Abstract
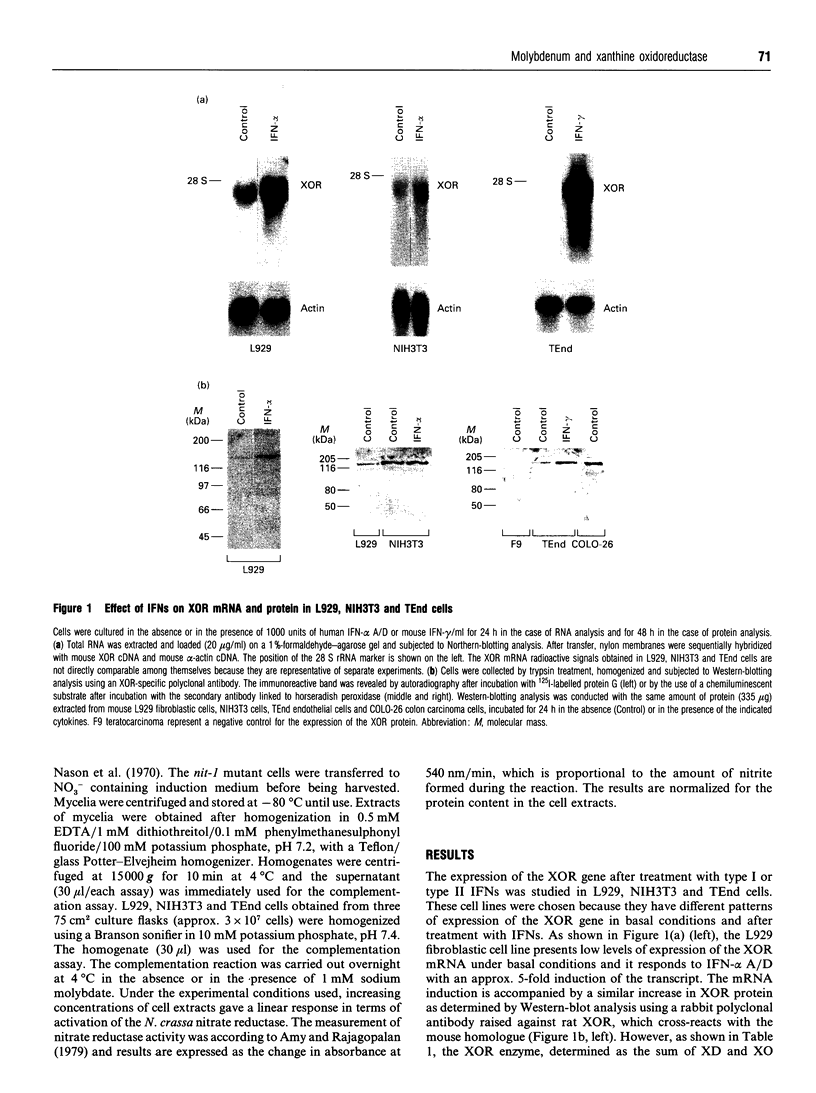
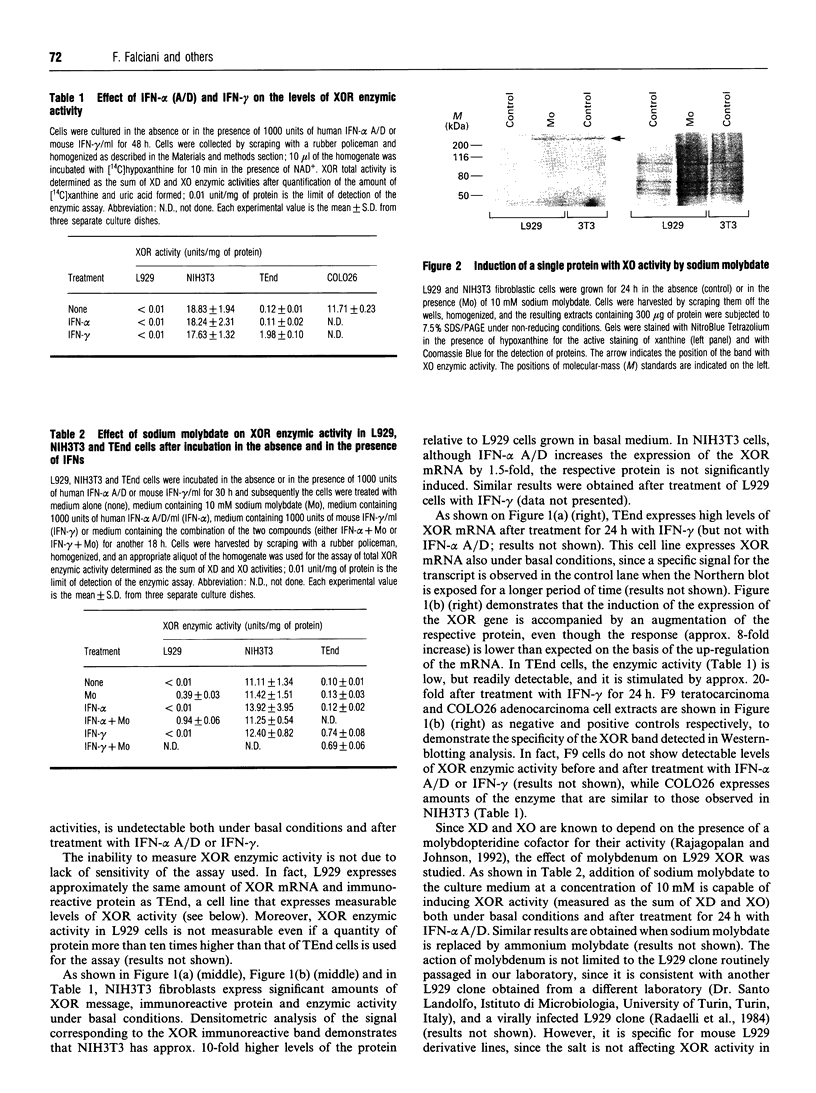
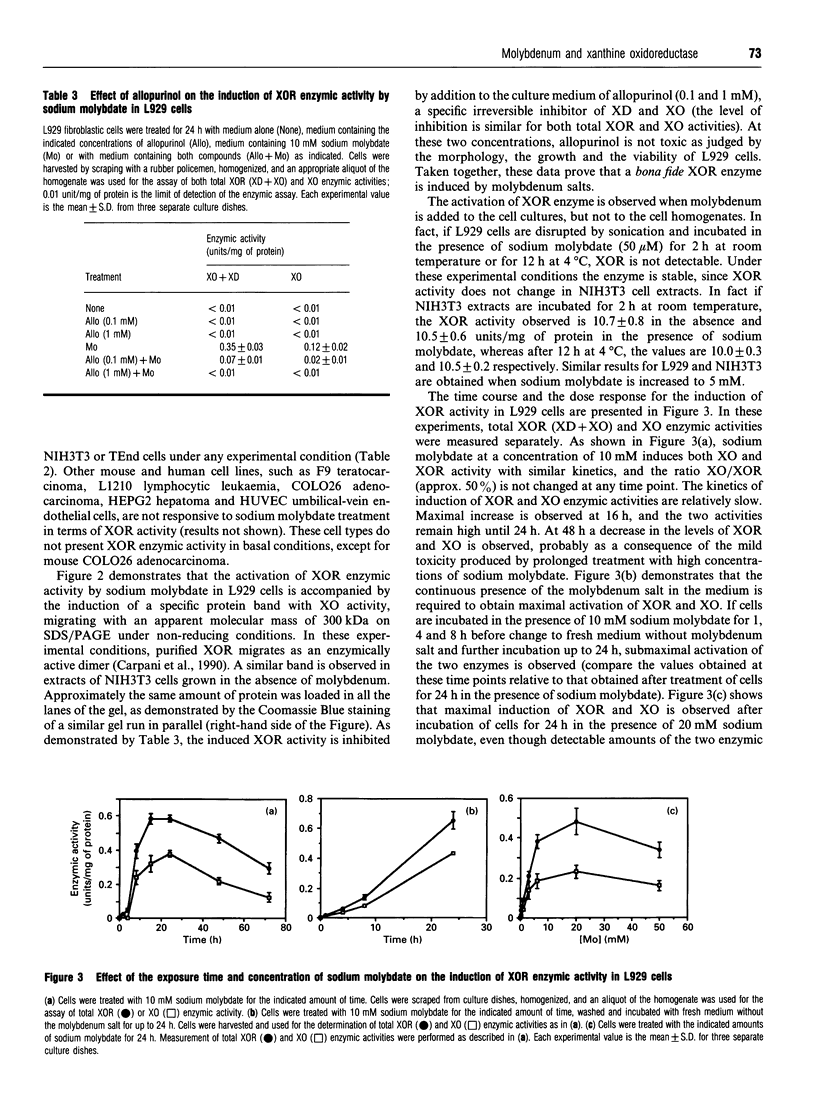
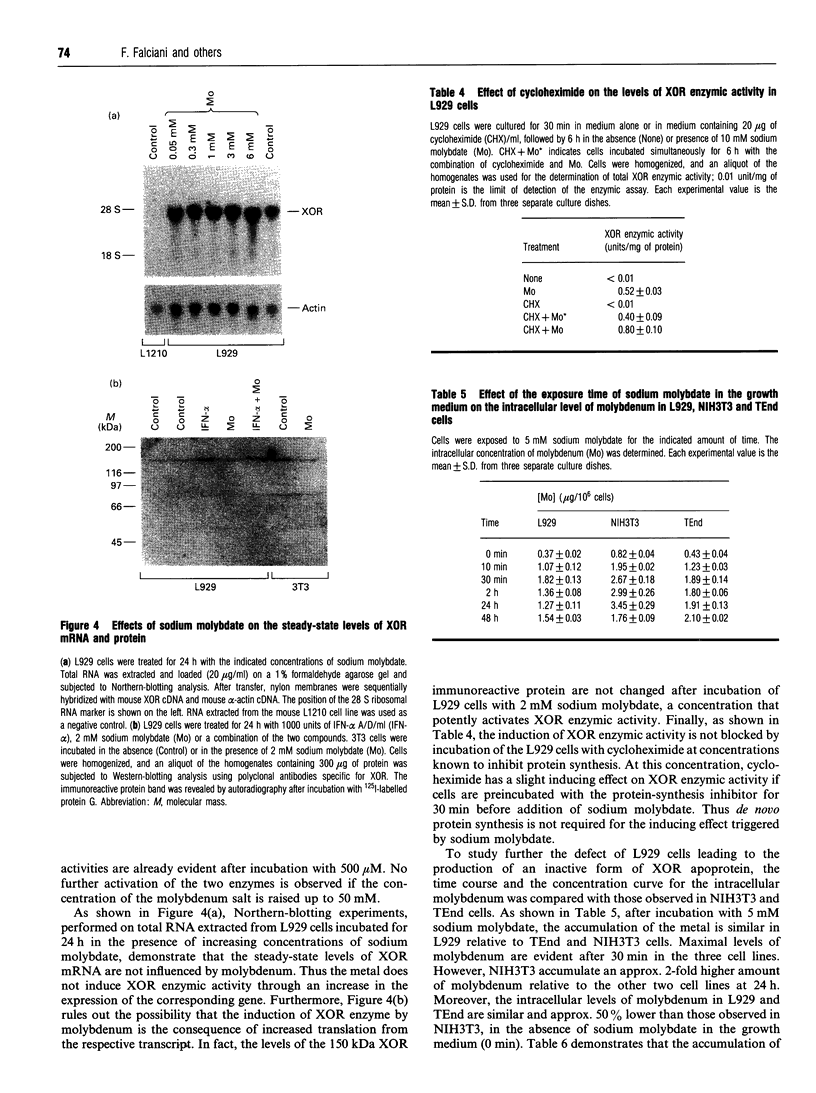
The mouse L929 fibroblastic cell line presents low, but detectable, levels of the mRNA encoding xanthine oxidoreductase under basal conditions, and it responds to type I and type II interferons by inducing the expression of the transcript [Falciani, Ghezzi, Terao, Cazzaniga, and Garattini (1992) Biochem. J. 285, 1001-1008]. This cell line, however, does not show any detectable amount of xanthine oxidoreductase enzymic activity, either before or after treatment with the cytokines. Molybdenum(VI) salts, in the millimolar range, are capable of activating xanthine oxidoreductase in L929 cells both under basal conditions and after treatment with interferon-alpha. The increase is observed in mouse L929 as well as in clones derived from it, but not in many other human and mouse cell lines. The induction observed in L929 cells is post-translational in nature and it is insensitive to cycloheximide, indicating that the molybdenum ion converts a pool of inactive xanthine oxidoreductase apoenzyme into its holoenzymic form. When grown in the absence of sodium molybdate, the L929 cell line has undetectable intracellular levels of the molybdenum cofactor, since the cell extracts are unable to complement the nitrate reductase defect of the nit-1 mutant of Neurospora crassa. L929 cells grown in the presence of millimolar concentrations of sodium molybdate, however, become competent to complement the nit-1 defect. L929 cells accumulate molybdenum ion inside the intracellular compartment as efficiently as TEnd cells, a mouse endothelial cell line that expresses xanthine oxidoreductase activity both under basal conditions and after treatment with interferon-gamma, suggesting that L929 cells have a defect in one or more of the metabolic steps leading to the synthesis of the molybdenum cofactor.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amaya Y., Yamazaki K., Sato M., Noda K., Nishino T., Nishino T. Proteolytic conversion of xanthine dehydrogenase from the NAD-dependent type to the O2-dependent type. Amino acid sequence of rat liver xanthine dehydrogenase and identification of the cleavage sites of the enzyme protein during irreversible conversion by trypsin. J Biol Chem. 1990 Aug 25;265(24):14170–14175. [PubMed] [Google Scholar]
- Amy N. K., Rajagopalan K. V. Characterization of molybdenum cofactor from Escherichia coli. J Bacteriol. 1979 Oct;140(1):114–124. doi: 10.1128/jb.140.1.114-124.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aukett A., Bennett M. J., Hosking G. P. Molybdenum co-factor deficiency: an easily missed inborn error of metabolism. Dev Med Child Neurol. 1988 Aug;30(4):531–535. doi: 10.1111/j.1469-8749.1988.tb04781.x. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Bray R. C. The inorganic biochemistry of molybdoenzymes. Q Rev Biophys. 1988 Aug;21(3):299–329. doi: 10.1017/s0033583500004479. [DOI] [PubMed] [Google Scholar]
- Carpani G., Racchi M., Ghezzi P., Terao M., Garattini E. Purification and characterization of mouse liver xanthine oxidase. Arch Biochem Biophys. 1990 Jun;279(2):237–241. doi: 10.1016/0003-9861(90)90487-j. [DOI] [PubMed] [Google Scholar]
- Corte E. D., Stirpe F. Regulation of xanthine oxidase in rat liver: modifications of the enzyme activity of rat liver supernatant on storage at 20 degrees. Biochem J. 1968 Jun;108(2):349–351. doi: 10.1042/bj1080349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corte E. D., Stirpe F. The regulation of rat liver xanthine oxidase. Involvement of thiol groups in the conversion of the enzyme activity from dehydrogenase (type D) into oxidase (type O) and purification of the enzyme. Biochem J. 1972 Feb;126(3):739–745. doi: 10.1042/bj1260739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endres W., Shin Y. S., Günther R., Ibel H., Duran M., Wadman S. K. Report on a new patient with combined deficiencies of sulphite oxidase and xanthine dehydrogenase due to molybdenum cofactor deficiency. Eur J Pediatr. 1988 Dec;148(3):246–249. doi: 10.1007/BF00441412. [DOI] [PubMed] [Google Scholar]
- Falciani F., Ghezzi P., Terao M., Cazzaniga G., Garattini E. Interferons induce xanthine dehydrogenase gene expression in L929 cells. Biochem J. 1992 Aug 1;285(Pt 3):1001–1008. doi: 10.1042/bj2851001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Fridovich I. Quantitative aspects of the production of superoxide anion radical by milk xanthine oxidase. J Biol Chem. 1970 Aug 25;245(16):4053–4057. [PubMed] [Google Scholar]
- Gardlik S., Rajagopalan K. V. The state of reduction of molybdopterin in xanthine oxidase and sulfite oxidase. J Biol Chem. 1990 Aug 5;265(22):13047–13054. [PubMed] [Google Scholar]
- Glaser J. H., DeMoss J. A. Phenotypic restoration by molybdate of nitrate reductase activity in chlD mutants of Escherichia coli. J Bacteriol. 1971 Nov;108(2):854–860. doi: 10.1128/jb.108.2.854-860.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes R. S., Leijten L. R., Duley J. A. Liver aldehyde oxidase and xanthine oxidase genetics in the mouse. Anim Blood Groups Biochem Genet. 1981;12(3):193–199. doi: 10.1111/j.1365-2052.1981.tb01550.x. [DOI] [PubMed] [Google Scholar]
- Huff S. D., Chaykin S. Genetic and androgenic control of N1-methylnicotinamide oxidase activity in mice. J Biol Chem. 1967 Mar 25;242(6):1265–1270. [PubMed] [Google Scholar]
- Imperial J., Ugalde R. A., Shah V. K., Brill W. J. Role of the nifQ gene product in the incorporation of molybdenum into nitrogenase in Klebsiella pneumoniae. J Bacteriol. 1984 Apr;158(1):187–194. doi: 10.1128/jb.158.1.187-194.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. L., Hainline B. E., Rajagopalan K. V., Arison B. H. The pterin component of the molybdenum cofactor. Structural characterization of two fluorescent derivatives. J Biol Chem. 1984 May 10;259(9):5414–5422. [PubMed] [Google Scholar]
- Johnson J. L., Hainline B. E., Rajagopalan K. V. Characterization of the molybdenum cofactor of sulfite oxidase, xanthine, oxidase, and nitrate reductase. Identification of a pteridine as a structural component. J Biol Chem. 1980 Mar 10;255(5):1783–1786. [PubMed] [Google Scholar]
- Johnson J. L., Rajagopalan K. V. Structural and metabolic relationship between the molybdenum cofactor and urothione. Proc Natl Acad Sci U S A. 1982 Nov;79(22):6856–6860. doi: 10.1073/pnas.79.22.6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. L., Waud W. R., Rajagopalan K. V., Duran M., Beemer F. A., Wadman S. K. Inborn errors of molybdenum metabolism: combined deficiencies of sulfite oxidase and xanthine dehydrogenase in a patient lacking the molybdenum cofactor. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3715–3719. doi: 10.1073/pnas.77.6.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. L., Wuebbens M. M., Mandell R., Shih V. E. Molybdenum cofactor biosynthesis in humans. Identification of two complementation groups of cofactor-deficient patients and preliminary characterization of a diffusible molybdopterin precursor. J Clin Invest. 1989 Mar;83(3):897–903. doi: 10.1172/JCI113974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketchum P. A., Swarin R. S. In vitro formation of assimilatory nitrate reductase: presence of the constitutive component in bacteria. Biochem Biophys Res Commun. 1973 Jun 19;52(4):1450–1456. doi: 10.1016/0006-291x(73)90663-3. [DOI] [PubMed] [Google Scholar]
- Kramer S. P., Johnson J. L., Ribeiro A. A., Millington D. S., Rajagopalan K. V. The structure of the molybdenum cofactor. Characterization of di-(carboxamidomethyl)molybdopterin from sulfite oxidase and xanthine oxidase. J Biol Chem. 1987 Dec 5;262(34):16357–16363. [PubMed] [Google Scholar]
- McCord J. M. Oxygen-derived free radicals in postischemic tissue injury. N Engl J Med. 1985 Jan 17;312(3):159–163. doi: 10.1056/NEJM198501173120305. [DOI] [PubMed] [Google Scholar]
- Minty A. J., Caravatti M., Robert B., Cohen A., Daubas P., Weydert A., Gros F., Buckingham M. E. Mouse actin messenger RNAs. Construction and characterization of a recombinant plasmid molecule containing a complementary DNA transcript of mouse alpha-actin mRNA. J Biol Chem. 1981 Jan 25;256(2):1008–1014. [PubMed] [Google Scholar]
- Nason A., Antoine A. D., Ketchum P. A., Frazier W. A., 3rd, Lee D. K. Formation of assimilatory nitrate reductase by in vitro inter-cistronic complementation in Neurospora crassa. Proc Natl Acad Sci U S A. 1970 Jan;65(1):137–144. doi: 10.1073/pnas.65.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino T., Nishino T., Schopfer L. M., Massey V. The reactivity of chicken liver xanthine dehydrogenase with molecular oxygen. J Biol Chem. 1989 Feb 15;264(5):2518–2527. [PubMed] [Google Scholar]
- Parks D. A., Granger D. N. Ischemia-induced vascular changes: role of xanthine oxidase and hydroxyl radicals. Am J Physiol. 1983 Aug;245(2):G285–G289. doi: 10.1152/ajpgi.1983.245.2.G285. [DOI] [PubMed] [Google Scholar]
- Pitterle D. M., Rajagopalan K. V. Two proteins encoded at the chlA locus constitute the converting factor of Escherichia coli chlA1. J Bacteriol. 1989 Jun;171(6):3373–3378. doi: 10.1128/jb.171.6.3373-3378.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radaelli A., Righi M., Liboi E., De Giuli Morghen C. Ultrastructural and biochemical evidence that the L929 cell retrovirus lacks the env gene translation product. J Gen Virol. 1984 Feb;65(Pt 2):295–307. doi: 10.1099/0022-1317-65-2-295. [DOI] [PubMed] [Google Scholar]
- Rajagopalan K. V., Handler P. Purification and properties of chicken liver xanthine dehydrogenase. J Biol Chem. 1967 Sep 25;242(18):4097–4107. [PubMed] [Google Scholar]
- Rajagopalan K. V., Johnson J. L. The pterin molybdenum cofactors. J Biol Chem. 1992 May 25;267(15):10199–10202. [PubMed] [Google Scholar]
- Rajagopalan K. V. Molybdenum: an essential trace element in human nutrition. Annu Rev Nutr. 1988;8:401–427. doi: 10.1146/annurev.nu.08.070188.002153. [DOI] [PubMed] [Google Scholar]
- Rambaldi A., Young D. C., Griffin J. D. Expression of the M-CSF (CSF-1) gene by human monocytes. Blood. 1987 May;69(5):1409–1413. [PubMed] [Google Scholar]
- Reiners J. J., Jr, Pence B. C., Barcus M. C., Cantu A. R. 12-O-tetradecanoylphorbol-13-acetate-dependent induction of xanthine dehydrogenase and conversion to xanthine oxidase in murine epidermis. Cancer Res. 1987 Apr 1;47(7):1775–1779. [PubMed] [Google Scholar]
- Reiss J., Kleinhofs A., Klingmüller W. Cloning of seven differently complementing DNA fragments with chl functions from Escherichia coli K12. Mol Gen Genet. 1987 Feb;206(2):352–355. doi: 10.1007/BF00333594. [DOI] [PubMed] [Google Scholar]
- Saito T., Nishino T. Differences in redox and kinetic properties between NAD-dependent and O2-dependent types of rat liver xanthine dehydrogenase. J Biol Chem. 1989 Jun 15;264(17):10015–10022. [PubMed] [Google Scholar]
- Stewart V. Nitrate respiration in relation to facultative metabolism in enterobacteria. Microbiol Rev. 1988 Jun;52(2):190–232. doi: 10.1128/mr.52.2.190-232.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada L. S., Rubinstein J. D., Lesnefsky E. J., Horwitz L. D., Leff J. A., Repine J. E. Existence and participation of xanthine oxidase in reperfusion injury of ischemic rabbit myocardium. Am J Physiol. 1991 Mar;260(3 Pt 2):H805–H810. doi: 10.1152/ajpheart.1991.260.3.H805. [DOI] [PubMed] [Google Scholar]
- Terao M., Cazzaniga G., Ghezzi P., Bianchi M., Falciani F., Perani P., Garattini E. Molecular cloning of a cDNA coding for mouse liver xanthine dehydrogenase. Regulation of its transcript by interferons in vivo. Biochem J. 1992 May 1;283(Pt 3):863–870. doi: 10.1042/bj2830863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waud W. R., Rajagopalan K. V. Purification and properties of the NAD+-dependent (type D) and O2-dependent (type O) forms of rat liver xanthine dehydrogenase. Arch Biochem Biophys. 1976 Feb;172(2):354–364. doi: 10.1016/0003-9861(76)90087-4. [DOI] [PubMed] [Google Scholar]
- de Groot H., Littauer A. Reoxygenation injury in isolated hepatocytes: cell death precedes conversion of xanthine dehydrogenase to xanthine oxidase. Biochem Biophys Res Commun. 1988 Aug 30;155(1):278–282. doi: 10.1016/s0006-291x(88)81080-5. [DOI] [PubMed] [Google Scholar]