence of amine and methylene groups together in the structure of PEI, supports the proton sponge hypothesis [80,82]. The role of PEI in this hypothesis includes not only inducing high osmotic pressure and endosomal rupture but also interacting with the phospholipid membrane of the endosome. This interaction permeabilizes the membrane, facilitating the release of the polyplex from the endosome [80,83,84]. These unique physical and chemical properties of PEI make it an effective and valuable polymer for gene delivery in TRAIL research.
For instance, PEI, a frequently used polymer in TRAIL research, was used in one study for modifying a novel biodegradable copolymer, TPGS-b-(PCL-ran-PGA) to deliver the TRAIL and/or endostatin genes to HeLa cells, both in-vitro and in-vivo [85]. By reducing the expression of P-glycoprotein, TPGS facilitate the entrance of drug into cells. benefiting the unique properties of TPGS, the authors expected the synergistic anticancer effect of TPGS with other therapeutic agents like TRAIL [85]. Another study used PEI-PLGA decoration on hyaluronic acid to create a nanoparticle system for the targeted co-delivery of gambogic acid (GA) and the TRAIL plasmid (pTRAIL) to MCF-7 and MDA-MB-231 cell lines (in-vitro) and 4T1-bearing mice (in-vivo) [78]. In 2018, researchers designed a biocompatible nanoplatform to co-deliver TRAIL DNA and monensin, a death receptor sensitizer, using PEI and cyclodextrin. The guest-host complexation facilitates the loading process of hydrophobic monensin on cyclodextrin. The polyplex core was further modified by Poly-γ-glutamic acid (γ-PGA) or RGD-γ-PGA. This specific design enabled the nanocomplex to have both dual ligand-targeting and dual stimuli-responsive activity [86]. The TRAIL molecule was effectively transported to the HCT8/ADR colon cell line, both in-vitro and in the HCT8/ADR tumor-bearing BALB/c nude mice (in-vivo), resulting in a tumor suppression rate of 83 % [86]. PEI-based carrier utilization in TRAIL gene delivery continued, as in 2020 a further study was conducted to deliver TRAIL gene to brain metastatic cancer cells. PEGylated PEI(PPR) modified with R6dGR peptide was designed as the carrier of plasmid encoding TRAIL protein (PPR/pTRAIL). To inhibit the degradation of their nanocarrier by brain capillary endothelial cells through the autophagy process, they employed wortmannin (Wtmn), an autophagy inhibitor, and encapsulated it into liposomes modified with R6dGR peptide (Wtmn-Lip). Wtmn was expected to inhibit autophagy as well as create a synergistic anti-cancer effect in combination with TRAIL protein. A similar physiological condition was obtained by using MDA-MB-231 and bEnd.3 cells as brain metastatic breast cancer cells and brain endothelial cells, respectively [87].
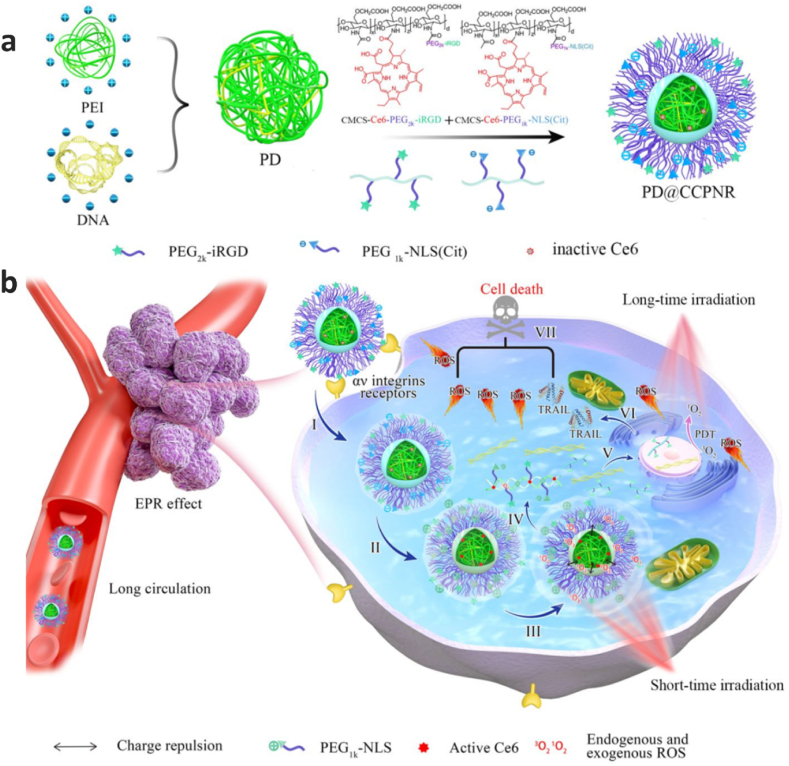
The effect of anticancer activity of TRAIL combined with photodynamic therapy was assessed in one research by designing a virus-inspired nanosystem (Fig. 3). In this novel complex nanoplatform a PEI/DNA polyplex core (PD) is coated with a dual pH responsive envelope. The envelop comprises and CCPR which refer to NLS(Cit)-modified O-carboxymethyl chitosan-Ce6 and iRGD-modified O-carboxymethyl chitosan-Ce6 conjugates, respectively. iRGD (internalizing RGD peptide) facilitate targeting delivery and cellular uptake, NLS is a nuclear localization signal enabling nucleus translocation of TRAIL gene and chlorin e6 (Ce6), and finally Ce6 serves as a photosensitizer for both Photochemical Internalization and Photodynamic Therapy through generating reactive oxygen species (ROS). The efficacy of the nanoplatform was assessed by delivering pTRAIL to the B16 cells [88].
Fig. 3.
Schematic illustration of a virus-inspired mimic for programmable TRAIL plasmid delivery without DNA damage. a. Preparation of virus-inspired PD@CCPNR. b. Diagram for cooperative anti-tumor therapy of PD@CCPNR. Reprinted with permission from [88].
In 2021, another study used a similar concept and designed a DNA-ejecting vector inspired by viruses using polymers. The utilization of quaternized linear PEI (QPEI) as a cationic polymer with long acyl chains facilitates DNA condensation and attachment to the cell membrane by determining the appropriate length for acyl chains. poly(γ-glutamic acid) was used for coating the polyplex to enhance in-vivo gene delivery efficiency to cervical carcinoma-bearing BALB/c-nude mice [89].
One another class of polymers that has garnered much interest in gene transfer, particularly in recent years, is poly (β-amino ester). The utilization of combined PEI and poly (β-amino ester) has performed in one study by Xu et al. PEI and PBAE, a pH sensitive poly (β-amino Ester), were coated with hyaluronic acid (HA) to form a nanosystem for co-delivery of TRAIL plasmid and embelin (EMB) to MDA-MB-231 cells [79]. Poly (β-amino ester) was utilized in another study to deliver sTRAIL (secretable TRAIL) to HepG2 cancer cells [90]. In order to overcome resistance to TRAIL, they conducted in-vitro experiments where they coupled treatment with histone deacetylase (HDAC) inhibitors (vorinostat, sodium butyrate, and MS-275). The findings demonstrated that the induction of apoptosis by TRAIL is directly influenced by the quantity of HDAC inhibitors. Additionally, it was observed that sTRAIL has a bystander effect on cells that have not been transfected. The researchers employed co-delivery of sTRAIL nanoparticles with intravenous vorinostat for in-vivo investigation. The tumor assessments during the initial 4 days of treatment revealed that the administration of sTRAIL nanoparticles combined with vorinostat resulted in a notable deceleration in tumor growth, compared to mice receiving control nanoparticles [90]. Recently, a research conducted by Zhao et al. designed a new structure of a highly branched-linear poly(β-amino ester)s, which demonstrated exceptional DNA condensation capability for gene delivery purposes [91]. Using mice with HepG2 tumors, the researchers evaluate the effectiveness of their nanosystems. By co-delivering TRAIL DNA and vorinostat, tumor development was significantly slowed compared to the control group [91].
HDAC inhibitors received significant attention due to their potent capacity to enhance non-viral gene transfer [92]. In another study, the effect of SAHA, a histone deacetylase inhibitor, on enhancing the gene delivery efficacy of a ROS-responsive charge-reversal cationic polymer (B-PDEAEA) and its synergistic effect with TRAIL gene therapy for cancer treatment was evaluated [93]. Upon oxidation of the boronic acid group by cellular ROS, the charge-reversal cationic polymer, B-PDEAEA, becomes ultimately negatively charged and releases the contained DNA. Thus, the amount of intracellular ROS dramatically influenced the effectiveness of gene transfer by B-PDEAEA/pDNA polyplexes. At a subtoxic concentration, SAHA causes an accumulation of ROS in cancer cells and improves transfection efficacy. SAHA has also the ability to enhance the responsiveness of cancer cells to TRAIL by increasing the expression of TRAIL death receptors and overcoming TRAIL resistance. Therefore, SAHA and B-PDEAEA collaborated to enhance cancer cell apoptosis by increasing TRAIL gene expression and potentially improving TRAIL sensitivity in resistant cancer cells [93].
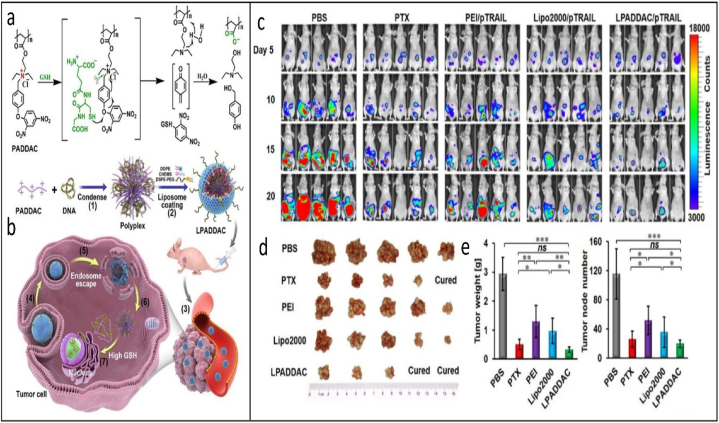
A highly selective glutathione (GSH)-responsive charge reversal cationic polymer has been employed in another research for intracellular gene delivery (Fig. 4a–b). Upon internalization in cancer cells, the cationic polymer undergoes GSH-induced thiolysis and degrades into negatively charged poly (acrylic acid), releasing the carried DNA and efficacious gene expression. The researchers coated the polymer with a PEGylated lipid to enhance the polyplexes' stability and extend the duration of their presence in the bloodstream, hence facilitating gene transfection in-vivo. TRAIL, as a therapeutic gene, was delivered to HeLa tumor cells in mice through the developed polymer. As showed in Fig. 4c–e, By the time the treatment was over, two mice had fully recovered from their tumors thanks to the more effective anticancer activity of LPADDAC/pTRAIL [94].
Fig. 4.
a. The structure of PADDAC and its GSH-triggered charge reversal. b. The formation of LPADDAC/DNA and its intracellular gene delivery cascade. c. Bioluminescence imaging of all nude mice bearing i.p. HeLa-Luci tumors measured by a Caliper IVIS Lumina II system. d. Images of the harvested tumor nodes in each group. e. left: Average tumor weight and right: node number per mouse in each group Reprinted with permission from [94].
1.7. Dendrimers
As a desirable class of drugs and gene delivery vector, dendrimers are well-defined three-dimensional nanopolymeric structures. Due to their abundant peripheral functional groups, Hyperbranched dendrimers, enable effectual attachment of targeting ligands and biomarkers [95]. These potential carriers, which have several polymeric branches, can be easily subjected to various structural changes and modifications and possess the ability to effectively entrap and transport high-molecular-weight hydrophobic or hydrophilic drugs or therapeutic genes, making them versatile and efficient nanocarriers [96].
Using dendrimers has been considered for TRAIL-related studies as well. Pishavar et al. used PAMAM one of the most widely utilized vectors in gene delivery for this purpose. Their study's goal was to modify polyamidoamine (PAMAM) dendrimers (G4 and G5) with hydrophobic compounds to increase their hydrophobicity and, as a result, their transfection efficiency while lowering their cytotoxicity. To this end, they used 10-bromodecanoic, cholesteryl chloroformate, and PEG, which promote lipophilicity–hydrophilicity balance. The in vitro and in vivo assessment was conducted to evaluate the cytotoxicity and effectiveness of the synthesized vectors in delivering the TRAIL gene to colon cancer (C26) cells [97].
A further study for TRAIL gene delivery focused on modifying generation 5 (G5) PAMAM dendrimers employing triazine moieties. Triazine exhibits the capability to form hydrogen bonds with DNA, enabling triazine-modified dendrimer () to possess favorable solubility in water, effective gene transport efficiency, and negligible toxicity towards osteosarcoma cells. Additionally, they demonstrated that /pTRAIL has anti-tumor effects in 3D cell cultures and in vivo mice models of osteosarcoma [98].
1.8. Lipid-based nanoparticles
Lipid-based nanoparticles (LNPs) have been widely employed as nonviral carriers for the delivery of drugs and genes [99]. They have demonstrated a robust capacity to compact and transport diverse nucleic acid molecules of varying sizes into cells, ranging from a few nucleotides to large ones. They additionally can be readily synthesized and conveniently modified [[99], [100], [101]]. Currently, cationic-lipid-based nanoplatforms are the most utilized nonviral carriers for delivering genes in therapeutic applications. The reason is that combining cationic lipids with DNA results in the formation of a positive charge complex, which enhances the gene transfection efficiency into cells [102,103].
Cationic lipid nanoparticles have been extensively used in TRAIL gene delivery. In one study, the researchers developed a multifunctional unique nanocarrier, LCPP, composed of Lipid/Calcium/Phosphate/Protamine, to transport TRAIL plasmid to hepatocellular carcinoma (HCC) cells [12]. The core of this nanocarrier is composed of a pH responsive calcium phosphate (CaP) that contains TRAIL plasmid and Protamine. Protamine served as a nucleus localization signal for intracellular delivery. The lipid bilayer is modified with an HCC- targeting peptide (SP94), improving the specific absorption of the nanoparticles by cancer cells. The release of from the CaP core could help overcom TRAIL resistance in HCC cells by upregulating the death receptor DR5 via CaMKII activation. The SP94-LCPP nanoparticles enhanced the expression of TRAIL in both human (Hep3B, JHH-7) and murine (HCA-1) hepatocellular carcinoma (HCC) cells under laboratory conditions (in-vitro) and in HCA-1 tumor-bearing mice (in-vivo). Enhancing its anticancer activity and clinic utilization, the researchers conducted additional investigations on the cytotoxic effects of SP94-LCPP nanoparticles through combination therapy using sorafenib. Interestingly, they could reduce liver fibrosis by specifically focusing on activated hepatic stellate cells (HSCs) in addition to suppressing tumor progression [12]. In 2021, Ravula et al. conducted a study to assess how arginine may affect TRAIL gene delivery to cancer cells. To accomplish this purpose, they synthesized a cationic lipid in which arginine serves as the cationic head group and vitamin E as the hydrophobic part. They also synthesized a glycine-based cationic lipid as the control with the same approach. The plasmid expressing TRAIL protein was effectively delivered to glioma cell lines, achieving a high transfection rate [104].
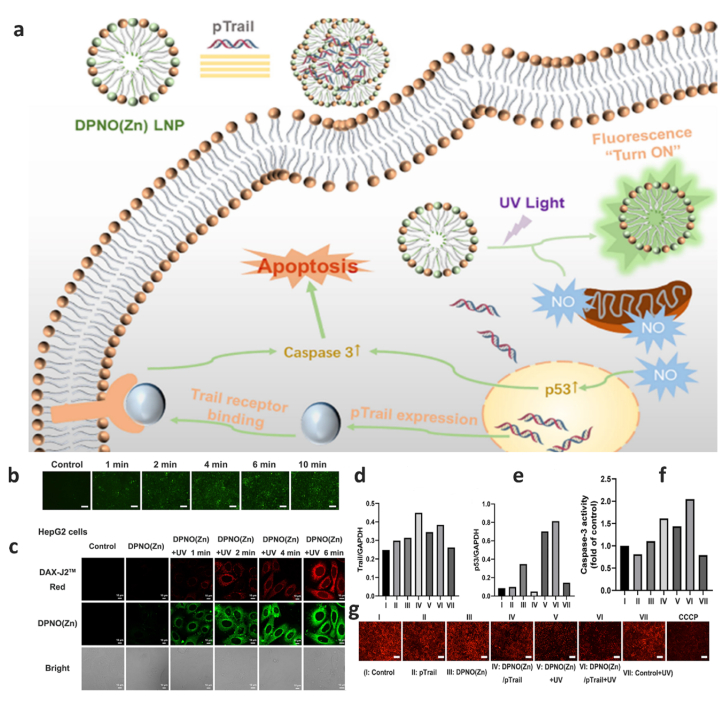
In one of the newest research performed, Yang et al. developed a novel light-responsive nanoplatform named DPNO(Zn) for gas/gene combination therapy [105]. Their designed multifunctional nanocomplex consisting a cationic lipid serving as the linking part and a Zn-DPA complex as the hydrophilic head group, utilized to simultaneously deliver nitric oxide (NO) and TRAIL as the therapeutic gene (Fig. 5a). The accumulation of p53 protein in cancer cells could occur through release of NO by exposure to UV light irradiation. This in turn leads to the activation of caspase-3 and mitochondrial damage. Interestingly, by UV irradiation, DPNO(Zn) LNPs undergo structural changes leading to different fluorescence emission wavelength. This phenomenon makes it possible to easily track the release of NO (Fig. 5b and c). Co-delivery of TRAIL gene with NO create a synergistic impact on HepG2 cells (Fig. 5d–g) [105].
Fig. 5.
a. schematic illustration of mechanism of light-responsive nanoplatform. b. Fluorescence microscopy images of DPNO(Zn) in HepG2 cells after UV light (3.0 mW/cm2) irradiation. Scale bar: 100 μm. c. CLSM images of DAX-J2 Red (15 μM) and DPNO(Zn) LNPs (15 μM) after being irradiated by UV light (3.0 mW/cm2). d and e. Relative Trail and p53 protein expression as the ratio of Trail (or p53 protein) to GAPDH from Western blot density analysis. f. Relative caspase-3 activity in HepG2 cells incubated with different formulations. (Dose was 20 μM for DPNO(Zn), 3.09 mg/L for pTrail). g. Detection of mitochondrial membrane potential under different treatments in HepG2 cells. CCCP was used as a positive control. Scale bar: 200 μm. Reprinted with permission from Ref. [105].
Researchers have utilized the potential characteristics of liposomes to transport drugs and genes to target cells, simultaneously. In one study, by taking advantage of the unique characteristics of cationic liposomes (CLPs) and angiopep-2, the researchers developed a nanosystem (ANG-CLP) for co-delivery of the pEGFP-hTRAIL therapeutic gene and the chemotherapy drug paclitaxel (PTX) to glioma cells. Modification with angiopep-2 endowed the CLPs with a dual targeting effect, improving BBB penetration as well as glioma targeted therapy [106]. In another research effort, a cationic nanoliposome was developed for the delivery of both TRAIL gene and the tyrosine kinase receptor 3 ligand (FL). Evaluating the combination therapy synergistic effect demonstrated enhanced apoptosis and reduced proliferation of Lovo cells [107].
Cancer stem-like cells (CSCs) exhibit deficient or lower expression of death receptors (DR), rendering them highly resistant to TRAIL-mediated apoptosis and limiting the therapeutic efficacy of TRAIL-based treatments. The liposomal assemblies co-encapsulating plasmid DNA encoding TRAIL and salinomycin enable cancer cells to act as protein generators expressing TRAIL. More importantly, this approach can acclimatize resistant CSCs, sensitizing them to TRAIL-triggered apoptosis through salinomycin-induced upregulation of DR expression on CSCs. This programmable liposome-based drug co-delivery system shows significant potential to efficiently eliminate CSCs and inhibit CSC-enriched tumor growth in an orthotopic colon tumor mouse model [108,109].
1.9. Carbon-based nanoparticles
Gene therapy can benefit greatly from the use of carbon-based nanoparticles because to their biocompatibility, high surface-to-volume ratios, simplicity of functionalization, and appropriate optical characteristics [110,111]. Carbon nanostructures are capable of delivering different DNA fragments to a specific target, ranging from large DNA molecules (plasmid DNA) to short DNA fragments like microRNA (miRNA) and small interfering RNA (siRNA) [110]. In this regard, several investigations have been conducted to develop carbon-based materials including fullerene, carbon nanotube, graphene, carbon dots, and diamonds for therapeutic purposes such as drug/gene delivery [112]. Carbon dots, a novel class of zero-dimensional sp2 carbon-based nanomaterials, have garnered significant interest as carriers. This is primarily due to their stable photoluminescence, broad excitation spectra, and excellent biocompatibility, making them suitable for delivery and imaging applications [112,113].
Zhao et al. utilized this potential and fabricated a multifunctional theranostic nanoplatform to simultaneously deliver and monitor the TRAIL gene to cancerous cells [114]. Carbon dots as the core were functionalized with HPAP polymer, increasing fluorescence quantum yield and GSH-triggered degradability. The core was coated by a pH-sensitive shell (mPEG-PEI-DMMA) named PPD. They investigated the delivery of the TRAIL gene using their theranostic nanoplatform in both in-vitro (MCF-7 cells and HUVECs cells) and in-vivo animal models (MCF-7 tumor-bearing mice) systems. The findings demonstrated that the growth of the tumor was suppressed by 70.84 % following a 16-day course of treatment [114].
In another study, human mesenchymal stem cells (hMSCs) were transfected by branched PEI carbon dots (bPEI25k CDs) TRAIL–GFP gene (pTRAIL–GFP) [115]. The therapeutic efficacy of TRAIL protein was evaluated on lung cancer cells (A549) through hMSCs [115].
1.10. Magnetic nanoparticles
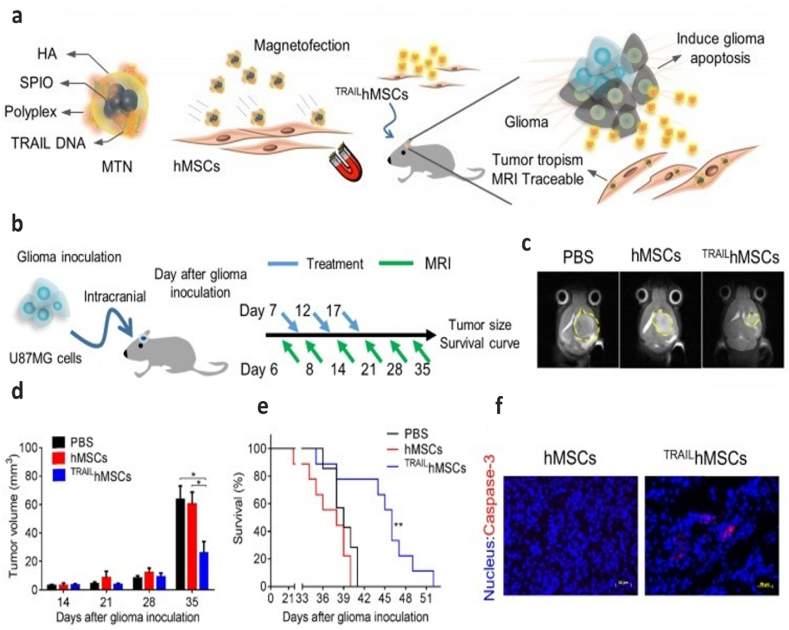
Having the capability to respond to an external magnetic field has made magnetic nanoparticles (MNPs) one of the most attractive nanoparticles in the realm of gene and drug delivery. Additionally, biocompatibility is another advantage these nanoparticles, making them more beneficial for biological applications. These nanoparticles are able to precisely deliver their cargo to the target cells when subjected to an external magnetic field, enhancing the gene delivery efficiency [[116], [117], [118]]. In order to leverage the fool potential of MNPs for TRAIL gene delivery, in one study, researchers developed a novel nanosystem compromising MNP, chitosan, and plasmid. The utilized plasmid (pCEM-TRAIL) contained a TRAIL protein-encoding gene regulated with Hsp70 promoter, which is able to respond to electromagnetic field (CEM). Thereby, employing a magnetic field not only guides MNPs to the target cells but also induces the gene expression by activating the promoter. The findings indicated that the nanosystem developed in this study effectively triggered apoptosis and inhibited the proliferation of melanoma B16F10 in the lungs [119]. A further study involved the creation of a ternary nanohybrid (MTN) system consisting of biodegradable cationic materials (PAE, bPEI, LPEI, SPEI, and lipofectamine), nucleic acids, and superparamagnetic iron oxide nanoparticles decorated with hyaluronic acid (HA-SPIONs) (Fig. 6a). HA-SPIO enhanced the transfection efficiency through the combined effects of CD44 receptors and magnetic force targeting. Even though they employed their nanosystem to transfer the TRAIL gene to human mesenchymal stem cells (hMSCs), the MTN-transfected TRAIL-expressing hMSCs were utilized to inhibit the growth of human glioma cells (U87MG), both in-vitro and in-vivo. They were monitored using magnetic resonance imaging (MRI) techniques (Fig. 6b–f) [120].
Fig. 6.
Physicochemical characterization of magnetic ternary nanohybrids and In vivo anti-cancer effects of TRAILhMSCs on orthotopic glioma-bearing xenograft animal model. a. Illustration of MTN formation and its applications on glioma treatment. b. Scheme of therapeutic plan for orthotopic glioma treatment. c. In vivo T2-weighted MR imaging of brain coronal sections on glioma-bearing mice at 35 days after the glioma inoculation. d. The tumor volume evolution in glioma-bearing mice, *P < 0.05 by two-way ANOVA with Tukey's multiple comparisons test and e. survival curve, n = 7 for PBS group and n = 9 for both hMSCs and TRAILhMSCs group, **P < 0.01 by exact log-rank test. f. Immunofluorescence staining of active caspase-3 on the brain tumor sections from mice received hMSCs or TRAILhMSCs treatments, Scale bar: 50 μm. Reprinted with permission from Ref. [120].
1.11. Protein-based nanoparticles
Protein-based nanoparticles offer several benefits for drug and gene delivery, including biocompatibility, biodegradability, abundant natural availability, simple synthesis process, cost-effectiveness, capacity to be modified with targeting molecules, high ability to bind drugs, and efficient uptake by specific cells. Furthermore, protein-based nanoparticles can modify their surface by attaching additional proteins and ligands through conjugation [[121], [122], [123]]. In one research, the researchers utilized Zein, a biocompatible prolamine-rich protein derived from maize with the ability to form protein nanoparticles by self-assembly. The objective of this study was to load the PTEN and TRAIL genes into Zein nanoparticles and evaluate their anti-tumor properties on liver tumor cells (HepG2) and rats with hepatocellular carcinoma (HCC) [124].
1.12. Metallic nanoparticles
Metallic nanoparticles have remarkable features, such as excellent stability and biocompatibility, easily conjugation with other molecules, and unique optical properties for imaging purposes make them appropriate candidates for gene and drug delivery [125,126]. In addition, the surface decoration of metal-organic frameworks (MOFs) with biomolecules has emerged as a promising approach to enhance their biocompatibility and targeting ability including DNA carrier [52].
Despite all of these great properties, they caught the attention to be used as TRAIL gene delivery. In one study, Chen and coworkers developed a sandwich-type nanocomplex based on gold nanoparticles (AuNPs) and PEI with the ability to target the nucleus and enhance the delivery of DNA with higher efficiency [127]. The researchers employed their nanocomplex (Au-PEI/DNA/PEI-Dexa) to transport the pTARIL to Hep3B tumor cells in mice. They successfully suppressed tumor formation and reduced the Hep3B tumor cells proliferation by 60 % compared to cells treated with pTRAIL, only five days after gene transfection [127].
2. Conclusion and prospective
TRAIL has shown excellent potential as a pharmaceutical agent for the therapeutic intervention of several cancer types. TRAIL holds promise as a candidate for cancer treatment, with clinical trials primarily focusing on recombinant TRAIL proteins and anti-TRAIL antibodies. However, challenges in delivery and drug resistance hinder successful translation of these findings, resulting in limited progress in clinical trials. To overcome these obstacles, efforts should prioritize the rational design of TRAIL formulations to enhance pharmacokinetic profiles and explore biomarkers specific to TRAIL-based therapy. Additionally, understanding TRAIL's anticancer mechanisms and combining it with TRAIL sensitizers could improve future applications. As precision medicine advances, a deeper understanding of TRAIL's antitumor functions may lead to the development of precise and effective treatments for patients. Various clinical trials have been conducted on TRAIL therapy for treating cancers such as advanced hepatocellular carcinoma, multiple myeloma, advanced solid tumors, and triple-negative breast cancer (TNBC). Many of these trials combine TRAIL with other therapeutic agents and utilize targeted formulations to enhance efficacy [30].
In recent decades, extensive research has explored various strategies to advance this therapy and promote its clinical application. These strategies include but are not limited to designing novel recombinant TRAIL and developing different expression techniques [[128], [129], [130]] employing other elements for conjugation purposes [131,132], and benefiting unique characteristics of nanoparticles [133,134], leading to increasing TRAIL's stability and bioactivity. In this regard, TRAIL gene delivery has emerged as an influential approach for overcoming the limitations associated with TRAIL stability and targeting, as well as combating TRAIL resistance in cancer cells.
Furthermore, new technologies like CRISPR-Cas9 and RNA interference hold promise in addressing TRAIL resistance by targeting and modifying genes involved in resistance pathways within cancer cells. CRISPR-Cas9 enables precise genome editing to enhance cancer cell sensitivity to TRAIL-induced apoptosis, while RNA interference techniques offer targeted gene silencing to sensitize cancer cells to TRAIL therapy, or, for example, by using some siRNAs that can downregulate c-FLIP expression, are able to overcome TRAIL resistance [47,135]. Integration of these technologies into TRAIL-based treatments has the potential to revolutionize cancer therapy by overcoming resistance mechanisms and improving treatment efficacy.
Nanoparticles have been extensively applied for TRAIL targeted therapy, providing a biocompatible surface for delivering genes to the desired cells. They have opened new opportunities for alternative therapeutic strategies such as combination therapy with other genes, drugs, or even photodynamic therapy (PDT) and photothermal therapy (PTT).
Currently, different designs of recombinant TRAIL either alone or in combination with other drugs are undergoing different phases of clinical trial experiments [[136], [137], [138]]. However, the use of TRAIL for clinical purposes continues to encounter different obstacles, mostly due to TRAIL resistance in cancer cells [34]. As a result, more profound comprehension of TRAIL's anticancer mechanism and the various ways through which cancer cells develop resistance to TRAIL treatment therapy would be needed to apply TRAIL in the future. Next research should concentrate on the development of more precise delivery systems, as well as the exploration of more effective strategies to overcome cancer cells' resistance to TRAIL. Based on all available results, it is expected that the remarkable properties of nanoparticles in TRAIL gene delivery would enable more reliable and efficient TRAIL-based therapies for patients.
Funding
This work was supported by Student Research Committee in Kermanshah University of Medical Sciences, Kermanshah, Iran (Grant numbers [4030036])
Ethical approval
This declaration is “not applicable”.
Data availability statement
No Data associated in the manuscript.
CRediT authorship contribution statement
Mina Habibizadeh: Writing – original draft. Shima Lotfollahzadeh: Writing – original draft. Parisa Mahdavi: Visualization. Soheila Mohammadi: Writing – review & editing. Omid Tavallaei: Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This research was supported partially by Student Research Committee, Kermanshah University of Medical Sciences, Kermanshah, Iran and the authors wish to acknowledge the financial support (Grant Number: 4030036) from Student Research Committee, Kermanshah University of Medical Sciences, Kermanshah, Iran.
References
- 1.Fesik S.W. Promoting apoptosis as a strategy for cancer drug discovery. Nat. Rev. Cancer. 2005;5(11):876–885. doi: 10.1038/nrc1736. [DOI] [PubMed] [Google Scholar]
- 2.Lemke J., et al. Getting TRAIL back on track for cancer therapy. Cell Death Differ. 2014;21(9):1350–1364. doi: 10.1038/cdd.2014.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galligan L., et al. Chemotherapy and TRAIL-mediated colon cancer cell death: the roles of p53, TRAIL receptors, and c-FLIP. Mol. Cancer Therapeut. 2005;4(12):2026–2036. doi: 10.1158/1535-7163.MCT-05-0262. [DOI] [PubMed] [Google Scholar]
- 4.Rahman M., et al. TRAIL induces apoptosis in triple-negative breast cancer cells with a mesenchymal phenotype. Breast Cancer Res. Treat. 2009;113:217–230. doi: 10.1007/s10549-008-9924-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ortiz-Otero N., et al. TRAIL-coated leukocytes to kill circulating tumor cells in the flowing blood from prostate cancer patients. BMC Cancer. 2021;21:1–12. doi: 10.1186/s12885-021-08589-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Favaro F., et al. TRAIL receptors promote constitutive and inducible IL-8 secretion in non-small cell lung carcinoma. Cell Death Dis. 2022;13(12):1046. doi: 10.1038/s41419-022-05495-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guimarães P.P., et al. Nanoparticles for immune cytokine TRAIL-based cancer therapy. ACS Nano. 2018;12(2):912–931. doi: 10.1021/acsnano.7b05876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lim S.M., et al. Improved biological half-life and anti-tumor activity of TNF-related apoptosis-inducing ligand (TRAIL) using PEG-exposed nanoparticles. Biomaterials. 2011;32(13):3538–3546. doi: 10.1016/j.biomaterials.2011.01.054. [DOI] [PubMed] [Google Scholar]
- 9.Jane E.P., Premkumar D.R., Pollack I.F. Bortezomib sensitizes malignant human glioma cells to TRAIL, mediated by inhibition of the NF-κB signaling pathway. Mol. Cancer Therapeut. 2011;10(1):198–208. doi: 10.1158/1535-7163.MCT-10-0725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koschny R., et al. TRAIL/bortezomib cotreatment is potentially hepatotoxic but induces cancer‐specific apoptosis within a therapeutic window. Hepatology. 2007;45(3):649–658. doi: 10.1002/hep.21555. [DOI] [PubMed] [Google Scholar]
- 11.Voortman J., et al. TRAIL therapy in non–small cell lung cancer cells: sensitization to death receptor–mediated apoptosis by proteasome inhibitor bortezomib. Mol. Cancer Therapeut. 2007;6(7):2103–2112. doi: 10.1158/1535-7163.MCT-07-0167. [DOI] [PubMed] [Google Scholar]
- 12.Liu C.H., et al. A multifunctional nanocarrier for efficient TRAIL‐based gene therapy against hepatocellular carcinoma with desmoplasia in mice. Hepatology. 2018;67(3):899–913. doi: 10.1002/hep.29513. [DOI] [PubMed] [Google Scholar]
- 13.Stuckey D.W., Shah K. TRAIL on trial: preclinical advances in cancer therapy. Trends Mol. Med. 2013;19(11):685–694. doi: 10.1016/j.molmed.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Roosmalen I.A., Quax W.J., Kruyt F.A. Two death-inducing human TRAIL receptors to target in cancer: similar or distinct regulation and function? Biochem. Pharmacol. 2014;91(4):447–456. doi: 10.1016/j.bcp.2014.08.010. [DOI] [PubMed] [Google Scholar]
- 15.Kruyt F.A. TRAIL and cancer therapy. Cancer letters. 2008;263(1):14–25. doi: 10.1016/j.canlet.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 16.Lacour S., et al. Chemotherapy enhances TNF-related apoptosis-inducing ligand DISC assembly in HT29 human colon cancer cells. Oncogene. 2003;22(12):1807–1816. doi: 10.1038/sj.onc.1206127. [DOI] [PubMed] [Google Scholar]
- 17.Kantari C., Walczak H. Caspase-8 and bid: caught in the act between death receptors and mitochondria. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research. 2011;1813(4):558–563. doi: 10.1016/j.bbamcr.2011.01.026. [DOI] [PubMed] [Google Scholar]
- 18.Dickens L.S., et al. A death effector domain chain DISC model reveals a crucial role for caspase-8 chain assembly in mediating apoptotic cell death. Molecular cell. 2012;47(2):291–305. doi: 10.1016/j.molcel.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spierings D.C., et al. Expression of TRAIL and TRAIL death receptors in stage III non-small cell lung cancer tumors. Clin. Cancer Res. 2003;9(9):3397–3405. [PubMed] [Google Scholar]
- 20.Kurbanov B.M., et al. Efficient TRAIL-R1/DR4-mediated apoptosis in melanoma cells by tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) J. Invest. Dermatol. 2005;125(5):1010–1019. doi: 10.1111/j.0022-202X.2005.23900.x. [DOI] [PubMed] [Google Scholar]
- 21.Fulda S., et al. Smac agonists sensitize for Apo2L/TRAIL-or anticancer drug-induced apoptosis and induce regression of malignant glioma in vivo. Nat. Med. 2002;8(8):808–815. doi: 10.1038/nm735. [DOI] [PubMed] [Google Scholar]
- 22.Kelley S.K., et al. Preclinical studies to predict the disposition of Apo2L/tumor necrosis factor-related apoptosis-inducing ligand in humans: characterization of in vivo efficacy, pharmacokinetics, and safety. J. Pharmacol. Exp. Therapeut. 2001;299(1):31–38. [PubMed] [Google Scholar]
- 23.Belkahla H., et al. TRAIL–NP hybrids for cancer therapy: a review. Nanoscale. 2017;9(18):5755–5768. doi: 10.1039/c7nr01469d. [DOI] [PubMed] [Google Scholar]
- 24.Alizadeh Zeinabad H., Szegezdi E. TRAIL in the treatment of cancer: from soluble cytokine to nanosystems. Cancers. 2022;14(20):5125. doi: 10.3390/cancers14205125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ichikawa K., et al. Tumoricidal activity of a novel anti-human DR5 monoclonal antibody without hepatocyte cytotoxicity. Nat. Med. 2001;7(8):954–960. doi: 10.1038/91000. [DOI] [PubMed] [Google Scholar]
- 26.Takeda K., Okumura K., Smyth M.J. Combination antibody‐based cancer immunotherapy. Cancer Sci. 2007;98(9):1297–1302. doi: 10.1111/j.1349-7006.2007.00529.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wajant H., Gerspach J., Pfizenmaier K. Engineering death receptor ligands for cancer therapy. Cancer letters. 2013;332(2):163–174. doi: 10.1016/j.canlet.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 28.de Miguel D., et al. Onto better TRAILs for cancer treatment. Cell Death Differ. 2016;23(5):733–747. doi: 10.1038/cdd.2015.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang K., Kievit F.M., Zhang M. Nanoparticles for cancer gene therapy: recent advances, challenges, and strategies. Pharmacol. Res. 2016;114:56–66. doi: 10.1016/j.phrs.2016.10.016. [DOI] [PubMed] [Google Scholar]
- 30.Zhong H.-h., et al. TRAIL-based gene delivery and therapeutic strategies. Acta Pharmacol. Sin. 2019;40(11):1373–1385. doi: 10.1038/s41401-019-0287-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kagawa S., et al. Antitumor activity and bystander effects of the tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) gene. Cancer Res. 2001;61(8):3330–3338. [PubMed] [Google Scholar]
- 32.Naoum G.E., et al. Role of nanotechnology and gene delivery systems in TRAIL-based therapies. Ecancermedicalscience. 2016;10 doi: 10.3332/ecancer.2016.660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang L., Fang B. Mechanisms of resistance to TRAIL-induced apoptosis in cancer. Cancer Gene Ther. 2005;12(3):228–237. doi: 10.1038/sj.cgt.7700792. [DOI] [PubMed] [Google Scholar]
- 34.Gampa S.C., Garimella S.V., Pandrangi S. Nano-TRAIL: a promising path to cancer therapy. Cancer Drug Resistance. 2023;6(1):78. doi: 10.20517/cdr.2022.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ozören N., et al. Homozygous deletion of the death receptor DR4 gene in a nasopharyngeal cancer cell line is associated with TRAIL resistance. Int. J. Oncol. 2000;16(5):917–942. doi: 10.3892/ijo.16.5.917. [DOI] [PubMed] [Google Scholar]
- 36.Locklin R.M., et al. Selective targeting of death receptor 5 circumvents resistance of MG-63 osteosarcoma cells to TRAIL-induced apoptosis. Mol. Cancer Therapeut. 2007;6(12):3219–3228. doi: 10.1158/1535-7163.MCT-07-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y., Zhang B. TRAIL resistance of breast cancer cells is associated with constitutive endocytosis of death receptors 4 and 5. Mol. Cancer Res. 2008;6(12):1861–1871. doi: 10.1158/1541-7786.MCR-08-0313. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Y., Yoshida T., Zhang B. TRAIL induces endocytosis of its death receptors in MDA-MB-231 breast cancer cells. Cancer Biol. Ther. 2009;8(10):917–922. doi: 10.4161/cbt.8.10.8141. [DOI] [PubMed] [Google Scholar]
- 39.Deng D., Shah K. TRAIL of hope meeting resistance in cancer. Trends in cancer. 2020;6(12):989–1001. doi: 10.1016/j.trecan.2020.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maksimovic-Ivanic D., et al. Resistance to TRAIL and how to surmount it. Immunol. Res. 2012;52:157–168. doi: 10.1007/s12026-012-8284-8. [DOI] [PubMed] [Google Scholar]
- 41.Hassanzadeh A., et al. Down‐regulation of intracellular anti‐apoptotic proteins, particularly c‐FLIP by therapeutic agents; the novel view to overcome resistance to TRAIL. J. Cell. Physiol. 2018;233(10):6470–6485. doi: 10.1002/jcp.26585. [DOI] [PubMed] [Google Scholar]
- 42.Von Karstedt S., Montinaro A., Walczak H. Exploring the TRAILs less travelled: TRAIL in cancer biology and therapy. Nat. Rev. Cancer. 2017;17(6):352–366. doi: 10.1038/nrc.2017.28. [DOI] [PubMed] [Google Scholar]
- 43.Thorburn A. Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) pathway signaling. J. Thorac. Oncol. 2007;2(6):461–465. doi: 10.1097/JTO.0b013e31805fea64. [DOI] [PubMed] [Google Scholar]
- 44.Takahashi K., et al. Functional roles of tumor necrosis factor‐related apoptosis‐inducing ligand–DR 5 interaction in B 16 F 10 cells by activating the nuclear factor‐κ B pathway to induce metastatic potential. Cancer Sci. 2013;104(5):558–562. doi: 10.1111/cas.12112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang L., et al. Mechanisms involved in development of resistance to adenovirus-mediated proapoptotic gene therapy in DLD1 human colon cancer cell line. Gene Ther. 2002;9(18):1262–1270. doi: 10.1038/sj.gt.3301797. [DOI] [PubMed] [Google Scholar]
- 46.De Looff M., De Jong S., Kruyt F.A. Multiple interactions between cancer cells and the tumor microenvironment modulate TRAIL signaling: implications for TRAIL receptor targeted therapy. Front. Immunol. 2019;10:1530. doi: 10.3389/fimmu.2019.01530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang Y., et al. Overcoming resistance to TRAIL-induced apoptosis in solid tumor cells by simultaneously targeting death receptors, c-FLIP and IAPs. Int. J. Oncol. 2016;49(1):153–163. doi: 10.3892/ijo.2016.3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chandrasekaran A.P., et al. YM155 sensitizes HeLa cells to TRAIL-mediated apoptosis via cFLIP and survivin downregulation. Oncol. Lett. 2020;20(4) doi: 10.3892/ol.2020.11933. 1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sophonnithiprasert T., Mahabusarakam W., Watanapokasin R. Artonin E sensitizes TRAIL-induced apoptosis by DR5 upregulation and cFLIP downregulation in TRAIL-refractory colorectal cancer LoVo cells. J. Gastrointest. Oncol. 2019;10(2):209. doi: 10.21037/jgo.2018.12.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sophonnithiprasert T., et al. Goniothalamin enhances TRAIL-induced apoptosis in colorectal cancer cells through DR5 upregulation and cFLIP downregulation. Int. J. Oncol. 2015;47(6):2188–2196. doi: 10.3892/ijo.2015.3204. [DOI] [PubMed] [Google Scholar]
- 51.Singh V., Khan N., Jayandharan G.R. Vector engineering, strategies and targets in cancer gene therapy. Cancer Gene Ther. 2022;29(5):402–417. doi: 10.1038/s41417-021-00331-7. [DOI] [PubMed] [Google Scholar]
- 52.Babaeenezhad E., et al. Cytotoxic and epigenetic effects of berberine-loaded chitosan/pectin nanoparticles on AGS gastric cancer cells: role of the miR-185-5p/KLF7 axis, DNMTs, and global DNA methylation. Int. J. Biol. Macromol. 2024;260 doi: 10.1016/j.ijbiomac.2024.129618. [DOI] [PubMed] [Google Scholar]
- 53.Yahya E.B., Alqadhi A.M. Recent trends in cancer therapy: a review on the current state of gene delivery. Life Sci. 2021;269 doi: 10.1016/j.lfs.2021.119087. [DOI] [PubMed] [Google Scholar]
- 54.Wang C., et al. Emerging non-viral vectors for gene delivery. J. Nanobiotechnol. 2023;21(1):272. doi: 10.1186/s12951-023-02044-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sung Y.K., Kim S. Recent advances in the development of gene delivery systems. Biomater. Res. 2019;23:1–7. doi: 10.1186/s40824-019-0156-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen Y.H., Keiser M.S., Davidson B.L. Viral vectors for gene transfer. Current protocols in mouse biology. 2018;8(4):e58. doi: 10.1002/cpmo.58. [DOI] [PubMed] [Google Scholar]
- 57.Norian L.A., James B.R., Griffith T.S. Advances in viral vector-based TRAIL gene therapy for cancer. Cancers. 2011;3(1):603–620. doi: 10.3390/cancers3010603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim C., et al. Cancer gene therapy using a novel secretable trimeric TRAIL. Gene Ther. 2006;13(4):330–338. doi: 10.1038/sj.gt.3302658. [DOI] [PubMed] [Google Scholar]
- 59.Lin T., et al. Combination of TRAIL gene therapy and chemotherapy enhances antitumor and antimetastasis effects in chemosensitive and chemoresistant breast cancers. Mol. Ther. 2003;8(3):441–448. doi: 10.1016/s1525-0016(03)00203-x. [DOI] [PubMed] [Google Scholar]
- 60.Jacob D., et al. Gene therapy in colon cancer cells with a fiber-modified adenovector expressing the TRAIL gene driven by the hTERT promoter. Anticancer research. 2004;24(5A):3075–3080. [PubMed] [Google Scholar]
- 61.Kim C.-Y., et al. Preclinical studies for pharmacokinetics and biodistribution of Ad-stTRAIL, an adenovirus delivering secretable trimeric TRAIL for gene therapy. Experimental & molecular medicine. 2011;43(10):580–586. doi: 10.3858/emm.2011.43.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang H., et al. Adenovirus-mediated TRAIL expression and downregulation of Bcl-2 expression suppresses non-small cell lung cancer growth in vitro and in vivo. Int. J. Mol. Med. 2012;30(2):358–364. doi: 10.3892/ijmm.2012.998. [DOI] [PubMed] [Google Scholar]
- 63.Voelkel-Johnson C., King D.L., Norris J.S. Resistance of prostate cancer cells to soluble TNF-related apoptosis-inducing ligand (TRAIL/Apo2L) can be overcome by doxorubicin or adenoviral delivery of full-length TRAIL. Cancer Gene Ther. 2002;9(2):164–172. doi: 10.1038/sj.cgt.7700420. [DOI] [PubMed] [Google Scholar]
- 64.Ren S., et al. Application of non-viral vectors in drug delivery and gene therapy. Polymers. 2021;13(19):3307. doi: 10.3390/polym13193307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tomanin R., Scarpa M. Why do we need new gene therapy viral vectors? Characteristics, limitations and future perspectives of viral vector transduction. Curr. Gene Ther. 2004;4(4):357–372. doi: 10.2174/1566523043346011. [DOI] [PubMed] [Google Scholar]
- 66.Mohammadinejad R., et al. In vivo gene delivery mediated by non-viral vectors for cancer therapy. J. Contr. Release. 2020;325:249–275. doi: 10.1016/j.jconrel.2020.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu M., Hu Y., Chen G. The antitumor effect of gene-engineered exosomes in the treatment of brain metastasis of breast cancer. Front. Oncol. 2020;10:1453. doi: 10.3389/fonc.2020.01453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jiang L., et al. Engineering exosomes endowed with targeted delivery of triptolide for malignant melanoma therapy. ACS applied materials & interfaces. 2021;13(36):42411–42428. doi: 10.1021/acsami.1c10325. [DOI] [PubMed] [Google Scholar]
- 69.Chen K., et al. A TRAIL-delivered lipoprotein-bioinspired nanovector engineering stem cell-based platform for inhibition of lung metastasis of melanoma. Theranostics. 2019;9(10):2984. doi: 10.7150/thno.31157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hu Y.-L., et al. Mesenchymal stem cells as a novel carrier for targeted delivery of gene in cancer therapy based on nonviral transfection. Mol. Pharm. 2012;9(9):2698–2709. doi: 10.1021/mp300254s. [DOI] [PubMed] [Google Scholar]
- 71.Rinaldi L., et al. Sonoporation by microbubbles as gene therapy approach against liver cancer. Oncotarget. 2018;9(63) doi: 10.18632/oncotarget.25875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Aied A., et al. Polymer gene delivery: overcoming the obstacles. Drug Discov. Today. 2013;18(21–22):1090–1098. doi: 10.1016/j.drudis.2013.06.014. [DOI] [PubMed] [Google Scholar]
- 73.Amreddy N., et al. Polymeric nanoparticle-mediated gene delivery for lung cancer treatment. Polymeric Gene Delivery Systems. 2018:233–255. doi: 10.1007/s41061-017-0128-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Eliyahu H., Barenholz Y., Domb A. Polymers for DNA delivery. Molecules. 2005;10(1):34–64. doi: 10.3390/10010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gary D.J., Puri N., Won Y.-Y. Polymer-based siRNA delivery: perspectives on the fundamental and phenomenological distinctions from polymer-based DNA delivery. J. Contr. Release. 2007;121(1–2):64–73. doi: 10.1016/j.jconrel.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 76.Vasir J.K., Labhasetwar V. Polymeric nanoparticles for gene delivery. Expet Opin. Drug Deliv. 2006;3(3):325–344. doi: 10.1517/17425247.3.3.325. [DOI] [PubMed] [Google Scholar]
- 77.Allensworth J.L., et al. XIAP inhibition and generation of reactive oxygen species enhances TRAIL sensitivity in inflammatory breast cancer cells. Mol. Cancer Therapeut. 2012;11(7):1518–1527. doi: 10.1158/1535-7163.MCT-11-0787. [DOI] [PubMed] [Google Scholar]
- 78.Wang S., et al. Co-delivery of gambogic acid and TRAIL plasmid by hyaluronic acid grafted PEI-PLGA nanoparticles for the treatment of triple negative breast cancer. Drug Deliv. 2017;24(1):1791–1800. doi: 10.1080/10717544.2017.1406558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xu Y., et al. Hyaluronic acid-coated pH sensitive poly (β-amino ester) nanoparticles for co-delivery of embelin and TRAIL plasmid for triple negative breast cancer treatment. Int. J. Pharm. 2020;573 doi: 10.1016/j.ijpharm.2019.118637. [DOI] [PubMed] [Google Scholar]
- 80.Gallops C., Ziebarth J., Wang Y. Polymers in Therapeutic Delivery. ACS Publications; 2020. A polymer physics perspective on why PEI is an effective nonviral gene delivery vector; pp. 1–12. [Google Scholar]
- 81.Jäger M., et al. Branched and linear poly (ethylene imine)-based conjugates: synthetic modification, characterization, and application. Chem. Soc. Rev. 2012;41(13):4755–4767. doi: 10.1039/c2cs35146c. [DOI] [PubMed] [Google Scholar]
- 82.Boussif O., et al. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc. Natl. Acad. Sci. USA. 1995;92(16):7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vermeulen L.M., et al. The proton sponge hypothesis: fable or fact? Eur. J. Pharm. Biopharm. 2018;129:184–190. doi: 10.1016/j.ejpb.2018.05.034. [DOI] [PubMed] [Google Scholar]
- 84.Behr J.-P. The proton sponge: a trick to enter cells the viruses did not exploit. Chimia. 1997;51(1–2) 34-34. [Google Scholar]
- 85.Zheng Y., et al. Surface modification of TPGS-b-(PCL-ran-PGA) nanoparticles with polyethyleneimine as a co-delivery system of TRAIL and endostatin for cervical cancer gene therapy. Nanoscale Res. Lett. 2013;8:1–12. doi: 10.1186/1556-276X-8-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xu F., et al. Targeting death receptors for drug-resistant cancer therapy: codelivery of pTRAIL and monensin using dual-targeting and stimuli-responsive self-assembling nanocomposites. Biomaterials. 2018;158:56–73. doi: 10.1016/j.biomaterials.2017.12.018. [DOI] [PubMed] [Google Scholar]
- 87.Wang X., et al. Autophagy inhibition changes the disposition of non-viral gene carriers during blood-brain barrier penetration and enhances TRAIL-induced apoptosis in brain metastatic tumor. J. Contr. Release. 2020;321:497–508. doi: 10.1016/j.jconrel.2020.02.042. [DOI] [PubMed] [Google Scholar]
- 88.Zhou J., et al. Virus-inspired mimics: dual-pH-responsive modular nanoplatforms for programmable gene delivery without DNA damage with the assistance of light. ACS applied materials & interfaces. 2020;12(20):22519–22533. doi: 10.1021/acsami.0c03486. [DOI] [PubMed] [Google Scholar]
- 89.Wang G., et al. Virus-mimetic DNA-ejecting polyplexes for efficient intracellular cancer gene delivery. Nano Today. 2021;39 [Google Scholar]
- 90.Vaughan H.J., et al. Poly (beta-amino ester) nanoparticles enable tumor-specific TRAIL secretion and a bystander effect to treat liver cancer. Molecular Therapy-Oncolytics. 2021;21:377–388. doi: 10.1016/j.omto.2021.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhao Y., et al. Superior TRAIL gene expression and cancer cell apoptosis mediated by highly branched-linear poly (β-amino ester) s. J. Nanobiotechnol. 2023;21(1):394. doi: 10.1186/s12951-023-02169-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.De Souza C., et al. Nanomaterials as potential transporters of HDAC inhibitors. Medicine in Drug Discovery. 2020;6 [Google Scholar]
- 93.Zhou X., et al. SAHA (vorinostat) facilitates functional polymer-based gene transfection via upregulation of ROS and synergizes with TRAIL gene delivery for cancer therapy. J. Drug Target. 2019;27(3):306–314. doi: 10.1080/1061186X.2018.1519028. [DOI] [PubMed] [Google Scholar]
- 94.Wang G., et al. Glutathione-specific and intracellularly labile polymeric Nanocarrier for efficient and safe cancer gene delivery. ACS applied materials & interfaces. 2020;12(13):14825–14838. doi: 10.1021/acsami.9b22394. [DOI] [PubMed] [Google Scholar]
- 95.Kesharwani P., Iyer A.K. Recent advances in dendrimer-based nanovectors for tumor-targeted drug and gene delivery. Drug Discov. Today. 2015;20(5):536–547. doi: 10.1016/j.drudis.2014.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Singh V., Sahebkar A., Kesharwani P. Poly (propylene imine) dendrimer as an emerging polymeric nanocarrier for anticancer drug and gene delivery. Eur. Polym. J. 2021;158 [Google Scholar]
- 97.Pishavar E., et al. Modified PAMAM vehicles for effective TRAIL gene delivery to colon adenocarcinoma: in vitro and in vivo evaluation. Artificial cells, nanomedicine, and biotechnology. 2018;46(sup3):503–513. doi: 10.1080/21691401.2018.1500372. [DOI] [PubMed] [Google Scholar]
- 98.Wang Y., et al. Triazine-modified dendrimer for efficient TRAIL gene therapy in osteosarcoma. Acta Biomater. 2015;17:115–124. doi: 10.1016/j.actbio.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 99.Zhao Y., Huang L. Lipid nanoparticles for gene delivery. Adv. Genet. 2014;88:13–36. doi: 10.1016/B978-0-12-800148-6.00002-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hashida M., Kawakami S., Yamashita F. Lipid carrier systems for targeted drug and gene delivery. Chem. Pharmaceut. Bull. 2005;53(8):871–880. doi: 10.1248/cpb.53.871. [DOI] [PubMed] [Google Scholar]
- 101.Dissanayake T., et al. Recent advances in lipid-protein conjugate-based delivery systems in nutraceutical, drug, and gene delivery. Food Hydrocolloids for Health. 2022;2 [Google Scholar]
- 102.Safinya C.R. Structures of lipid–DNA complexes: supramolecular assembly and gene delivery. Curr. Opin. Struct. Biol. 2001;11(4):440–448. doi: 10.1016/s0959-440x(00)00230-x. [DOI] [PubMed] [Google Scholar]
- 103.Zuhorn I.S., Engberts J.B., Hoekstra D. Gene delivery by cationic lipid vectors: overcoming cellular barriers. Eur. Biophys. J. 2007;36:349–362. doi: 10.1007/s00249-006-0092-4. [DOI] [PubMed] [Google Scholar]
- 104.Ravula V., et al. Arginine-tocopherol bioconjugated lipid vesicles for selective pTRAIL delivery and subsequent apoptosis induction in glioblastoma cells. Mater. Sci. Eng. C. 2021;126 doi: 10.1016/j.msec.2021.112189. [DOI] [PubMed] [Google Scholar]
- 105.Yang H.-Z., et al. Fluorescent self-reporting lipid nanoparticles for nitric oxide/gene Co-delivery and combination therapy. Mol. Pharm. 2023;20(2):1404–1414. doi: 10.1021/acs.molpharmaceut.2c00973. [DOI] [PubMed] [Google Scholar]
- 106.Sun X., et al. Co-delivery of pEGFP-hTRAIL and paclitaxel to brain glioma mediated by an angiopep-conjugated liposome. Biomaterials. 2012;33(3):916–924. doi: 10.1016/j.biomaterials.2011.10.035. [DOI] [PubMed] [Google Scholar]
- 107.Sun N.-f., et al. Nanoliposome-mediated FL/TRAIL double-gene therapy for colon cancer: in vitro and in vivo evaluation. Cancer letters. 2012;315(1):69–77. doi: 10.1016/j.canlet.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 108.Shen S., et al. Combating cancer stem-like cell-derived resistance to anticancer protein by liposome-mediated acclimatization strategy. Nano Lett. 2022;22(6):2419–2428. doi: 10.1021/acs.nanolett.2c00004. [DOI] [PubMed] [Google Scholar]
- 109.Mohajer F., et al. Advanced nanosystems for cancer therapeutics: a review. ACS Appl. Nano Mater. 2023;6(9):7123–7149. [Google Scholar]
- 110.Mostafavi E., Zare H. Carbon-based nanomaterials in gene therapy. OpenNano. 2022 [Google Scholar]
- 111.Mohajeri M., Behnam B., Sahebkar A. Biomedical applications of carbon nanomaterials: drug and gene delivery potentials. J. Cell. Physiol. 2019;234(1):298–319. doi: 10.1002/jcp.26899. [DOI] [PubMed] [Google Scholar]
- 112.Taghavi S., et al. Hybrid carbon-based materials for gene delivery in cancer therapy. J. Contr. Release. 2020;318:158–175. doi: 10.1016/j.jconrel.2019.12.030. [DOI] [PubMed] [Google Scholar]
- 113.Kailasa S.K., et al. Carbon dots as carriers for the development of controlled drug and gene delivery systems. Biomedical applications of nanoparticles. 2019:295–317. [Google Scholar]
- 114.Zhao H., et al. Microenvironment-driven cascaded responsive hybrid carbon dots as a multifunctional theranostic nanoplatform for imaging-traceable gene precise delivery. Chem. Mater. 2018;30(10):3438–3453. [Google Scholar]
- 115.Han J., Na K. Transfection of the TRAIL gene into human mesenchymal stem cells using biocompatible polyethyleneimine carbon dots for cancer gene therapy. J. Ind. Eng. Chem. 2019;80:722–728. [Google Scholar]
- 116.Ghosal K., et al. A detailed review on synthesis, functionalization, application, challenges, and current status of magnetic nanoparticles in the field of drug delivery and gene delivery system. AAPS PharmSciTech. 2022;24(1):25. doi: 10.1208/s12249-022-02485-5. [DOI] [PubMed] [Google Scholar]
- 117.Huang R.-Y., et al. Magnetic nanocomplexes for gene delivery applications. J. Mater. Chem. B. 2021;9(21):4267–4286. doi: 10.1039/d0tb02713h. [DOI] [PubMed] [Google Scholar]
- 118.Bi Q., et al. Magnetofection: magic magnetic nanoparticles for efficient gene delivery. Chin. Chem. Lett. 2020;31(12):3041–3046. [Google Scholar]
- 119.Alvizo-Baez C.A., et al. Systemic delivery and activation of the TRAIL gene in lungs, with magnetic nanoparticles of chitosan controlled by an external magnetic field. International journal of nanomedicine. 2016:6449–6458. doi: 10.2147/IJN.S118343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Huang R.-Y., et al. Magnetic ternary nanohybrids for nonviral gene delivery of stem cells and applications on cancer therapy. Theranostics. 2019;9(8):2411. doi: 10.7150/thno.29326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Elzoghby A.O., Samy W.M., Elgindy N.A. Protein-based nanocarriers as promising drug and gene delivery systems. J. Contr. Release. 2012;161(1):38–49. doi: 10.1016/j.jconrel.2012.04.036. [DOI] [PubMed] [Google Scholar]
- 122.Salatin S., et al. A sight on protein-based nanoparticles as drug/gene delivery systems. Ther. Deliv. 2015;6(8):1017–1029. doi: 10.4155/tde.15.28. [DOI] [PubMed] [Google Scholar]
- 123.Jain A., et al. Protein nanoparticles: promising platforms for drug delivery applications. ACS Biomater. Sci. Eng. 2018;4(12):3939–3961. doi: 10.1021/acsbiomaterials.8b01098. [DOI] [PubMed] [Google Scholar]
- 124.El Sharkawi F.Z., et al. PTEN and TRAIL genes loaded zein nanoparticles as potential therapy for hepatocellular carcinoma. J. Drug Target. 2017;25(6):513–522. doi: 10.1080/1061186X.2017.1289536. [DOI] [PubMed] [Google Scholar]
- 125.Mody V.V., et al. Introduction to metallic nanoparticles. J. Pharm. BioAllied Sci. 2010;2(4):282. doi: 10.4103/0975-7406.72127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bahadur K.C.R., Thapa B., Bhattarai N. Gold nanoparticle-based gene delivery: promises and challenges. Nanotechnol. Rev. 2014;3(3):269–280. [Google Scholar]
- 127.Chen Z., et al. Sandwich-type Au-PEI/DNA/PEI-Dexa nanocomplex for nucleus-targeted gene delivery in vitro and in vivo. ACS applied materials & interfaces. 2014;6(16):14196–14206. doi: 10.1021/am503483w. [DOI] [PubMed] [Google Scholar]
- 128.Tavallaei O., et al. Periplasmic expression of TNF related apoptosis inducing ligand (TRAIL) in E. coli. Iran. J. Pharm. Res. (IJPR): IJPR. 2015;14(2):617. [PMC free article] [PubMed] [Google Scholar]
- 129.Tavallaei O., et al. Production and secretion of TNF related apoptosis inducing ligand (TRAIL/Apo2L) in the Escherichia coli periplasm using PhoA signal peptide. Journal of Reports in Pharmaceutical Sciences. 2014;3(1):90–98. [Google Scholar]
- 130.Rasekhian M., et al. Assessment of prokaryotic signal peptides for secretion of tumor necrosis factor related apoptosis inducing ligand (trail) in E. Coli: an in silico approach. J. Pure Appl. Microbiol. 2016;10(4):2647–2653. [Google Scholar]
- 131.Lotfollahzadeh S., et al. TRAIL/S-layer/graphene quantum dot nanohybrid enhanced stability and anticancer activity of TRAIL on colon cancer cells. Sci. Rep. 2022;12(1):5851. doi: 10.1038/s41598-022-09660-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Siegemund M., et al. IgG-single-chain TRAIL fusion proteins for tumour therapy. Sci. Rep. 2018;8(1):7808. doi: 10.1038/s41598-018-24450-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Li H., et al. Supramolecular assembly of protein-based nanoparticles based on tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) for cancer therapy. Colloids Surf. A Physicochem. Eng. Asp. 2020;590 [Google Scholar]
- 134.Na S.J., et al. Stability and bioactivity of nanocomplex of TNF-related apoptosis-inducing ligand. International journal of pharmaceutics. 2008;363(1–2):149–154. doi: 10.1016/j.ijpharm.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 135.Gunes S., et al. Enhancement of Apo2L/TRAIL signaling pathway receptors by the activation of Klotho gene with CRISPR/Cas9 in Caco-2 colon cancer cells. Med. Oncol. 2021;38(12):146. doi: 10.1007/s12032-021-01595-7. [DOI] [PubMed] [Google Scholar]
- 136.ClinicalTrials.gov Phase 1b/2 study of AMG 655 with mFOLFOX6 and bevacizumab for first-line metastatic colorectal cancer. https://clinicaltrials.gov/ct2/show/NCT00625651 Available from:
- 137.ClinicalTrials.gov Open-label study of CS-1008 for subjects with untreated and unresectable pancreatic cancer. https://clinicaltrials.gov/ct2/show/NCT00521404 Available from:
- 138.ClinicalTrials.gov A phase III trial of recombinant human Apo-2 ligand for injection. https://clinicaltrials.gov/ct2/show/NCT03083743 Available from:
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No Data associated in the manuscript.