Abstract
Ferroptosis is an important pathological mechanism of chronic heart failure (CHF). This study aimed to investigate the protective mechanism of Astragaloside IV (AS-IV) on CHF rats by integrating bioinformatics and ferroptosis. CHF-related targets and ferroptosis-related targets were collected. After the intersection, the common targets were obtained. The PPI network of the common targets was constructed, and topological analysis of the network was carried out. The target with the highest topological parameter values was selected as the key target. The key target p53 was obtained through bioinformatics analysis, and its molecular docking model with AS-IV was obtained, as well as molecular dynamics simulation analysis. The rat models of CHF after myocardial infarction were established by ligation of left coronary artery and treated with AS-IV for 4 weeks. AS-IV treatment significantly improved cardiac function in CHF rats, improved cardiomyocyte morphology and myocardial fibrosis, reduced mitochondrial damage, decreased myocardial MDA and Fe2+ content, increased GSH content, inhibited the expression of p53 and p-p53, and up-regulated the expression of SLC7A11 and GPX4. In conclusion, AS-IV improved cardiac function in CHF rats, presumably by regulating p53/SLC7A11/GPX4 signaling pathway and inhibiting myocardial ferroptosis.
Subject terms: Cardiology, Medical research
Introduction
Chronic heart failure (CHF) is a complex group of clinical syndromes, whose symptoms and signs are caused by various factors leading to cardiac diastolic dysfunction and impaired blood pumping ability. CHF is the ultimate destination and leading cause of death in the development of most cardiovascular diseases1. Ferroptosis is a distinct regulated form of cell death driven by iron-dependent lipid peroxidation, and its main characteristics are iron overload and lipid peroxidation2. Ferroptosis plays an important role in a variety of diseases, such as tumors, gastrointestinal diseases, neurodegenerative diseases, cardiovascular and cerebrovascular diseases, etc3. In recent years, more and more studies have found that ferroptosis is closely related to myocardial injury in CHF, and the regulation of ferroptosis in myocardium has also become a hot research field on the prevention and treatment of CHF4.
With thousands of years of application experience and good clinical efficacy, traditional Chinese medicine has received extensive attention in the prevention and treatment of CHF, and relevant mechanism studies have been increasing year by year. Moreover, a variety of traditional Chinese medicine and natural active ingredients have been confirmed to regulate the pathways related to ferroptosis, improve myocardial injury and inhibit the progression of CHF effectively5. Radix Astragali is a commonly used traditional Chinese medicine for the treatment of CHF, and Astragaloside IV (AS-IV) is one of its main active components, which has various pharmacological effects on the prevention and treatment of CHF6, however, there are few studies on AS-IV in regulating the mechanism of ferroptosis so far.
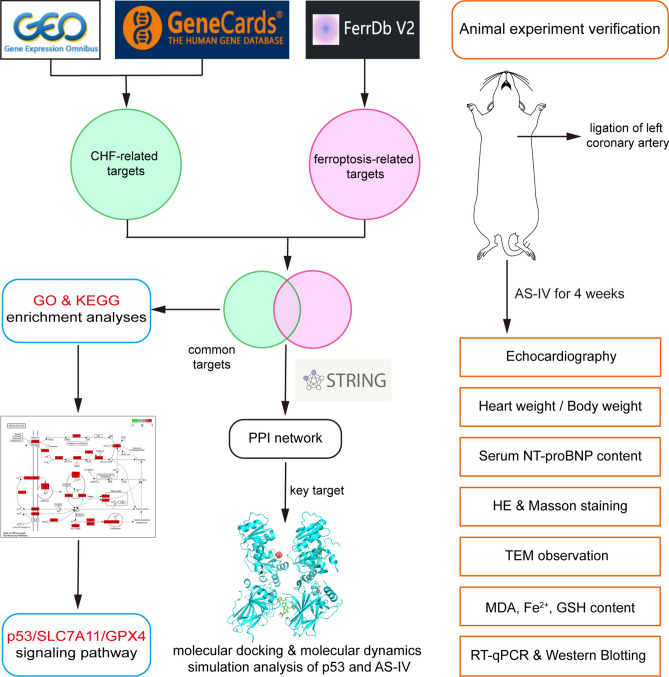
In this study, the potential targets and pathways of CHF treatment were analyzed from the perspective of ferroptosis through bioinformatics, and the key target “p53” was screened. Molecular docking and molecular dynamics (MD) simulation analysis of p53 and AS-IV were performed. The rat models of CHF after myocardial infarction were established by ligation of left coronary artery. Then, AS-IV was used to treat CHF rats for 4 weeks, and its effects on cardiac function and ferroptosis indicators, such as Malondialdehyde (MDA), Fe2+, Glutathione (GSH) and p53/Solute carrier family 7 member 11 (SLC7A11)/Glutathione peroxidase 4 (GPX4) signaling pathway, were observed to further explore the mechanism of AS-IV in treating CHF. The workflow chart of the research process is displayed in (Fig. 1).
Fig. 1.
The workflow chart of bioinformatics analysis and animal experiment.
Materials and methods
Bioinformatics analysis
Collection of CHF-related targets
First, data sets related to CHF were retrieved through GEO database (https://www.ncbi.nlm.nih.gov/gds/), and the screening criteria was “Homo sapiens, study type of expression profiling by array, sample size greater than 50 and containing both normal and disease samples”. The screened data sets were annotated based on the corresponding platform file and merged by Perl, and batch normalized through the “sva” package of R software. Then the difference analysis of genes obtained after batch normalization was carried out through the “limma” package of R software. Then, “adjusted P-value < 0.05 and absolute value of logFC > 1” were set as the criteria to screen differentially expressed genes. The volcano plot and the heat map of differentially expressed genes were drawn by “ggplot2” and “pheatmap” packages of R software. Second, gene targets related to CHF were searched through GeneCards database (https://www.genecards.org/), and targets with relevance score > 10 were selected. Third, the targets retrieved from the above two databases were combined and the duplicate genes were removed to obtain CHF-related targets.
Collection of ferroptosis-related targets
The gene expression data for drivers, suppressors, and markers of ferroptosis were downloaded from FerrDb V2 platform7 (http://www.zhounan.org/ferrdb/). The three kinds of gene expression data were combined, and ferroptosis-related targets were obtained after removing duplicate genes.
PPI network of common targets and screening of core targets
The common targets between CHF-related targets and ferroptosis-related targets were obtained by intersecting them two using a Venn diagram8. The common targets were imported into STRING database (https://string-db.org/), and the analysis mode was set as “Multiple proteins” and species as “Homo sapiens” to obtain the protein–protein interaction (PPI) network of them. Then the PPI network was imported into Cytoscape3.9.0 software to build a visual network and execute topological analysis. With “Betweenness, Closeness and Degree all greater than the median” as the screening criterion, the network of core targets was acquired after two rounds of screenings.
GO and KEGG enrichment analyses of common targets
In order to understand the main gene functions and related signaling pathways, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were conducted for the common targets obtained in 2.1.3 with the help of R software, using packages including “colorspace”, “stringi”, “ggplot2”, “DOSE”, “clusterProfiler”, “enrichplot” and “pathview”.
Molecular docking of the key target with AS-IV
Through the above bioinformatics analysis, the target with the highest Betweenness, Closeness and Degree among the core targets was taken as the key target. The protein ID of the key target was retrieved through UniProt database (https://www.uniprot.org/), then the receptor file of the protein was downloaded through PDB database (https://www.rcsb.org/), and the water molecule and small molecule ligand were removed by PyMOL software. The 2D structure of AS-IV was downloaded from PubChem database (https://pubchem.ncbi.nlm.nih.gov/) and imported into ChemBio3D software to generate 3D structure and optimize modification to obtain AS-IV ligand file. The protein receptor file and AS-IV ligand file were imported into AutoDockTools9, then the protein receptor was hydrogenated, and the above two files were saved as pdbqt files. The active pocket of the protein receptor was determined and re-output as a gpf file after saving parameters. AutoDockVina10 software was used to execute molecular docking between the protein receptor and AS-IV ligand. The energy difference was set to 5 and the number of models was set to 20. The obtained models were visualized with PyMOL software.
MD simulation analysis of the key target-AS-IV complex
Gromacs2022.3 software11,12 was used for MD simulation. For the small molecule preprocessing, AmberTools22 was used to add GAFF force field to AS-IV ligand, while Gaussian 16W was used to hydrogenate AS-IV ligand and calculate RESP potential. Potential data were added to the topology file of MD system. The simulation conditions were carried out at static temperature of 300 K and atmospheric pressure of 1 Bar. Amber99sb-ildn was used as force field and water molecules were used as solvent (Tip3p water model). The total charge of the simulation system was neutralized by adding an appropriate number of Na+ ions. The simulation system adopted the steepest descent method to minimize the energy, and then carried out the isothermal isovolumic ensemble (NVT) equilibrium and isothermal isobaric ensemble (NPT) equilibrium for 100,000 steps, respectively, with the coupling constant of 0.1 ps and the duration of 100 ps. Finally, the free MD simulation was performed. The process consisted of 5,000,000 steps, with a step length of 2 fs and a total duration of 100 ns. After the simulation was completed, the built-in tool of the software was used to analyze the trajectory of each amino acid, and the root-mean-square deviation (RMSD), root-mean-square fluctuation (RMSF), the radius of gyration (Rg) value, solvent accessible surface area (SASA), hydrogen bonds (H-bonds), and binding free energy (MM/GBSA) were calculated.
Animal experiment verification
Drugs and reagents
AS-IV (Cat#SA8640) was obtained from Beijing Suolaibao Technology Co., Ltd. (Beijing, China) and prepared into suspension with a concentration of 2 mg/mL with normal saline. Pentobarbital sodium (Cat#P3761) came from Merck KgaA (Germany). Penicillin sodium for injection (Cat#H13020657) came from North China Pharmaceutical Co., Ltd. (Shijiazhuang, China). Isoflurane (Cat#R510-22-10) was obtained from RWD Life Science Co., Ltd. (Shenzhen, China). Rat N-Terminal Pro-Brain Natriuretic Peptide (NT-proBNP) ELISA kit (Cat#JM-01736R1) was obtained from Jiangsu Jingmei Biotechnology Co., Ltd. (Yancheng, China). Hematoxylin–eosin (HE) dye solution set (Cat#G1003), Masson dye solution set (Cat#G1006), RNA extraction solution (Cat#G3013), RIPA lysate (Cat# G2002-100ML), BCA protein assay kit (Cat#G2026), sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel preparation kit (Cat#G2003-50T), GAPDH antibody (Cat#B11002), p53 antibody (Cat#GB11626), GPX4 antibody (Cat#GB115275) and HRP-conjugated goat anti rabbit (Cat#GB23303) secondary antibodies were all purchased from Servicebio Technology Co., Ltd. (Wuhan, China). SLC7A11 antibody (Cat#ab307601) came from Abcam (UK). Phospho-p53 (p-p53) antibody (Cat#82530) came from Cell Signaling Technology (USA). MDA (Cat#E-BC-K025-M), Fe2+ (Cat#E-BC-K773-M) and GSH (Cat#E-BC-K030-M) colorimetric assay kits were all obtained from Elabscience Biotechnology Co., Ltd. (Wuhan, China).
Main instruments
Small animal ventilator (RWD/R407, China); Electrocardiograph (3RAY/VECG-2303B, China); Color doppler ultrasound imaging system for small animals (VINNO/6LAB, China); Enzyme labeling instrument (Rayto/RT-6100, China); Dehydrator (DIAPATH/Donatello, Italy); Embedding machine (Wuhan Junjie Electronics Co., LTD./JB-P5, China); Pathological microtome (Shanghai Leica Instrument Co., LTD./RM2016, China); Dyeing machine (DIAPATH/Giotto, Italy); Upright optical microscope (Nikon/Eclipse E100, Japan); Transmission electron microscope (HITACHI/HT7700, Japan); Fluorescent quantitative PCR instrument (Bio-rad/CFX Connect, USA); Ultramicrospectrophotometer (Thermo/NanoDrop2000, USA); Chemiluminescence instrument (CLINX/6100, China).
Animals and experiment protocols
A total of 30 SPF-grade Sprague Dawley male rats (No. 430727231100274116) (aged eight weeks, weighing 200–220 g) were purchased from Hunan Silaikejingda Experimental Animal Co., Ltd. with license No. SCXK (Xiang) 2019-0004. Based on the Resource Equation Approach13 (n = degrees of freedom/number of groups + 1) and taking into account 10–30% mortality due to modeling (according to our previous experience), the maximum sample size of n = 10 was selected. These rats were raised in the Experimental Animal Center of Hunan University of Chinese Medicine at a room temperature of (23 ± 2) °C and a humidity of (56 ± 4) % with free access to food and water. After feeding 30 rats for 1 week, they were randomly divided into sham operation group (Sham) (10 rats) and modeling group (20 rats). Random numbers were generated using the standard = RAND() function in Microsoft Excel. The left coronary artery ligation was performed on 20 rats in modeling group to establish CHF models after myocardial infarction14. Rats were anesthetized with 2% pentobarbital sodium (50 mg/kg). When the righting reflex and paw withdraw reflex disappeared, the breathing became deep and slow and regular, the rats were considered to be in a complete state of anesthesia. Then the rats were supine on a thermostatic pad and their limbs were fixed. After breast coat removal, skin disinfection, and mechanical ventilation, the skin on the left side of the rat breast was incised, and the 3–4 intercostal muscles and pericardium were separated by blunt dissection to fully expose the heart. Ligation using a 6-0 suture was performed approximately 2.5–3.5 mm below the root of the left coronary artery, and the thorax was closed with 3-0 sutures at the end. The operation was completed within 30 min. Electrocardiogram (ECG) detection was performed before and after operation. When the ECG showed ST segment elevation and the myocardial whitening at the ligation site was observed during operation, the myocardial infarction model could be judged successful. The rats were intramuscularly injected with penicillin (80,000 U/rat) for 3 days after operation to prevent infection. Rats in the Sham group underwent the same operation except ligation.
After 4 weeks of modeling, echocardiography was performed to detect the cardiac function of each group. The CHF model was considered successful when the left ventricular fractional shortening (LVFS) of modeling group was significantly lower than that of the Sham group. Then 20 rats in modeling group were randomly divided into model group (Model) and AS-IV group (AS-IV), with 10 rats in each group. The rats in AS-IV group were given AS-IV once a day (20 mg/kg by gavage) for 4 weeks with a volume of 10 mL/kg. The Model group and Sham group were given the same amount of normal saline. All researchers involved in the experiment were aware of the group allocation. While during the data analysis, the data was coded prior to analysis so that the treatment group couldn't be identified by the people who contributed to formal analysis. There were 9 rats in each group survived after 4 weeks of administration. All rats underwent echocardiography, serum assay, and heart weight index calculation. Within each group, 3 rats were randomly selected and subjected to HE and Masson staining, TEM observation, RT-qPCR and Western Blotting, while the other 6 rats were used for the detection of MDA, Fe2+ and GSH content. No animal was excluded for analysis. All authors complied with the ARRIVE guideline. All animal experiments were performed in accordance with the guidelines of Animal Research Ethics Committee of the Hunan University of Chinese Medicine, and approved by the Animal Research Ethics Committee of the Hunan University of Chinese Medicine (Permission Number: LL2022030207).
Echocardiography
After 4 weeks of AS-IV administration, the rats in each group were given respiratory anesthesia with 3% isoflurane. After completely anesthetized (the same criteria as above), the rats were supine and fixed on the dissecting table with 1% isoflurane to maintain anesthesia. Then, the breast coat was removed with hair removal cream to ensure the exposure of the skin from the neck to the abdomen, and ultrasound coupling agent was applied on to help observe the heart structure. Specifically, small animal high-frequency ultrasound probe (17–18 MHz) was used to detect the left ventricle, left atrium, right atrium, left ventricular outflow tract and other structures from the position parallel to the long axis of the heart. Left ventricular apical dynamic 2D and M-ultrasound images were collected at the lower margin of mitral papillary muscle for no less than 3 cardiac cycles, and LVFS, left ventricular end systolic dimension (LVIDs), left ventricular end diastolic dimension (LVIDd), left ventricular end systolic volume (LVESV) and left ventricular end diastolic volume (LVEDV) were measured.
Serum assay and heart isolation
After echocardiography, the rats were kept under anesthesia and blood was collected from the abdominal aorta. The collected blood was placed at room temperature for 2 h and centrifuged at 3000 rpm for 15 min. The serum obtained after centrifugation was packaged and stored in the refrigerator at − 80 °C, and the serum NT-proBNP content was determined by ELISA method according to the instruction of rat NT-proBNP ELISA kit. After collecting blood, the rats were sacrificed by cervical dislocation. Then the heart was isolated from the thorax, rinsed in cold normal saline to remove the blood, and dried with filter paper. The heart weight index (heart weight/body weight) was further calculated by weighing the heart.
HE and Masson staining
The ventricular part of the heart was taken by a ophthalmic scissor and fixed in 4% paraformaldehyde solution for 72 h, then embedded with paraffin and sliced transversely from the apex to the basal part of the left ventricular with the interval of 500 μm between each section. HE staining procedures: Paraffin slices were dewaxed to water, stained with hematoxylin for 3–5 min, differentiated with differentiation solution, reverted blue with reverted blue solution, stained with eosin for 5 min, sealed with neutral gum after dehydration, and examined under microscope to observe myocardial histomorphological changes and size of cardiomyocyte cross-section. Masson staining procedures: Paraffin slices were dewaxed to water, soaked overnight in Masson A solution, soaked in an equal mixture of Masson B and Masson C solution for 1 min, differentiated by differentiation solution, and soaked in Masson D solution for 6 min, Masson E solution for 1 min and Masson F solution for 20 s. After rinsing and differentiation with 1% acetic acid and following dehydration, the slice was sealed with neutral gum, and the myocardial histopathological changes including infarct size and myocardial fibrosis were observed under microscope. The images were captured and then analyzed using the ImageJ software. The infarct size was measured by area-based approaches15.
TEM observation
The heart tissue of the infarct area was taken with a ophthalmic scissor and placed in 4 °C fixative specially used for electron microscope. The tissue was cut into small pieces by scalpel in the fixative and then stained with uranium acetate and lead nitrate after resin embedding. The morphological and structural pathological changes of myocardial mitochondria were observed by transmission electron microscopy (TEM) and mitochondrial density and average size were measured using the ImageJ software. The mitochondrial profiles were manually drawn and threshold segmentation was used to identify mitochondria in TEM images. The number of mitochondria, total mitochondrial area and actual image area were calculated, and then the mitochondrial density (mitochondrial number/actual image area) and average size (total mitochondrial area/number of mitochondria) could be obtained.
MDA, Fe2+ and GSH content
After homogenization, the myocardium was centrifuged at 12,000 rpm and 4 °C for 10 min, and the supernatant was placed on ice waiting for detection. Part of the supernatant was reserved for determination of protein concentration. The contents of MDA, Fe2+ and GSH in myocardium were determined by colorimetry according to the instructions of MDA, Fe2+ and GSH colorimetric assay kits.
RT-qPCR
Total RNA was extracted from myocardial tissue and reverse transcribed into cDNA for RT-qPCR analysis. The amplification procedures included pre-denaturation at 95 °C for 10 min, 40 cycles of denaturation at 95 °C for 15 s and annealing at 60 °C for 30 s. The dissolution curve ranged from 65 to 95 °C, and the fluorescence signal was collected every time the temperature rose 0.5 °C. The data were processed by 2−ΔΔCT formula, and the mRNA expression of p53, SLC7A11 and GPX4 were determined. The primers, as shown in Table 1, were synthesized by Servicebio Technology Co., Ltd. (Wuhan, China).
Table 1.
Primer information for RT-qPCR.
| Gene | Primer sequence (5′–3′) | Length (bp) |
|---|---|---|
| GAPDH | Forward: CTGGAGAAACCTGCCAAGTATG | 138 |
| Reverse: GGTGGAAGAATGGGAGTTGCT | ||
| p53 | Forward: GGAGGATTCACAGTCGGATATG | 291 |
| Reverse: TGAGAAGGGACGGAAGATGAC | ||
| GPX4 | Forward: AGGCAGGAGCCAGGAAGTAATC | 212 |
| Reverse: ACCACGCAGCCGTTCTTATC | ||
| SLC7A11 | Forward: TATGCTGAATTGGGTACGAGC | 114 |
| Reverse: TATTACCAGCAGTTCCACCCA |
Western blotting
Protein concentration of myocardial tissue was determined by BCA assay, and the extracted proteins were added to the loading buffer and boiled. Proteins were Separated by SDS-PAGE and transferred onto polyvinyl difluoride membranes. The membranes were blocked with milk at room temperature for 2 h and then immunoblotted with primary antibodies of p53, p-p53, SLC7A11 and GPX4 (1:1000) overnight at 4 °C, and incubated with the secondary antibody (1:3000) for 1 h at room temperature. Finally, the membranes were exposed and photographed after adding ECL chemiluminescence reagent. The gray values of protein bands were analyzed by Image J software and the relative expressions of proteins (gray value of target protein/gray value of reference protein) were calculated.
Statistical analysis
All the measurement data were analyzed by statistical software SPSS 27 and expressed as mean ± standard deviation (). Shapiro–Wilk test was used to verify the normality of the data. Levene's Test verifies the homogeneity of the variance. Unpaired t-test was applied to the comparison between two groups. One-way ANOVA was used for comparison of more than two groups, and the LSD method was used when the data conformed to the homogeneity of variance; otherwise the Games-Howell method was used. When the data didn't conform to the normal distribution, the non-parametric tests including the Mann–Whitney U test (for comparing two groups) and the Kruskal–Wallis test (for comparing multiple groups) were used. Statistical significance was defined as P < 0.05.
Results
CHF-related targets and ferroptosis-related targets
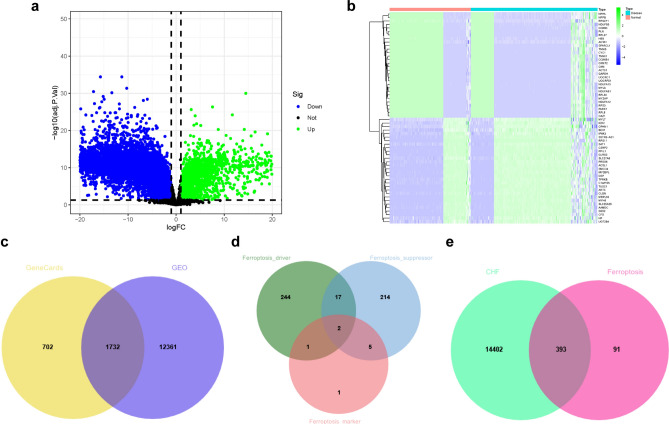
Two data sets GSE120895 and GSE57338 were obtained by retrieving and screening in GEO database. The GSE120895 dataset, whose title is “Correlation of clinical parameters of heart failure with myocardial gene expression indilated cardiomyopathy”, contains 47 disease samples and 8 normal samples. The GSE57338 dataset, whose title is “RNA-Seq Identifies Novel Myocardial Gene Expression Signatures of Heart Failure”, contains 177 disease samples and 136 normal samples. The two data sets were annotated respectively according to the platform files GPL570 and GPL11532. Then they were merged and batch normalized, and the “limma” difference analysis was executed. The number of differentially expressed genes was 14,093 and the volcano plot was shown as (Fig. 2a). Also, the heat map of differentially expressed genes was obtained by setting the gene number as 30 (Fig. 2b). A total of 12,796 CHF-related gene targets were retrieved from GeneCards database, and there were 2434 targets screened with relevance score greater than 10. A total of 14,795 CHF-related targets were obtained by combining the targets retrieved from the above two databases and removing duplicate genes (Fig. 2c). There were 484 ferroptosis-related targets collected by FerrDb V2 platform (Fig. 2d).
Fig. 2.
CHF-related targets and ferroptosis-related targets. (a) The volcano plot of differentially expressed genes. (b) The heat map of partial differentially expressed genes. (c) Venn diagram of CHF-related targets. (d) Venn diagram of ferroptosis-related targets. (e) Venn diagram of CHF-related targets and ferroptosis-related targets.
PPI network of common targets and network of core targets
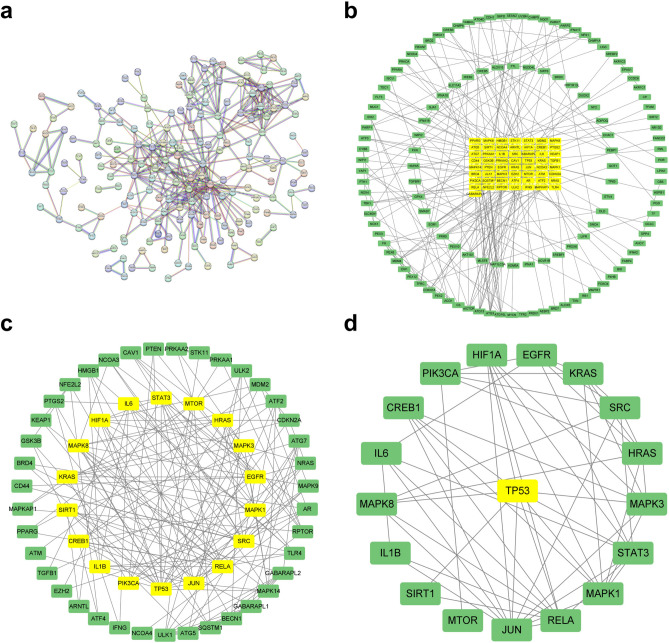
A total of 393 common targets were obtained by taking the intersection of CHF-related targets and ferroptosis-related targets (Fig. 2e). The common targets were imported into the STRING database, and the PPI network was obtained by setting the confidence level to 0.95 and hiding the free nodes (Fig. 3a). Then it was imported into Cytoscape3.9.0 software to build a visual network and execute topological analysis. After two screenings, the network of core targets was acquired (Fig. 3b–d, Table 2).
Fig. 3.
PPI network of common targets and network of core targets. (a) PPI network of common targets. (b) First screening of common targets. (c) Second screening of core targets. (d) Screening of key target p53 through topological analysis.
Table 2.
Topological analysis of the core targets.
| Gene | Betweenness | Closeness | Degree |
|---|---|---|---|
| TP53 | 865.0016346 | 0.5436893 | 20 |
| STAT3 | 147.1061907 | 0.4590164 | 9 |
| SRC | 247.9310858 | 0.4375 | 13 |
| SIRT1 | 359.7111989 | 0.4444444 | 8 |
| RELA | 466.2387138 | 0.5137615 | 14 |
| PIK3CA | 185.9385613 | 0.4210526 | 7 |
| MTOR | 283.2259574 | 0.3943662 | 9 |
| MAPK8 | 78.08196023 | 0.4444444 | 9 |
| MAPK3 | 58.0740529 | 0.4341085 | 9 |
| MAPK1 | 90.56457153 | 0.4444444 | 11 |
| KRAS | 118.0997267 | 0.4028777 | 8 |
| JUN | 154.5255859 | 0.4827586 | 14 |
| IL6 | 153.3786971 | 0.4210526 | 9 |
| IL1B | 67.20881341 | 0.4028777 | 7 |
| HRAS | 52.78975995 | 0.4117647 | 9 |
| HIF1A | 91.0314127 | 0.4375 | 9 |
| EGFR | 161.5513198 | 0.4179104 | 11 |
| CREB1 | 119.9051715 | 0.4242424 | 8 |
GO enrichment analysis of common targets
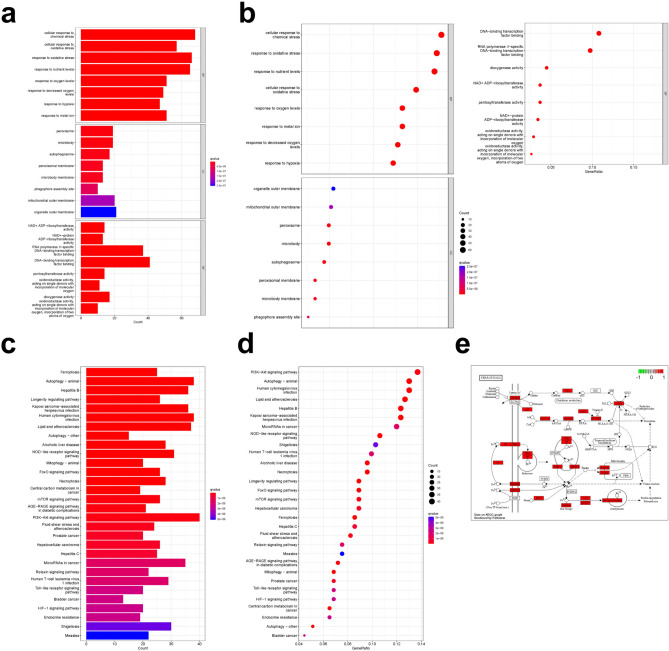
The result of GO enrichment analysis showed that the total number was 2666, including 2438 biological processes (BP), which mainly involved cellular response to chemical stress, response to oxidative stress, response to metal ion, etc. There were 81 cellular components (CC), which mainly involved mitochondrial outer membrane, peroxisome, autophagosome, etc. There were 147 molecular functions (MF), which were mainly related to DNA-binding transcription factor binding, dioxygenase activity, oxidoreductase activity, etc. Take the top 8 items to plot barplot and bubble chart (Fig. 4a,b).
Fig. 4.
GO and KEGG enrichment analyses of common targets. (a) The barplot of GO enrichment analysis. (b) The bubble chart of GO enrichment analysis. (c) The barplot of KEGG enrichment analysis. (d) The bubble chart of KEGG enrichment analysis. (e) The map of ferroptosis pathways.
KEGG enrichment analysis of common targets
A total of 165 pathways were identified by KEGG enrichment analysis, including ferroptosis, PI3K-Akt signaling pathway, autophagy, lipid and atherosclerosis, necroptosis, FoxO signaling pathway, NOD-like receptor signaling pathway, etc. Take the top 30 pathways to plot barplot and bubble chart (Fig. 4c,d). The map of ferroptosis pathways was drawn by R software as shown in (Fig. 4e).
Molecular docking of p53 with AS-IV
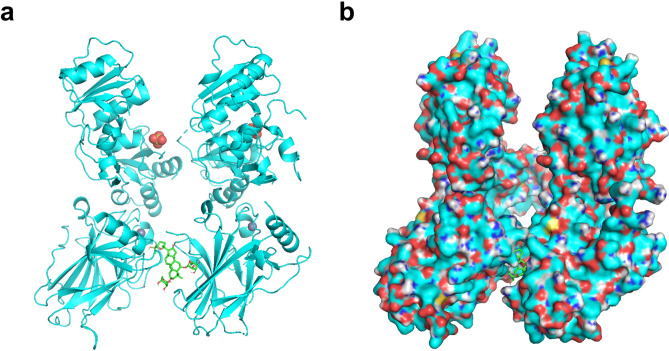
According to the above bioinformatics analysis, the key target p53 was obtained. The protein ID of p53 retrieved from the UniProt database was P04637, and the protein receptor file “1GZH” was downloaded from the PDB database. Molecular docking of p53 protein receptor with AS-IV ligand showed that the lowest binding free energy was − 9.2 kcal/mol, indicating a good affinity. The optimal model was shown in (Fig. 5a,b).
Fig. 5.
Molecular docking model of p53 with AS-IV. (a) Molecular docking model without surface of receptor. (b) Molecular docking model showing surface of receptor.
MD simulation analysis of p53-AS-IV complex
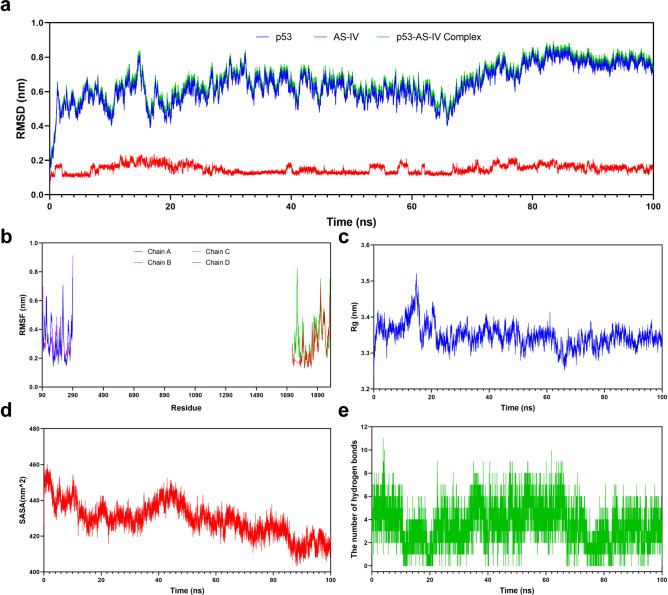
A 100 ns MD simulation was conducted to investigate the dynamic properties of p53-AS-IV complex obtained by molecular docking. The results of the MD simulations provided a crucial role16 in determining the stability of p53-AS-IV complex, and in evaluating the structural stability, hydrophobicity of amino acid residues and other relevant factors of p53 protein after binding with AS-IV. Among them, the RMSD showed the position change between the conformations of p53 and AS-IV in the simulation process and their initial conformations. The change trend was an important characterization to judge whether the simulation has reached stability. In this MD simulation, the RMSD curve of AS-IV was stabilized at 0.1–0.2 nm. The RMSD curve of p53-AS-IV complex maintained the same fluctuation trend as p53 in the whole MD simulation, and both of them were relatively stable at 0.6–0.8 nm for 24–100 ns. Overall, the RMSD value remained in a small range, indicating that p53 could generate a stable complex system with AS-IV (Fig. 6a). The RMSF is an index to evaluate the flexibility of protein amino acids during the whole simulation process, and the greater RMSF value indicates the greater flexibility of amino acid residues, which can be used to explain the change of complex at the residue level. As can be seen in Fig. 5, p53 protein has 4 amino acid chains. In this MD simulation, RMSF curve showed that the fluctuation range of most amino acid residues in the amino acid chains of p53 was stable within 0.5 nm except for a few residues with a greater flexibility, which meant that the flexibility variation of the protein was within a reasonable range throughout the MD simulation (Fig. 6b). The Rg value is used to evaluate the tightness of protein structure. The higher the Rg value is, the looser the peptide chains are and the more unstable the protein structure becomes. On the contrary, the lower the Rg value is, the more dense and stable the protein structure becomes. In this MD simulation, the Rg value increased briefly at the beginning of MD simulation, but gradually decreased and stabilized after 20 ns with a fluctuation range of 3.3–3.4 nm, indicating good tightness of p53 protein structure (Fig. 6c). The SASA is utilized to assess protein surface area. The results of MD simulation showed that the SASA value of p53 protein was large before binding with AS-IV, but gradually became smaller during MD simulation, suggesting that the area of the protein exposed to the solution gradually decreased. It meant that the folding of p53 was more tightly after binding with AS-IV (Fig. 6d). H-bonds between the small molecule ligand and the protein receptor are conducive to the stabilization of complex. The results demonstrated that the complex systems of p53-AS-IV with stable H-bonds were shown to exist stably throughout the MD simulation, with the maximum H-bonds number of 11 (Fig. 6e). Furthermore, in order to determine whether the combination between protein and small molecule is reasonable, some other indicators need to be evaluated. One of the most commonly used indicators is the MM/PBSA analysis. By calculating the binding energy of protein and small molecule, the total binding free energy of complex can be obtained. The lower binding free energy indicates a more stable binding. Here, the binding free energy was calculated by MM/GBSA method and the average binding free energy of − 47.52 kcal/mol was obtained, indicating a good stability of p53-AS-IV complex system (Table 3).
Fig. 6.
MD simulation analysis of p53-AS-IV complex. (a) RMSD curve of p53, AS-IV and p53-AS-IV complex. (b) RMSF curve of p53’s amino acid chains. (c) Rg curve of p53. (d) SASA curve of p53. (e) H-bonds of p53-AS-IV complex.
Table 3.
Binding free energy calculation of p53-AS-IV complex.
| Energy component | Energy (kcal/mol) |
|---|---|
| ΔVDWAALS | − 61.81 ± 2.23 |
| ΔEEL | − 39.06 ± 4.8 |
| ΔEGB | 62.18 ± 1.34 |
| ΔESURF | − 8.82 ± 0.04 |
| ΔGGAS | − 100.87 ± 5.3 |
| ΔGSOLV | 53.35 ± 1.34 |
| ΔTOTAL | − 47.52 ± 5.47 |
ΔVDWAALS Van der Waals energy, ΔEEL Electrostatic energy, ΔEGB polar solvation free energy, ΔESURF non-polar solvation energy, ΔGGAS energy in the gas phase = ΔVDWAALS + ΔEEL, ΔGSOLV solvation free energy = ΔEGB + ΔESURF, ΔTOTAL total binding free energy = ΔGGAS + ΔGSOLV.
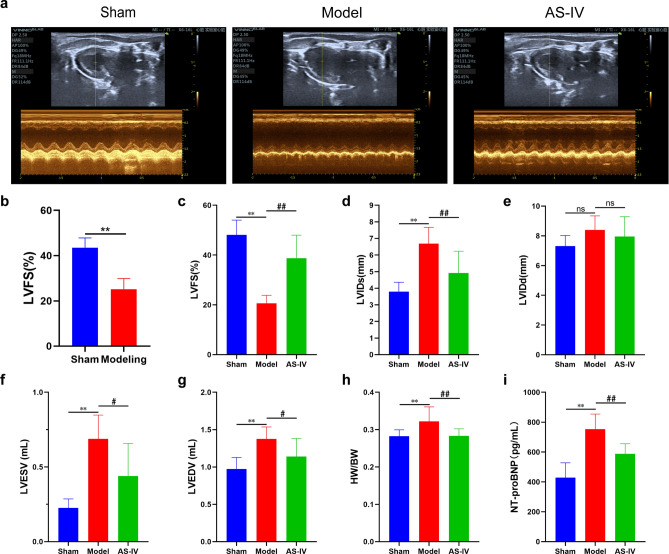
AS-IV improved cardiac function in CHF rats
After 4 weeks of modeling, the results of echocardiography showed that the LVFS of the Modeling group was significantly lower than that of the Sham group, indicating that CHF models were successfully induced (Fig. 7b). After 4 weeks of AS-IV administration, compared with the Sham group, LVFS in the Model group decreased, and LVIDs, LVIDd, LVESV, LVEDV, heart weight index and NT-proBNP level increased. These results suggested that the heart of the Model group was dilated, the myocardium in the non-infarcted zone was hypertrophic, and the cardiac function was significantly decreased. While, compared with the Model group, AS-IV treatment increased LVFS and decreased LVIDs, LVIDd, LVESV, LVEDV, heart weight index and NT-proBNP level (Fig. 7a,c–i).
Fig. 7.
AS-IV improved cardiac function in CHF rats. (a) Representative echocardiographic images of rats after 4 weeks of AS-IV administration. (b) The result of LVFS at 4 weeks after coronary ligation, Sham (n = 9), Modeling (n = 20). (c–i) The results of LVFS, LVIDs, LVIDd, LVESV, LVEDV, heart weight index and serum NT-proBNP content (n = 9). **P < 0.01 vs. Sham group; ##P < 0.01, #P < 0.05 vs. Model group.
AS-IV alleviated myocardial histomorphological changes in CHF rats
HE staining of the Sham group showed that the myocardial fiber lines were clear, the morphology of cardiomyocyte was normal and arranged neatly, and there was no swelling and inflammatory infiltration in the interstitial area. While in the Model group, the myocardial fibers in the infarcted border zone were swollen, broken, blurred or disappeared, with swelling and inflammatory infiltration observed in the interstitial area. The cardiomyocyte in the non-infarcted zone also showed hypertrophy and disordered arrangement. Compared with the Model group, AS-IV treatment significantly alleviated swelling and inflammatory infiltration in the interstitial area, and there was a certain degree of morphological recovery in both the myocardial fibers of the infarcted border zone and the cardiomyocytes in the non-infarcted zone (Fig. 8a–c,g). Masson staining showed that there was no myocardial infarction in the Sham group, and there was no statistically significant difference in myocardial infarct size between the Model and AS-IV group, indicating that the myocardial infarct size of rats in the two groups was comparable before administration (Fig. 8d,h). Additionally, a small amount of scattered filamentous collagen fibers could be seen in the myocardial surgical area of the Sham group rats. In the model group, there was extensive collagen fiber deposition at the infarcted border zone, significant proliferation of fibrous connective tissue in the interstitial area, and a large area of myocardial fibrosis. Nevertheless, AS-IV treatment reduced myocardial fibrosis in the infarcted border zone (Fig. 8e,i). In the non-infarcted zone, fibrosis was also significantly increased in the Model group, but it was markedly reduced in the AS-IV group (Fig. 8f,j).
Fig. 8.
AS-IV alleviated myocardial histomorphological changes in CHF rats. (a,b) Representative images of HE staining. (c,g) Representative images and quantification of the cardiomyocyte cross-sectional area in the non-infarcted zone (n = 3). (d,h) Representative images of Masson staining and quantification of the left ventricular infarct size (n = 3). (e,i) Representative images and quantification of the fibrosis in the infarcted border zone (surgical area for the Sham group, n = 3). (f,j) Representative images and quantification of the fibrosis in the non-infarcted zone (n = 3). **P < 0.01 vs. Sham group; ##P < 0.01 vs. Model group.
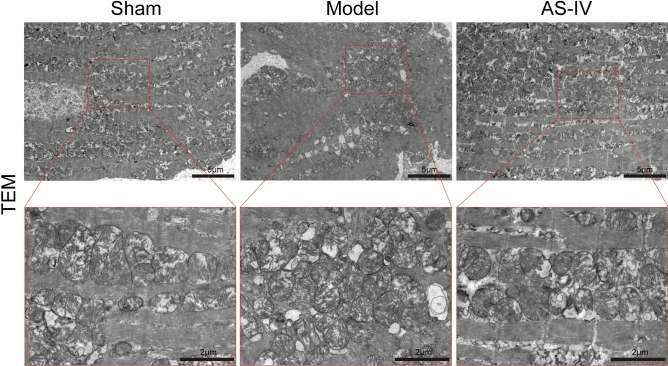
AS-IV alleviated myocardial mitochondrial damages in CHF rats
TEM showed that myocardial mitochondria in the Sham group were dense and evenly distributed, with clear ridge structure, small vacuoles and few damaged mitochondria. The mitochondrial density was 38 per 100 μm2 and the average size was 1.003 μm2. Compared with the Sham group, mitochondria in the Model group were sparse in number, smaller in volume, and disordered in arrangement, with membrane rupture, ridge fracture, large vacuoles, and more damaged mitochondria. The mitochondrial density was 33.55 per 100 μm2 and the average size was 0.796 μm2. However, compared with the Model group, the number of mitochondria in the AS-IV group increased, and the arrangement and distribution of mitochondria were more uniform and dense. Most of the mitochondria had normal morphology and more complete structure, and fewer damaged mitochondria could be seen. The mitochondrial density was 42.32 per 100 μm2 and the average size was 1.846 μm2 (Fig. 9).
Fig. 9.
AS-IV alleviated myocardial mitochondrial damage in CHF rats. Representative images of TEM.
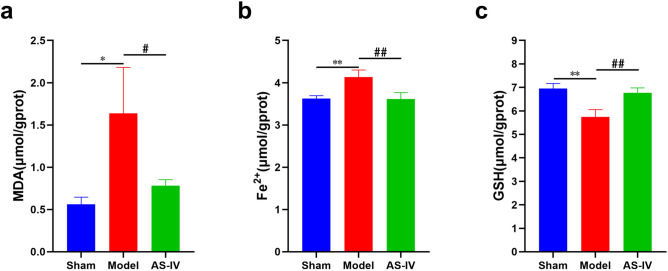
AS-IV decreased MDA content and Fe2+ accumulation and increased GSH content in CHF rats
Compared with the Sham group, MDA content and Fe2+ accumulation increased, while GSH content decreased in myocardial tissue of the Model group. Compared with the Model group, using AS-IV reduced MDA content and Fe2+ accumulation, and increased GSH content in myocardial tissue (Fig. 10a–c).
Fig. 10.
AS-IV decreased MDA content and Fe2+ accumulation and increased GSH content in CHF rats. (a) The result of MDA content in myocardium (n = 6). (b) The result of Fe2+ content in myocardium (n = 6). (c) The result of GSH content in myocardium (n = 6). **P < 0.01, *P < 0.05 vs. Sham group; ##P < 0.01, #P < 0.05 vs. Model group.
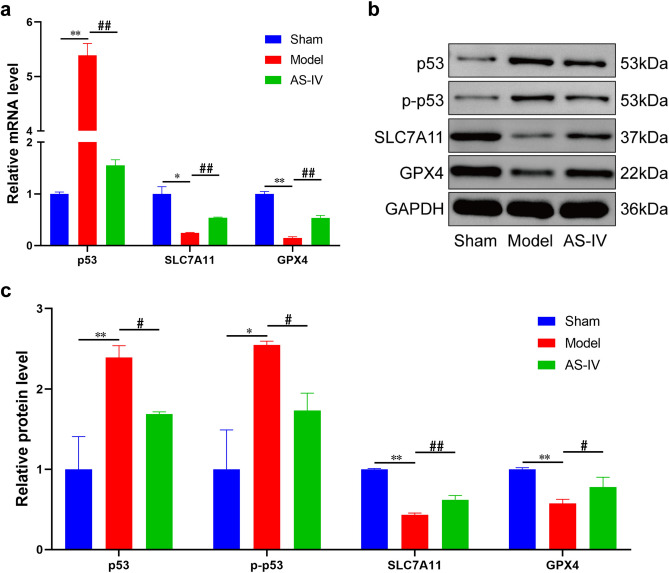
AS-IV regulated the expression of p53/SLC7A11/GPX4 signaling pathway
Results of RT-qPCR and Western Blotting showed that compared with the Sham group, expression of p53 and p-p53 increased, while expression of SLC7A11 and GPX4 decreased in myocardial tissue of the Model group. However, compared with the Model group, expression of p53 and p-p53 in the AS-IV group was inhibited, while expression of SLC7A11 and GPX4 was up-regulated. It was indicated that the expression of p53/SLC7A11/GPX4 signaling pathway was significantly regulated by AS-IV (Fig. 11a–c).
Fig. 11.
AS-IV regulated the expression of p53/SLC7A11/GPX4 signaling pathway. (a) The mRNA expression of p53/SLC7A11/GPX4 signaling pathway (n = 3). (b) The representative Western Blotting bands of p53/SLC7A11/GPX4 signaling pathway. The original blots/gels were presented in Supplementary Figure. (c) The protein expression of p53/SLC7A11/GPX4 signaling pathway (n = 3). **P < 0.01, *P < 0.05 vs. Sham group; ##P < 0.01, #P < 0.05 vs. Model group.
Discussion
It’s reported that the total number of worldwide patients with CHF is gradually increasing, posing a serious threat to human life and health17. At present, researches on new drugs for CHF mainly focuses on Angiotensin II Receptor/Neprilysin Inhibitor (ARNI), Sodium-Glucose Co-Transporter-2 Inhibitors (SGLT2i), Soluble Guanylyl Cyclase (sGC) Stimulator and Cardiac Myosin Activator, which has been effective in improving cardiac function and relieving symptoms, but there are still some problems exist such as high cost and many side effects of drug treatment18. Therefore, it is particularly important to find better treatment ideas and develop new drugs for CHF. In recent years, bioinformatics, as an interdisciplinary subject integrating molecular biology, genetic engineering and computer science, has developed rapidly and occupied a vital position in the field of medical research. It can effectively provide biological evidences for clarifying the complicated mechanism of action between drugs and diseases from abundant biological data, which is beneficial to the implementation of precision medicine and the speed of new drug research and development19.
In this study, the key target p53 was obtained through bioinformatics analysis and screening from the perspective of ferroptosis, and it could be concluded that there was a good affinity between p53 and AS-IV through molecular docking and MD simulation analysis. Furthermore, the map of ferroptosis pathways showed that the p53/SLC7A11/GPX4 pathway is one of the main pathways regulating ferroptosis. Then the results of animal experiment showed that AS-IV treatment significantly mitigated cardiac hypertrophy and myocardial remodeling in CHF rats, improved cardiac function, reduced mitochondrial damage, decreased myocardial MDA content and Fe2+ accumulation, increased GSH content, inhibited myocardial p53 and p-p53 expression, and up-regulated SLC7A11 and GPX4 expression.
Ferroptosis was first proposed in 2012 as a form of regulatory cell death driven by iron accumulation and lipid peroxidation20, so Fe2+ and MDA are both important molecular markers for the detection of ferroptosis and lipid peroxidation21. The morphological characteristics of ferroptosis are mainly reflected in the decreased number and smaller volume of mitochondria, also including reduced ridge, increased membrane density and membrane rupture22. In addition, from biologically speaking, the increase of lipid peroxides induced by depletion of intracellular antioxidant GSH and decreased activity of GPX4 is the most critical step leading to ferroptosis23. The p53/SLC7A11/GPX4 pathway is crucial for regulating ferroptosis24. SLC7A11 is an important component of cystine/glutamate antitransporter (System Xc−), which mediates the exchange of intracellular glutamic acid and extracellular cystine on the plasma membrane so as to complete the cell uptake of cystine25. After entering the cell, cystine is reduced to cysteine, which is involved in the synthesis of GSH as one of the essential components26. GPX4 can detoxify oxidative free radicals assisted with the activity of multiple reducing agents, and GSH is the main reducing substrate27. The cooperation of GPX4 and GSH can effectively remove the excessive accumulation of lipid peroxides, reduce oxidative stress reaction, and protect cell membrane from lipid peroxidation28,29. It has been reported that inhibiting the synthesis of cysteine and GSH in cells by the regulation of System Xc− or inhibiting GPX4 activity directly can both induce ferroptosis, for instance, using ferroptosis inducers such as erastin, sulfasalazine, RSL3, ML162, DPI10, FIN56, and so on30,31. The mechanism of p53 promoting ferroptosis is the same as above32. Jiang et al. reported for the first time that p53 affected the cystine transport function of System Xc− through inhibiting SLC7A11 expression, thereby reduced cystine uptake and inhibited GPX4 activity, ultimately led to the occurrence of ferroptosis33,34.
In recent years, more and more studies have reported that ferroptosis plays an important role in CHF pathogenesis35. CHEN et al. observed iron overload and increased lipid peroxide content in cardiomyocytes in CHF rat models induced by descending aortic banding procedure, suggesting that ferroptosis was involved in the occurrence and development of CHF36. In pressure-loaded CHF mouse models constructed by transverse aortic constriction, Wang et al. found that mitochondria of cardiomyocytes decreased and became obviously smaller, and the mitochondrial membrane density increased, accompanied by the outer membrane rupture. At the same time, it was also found that MDA content increased, GSH content decreased, SLC7A11, GPX4 expression decreased, and p53 expression increased, suggesting that ferroptosis was a critical pathological process of myocardial hypertrophy37. In CHF mouse models induced by isoproterenol, iron accumulation, decreased GSH level, increased MDA content, and down-regulated GPX4 and SLC7A11 protein expression were observed in damaged myocardial tissue. However, using ferrostatin-1 and other ferroptosis inhibitors effectively rescued isoproterenol-induced myocardial injury and improved CHF symptoms, indicating that ferroptosis was involved in CHF process and inhibiting ferroptosis could effectively prevent the progression of CHF38. There are also studies have pointed that ferritin deficient mice showed severe myocardial hypertrophy and decreased cardiac function when given a high iron diet, but SLC7A11 overexpression in the mice could effectively inhibit ferroptosis and improve ventricular remodeling39. In this study, rat models of CHF induced by left coronary artery ligation exhibited significant cardiac dysfunction and myocardial remodeling, including cardiomyocyte damage and hypertrophy, obvious myocardial fibrosis, severe mitochondrial damage with decreased amount, iron overload, lipid peroxide accumulation, decreased antioxidant capacity, increased p53 and p-p53 expression, and down-regulated SLC7A11 and GPX4 expression. These results suggested that ferroptosis was an important pathological mechanism of CHF rats in this study.
Radix Astragali is prepared from the dry roots of Astragalus memeranaceus (Fisch.) Bge. Var. mongholicus (Bge.) Hsiao or Astragalus membranaceus (Fisch.) Bge. In traditional Chinese medicine, it has several efficacies such as fortifying the spleen and replenishing qi, boosting defense and securing exterior, engendering fluid and nourishing blood, inducing diuresis to alleviating edema, moving stagnation and unblocking impediment, etc40. Traditional Chinese medicine believes that the main location of CHF is in the heart, and its basic pathological characteristic is qi deficiency with blood stasis, which runs through the beginning and end of CHF. Therefore, Radix Astragali is a commonly used Chinese medicine for the treatment of CHF41. AS-IV is the principal active ingredient of Radix Astragali, and many studies have supported its efficacy and safety in both in-vivo and in-vitro CHF models6,42. AS-IV has a wide range of pharmacological actions for the treatment of CHF, including improving oxidative stress, regulating energy metabolism, reducing cardiomyocyte apoptosis, restraining inflammatory response, adjusting calcium homeostasis, inhibiting cardiomyocyte autophagy, alleviating myocardial fibrosis, and so on43. Our study demonstrated that AS-IV significantly improved the cardiac function of CHF rats, increased the quantity of mitochondria, reduced the accumulation of Fe2+ and lipid peroxide, increased the content of GSH to enhance the antioxidant capacity, suppressing the expression of p53 and p-p53 in myocardium, and promote the expression of SLC7A11 and GPX4. These results all indicated that AS-IV effectively inhibited cardiac ferroptosis in CHF rats. The study revealed the protective effect of AS-IV on CHF rats from the perspective of ferroptosis. Its ability to regulate the p53/SLC7A11/GPX4 pathway suggested a new mechanism of AS-IV as a potential therapeutic agent for CHF and provide a potential therapeutic strategy for human CHF treatment.
Some limitations still exist in this study, which should be discussed. Firstly, the etiology of CHF is diverse, but we only used one method and one animal species to induce the CHF model. Moreover, the establishment of the CHF model requires a certain level of technical expertise from researchers, which could lead to variability across different laboratories. Future studies should consider using multiple models to verify the effects of AS-IV, thereby enhancing the generalizability and reliability of the results. Secondly, we only studied one dosage form and one administration mode of AS-IV, and the pharmacokinetics of AS-IV has not been studied. This limits our understanding of the optimal dosage and administration regimen for AS-IV to some extent. So the pharmacological research of AS-IV targeting the mechanism of ferroptosis in CHF still need further development in the future. Thirdly, other types of cell death also occur in CHF cardiomyocytes. Various forms of cell death often interact with each other, and cell death signaling pathways cross each other. In addition to inhibiting ferroptosis, AS-IV may also interfere with a variety of other forms of cell death. However, our study focused only on ferroptosis and did not jointly analyze other protective functions of AS-IV. In the future, exploring multiple cell death pathways targeting CHF may be more helpful in fully revealing the protective mechanism of AS-IV in CHF. Fourthly, although our study achieved statistically significant results, studies with larger sample sizes would be more helpful in verifying the reliability of these findings and further eliminating the influence of random factors. Fifthly, bioinformatics analysis relies on existing public databases, whose update speed and data quality may also affect the accuracy of the analysis results. Therefore, future research should include more data set validation and experimental verification to reduce the impact of these potential biases. Sixthly, this study only measured cardiac function and molecular indicators at specific time points, which may not fully reflect the dynamic impact of AS-IV on CHF progression over time. CHF is a complex, progressive process, and different stages may involve different pathological mechanisms. Therefore, future studies should consider more systematic follow-up observations at various time points to comprehensively understand the mechanism of AS-IV on CHF.
Conclusion
In this study, p53 was the key target for the treatment of CHF from the perspective of ferroptosis and possessed a good affinity with AS-IV, which was obtained through bioinformatics analysis. The animal experiment proved that AS-IV improved the cardiac function of CHF rats, and its mechanism may be related to the regulation of p53/SLC7A11/GPX4 signaling pathway and the inhibition of myocardial ferroptosis. The results not only provided theoretical and experimental evidences for the clinical application of AS-IV, but also further enriched the pharmacological effects of AS-IV in the prevention and treatment of CHF. In the future, based on the results of this study, we will conduct gene silencing experiments or other experiments to further verify our conclusion. We will also investigate the effects of AS-IV in other CHF models to determine whether its therapeutic effects are consistent.
Supplementary Information
Abbreviations
- CHF
Chronic heart failure
- AS-IV
Astragaloside IV
- PPI
Protein–protein interaction
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- HE
Hematoxylin–eosin
- TEM
Transmission electron microscopy
- MDA
Malondialdehyde
- GSH
Glutathione
- SLC7A11
Solute vector family 7 member 11
- GPX4
Glutathione peroxidase 4
- LVFS
Left ventricular fractional shortening
- LVIDs
Left ventricular end systolic dimension
- LVIDd
Left ventricular end diastolic dimension
- NT-proBNP
N-terminal pro-brain natriuretic peptide
- SDS-PAGE
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- p-p53
Phospho-p53
- System Xc-
Cystine/glutamate antitransporter
Author contributions
H.Y. and M.S. contributed to conceptualization, investigation, project administration, formal analysis and writing—original draft. J.W., C.L. and Z.W. contributed to investigation and writing—original draft. Y.L. contributed to conceptualization, funding acquisition and writing—review and editing. Z.G. contributed to conceptualization, funding acquisition, supervision and writing—review and editing. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by National Natural Science Foundation of China [grant number 82174343 and 81673955], Key Scientific Research Project of Hunan Province [grant number 2022SK2012], “Yi-Fang” Graduate Innovation Project of Hunan University of Chinese Medicine [grant number 2023YF10], and Graduate Research Innovation Project of Hunan Province [grant number QL20230197 and CX20230832].
Data availability
Data is provided within the manuscript or supplementary information files.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Hui Yuan and Min Shi.
Contributor Information
Ya Li, Email: 003872@hnucm.edu.cn.
Zhihua Guo, Email: 004294@hnucm.edu.cn.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-72011-z.
References
- 1.Heidenreich, P. A. et al. 2022 AHA/ACC/HFSA Guideline for the Management of Heart Failure: Executive Summary: A Report of the American College of Cardiology/American Heart Association Joint Committee on Clinical Practice Guidelines. Circulation145(18), e876–e894 (2022). [DOI] [PubMed] [Google Scholar]
- 2.Yu, Y. et al. Ferroptosis: A cell death connecting oxidative stress, inflammation and cardiovascular diseases. Cell Death Discov.7(1), 193 (2021). 10.1038/s41420-021-00579-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang, X. D. et al. Mechanisms and regulations of ferroptosis. Front. Immunol.14, 1269451 (2023). 10.3389/fimmu.2023.1269451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang, X. et al. Ferroptosis in heart failure. J. Mol. Cell. Cardiol.173, 141–153 (2022). 10.1016/j.yjmcc.2022.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dong, L. et al. Research progress of Chinese medicine in the treatment of myocardial ischemia–reperfusion injury. Am. J. Chin. Med.51(1), 1–17 (2023). 10.1142/S0192415X23500015 [DOI] [PubMed] [Google Scholar]
- 6.Zang, Y. et al. An updated role of astragaloside IV in heart failure. Biomed. Pharmacother.126, 110012 (2020). 10.1016/j.biopha.2020.110012 [DOI] [PubMed] [Google Scholar]
- 7.Zhou, N. et al. FerrDb V2: Update of the manually curated database of ferroptosis regulators and ferroptosis-disease associations. Nucleic Acids Res.51(D1), D571–D582 (2023). 10.1093/nar/gkac935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bardou, P. et al. jvenn: An interactive Venn diagram viewer. BMC Bioinform.15(1), 293 (2014). 10.1186/1471-2105-15-293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanner, M. F. Python: A programming language for software integration and development. J. Mol. Graph. Model.17(1), 57–61 (1999). [PubMed] [Google Scholar]
- 10.Trott, O. & Olson, A. J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem.31(2), 455–461 (2010). 10.1002/jcc.21334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van der Spoel, D. et al. GROMACS: Fast, flexible, and free. J. Comput. Chem.26(16), 1701–1718 (2005). 10.1002/jcc.20291 [DOI] [PubMed] [Google Scholar]
- 12.Abraham, M. J. et al. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX1–2, 19–25 (2015). 10.1016/j.softx.2015.06.001 [DOI] [Google Scholar]
- 13.Arifin, W. N. & Zahiruddin, W. M. Sample size calculation in animal studies using resource equation approach. Malays. J. Med. Sci.24(5), 101–105 (2017). 10.21315/mjms2017.24.5.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeng, Z. et al. Qishen granule attenuates cardiac fibrosis by regulating TGF-β/Smad3 and GSK-3β pathway. Phytomedicine62, 152949 (2019). 10.1016/j.phymed.2019.152949 [DOI] [PubMed] [Google Scholar]
- 15.Takagawa, J. et al. Myocardial infarct size measurement in the mouse chronic infarction model: Comparison of area- and length-based approaches. J. Appl. Physiol.102(6), 2104–2111 (2007). 10.1152/japplphysiol.00033.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pan, Y. et al. Screening of active substances regulating Alzheimer’s disease in ginger and visualization of the effectiveness on 6-gingerol pathway targets. Foods13(4), 612 (2024). 10.3390/foods13040612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Groenewegen, A. et al. Epidemiology of heart failure. Eur. J. Heart Fail.22(8), 1342–1356 (2020). 10.1002/ejhf.1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsutsui, H. Recent advances in the pharmacological therapy of chronic heart failure: Evidence and guidelines. Pharmacol. Ther.238, 108185 (2022). 10.1016/j.pharmthera.2022.108185 [DOI] [PubMed] [Google Scholar]
- 19.Gómez-López, G. et al. Precision medicine needs pioneering clinical bioinformaticians. Brief. Bioinform.20(3), 752–766 (2019). 10.1093/bib/bbx144 [DOI] [PubMed] [Google Scholar]
- 20.Stockwell, B. R. Ferroptosis turns 10: Emerging mechanisms, physiological functions, and therapeutic applications. Cell185(14), 2401–2421 (2022). 10.1016/j.cell.2022.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang, D., Kroemer, G. & Kang, R. Ferroptosis in immunostimulation and immunosuppression. Immunol. Rev.321(1), 199–210 (2024). 10.1111/imr.13235 [DOI] [PubMed] [Google Scholar]
- 22.Li, J. et al. Ferroptosis: Past, present and future. Cell Death Dis.11(2), 88 (2020). 10.1038/s41419-020-2298-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu, M. et al. The critical role and molecular mechanisms of ferroptosis in antioxidant systems: A narrative review. Ann. Transl. Med.10(6), 368 (2022). 10.21037/atm-21-6942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang, R., Kroemer, G. & Tang, D. The tumor suppressor protein p53 and the ferroptosis network. Free Radic. Biol. Med.133, 162–168 (2019). 10.1016/j.freeradbiomed.2018.05.074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo, Y. et al. Ferroptosis in cardiovascular diseases: Current status, challenges, and future perspectives. Biomolecules12(3), 390 (2022). 10.3390/biom12030390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, Q. et al. Ferroptosis: The potential target in heart failure with preserved ejection fraction. Cells11(18), 2842 (2022). 10.3390/cells11182842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie, L., Fang, B. & Zhang, C. The role of ferroptosis in metabolic diseases. Biochim. Biophys. Acta Mol. Cell Res.1870(6), 119480 (2023). 10.1016/j.bbamcr.2023.119480 [DOI] [PubMed] [Google Scholar]
- 28.Liu, Y. et al. The diversified role of mitochondria in ferroptosis in cancer. Cell Death Dis.14(8), 519 (2023). 10.1038/s41419-023-06045-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang, X., Stockwell, B. R. & Conrad, M. Ferroptosis: Mechanisms, biology and role in disease. Nat. Rev. Mol. Cell Biol.22(4), 266–282 (2021). 10.1038/s41580-020-00324-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dixon, S. J. et al. Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell149(5), 1060–1072 (2012). 10.1016/j.cell.2012.03.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang, M. et al. Relationship between ferroptosis and mitophagy in renal fibrosis: A systematic review. J. Drug Target.31(8), 858–866 (2023). 10.1080/1061186X.2023.2250574 [DOI] [PubMed] [Google Scholar]
- 32.Xu, R., Wang, W. & Zhang, W. Ferroptosis and the bidirectional regulatory factor p53. Cell Death Discov.9(1), 197 (2023). 10.1038/s41420-023-01517-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang, L. et al. Ferroptosis as a p53-mediated activity during tumour suppression. Nature520(7545), 57–62 (2015). 10.1038/nature14344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu, S., Li, X. & Wang, Y. Regulation of the p53-mediated ferroptosis signaling pathway in cerebral ischemia stroke (Review). Exp. Ther. Med.25(3), 113 (2023). 10.3892/etm.2023.11812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.del Re, D. P. et al. Fundamental mechanisms of regulated cell death and implications for heart disease. Physiol. Rev.99(4), 1765–1817 (2019). 10.1152/physrev.00022.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen, X. et al. Role of TLR4/NADPH oxidase 4 pathway in promoting cell death through autophagy and ferroptosis during heart failure. Biochem. Biophys. Res. Commun.516(1), 37–43 (2019). 10.1016/j.bbrc.2019.06.015 [DOI] [PubMed] [Google Scholar]
- 37.Wang, J. et al. Pyroptosis and ferroptosis induced by mixed lineage kinase 3 (MLK3) signaling in cardiomyocytes are essential for myocardial fibrosis in response to pressure overload. Cell Death Dis.11(7), 574 (2020). 10.1038/s41419-020-02777-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen, Y. et al. Ferroptosis contributes to catecholamine-induced cardiotoxicity and pathological remodeling. Free Radic. Biol. Med.207, 227–238 (2023). 10.1016/j.freeradbiomed.2023.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fang, X. et al. Loss of cardiac ferritin H facilitates cardiomyopathy via Slc7a11-mediated ferroptosis. Circ. Res.127(4), 486–501 (2020). 10.1161/CIRCRESAHA.120.316509 [DOI] [PubMed] [Google Scholar]
- 40.Zhang, C. H. et al. Ethnopharmacology, phytochemistry, pharmacology, toxicology and clinical applications of radix astragali. Chin. J. Integr. Med.27(3), 229–240 (2021). 10.1007/s11655-019-3032-8 [DOI] [PubMed] [Google Scholar]
- 41.Wang, Y. et al. A review of Chinese herbal medicine for the treatment of chronic heart failure. Curr. Pharm. Des.23(34), 5115–5124 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang, C. et al. Review on the protective mechanism of astragaloside IV against cardiovascular diseases. Front. Pharmacol.14, 1187910 (2023). 10.3389/fphar.2023.1187910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liang, Y. et al. Pharmacological effects of astragaloside IV: A review. Molecules28(16), 6118 (2023). 10.3390/molecules28166118 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is provided within the manuscript or supplementary information files.