Abstract
In Candida albicans, Cdr1 pumps azole drugs out of the cells to reduce intracellular accumulation at detrimental concentrations, leading to azole-drug resistance. Milbemycin oxime, a veterinary anti-parasitic drug, strongly and specifically inhibits Cdr1. However, how Cdr1 recognizes and exports azole drugs, and how milbemycin oxime inhibits Cdr1 remain unclear. Here, we report three cryo-EM structures of Cdr1 in distinct states: the apo state (Cdr1Apo), fluconazole-bound state (Cdr1Flu), and milbemycin oxime-inhibited state (Cdr1Mil). Both the fluconazole substrate and the milbemycin oxime inhibitor are primarily recognized within the central cavity of Cdr1 through hydrophobic interactions. The fluconazole is suggested to be exported from the binding site into the environment through a lateral pathway driven by TM2, TM5, TM8 and TM11. Our findings uncover the inhibitory mechanism of milbemycin oxime, which inhibits Cdr1 through competition, hindering export, and obstructing substrate entry. These discoveries advance our understanding of Cdr1-mediated azole resistance in C. albicans and provide the foundation for the development of innovative antifungal drugs targeting Cdr1 to combat azole-drug resistance.
Subject terms: Cryoelectron microscopy, Antimicrobial resistance, Transporters, Small molecules
Candida albicans Cdr1 pumps azole antifungal drugs out of the cell, driving drug resistance. Here, the authors determine the mechanism of Cdr1-mediated fluconazole resistance and milbemycin oxime inhibition, providing a foundation for future drug discovery efforts.
Introduction
Globally, over one billion people suffer from fungal infections, causing over 1.5 million deaths annually1. Regrettably, fungal diseases are frequently overlooked by public health authorities, resulting in many avoidable deaths1. Azole drugs including triazoles and imidazoles, are a common class of antifungal drugs that disrupt fungal growth by inhibiting the production of ergosterol through the inhibition of P450-dependent lanosterol 14-α-demethylase2. They are widely used as first-line options for treatment of fungal infections, particularly systemic fungal infections3. However, prolonged therapeutic use of azole antifungals can lead to drug resistance, restricting treatment options and causing more deaths4,5.
A key strategy of azole-drug resistance for fungal pathogens is to reduce intracellular accumulation to non-lethal levels by actively pumping azole antifungals out of the cells6–9. In C. albicans, the most commonly encountered fungal pathogen10, efflux pumps, including ATP-binding cassette (ABC) transporters (Cdr1 and Cdr2) and the major facilitator superfamily (MFS) transporter Mdr1, are responsible for azole drug resistance5,11(Fig. 1a). Cdr1 and Cdr2 belong to the pleiotropic drug resistance (PDR) family, specifically the PDR5 subfamily12,13. They are capable of transporting azole antifungals such as fluconazole, itraconazole, and ketoconazole, as well as structurally and functionally unrelated compounds such as rhodamine 6 G, terbinafine, and cycloheximide14–18. Despite the high sequence similarity of 92% between Cdr1 and Cdr2, it is Cdr1 that plays a primary role in determining azole resistance17. Recently, the structures of Cdr1 homologue Pdr5 from non-pathogenic fungus Saccharomyces cerevisiae have provided the structural insights into the PDR5 subfamily19. However, owing to the critical absence of azole-bound structures, the mechanism by which the efflux pump Cdr1 recognizes and exports fluconazole remains unclear.
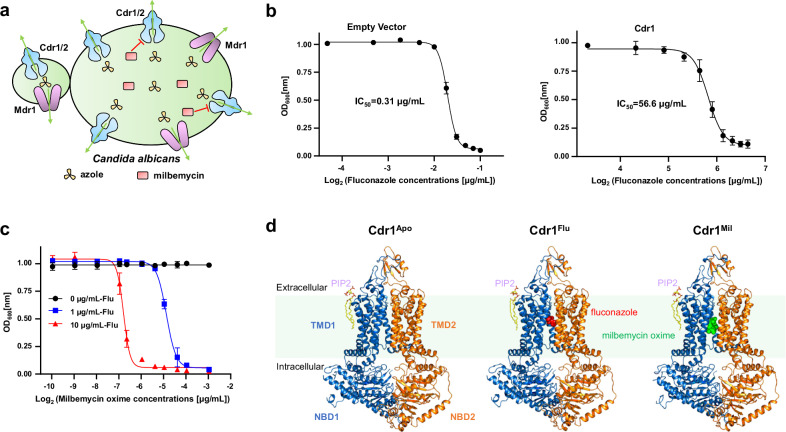
Fig. 1. Functional and cryo-EM studies of C. albicans Cdr1.
a A schematic diagram of efflux pumps in C. albicans. Cdr1, Cdr2 and Mdr1 can expel azole antifungal drugs. Milbemycin inhibits Cdr1 and Cdr2 efflux activity. The green arrows indicate the direction of azole drug efflux. b Fluconazole-sensitivity assay by measuring OD600nm under different fluconazole concentrations. The IC50 value for the empty vector is 0.31 µg/mL (left figure), while Cdr1 overexpression raises the IC50 value to 56.6 µg/mL (right figure). Data are presented as mean values ± SD; n = 3 independent experiments. c Milbemycin oxime-sensitivity assay measuring OD600nm under different milbemycin oxime concentrations, with fluconazole concentrations of 0 µg/mL, 1 µg/mL, and 10 µg/mL. The IC50 values for 1 µg/mL and 10 µg/mL fluconazole are 0.034 µg/mL and 0.009 µg/mL, respectively. Data are presented as mean values ± SD; n = 3 independent experiments. d Overall structures of Cdr1Apo, Cdr1Flu and Cdr1Mil. TMD1 and NBD1 are depicted in sky-blue, while TMD2 and NBD2 are in orange. PIP2, fluconazole, and milbemycin oxime are colored by yellow, red, and green, respectively. The green shading represents plasma membrane region.
Inhibiting the efflux pumps is one principal approach to combat drug resistance20,21. Several chemicals, such as FK506, enniatin B, beauvericin, D-octapeptide RC21v3, milbemycins and their oxime derivatives, have been approved to inhibit the pump activity of Cdr1, and thereby effectively combat drug resistance when combined with the widely used fluconazole21–23. Among them, milbemycin α25 exhibited potent inhibition for both C. albicans Cdr1 and Cdr2, except for Pdr521. Milbemycin A3/A4 oxime derivatives, an FDA-approved anti-parasite drug for animals (e.g., dogs and cats), have strong synergistic effects with fluconazole and reversed azole resistance in azole-resistant clinical C. albicans isolates23. However, the inhibitory mechanism of milbemycin oxime for the efflux pump Cdr1 remains unclear, hindering our ability to develop novel Cdr1 inhibitors.
In this work, we used single-particle cryoelectron microscopy (cryo-EM) to determine the structures of C. albicans Cdr1 in three distinct states: the apo state, the fluconazole-bound state, and the milbemycin oxime-inhibited state, with resolutions of 3.38 Å, 3.30 Å, and 3.08 Å, respectively. These structures reveal the mechanism of recognition and exportation of fluconazole, a representative azole antifungal drug, and elucidate the inhibitory mechanism of anti-parasite drug milbemycin oxime. This work provides the basis for comprehending Cdr1-mediated azole resistance in C. albicans and pioneering the development of innovative antifungal drugs that target the Cdr1 efflux pump to combat azole-drug resistance.
Results
Functional characterization and structural determination of C. albicans Cdr1
The full-length C. albicans Cdr1 was transformed into S. cerevisiae with a knockout of Pdr5 (Cdr1 homologue in S. cerevisiae) and the cell density was observed under different fluconazole concentrations. The overexpression of Cdr1 in Pdr5-deficient S. cerevisiae significantly conferred fluconazole resistance, resulting in an IC50 of 56.6 µg/mL (Fig. 1b). In contrast, the absence of Cdr1 made the cells highly sensitive to fluconazole, with a much lower IC50 of 0.31 µg/mL (Fig. 1b). The IC50 of overexpressed Cdr1 was approximately 180 times higher than that of cells lacking Cdr1, confirming that Cdr1 plays a significant role in conferring fluconazole resistance (Fig. 1a). Notably, the fluconazole resistance decreased dramatically in a dose-dependent manner with milbemycin oxime (Fig. 1c). As the fluconazole concentration increased from 1 µg/mL to 10 µg/mL, the IC50 of milbemycin oxime decreased from 0.034 µg/mL to 0.009 µg/mL (Fig. 1c), which is consistent with previous findings that milbemycin oxime exhibits a synergistic effect with fluconazole23.
To investigate the mechanism of Cdr1-mediated fluconazole resistance and its inhibition by milbemycin oxime, we next sought to determine the presentative Cdr1 structures in three distinct states, including the apo state, the fluconazole-bound state, and the milbemycin oxime-inhibited state. The purified wild-type Cdr1 in glyco-diosgenin (GDN) detergent exhibited homogeneity (Supplementary Fig. 1a) and displayed ATPase activity with a Vmax value of 37.13 nmol/min/mg (Supplementary Fig. 1b). Subsequently, the purified Cdr1 in GDN detergent was incubated with no ligand (apo), the triazole substrate (fluconazole), or the inhibitor (milbemycin oxime) before cryo-EM sample preparation. Following standard cryo-EM sample preparation and data processing procedures, we reconstructed cryo-EM maps for the apo state, the fluconazole-bound state, and the milbemycin oxime-inhibited state to an overall resolution of 3.38 Å, 3.30 Å, and 3.08 Å (Supplementary Figs. 1–4), respectively. For simplicity, the three structures were referred as Cdr1Apo, Cdr1Flu and Cdr1Mil (Apo for apo state, Flu for fluconazole, and Mil for milbemycin oxime) (Fig. 1d).
The three cryo-EM map densities exhibit high quality, enabling us to de novo build models for most regions in different states (Supplementary Figs. 1–5). In comparison to Cdr1Apo, both Cdr1Flu and Cdr1Mil present additional densities with high quality within the central cavity, corresponding well to fluconazole and milbemycin oxime A4 (Supplementary Fig. 5d, e), respectively. Furthermore, we observed a strong phospholipid-like density, consistent with a PIP2 molecule, in the exoplasmic membrane leaflet (Supplementary Fig. 6). This lipid-like density was not observed in Pdr519. Interestingly, quadruple mutation R624A/H706A/N758A/K761A did not significantly alter the OD600nm value (Supplementary Fig. 6d–f), suggesting that this lipid-like molecule might not be critical for fluconazole resistance. Cdr1Apo exhibits a significant structural similarity to Pdr5Apo, with a root-mean-square deviation of 0.814 Å (Supplementary Fig. 7a). In this work, our primary focus is to address two critical, as-yet-unsolved questions in the field: the mechanism of recognition and exportation of representative azole-substrate fluconazole, and the inhibitory mechanism of anti-parasite drug milbemycin oxime for the efflux pump Cdr1.
Specific recognition of fluconazole by Cdr1
To ensure the accurate identification of the fluconazole density, the protein sample for Cdr1Flu remained identical to that of Cdr1Apo, obtained from the same purification batch, except for the addition of the fluconazole ligand for Cdr1Flu. In the Cdr1Flu map, a distinctive shamrock-shaped density was identified and precisely matched with a fluconazole molecule (Supplementary Fig. 7b). In contrast, no corresponding density was observed at the same position in the Cdr1Apo map (Supplementary Fig. 7b). In the Cdr1Flu structure, transmembrane domain 1 and 2 (TMD1 and TMD2) enclose a cavity that opens toward the intracellular side, a conformation referred to as the inward-open state (Fig. 2a, b). Fluconazole, a member of the triazole family, binds at the top of this cavity, surrounded by TM1, TM2, TM5 of TMD1, and TM8, TM11 of TMD2 (Fig. 2b). This binding position is consistent with alanine scanning mutagenesis in these helices, particularly in TM2, which has been shown to increase susceptibility to azole drugs24. Notably, the binding position of fluconazole in Cdr1Flu differs from that of the dye molecule rhodamine 6 G (R6G) in Pdr519, where R6G is positioned beneath fluconazole (Supplementary Fig. 7c).
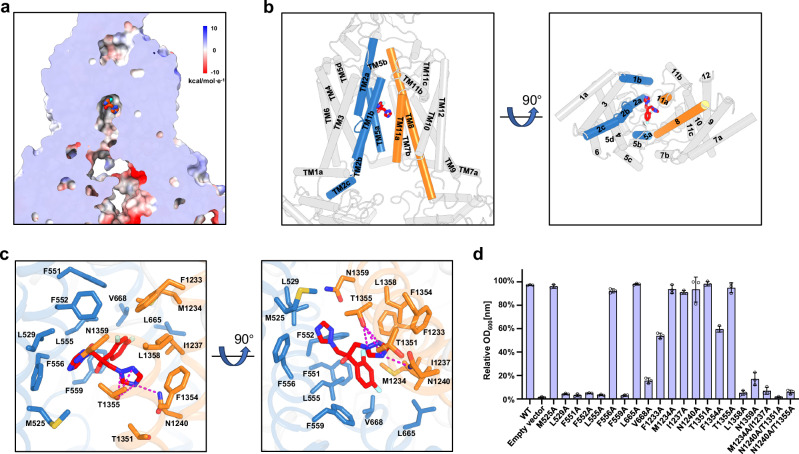
Fig. 2. Fluconazole recognition by TMDs of Cdr1.
a Sectional view displaying the electrostatic surface of Cdr1Flu. Red-colored fluconazole resides at the top of the hydrophobic central cavity. b Side (left figure) and bottom (right figure) views of Cdr1Flu. Fluconazole is surrounded by TM1b, TM2a and TM5a from TMD1(sky-blue), TM8 and TM11a from TMD2 (orange). c Coordination of fluconazole by TMDs of Cdr1Flu. Residues from TMD1 are colored by sky-blue, while those from TMD2 are colored by orange. Magenta dashes indicate hydrogen bonds. d Relative OD600nm values for wild type and mutants under a 10 µg/mL fluconazole concentration. The y-axis represents the relative OD600nm values as a percentage of the control (no drug). Three independent experiments were conducted for both wild type and mutants. Data are presented as mean values ± SD; n = 3 independent experiments.
The shamrock-shaped fluconazole is mainly surrounded by a large number of hydrophobic residues, forming a hydrophobic cavity (Fig. 2a, c). This hydrophobic cavity comprises Met525 and Leu529 from TM1, Phe551, Phe552, Leu555, Phe556, Phe559 from TM2, Leu665 and Val668 from TM5, Phe1233, Met1234, Ile1237 from TM8, and Thr1351, Phe1354, and Leu1358 from TM11 (Fig. 2c). In addition to these hydrophobic interactions, Asn1240 of TM8 and Thr1355 of TM11 contribute to three pairs of hydrogen bonds with one of 1,2,4-triazol-1-yl group, while Asn1359 of TM11 forms van der Waals interaction with the other 1,2,4-triazol-1-yl group (Fig. 2c). Single mutations within the cavity exhibit varying effects on fluconazole sensitivity and resistance. For instance, single mutations such as L529A, F551A, F552A, L555A, F559A, V668A, L1358A, and N1359A result in significant sensitivity to fluconazole (Fig. 2d). F1233A and F1354A decrease fluconazole resistance, while M525A, F556A, L665A, M1234A, I1237A, N1240A, T1351A, and T1355A maintain resistance under our experimental conditions (Fig. 2d). Notably, double mutations M1234A/I1237A, N1240A/T1351A and N1240A/T1355A lead to pronounced sensitivity to fluconazole (Fig. 2d).
To investigate whether drug sensitivity is influenced by protein expression levels, we conducted western blot analysis on various Cdr1 mutants (Supplementary Fig. 7e). The results showed that most of these mutations exhibited expression levels similar to the wild type, except for the N1240A/T1351A mutant (Supplementary Fig. 7e). This N1240A/T1351A double mutation resulted in nearly undetectable expression (Supplementary Fig. 7e), which likely contributes to its heightened drug sensitivity. We also assessed the ATP hydrolysis activity of mutants that exhibited decreased resistance to fluconazole (Supplementary Fig. 7f). Our study revealed that group 1 mutants (F551A, F552A, L555A, F559A, V668A, F1233A, F1354A, L1358A, M1234A/I1237A, and N1240A/T1355A) displayed decreased ATP hydrolysis activity (Supplementary Fig. 7f). In contrast, group 2 mutants (L529A and N1359A) did not show this reduction (Supplementary Fig. 7f). For group 1 mutants, the decrease in drug resistance may be attributed to impaired fluconazole recognition as well as reduced ATP hydrolysis activity. Conversely, for group 2 mutants, the decrease in drug resistance appears to be primarily due to impaired fluconazole recognition.
In addition to fluconazole, Cdr1 also transports other types of azole drugs, including short-tail azoles (such as miconazole and voriconazole) and long-tail azoles (such as ketoconazole and itraconazole)14,22,25. These azole drugs are hydrophobic in nature and have limited water solubility, making them compatible with the hydrophobic central cavity of Cdr1. When we docked these representative azoles into our fluconazole-bound Cdr1 map, we observed that voriconazole fit well, while miconazole and ketoconazole had minor clashes (Supplementary Fig. 8). In contrast, itraconazole exhibited severe steric hindrance due to its long-tail structure (Supplementary Fig. 8). This suggests that the recognition position of itraconazole differs from that of fluconazole, likely to accommodate the steric clash. Thus, we conclude that Cdr1 primarily recognizes azole drugs through hydrophobic interactions, and various azole drugs likely have specific recognition positions that match their individual structures.
Structural comparison of fluconazole-bound Cdr1 and vanadate-trapped Pdr5
The previously reported structure of vanadate-trapped Pdr5 (Pdr5AOV) adopts an outward-facing conformation, facilitating the release of the substrate19. The high structural and functional similarities between Cdr1 and Pdr5 suggest that the Pdr5AOV is a reliable representative of the outward-facing state of Cdr1. Structural alignment between Cdr1Flu and Pdr5AOV reveals that the two nucleotide-binding domains (NBDs) in Pdr5AOV come closer together upon binding ATP/ADP-Vi, thereby promoting an outward conformational change in their transmembrane domains (TMDs) (Fig. 3a). TM1b and TM11a draw closer and seal the substrate entry channel, thereby preventing the reverse passage of fluconazole into the inner membrane leaflet (Fig. 3b and Supplementary Fig. 9a). Similarly, the lower sections of TM1b, TM2b, TM8, and TM11 shift inward, thus obstructing the retrograde movement of fluconazole into the cytoplasm (Fig. 3c and Supplementary Fig. 9b).
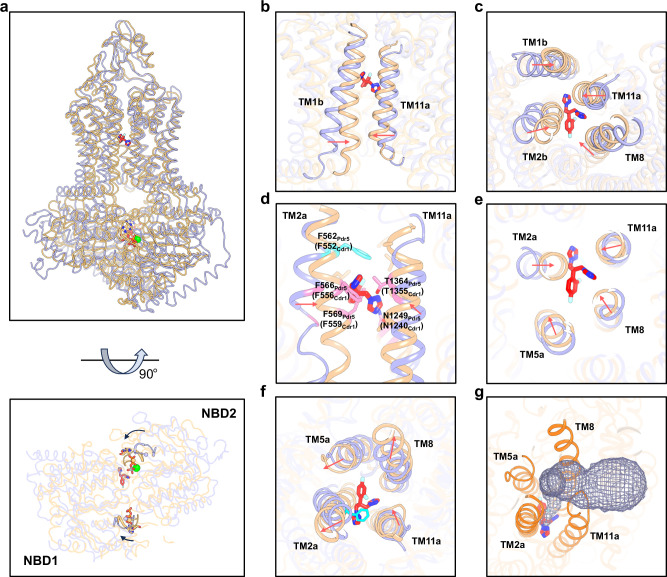
Fig. 3. Structural comparison of Cdr1Flu and Pdr5AOV.
a Side view (upper figure) and bottom view (lower figure) for structural alignment Cdr1Flu and Pdr5AOV (PDB: 7P06). Models of Cdr1Flu and Pdr5AOV are distinguished by using light-blue and wheat colors, respectively. b Zoomed-in view of TM1b and TM11a region of Cdr1Flu and Pdr5AOV. TM1b and TM11a move inward in Pdr5AOV. c, Bottom view of TM1b, TM2b, TM8 and TM11 regions of Cdr1Flu and Pdr5AOV. The lower regions of these helices move inward in Pdr5AOV. d Residues in TM2, TM8 and TM11 of Pdr5AOV exhibit clashes with fluconazole. Phe562 in Pdr5AOV prevents upward movement of fluconazole along the axis between TM2 and TM11. e Section view within the fluconazole binding region. TM2, TM5, TM8, and TM11 in Pdr5AOV undergo an inward movement within the fluconazole binding site. f Top view of TM2a, TM5a, TM8 and TM11 regions of Cdr1Flu and Pdr5AOV. The upper regions of TM2a, TM5a, and TM8 move outward, while that of TM11 shifts inward. g The light-blue grid mesh depicts the exit channel of fluconazole in Pdr5AOV. The red arrows indicate the movement direction of the helices.
Next, we carefully examined conformational changes within the vicinity of the fluconazole binding region. In the outward-facing conformation, the fluconazole binding cavity ceases to exist, as some residues that originally interacted with fluconazole in the inward-facing structure move inward (Fig. 3d and Supplementary Fig. 9c, d). Phe556, Phe559, Asn1240 and Thr1355 cause steric clash with fluconazole in outward-facing conformation (Fig. 3d). TM2, TM5, TM8, and TM11 undergo an inward movement within the fluconazole binding site (Fig. 3e), leading to the extrusion of fluconazole. In particular, TM2 undergoes a significant shift (Fig. 3d, e). Phe552 in TM2 remains at the top of fluconazole, thereby impeding the translocation pathway along the axis between TM2 and TM11 (Fig. 3d–f). However, the upper regions of TM2a, TM5a, and TM8 move outward, while that of TM11 shifts inward (Fig. 3f), collectively forming an exit channel that enables the release of fluconazole into the environment via a side pathway (Fig. 3g).
Specific recognition of milbemycin oxime by Cdr1
In the Cdr1Mil map, a strong density that was not observed in the Cdr1Apo map, was founded in the central cavity, which corresponded to the milbemycin oxime molecule (Fig. 4a and Supplementary Fig. 7d). The excellent density for milbemycin oxime enabled unambiguous assignment of the compound (Supplementary Figs. 5e and 7d). Like fluconazole, milbemycin oxime is also accommodated within the central cavity surrounded by TM1, TM2, TM5 of TMD1, and TM8, TM11 of TMD2 (Fig. 4a, b). Unlike fluconazole, which adheres to the top of the cavity, milbemycin oxime occupies the entire cavity (Figs. 2a and 4a).
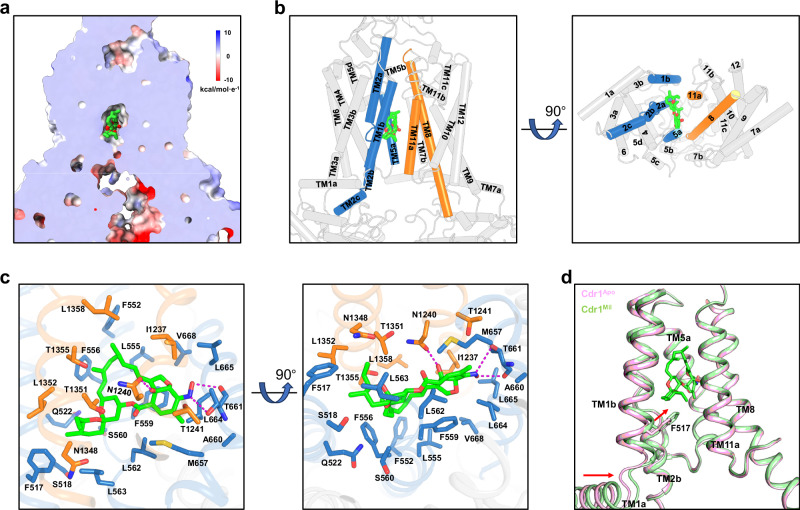
Fig. 4. Milbemycin oxime recognition by TMDs of Cdr1.
a Sectional view displaying the electrostatic surface of Cdr1Mil. Green-colored milbemycin oxime resides at the top of the hydrophobic central cavity. b Side (left figure) and bottom (right figure) views of Cdr1Mil. Milbemycin oxime is surrounded by TM1b, TM2a and TM5a from TMD1(sky-blue), TM8 and TM11a from TMD2 (orange). c Coordination of milbemycin oxime by TMDs of Cdr1Mil. Residues from TMD1 are colored by sky-blue, while those from TMD2 are colored by orange. Magenta dashes indicate hydrogen bonds. d Comparison of the TM1b region in Cdr1Mil and Cdr1Apo, with the lower portion of TM1b moving toward the central cavity due to hydrophobic interactions. The red arrows indicate the movement direction of TM1a and TM1b.
Milbemycin oxime, belonging to the 16-membered ring macrolide family, exhibits strong hydrophobic characteristics with poor water solubility. Consistent with this property, in the structure of Cdr1Mil, milbemycin oxime is primarily coordinated by a large number of hydrophobic residues from TM1, TM2, TM5, TM8 and TM11 (Fig. 4b, c). In addition to hydrophobic interactions, milbemycin oxime forms hydrogen bonds with the carbonyl oxygens of residues Thr661, as well as the side chains of Thr661 and Asn1240 (Fig. 4c). It is reported that the mutants of G521R in TM1 and T1355N in TM11 which are located in the central cavity, exhibited milbemycin resistance26. In our Cdr1Mil model, the G521R mutation could cause steric clashes, preventing milbemycin oxime from binding to the cavity, while the T1355N mutation could lead to conflicts between the hydrophilic side chain of asparagine and the hydrophobic backbone of milbemycin oxime (Supplementary Fig. 10a, b). Despite the high structural and functional similarities between Cdr1 and Pdr5, milbemycin oxime, much like milbemycin α2521, selectively inhibits Cdr1 rather than Pdr5 (Supplementary Fig. 10c). Structural comparison between Cdr1 and Pdr5 reveals that TM5a of Pdr5 moves toward the central cavity, causing steric clashes with milbemycin oxime (Supplementary Fig. 10d). Additionally, the side chain of Phe566 in Pdr5 is also involved in steric clash with milbemycin oxime (Supplementary Fig. 10d). These may explain why milbemycin oxime selectively inhibits Cdr1 instead of Pdr5.
Although the overall architecture of Cdr1Mil is similar to that of Cdr1Apo, with an RMSD of 0.511 Å over Cα atoms, conformational changes in local regions between Cdr1Mil and Cdr1Apo are evident (Fig. 4d and Supplementary Fig. 10e–g). The bottom region of TM1b moves toward the central cavity due to the attraction of milbemycin oxime (Fig. 4d). Compared to Cdr1Apo, Phe517 of Cdr1Mil moves closer to the spiroketal group with an ethyl substituent of milbemycin oxime, mainly mediated by hydrophobic interactions (Fig. 4d). This may explain the conformational changes in the bottom region of TM1b. The movement of TM1b induces nearly rigid movement in TM1a and the associated NBD1 (Fig. 4d and Supplementary Fig. 10e–g). Other than these regions, the rest parts remain almost unchanged between Cdr1Mil and Cdr1Apo (Supplementary Fig. 10e).
Inhibitory mechanism of milbemycin oxime
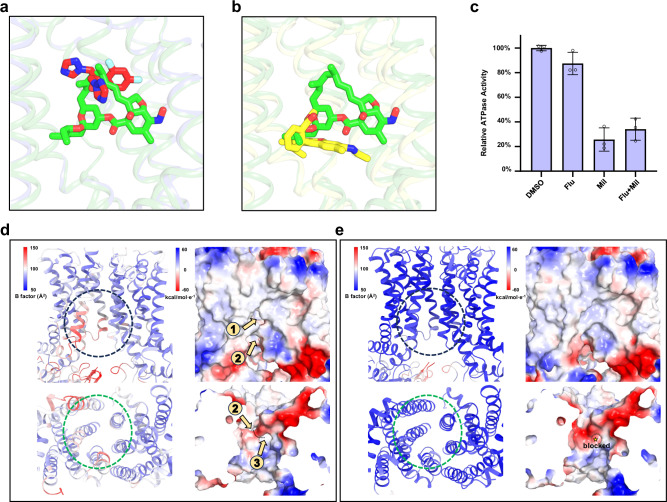
Structural comparison of Cdr1Mil and Cdr1Flu sheds light on our understanding of the inhibitory mechanism of milbemycin oxime for Cdr1. Milbemycin oxime partially overlaps with fluconazole, resulting in a steric clash between the two compounds (Fig. 5a). This incompatibility between milbemycin oxime and fluconazole may explain the inhibitory effect of milbemycin oxime, to some extent. Additionally, structural alignment of Cdr1Mil and Pdr5R6G also reveals a steric clash between milbemycin oxime and R6G (Fig. 5b). Given the substantial and expansive size of milbemycin oxime, which completely occupies the central cavity, it has the potential to act as a broad-spectrum inhibitor for Cdr1.
Fig. 5. Inhibitory mechanism of milbemycin oxime.
a Overlapped binding poses of milbemycin oxime and fluconazole. Steric clash explains their competition for binding to Cdr1. Fluconazole, and milbemycin oxime are colored by red and green, respectively. b Overlapped binding poses of milbemycin oxime in Cdr1Mil and R6G in Pdr5R6G (PDB: 7P05). Steric clash explains their competition for binding to Cdr1. Milbemycin oxime and R6G are colored by green and yellow, respectively. c Relative ATPase activity of wild type Cdr1 by adding DMSO, fluconazole (Flu), milbemycin oxime (Mil) or fluconazole with milbemycin oxime (Flu+Mil). The concentrations of fluconazole and/or milbemycin oxime were ten times greater than that of the purified Cdr1. These drugs are dissolved in DMSO. Data are presented as mean values ± SD; n = 3 independent experiments. Stability analysis of the cytoplasmic and inner-leaflet entrances of Cdr1Flu (d) and Cdr1Mil (e). The left panels in both figures are shown by B-factor analysis. Higher B-factors are depicted in red, indicating greater flexibility, while lower B-factors are shown in blue, signifying rigidity. The electrostatic potential is color-coded in the right panels, ranging from red (indicating negative charge) to blue (indicating positive charge). The dashed circle marks the cytoplasmic (black) and inner-leaflet (green) entrances in the panels. The orange arrows indicate the substrate entrance channel.
If milbemycin oxime acts solely as a competitor of fluconazole within the central cavity, it might be exported by Cdr1 as a substrate and, thus, may not exhibit potent inhibition23 (Fig. 1c). This suggests that milbemycin oxime may hinder exportation. Indeed, the ATPase activity assay revealed that milbemycin oxime inhibited ATP hydrolysis to 25.7% (Fig. 5c), indicating its potential to hinder the outward conformation required for exportation. As mentioned earlier, milbemycin oxime engages in numerous hydrophobic interactions with the residues in the central cavity surrounded by two TMDs of Cdr1, which suggests that milbemycin oxime may act as molecular glue to constrain conformational changes even after ATP binding. In contrast, ATPase activity decreases slightly upon the addition of fluconazole (Fig. 5c), which is consistent with the lack of the substantial conformational change following fluconazole binding (Supplementary Fig. 7a). Remarkably, in the presence of a mixture of fluconazole and milbemycin oxime, ATP hydrolysis decreases to 34.1%, slightly higher than that observed with milbemycin oxime alone (Fig. 5c). This suggests that milbemycin oxime exhibits a higher binding affinity to the central cavity compared to fluconazole. Indeed, the microscale thermophoresis (MST) assay showed that milbemycin oxime bound more strongly to Cdr1 than fluconazole, with dissociation constants (Kd) of 147 nM and 61.4 µM (Supplementary Fig. 11), respectively. Consequently, milbemycin oxime competes with substrate binding and inhibits the transport activity of Cdr1 by blocking ATP hydrolysis.
Like Pdr5, Cdr1Flu adopts an inward-facing conformation with the substrate entry channel opened towards the cytoplasm and inner leaflet of the membrane (Figs. 2a, 5d)19. Despite the apparent narrowness of the entrance region, it displays a certain degree of flexibility, as evidenced by the B factor analysis (Fig. 5d), which enables substrates of certain sizes to have the potential to pass through the channel. In contrast, when milbemycin oxime is introduced, conformational changes occur, leading to the closure of both the cytoplasmic and inner-leaflet entrances (Fig. 5e). Moreover, the entrance region of Cdr1Mil appears to be relatively rigid, as suggested by the B factor analysis (Fig. 5e). Consequently, milbemycin oxime effectively hinders substrates from entering the central cavity, thereby serving as another mechanism of inhibiting Cdr1. In summary, milbemycin oxime inhibits Cdr1 through a multifaceted mechanism involving competition, hindrance of exportation, and obstruction of substrate entry.
Discussion
In C. albicans, Cdr1 acts as an efflux pump to effectively remove azole compounds from fungal cells. This prevents their accumulation to harmful levels, ultimately leading to azole-drug resistance. Despite the molecular cloning of C. albicans Cdr1 dating back to 199527, the precise mechanism underlying how this efflux pump recognizes and transports azole drugs have remained elusive for nearly three decades. In this work, we have successfully determined the structural details of fluconazole-bound Cdr1. Complemented by structural, biochemical and functional analyses, our study provides a comprehensive understanding of how Cdr1 recognizes and transports fluconazole, a representative short-tail azole drug. This work shed lights on a 30-year-old puzzle regarding how Pdr5 subfamily (including Cdr1) mediates azole resistance.
Considering the limited availability of antifungal drugs to treat multi-drug resistant fungal infections, understanding the inhibitory mechanism for Cdr1 is crucial for the development of novel Cdr1 inhibitors aimed at addressing drug resistance. In this work, we report the high-resolution structure of milbemycin oxime-inhibited Cdr1, providing insight into Cdr1’s interaction with an inhibitor. Our findings suggest that milbemycin oxime inhibits Cdr1 through three distinct mechanisms: competitive binding with fluconazole, inhibition of ATPase activity, and closure of the entry channel. Furthermore, the structural insights gained from the milbemycin oxime-inhibited Cdr1 may offer valuable clues regarding the inhibitory mechanisms of other macrolide inhibitors, such as FK506, enniatin B, and beauvericin21–23. These findings open up new possibilities for the development of effective inhibitors targeting Cdr1, ultimately aiding in the fight against azole-drug-resistant fungal infections.
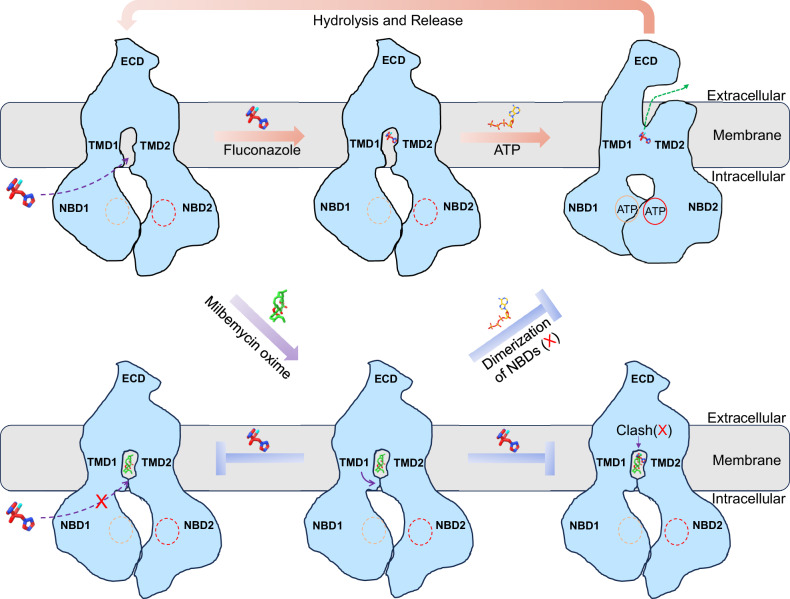
The structural and functional results mentioned above imply a model of fluconazole recognition, exportation and inhibition by milbemycin oxime (Fig. 6). In its resting state, Cdr1 maintains an inward-open conformation, allowing the entry of hydrophilic substrates from the cytoplasm or hydrophobic substrates from the inner membrane leaflet. As fluconazole enters the hydrophobic cavity, ATP molecules act as molecular glue to promote the dimerization of the two NBDs. This dimerization of two NBDs subsequently triggers outward conformational changes in the two TMDs, which, in turn, facilitate the expulsion of fluconazole. Following the hydrolysis of the nucleotide in NBD2, the transporter reverts back to its inward-facing conformation, initiating the next cycle of substrate transport. Milbemycin oxime, upon entering the hydrophobic cavity, induces a conformational change that closes the entrances. It competes for binding at the central hydrophobic cavity, obstructing fluconazole and other substrates from entering this central region. Additionally, the closure of the entrances effectively blocks the entry of fluconazole and other substrates into this central region. Furthermore, milbemycin oxime acts as a molecular glue to lock the two TMDs, thereby inhibiting ATPase activity and preventing the transportation process. The mechanisms of Cdr1-mediated fluconazole resistance and its inhibition by milbemycin oxime that we have revealed here will serve as a catalyst for future investigations to combat antifungal drug resistance.
Fig. 6. Model for fluconazole efflux and inhibition by milbemycin oxime.
In the resting state, Cdr1 maintains an inward-open conformation, allowing the entry of hydrophilic fluconazole from the cytoplasm (dash purple arrow). As fluconazole enters the hydrophobic cavity, ATP molecules act as molecular glue to induce dimerization of NBDs and trigger a conformational change from inward to outward, leading to the release of fluconazole (dash green arrow). Following the hydrolysis of the ATP in NBD2, Cdr1 returns to its inward-facing conformation, starting the next cycle of fluconazole transport. Milbemycin oxime, upon entering the hydrophobic cavity, induces a conformational change that closes the entrances (purple arrow). It competes for binding at the central hydrophobic cavity, obstructing fluconazole. Additionally, the closure of the entrances effectively blocks the entry of fluconazole into this region (blocked dash purple arrow). Furthermore, milbemycin oxime acts as a molecular glue, locking the two TMDs, inhibiting ATPase activity, and preventing the transportation process.
Methods
Cloning, expression and purification
The Cdr1 gene was amplified from the genome of C. albicans SC5314 strain and then cloned into the p416GAL1 vector using ClonExpress Ultra One Step Cloning Kit (Vazyme Biotech), incorporating a C-terminal tandem twinstrep II and flag tag. This same method was employed to generate all Cdr1 variants. Cdr1 was transformed into S. cerevisiae INVSc1 strain by the lithium acetate method28. Yeast cells were initially cultured in synthetic uracil-dropout medium (SD-Ura, Coolaber) at 30°C, with agitation at 200 rpm, for 24 h. Subsequently, they were added into YPG medium (containing 1% Yeast, 2% Peptone, and 2% D-Galactose) for an additional 24 h to induce protein over-expression. Yeast cells were harvested and suspended in a buffer consisting of 25 mM Tris-HCl at pH 8.0, 150 mM NaCl with protease inhibitors, and then disrupted for six rounds by cell disruptor (nano-1500, ATS). The cell debris was removed by low-speed centrifugation (4000 g) for 10 min. After centrifugation at 58000 g for 1 h, the membranes were collected and solubilized by 1% n-dodecyl-α-d-maltoside (DDM, Anatrace) and 0.2% cholesteryl hemisuccinate Tris salt (CHS, Anatrace) for 1.5 h at 4 °C. Insoluble materials were removed by centrifugation at 58000 g for 1 h and the detergent solubilized supernatant was then applied to Strep-Tactin resin (IBA). The resin was washed with buffer containing 25 mM Tris-HCl at pH 8.0, 150 mM NaCl, 0.02% glyco-diosgenin (GDN, Anatrace) and eluted with buffer containing 25 mM Tris-HCl at pH 8.0, 150 mM NaCl, 0.02% GDN, 2.5 mM desthiobiotin. The eluted protein was concentrated and subjected to further purification through size-exclusion chromatography (Superose 6 10/300 GL Increased, Cytiva). The peak fractions were analyzed by 15% SDS-PAGE. The purified sample was concentrated and stored at −80 °C for future use.
Cryo-EM sample preparation and data collection
The Cdr1 protein sample, at a concentration of about 24 mg/mL, was mixed with either 10 mM fluconazole or 200 μM milbemycin oxime and incubated on ice for 30 min. Aliquots of 3.5 μL of the sample were applied to a glow-discharged holey carbon grid (R1.2/1.3, 400 mesh, Quantifoil), and then incubated for 30 s and subsequently blotted for 4 s at 8 °C and 100% humidity. The grids were subsequently plunge-frozen in liquid ethane cooled by liquid nitrogen using FEI Vitrobot Mark IV, and then loaded into an FEI Titan Krios electron microscope operated at 300 kV. All images were recorded using a K3 direct electron detector (Gatan) at a nominal magnification of 105000x (physical pixel size: 0.837 Å/pixel) using AutoEMation software29 for Cdr1Apo and Cdr1Flu or 81000x (physical pixel size: 1.088 Å/pixel) using EPU software for Cdr1mil. For Cdr1Apo and Cdr1Flu, all micrographs were dose-fractionated to 32 frames with a total exposure time of 2.56 s and total dose of 50 e/Å2. For Cdr1Mil, all micrographs were dose-fractionated to 32 frames with a total exposure time of 3.2 s and total dose of 55 e/Å2. A defocus range of −1.5 to −2.1 µm was used for all micrographs.
Cryo-EM image processing
For the Cdr1Flu dataset, beam-induced motion of 2007 movies were corrected by MotionCor230. Defocus parameters were estimated using CTFFIND431. A total of 2,382,785 autopicked particles underwent multiple rounds of 2D and 3D classification within RELION32 to create a high-quality 3D reference. The original particles were divided into four datasets, and for each dataset, 3D classification was performed using the previously reconstructed 3D reference. The best class from each dataset was selected and merged, resulting in a total of 723,501 particles. Following 3D classification, 3D refinement, CTF refinement, and post-processing, a high-quality map was reconstructed at a resolution of 3.30 Å, based on the ‘gold standard’ Fourier shell correlation (FSC) = 0.143. Local-resolution estimations were performed by ResMap33.
For the data processing of Cdr1Apo and Cdr1Mil, 3950 and 3985 dosed-weighted micrographs were imported into cryoSPARC34 to determine CTF parameters through patch-CTF. Initial particles were picked based on the template from Cdr1Flu. Three rounds of 2D classification were performed to select good particles for ab-initio reconstruction, resulting in 690,501 and 652,297 particles for Cdr1Apo and Cdr1Mil, respectively. An initial map with distinct features was generated and served as a reference for subsequent heterogeneous refinement, leaving 206,300 and 329,525 particles for Cdr1Apo and Cdr1Mil, respectively. After applying non-uniform refinement, CTF refinement and local refinement, we eventually obtained the cryo-EM maps of Cdr1Apo and Cdr1Mil at resolutions of 3.38 Å and 3.08 Å, respectively, based on the ‘gold standard’ Fourier shell correlation (FSC) = 0.143.
Model building
The Cdr1 model, as predicted by Alphafold35, and fluconazole molecule were fitted into Cdr1Flu cryo-EM map in Chimera36 and adjusted manually by Coot37 based on the density. The resulting model then underwent real space refinement in Phenix38. For the Cdr1Apo and Cdr1Mil, the models were built in Coot using Cdr1Flu structure as the initial model and refined in real space by Phenix. The restraints files of milbemycin oxime and PIP2 were generated by AceDRG39 in CCP4 and eLBOW40 in Phenix. The geometries of all models were evaluated by MolProbity41. Cryo-EM data collection, refinement and validation statistics are presented in Supplementary Table 1.
Drug-sensitivity assay
Pdr5 in S. cerevisiae BY4741 strain was knocked out using LEU2 selection marker. Cdr1 wild-type and mutants were constructed using p416GAL1 plasmid and subsequently transformed into the ΔPdr5 strain. The yeast cells were initially inoculated in fresh SC-Ura medium with 2% glucose for 24 h and then resuspended at an OD600nm value of 0.01 in fresh SC-Ura medium with 2% galactose and varying drug concentrations. Drug susceptibility was assessed by measuring the OD600nm after culturing at 30°C for 48 h. Each result was analyzed using GraphPad Prism 8.0 software.
ATP activity assay
ATPase activity was assessed by measuring the release of inorganic phosphate (Pi) according to the Malachite Green Phosphate Detection Kit (Beyotime Biotechnology, China). Briefly, 4 μM of fluconazole or milbemycin oxime were incubated with 0.4 μM of purified Cdr1 in ice for 30 min. Reactions were carried out at 37 °C for 30 min in a 50-μL system containing 50 mM HEPES 7.4, 150 mM NaCl, 0.02% GDN, 2 mM ATP/MgCl2. For various Cdr1 mutants with deceased drug sensitivity, 1.0 μM of purified Cdr1 mutants were incubated with 2 mM ATP/MgCl2 at 37 °C for 30 min in a 50-μL system containing 50 mM HEPES (pH 7.4), 150 mM NaCl, and 0.02% GDN. The reaction mixture was then diluted with 50 μL of ddH2O before adding 35 μL of chromogenic reagent and stopped by adding 10 μL citric acid. The mixture was incubated for an additional 20 min at room temperature. Each data point was measured at 630 nm by BioTek Synergy H1 microplate reader. The data were subsequently analyzed using GraphPad Prism 8.0 software.
Western blotting analysis
Cdr1 wild-type and mutants were transformed into the S. cerevisiae BY4741 ΔPdr5 strain and induced with 2% galactose. The cells were normalized based on OD600nm, then disrupted and extracted using 1% DDM and 0.2% CHS for 2 h at 4 °C. The samples were loaded onto the 8% SDS-PAGE gels. After electrophoresis, the separated proteins were transferred onto a PVDF membrane. The membrane was blocked in 5% skimmed milk at 4 °C overnight and then incubated with an anti-Flag mouse monoclonal antibody (Abmart, catalog number M20008, 5000-fold dilution) for 1 h at room temperature. After three washes with TBST buffer (25 mM Tris at pH 7.5, 150 mM NaCl, and 0.1% Tween 20), the membrane was incubated with a goat anti-mouse secondary antibody (Abmart, catalog number M21001, 5000-fold dilution) for 1 h at room temperature. Following additional washes with TBST buffer and a final wash with TBS buffer (25 mM Tris at pH 7.5, and 150 mM NaCl), the membrane was detected using a ChemiDoc MP (Bio-Rad) with the enhanced chemiluminescence method.
Microscale thermophoresis (MST)
Cdr1 fused with EGFP at the C-terminus was purified in phosphate-buffered saline (PBS) with 0.02% DDM. The binding affinity of Cdr1 with fluconazole was assessed by incubating 50 nM Cdr1 with fluconazole at 16 different concentrations, ranging from 1 mM to 0.0000305 mM, in PBS buffer with 0.02% DDM for 20 min at 25 °C. Similarly, the binding affinity of Cdr1 with milbemycin oxime was assessed by incubating 100 nM Cdr1 with milbemycin oxime at 16 different concentrations, ranging from 10 µM to 0.000305 µM, in PBS buffer with 0.02% DDM for 20 min at 25 °C. Samples were loaded into capillaries, and MST analyses were conducted using a Monolith NT.115 (Nano-Temper Technologies GmbH) at 25 °C with 40% LED power and 40% MST power. Each assay was repeated three times, and Kd values were calculated using MO. Affinity Analysis v.2.3 software.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
This work was supported by the National Natural Science Foundation of China (32371279 and 32100972 to Zhaofeng Yan; 32200992 to Meng Yin), the Science and Technology Innovation Program of Hunan Province (2021RC3046 to Zhaofeng Yan), the Natural Science Foundation of Hunan Province (2023JJ20007 and 2022JJ40052 to Meng Yin; 2023SK2081 to Zhixiong Fang) and the Changsha Municipal Natural Science Foundation (kq2202151 to Meng Yin). We thank the Tsinghua University Branch of China National Center for Protein Sciences (Beijing) for the cryo-EM facility support and protein preparation (Fan Yang, Xiaomin Li and Bo Shen for technical support in EM data acquisition; Zi Yang for protein preparation). We thank the Shanxi Academy of Advanced Research and Innovation for the cryo-EM facility support, and Yuqian Mi, Erqi Pang, Qianqian Dong for technical support on the Cryo-EM.
Author contributions
Zhaofeng Yan conceived and supervised the project. Y.P., M.Y. and Zhaofeng Yan designed all the experiments. Y.P., X.H., Z.F., Y.Q., J.T. and T.Q. performed the molecular cloning and protein purification. Y.P. performed the biochemical and functional assays. Y.L., H.S., J.M. and X.L. collected the cryo-EM data. H.S. performed the cryo-EM data processing and model building. All authors contributed to the data analysis and manuscript preparation. H.S., Y.P., M.Y., and Zhaofeng Yan prepared the figures. Zhaofeng Yan wrote the manuscript.
Peer review
Peer review information
Nature Communications thanks Atsushi Kodan and the other, anonymous, reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
Density maps of the Cdr1Apo, Cdr1Flu and Cdr1Mil are available through the EMDB with entry codes EMD-60908, EMD-60909 and EMD-60910, respectively. Models of the Cdr1Apo, Cdr1Flu and Cdr1Mil are deposited in the Protein Data Bank (PDB) with entry codes 9IUK, 9IUL and 9IUM, respectively. Previously reported Pdr5 structures used in this study are: 7P03, 7P05, and 7P06. Sequences of mutagenesis primers in this study are included in Supplementary Data 1. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Ying Peng, Yan Lu, Hui Sun, Jinying Ma, Xiaomei Li, Xiaodan Han.
Contributor Information
Meng Yin, Email: yinm@hnu.edu.cn.
Zhaofeng Yan, Email: zhaofengyan@hnu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-52107-w.
References
- 1.Bongomin, F., Gago, S., Oladele, R. O. & Denning, D. W. Global and multi-national prevalence of fungal diseases-estimate precision. J. Fungi (Basel)3, 57 (2017). 10.3390/jof3040057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robbins, N., Caplan, T. & Cowen, L. E. Molecular evolution of antifungal drug resistance. Annu Rev. Microbiol.71, 753–775 (2017). 10.1146/annurev-micro-030117-020345 [DOI] [PubMed] [Google Scholar]
- 3.Benitez, L. L. & Carver, P. L. Adverse effects associated with long-term administration of azole antifungal agents. Drugs79, 833–853 (2019). 10.1007/s40265-019-01127-8 [DOI] [PubMed] [Google Scholar]
- 4.Kainz, K., Bauer, M. A., Madeo, F. & Carmona-Gutierrez, D. Fungal infections in humans: the silent crisis. Micro. Cell7, 143–145 (2020). 10.15698/mic2020.06.718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monk, B. C. & Goffeau, A. Outwitting multidrug resistance to antifungals. Science321, 367–369 (2008). 10.1126/science.1159746 [DOI] [PubMed] [Google Scholar]
- 6.Perea, S. et al. Prevalence of molecular mechanisms of resistance to azole antifungal agents in Candida albicans strains displaying high-level fluconazole resistance isolated from human immunodeficiency virus-infected patients. Antimicrob. Agents Chemother.45, 2676–2684 (2001). 10.1128/AAC.45.10.2676-2684.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.White, T. C., Marr, K. A. & Bowden, R. A. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin. Microbiol Rev.11, 382–402 (1998). 10.1128/CMR.11.2.382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Banerjee, A. et al. Directed mutational strategies reveal drug binding and transport by the MDR transporters of Candida albicans. J. Fungi (Basel)7, 68 (2021). 10.3390/jof7020068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fisher, M. C. et al. Tackling the emerging threat of antifungal resistance to human health. Nat. Rev. Microbiol20, 557–571 (2022). 10.1038/s41579-022-00720-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martins, N., Ferreira, I. C., Barros, L., Silva, S. & Henriques, M. Candidiasis: predisposing factors, prevention, diagnosis and alternative treatment. Mycopathologia177, 223–240 (2014). 10.1007/s11046-014-9749-1 [DOI] [PubMed] [Google Scholar]
- 11.Cannon, R. D. et al. Efflux-mediated antifungal drug resistance. Clin. Microbiol Rev.22, 291–321 (2009). 10.1128/CMR.00051-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lamping, E. et al. Fungal PDR transporters: Phylogeny, topology, motifs and function. Fungal Genet Biol.47, 127–142 (2010). 10.1016/j.fgb.2009.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Egner, R., Rosenthal, F. E., Kralli, A., Sanglard, D. & Kuchler, K. Genetic separation of FK506 susceptibility and drug transport in the yeast Pdr5 ATP-binding cassette multidrug resistance transporter. Mol. Biol. Cell9, 523–543 (1998). 10.1091/mbc.9.2.523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakamura, K. et al. Functional expression of Candida albicans drug efflux pump Cdr1p in a Saccharomyces cerevisiae strain deficient in membrane transporters. Antimicrob. Agents Chemother.45, 3366–3374 (2001). 10.1128/AAC.45.12.3366-3374.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shukla, S. et al. Functional characterization of Candida albicans ABC transporter Cdr1p. Eukaryot. Cell2, 1361–1375 (2003). 10.1128/EC.2.6.1361-1375.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao, P. H. et al. Drug susceptibilities of yeast cells are affected when expressing mutant Candida albicans drug resistance protein. Int J. Antimicrob. Agents28, 69–74 (2006). 10.1016/j.ijantimicag.2006.02.016 [DOI] [PubMed] [Google Scholar]
- 17.Tsao, S., Rahkhoodaee, F. & Raymond, M. Relative contributions of the Candida albicans ABC transporters Cdr1p and Cdr2p to clinical azole resistance. Antimicrob. Agents Chemother.53, 1344–1352 (2009). 10.1128/AAC.00926-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanglard, D., Ischer, F., Monod, M. & Bille, J. Cloning of Candida albicans genes conferring resistance to azole antifungal agents: characterization of CDR2, a new multidrug ABC transporter gene. Microbiol. (Read.)143, 405–416 (1997). 10.1099/00221287-143-2-405 [DOI] [PubMed] [Google Scholar]
- 19.Harris, A. et al. Structure and efflux mechanism of the yeast pleiotropic drug resistance transporter Pdr5. Nat. Commun.12, 5254 (2021). 10.1038/s41467-021-25574-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prasad, R., Banerjee, A., Khandelwal, N. K. & Dhamgaye, S. The ABCs of Candida albicans Multidrug Transporter Cdr1. Eukaryot. Cell14, 1154–1164 (2015). 10.1128/EC.00137-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanabe, K. et al. FK506 resistance of saccharomyces cerevisiae Pdr5 and Candida albicans Cdr1 involves mutations in the transmembrane domains and extracellular loops. Antimicrob. Agents Chemother.63, e01146–18 (2019). 10.1128/AAC.01146-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niimi, K. et al. Specific interactions between the Candida albicans ABC transporter Cdr1p ectodomain and a D-octapeptide derivative inhibitor. Mol. Microbiol.85, 747–767 (2012). 10.1111/j.1365-2958.2012.08140.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Silva, L. V. et al. Milbemycins: more than efflux inhibitors for fungal pathogens. Antimicrob. Agents Chemother.57, 873–886 (2013). 10.1128/AAC.02040-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rawal, M. K. et al. Insight into pleiotropic drug resistance ATP-binding cassette pump drug transport through mutagenesis of Cdr1p transmembrane domains. J. Biol. Chem.288, 24480–24493 (2013). 10.1074/jbc.M113.488353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szepinski, E., Martynow, D., Szweda, P., Milewska, M. J. & Milewski, S. Voriconazole-based salts are active against multidrug-resistant human pathogenic yeasts. Molecules24, 3635 (2019). 10.3390/molecules24203635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niimi, M., Niimi, K., Tanabe, K., Cannon, R. D. & Lamping, E. Inhibitor-resistant mutants give important insights into Candida albicans ABC Transporter Cdr1 substrate specificity and help elucidate efflux pump inhibition. Antimicrob. Agents Chemother.66, e0174821 (2022). 10.1128/AAC.01748-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prasad, R., De Wergifosse, P., Goffeau, A. & Balzi, E. Molecular cloning and characterization of a novel gene of Candida albicans, CDR1, conferring multiple resistance to drugs and antifungals. Curr. Genet27, 320–329 (1995). 10.1007/BF00352101 [DOI] [PubMed] [Google Scholar]
- 28.Gietz, R. D. & Woods, R. A. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol.350, 87–96 (2002). 10.1016/S0076-6879(02)50957-5 [DOI] [PubMed] [Google Scholar]
- 29.Lei, J. & Frank, J. Automated acquisition of cryo-electron micrographs for single particle reconstruction on an FEI Tecnai electron microscope. J. Struct. Biol.150, 69–80 (2005). 10.1016/j.jsb.2005.01.002 [DOI] [PubMed] [Google Scholar]
- 30.Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods14, 331–332 (2017). 10.1038/nmeth.4193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rohou, A. & Grigorieff, N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol.192, 216–221 (2015). 10.1016/j.jsb.2015.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol.180, 519–530 (2012). 10.1016/j.jsb.2012.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kucukelbir, A., Sigworth, F. J. & Tagare, H. D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods11, 63–65 (2014). 10.1038/nmeth.2727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods14, 290–296 (2017). 10.1038/nmeth.4169 [DOI] [PubMed] [Google Scholar]
- 35.Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature596, 583–589 (2021). 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pettersen, E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput Chem.25, 1605–1612 (2004). 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- 37.Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr.66, 486–501 (2010). 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr.66, 213–221 (2010). 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Long, F. et al. AceDRG: a stereochemical description generator for ligands. Acta Crystallogr. D. Struct. Biol.73, 112–122 (2017). 10.1107/S2059798317000067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moriarty, N. W., Grosse-Kunstleve, R. W. & Adams, P. D. Electronic Ligand Builder and Optimization Workbench (eLBOW): a tool for ligand coordinate and restraint generation. Acta Crystallogr. D. Biol. Crystallogr. 65, 1074–1080 (2009). 10.1107/S0907444909029436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. Biol. Crystallogr.66, 12–21 (2010). 10.1107/S0907444909042073 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
Density maps of the Cdr1Apo, Cdr1Flu and Cdr1Mil are available through the EMDB with entry codes EMD-60908, EMD-60909 and EMD-60910, respectively. Models of the Cdr1Apo, Cdr1Flu and Cdr1Mil are deposited in the Protein Data Bank (PDB) with entry codes 9IUK, 9IUL and 9IUM, respectively. Previously reported Pdr5 structures used in this study are: 7P03, 7P05, and 7P06. Sequences of mutagenesis primers in this study are included in Supplementary Data 1. Source data are provided with this paper.