Abstract
Background
Clear cell renal cell carcinoma (ccRCC) is a highly aggressive cancer associated with higher death rates. However, traditional anti-angiogenic therapies have limited effectiveness due to drug resistance. Vascular mimicry (VM) provides a different way for tumors to develop blood vessels without relying on endothelial cells or angiogenesis. However, the intricate mechanisms and interplay between it and the immune microenvironment in ccRCC remain unclear.
Methods
A PubMed and GeneCards literature review was conducted to identify VM-related genes (VMRGs). VMRGs expression profiles were obtained from The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO), developing a novel VM risk score model and nomogram for ccRCC. The EBI ArrayExpress database (the validation set) was obtained to validate the prognostic model. The relationship between VMRGs risk score clinical characteristics and immune infiltration was investigated. Finally, the expression of six model VMRGs was validated using single-cell analysis, GEPIA, Human Protein Atlas (HPA), and quantitative Real-time PCR (qRT-PCR).
Results
Cox regression analysis and nomogram identified L1CAM, TEK, CLDN4, EFNA1, SERPINF1, and MALAT1 as independent prognostic risk factors, which could be used to stratify the ccRCC population into two risk groups with distinct immune profiles and responsiveness to immunotherapy. The results of single-cell analysis, GEPIA, HPA, and qRT-PCR validated the model genes’ expression.
Conclusions
Our novel findings constructed a convenient and reliable 6 gene signatures as potential immunologic and prognostic biomarkers of VM in ccRCC.
Keywords: Vasculogenic mimicry, Tumor microenvironment, Clear cell renal cell carcinoma, Nomogram, Immune infiltration
Highlights
-
•
The intricate mechanisms and interplay between vascular mimicry (VM) and the tumor microenvironment in ccRCC remain unclear.
-
•
Integrated analyses on multi-omics characteristics of VM on tumor microenvironment, immunotherapy, and prognosis.
-
•
L1CAM, EFNA1, TEK, CLDN4, SERPINF1, and MALAT1 can serve as potential prognostic biomarkers associated with VM in ccRCC.
-
•
A nomogram combining the six-gene signature with clinicopathological parameters was constructed.
1. Introduction
Clear cell renal cell carcinoma (ccRCC) is a prevalent histological subtype of renal cell carcinoma (RCC). Approximately 33 % of patients display metastatic lesions as the disease advances [1].
The leading causes of mortality among patients with cancer are tumor recurrence and metastasis. Tyrosine kinase inhibitors (TKIs) like sunitinib, pazopanib, and axitinib, which are anti-angiogenic drugs, have shown effectiveness in most instances of metastatic RCC. Nevertheless, resistance and relapse often emerge in many patients over time, with some individuals inherently resistant to targeted therapy [2,3]. This raises the question of whether alternative sources of blood and nutrients exist.
Vascular mimicry (VM) is a novel pattern of tumor perfusion that differs from typical tumor angiogenesis by creating channels lined with cancer cells [4]. Due to its ability to circulate blood from vessels to tumor tissues and facilitate tumor cells into the extracellular matrix, VM is crucial in boosting tumor growth and promoting metastasis [[5], [6], [7], [8], [9]]. Nevertheless, no studies have reported the synergistic effects of these genes related to VM, known as VM-related genes (VMRGs). Whether VMRGs have the potential to act as prognostic markers for ccRCC remains unclear.
ccRCC development depends on a multifaceted tissue environment to support continuous growth, invasion, and metastasis. Additionally, in advanced ccRCC, the tumor microenvironment (TME) may induce drug resistance because it is highly dynamic, adaptable, and heterogeneous [10]. Hence, a comprehensive investigation and clarification of the specific communication mechanisms between TME and VM is required.
A thorough literature review of PubMed [[11], [12], [13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24]] and GeneCards was conducted to identify VMRGs. The investigation of VM in ccRCC used resources like The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO). Various VM subgroups were identified, and a prognostic scoring model was developed for ccRCC to highlight their significance in TME. This study emphasizes the functional importance of the VMRGs signature and provides novel insights into possible targets for therapy.
2. Material and methods
2.1. Dataset preparation
Overall, 294 VMRGs were extracted from GeneCards (https://www.genecards.org) and 24 genes from previous reviews. The duplicates were removed, and 300 VMRGs were obtained. RNA-Seq and clinical and survival data were obtained for 533 ccRCC samples and 81 tumor-adjacent normal samples from the TCGA database (http://portal.gdc.cancer.gov/). Furthermore, 39 ccRCC specimens were acquired from the GEO database (https//www.ncbi.nlm.nih.gov/geo) under the accession code GSE29609. The clinicopathological features of ccRCC patients are detailed in Supplementary Table 1. Differential gene expression analysis was performed with the R package ‘limma.’ This expression profiling of the 53 ccRCC samples with corresponding available clinical information about recurrence and prognosis was downloaded from the EBI ArrayExpress databases (https://www.ebi.ac.uk/arrayexpress/), which were used as a validation dataset.
2.2. Identification of robust and prognostic VMRGs
Screening criteria of p < 0.05 and fold change (FC) > 2 were used to identify genes that were expressed differently. Heatmaps were generated using the “pheatmap” R package, while volcano plots were created using the “Ggplot2″ package. Data from TCGA-KIRC and GSE29609 datasets were merged, and batch effect correction was performed using the “ComBat” algorithm from the “sva” package [25]. Survival analyses of 109 VMRGs, defined as differentially expressed genes, were performed using the “survival” package. The network of protein-protein interactions (PPI) created with the STRING platform exposed the connections between these VMRGs. The Gene Pattern platform [26] was utilized to evaluate the level of somatic mutations and changes in copy number variation (CNV) for the GDC TCGA Kidney Clear Cell Carcinoma (KIRC) cohort (https://xena.ucsc.edu/).
2.3. Formation of VMRGs cluster
To explore diverse patterns among VMRGs, the “ConsensusClusterPlus” algorithm was employed [27]. This method segregated patients into distinct molecular subgroups based on gene expression profiles. The main parameters of the function were defined as: max cluster number (maxK) set to 9; repetitions (reps) at 50; the proportion of items to sample (pItem) at 0.8; the proportion of features to sample (pFeature) at 1 [28]; cluster algorithm (clusterAlg) utilizing the k-means algorithm; and distance metric set to Euclidean. Subsequently, principal component analysis (PCA) was conducted with the “ggplot2″ R package. Survival analyses were performed to assess potential differences in patient survival times among the identified VMRGs subtypes. Differentially expressed VMRGs were screened for from these clusters and generated a heatmap.
2.4. Function enrichment analysis of VMRGs cluster
To further investigate deeper into the potential mechanism underlying the VMRGs pattern in TCGA-KIRC patients, enrichment function annotation analyses were conducted using gene set variation analysis (GSVA) and gene set enrichment analysis (GSEA). The “c5. go.symbols.gmt” and “c2. cp.kegg.v6.2. symbols.gmts” datasets from the MsigDB database [29] were utilized for the analysis.
2.5. Construction of prognostic prediction mode
The TCGA-KIRC and GSE29609 cohorts were divided into training and testing sets. The correlation between VMRGs and ccRCC survival was determined using a univariate Cox regression analysis in the training set. The testing set was used to validate the signature performance. LASSO regression analysis was performed using the R package ‘glmnet’ to discover the important prognostic genes [30]. The following formula was used to select candidate genes to build prognostic signatures based on VMRGs using multivariate Cox analyses: VM risk score = gene expression [1] × corresponding coefficient [1] + gene expression [n] × corresponding coefficient [n].
Patients were categorized into high-risk and low-risk groups based on the median risk score calculated with this formula. Differences in survival time between the two groups were estimated using the Kaplan–Meier method. Model accuracy was assessed using receiver operating characteristic (ROC) curves. Furthermore, multivariate Cox regression analyses were conducted to investigate whether the risk score was independent of the other available clinicopathological features.
2.6. Construction of predictive nomogram
A visual nomogram model incorporating risk, TNM stage, and VMRGs risk was developed using the “rms” package. The predictive power of this prognostic model was evaluated using the area under the ROC curve (AUC) and concordance index, while calibration curve analysis was employed to assess its accuracy. The nomogram model's clinical utility was evaluated using decision curve analysis (DCA).
2.7. Immune infiltration level and tumor microenvironment
The single-sample Gene Set Enrichment Analysis (ssGSEA) algorithm was applied to compute immune cell type scores, estimating the abundance of infiltrating immune cells using cell type-specific gene expression profiles from VMRGs clusters [31]. The CIBERSORT algorithm quantified levels of 22 immune cell subtypes and immune checkpoints (ICPs) in low- and high-risk groups [32]. To assess the Tumor Microenvironment (TME) of ccRCC, the “ESTIMATE” R package was utilized to obtain immune, stromal, and estimate scores [33].
2.8. Single-cell analysis
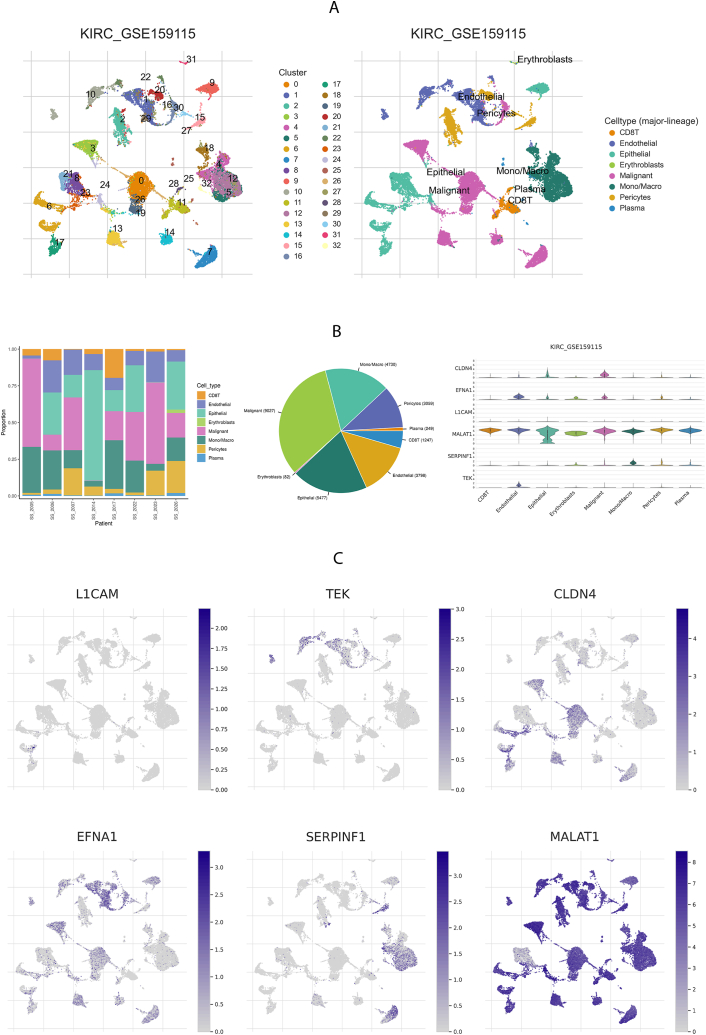
To determine the differential expression of the model genes in cells, scRNA-seq datasets (GSE159115) from TISCH (http://tisch.comp-genomics.org/) were used [34]. The expression of model genes in each of the seven cell populations was analyzed based on ccRCC marker gene expression, including CD8 T, endothelial, epithelial, erythroblast, malignant, monocyte (Mono)/macrophage (Macro), pericyte, and plasma cells.
2.9. Model gene expression examined by GEPIA and HPA
The “Expression Analysis- Box Plots” module of the GEPIA database(http://gepia2.cancer-pku.cn/#analysis) was used to further confirm differential model gene expression in the ccRCC tissues compared with the corresponding normal tissues from TCGA databases. The Human Protein Atlas (HPA) database includes information regarding the spatial distribution and expression of various proteins in human tissues and cells. The levels of the model gene protein expression in various tumor tissues and their corresponding normal tissues were analyzed based on the immunohistochemistry data in the HPA database [35]. Survival analysis of the model gene from the TCGA database was performed using GEPIA online software.
2.10. Real-time quantitative PCR (qRT-PCR)
The ccRCC cell lines, including caki-1 and 786-O, and the immortalized human kidney cell line HK-2, were procured from the American Type Culture Collection (ATCC). Cells were cultured in 1640 medium from Biological Industries supplemented with 10 % fetal bovine serum (FBS) also from Biological Industries. Authentication of all human cell lines was performed using STR profiling, and experiments were carried out using mycoplasma-free cells. Total RNA was extracted using a TRIZOL reagent (Cat# 9108; TaKaRa, Dalian, China). The ratio of absorbance at 260 and 280 nm (the A260/280 ratio) was used to evaluate the purity of RNA. Subsequently, cDNAs were synthesized using the Transcriptor First Strand cDNA Synthesis Kit from Tiangen (China) according to the manufacturer's instructions. Real-time PCR was conducted using the SYBR Green Master Mix from Yeasen (China) on a QuantStudio 3 Real-Time 170 PCR system (Thermo Fisher Scientific). Relative expression levels of mRNAs and lncRNAs were calculated using the 2–ΔΔ Ct method with GAPDH as the internal control. The primer sequences used for qRT-PCR were as follows:
L1CAM -forward: 5′- CACTATGGCCTTGTCTGGGA -3′
L1CAM - reverse: 5′- ACATACTGTGGCGAAAGGGA -3′
CLDN4-forward: 5′- AGAGTGGATGGACGGGTTT -3′
CLDN4- reverse: 5′- GAAGGGGCAGAGGACTCA -3′
TEK -forward: 5′- TAAACTTGGACACCATCCAAA -3′
TEK - reverse: 5′- CCAGATCCCTGTGGATAAACT -3′
EFNA1-forward: 5′- CCGCTCATCGTGCAACCTG -3′
EFNA1-reverse: 5′- ATAAGAAGGCATCAGATCG -3′
SERPINF1-forward: 5′- TGTCTCCAACTTCGGCTATG -3′
SERPINF1- reverse: 5′- AGTAGAGAGCCCGGTGAATG -3′
MALAT1-forward: 5′- GCAGGCGTTGTGCGTAGAG -3′,
MALAT1-reverse: 5′- TTGCCGACCTCACGGATT -3′.
2.11. Statistical analysis
Data processing, analysis, and presentation were conducted utilizing R software (version 4.3.1; http://www.Rproject.org). Survival correlations were assessed through Kaplan–Meier plots, log-rank tests, and univariate and multivariate analyses using the Cox proportional hazards method. Mann–Whitney-Wilcoxon Test or Student's t-test was executed to compare the difference between defined groups for continuous variables. Categorical clinical variables were assessed using Chi-square or Fisher's exact tests. Spearman's correlation analysis was used for correlation assessment. A significance level of P < 0.05 was set for statistical significance.
3. Results
3.1. Expression and mutation of VMRGs in ccRCC
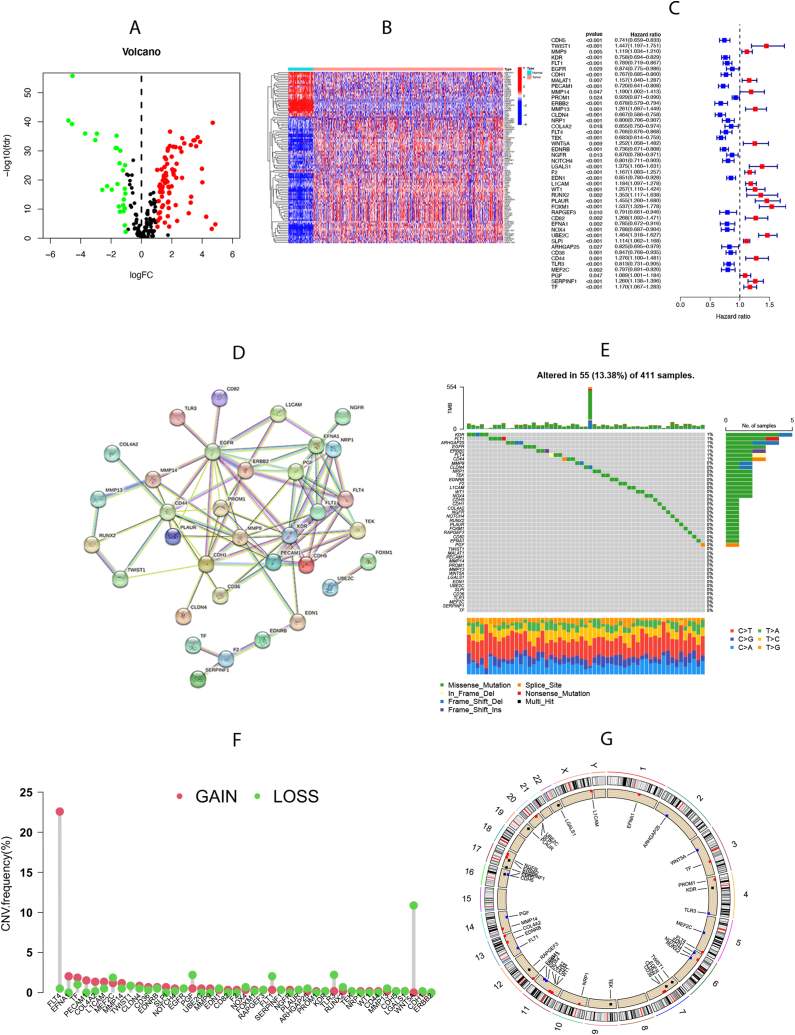
A total of 300 VMRGs were incorporated in this study based on a previous study and GeneCards. Among these, 109 genes with differential expression between normal and tumor tissues were identified (Fig. 1A and B). Samples from the TCGA-KIRC and GSE29609 cohorts were combined to explore the survival implications of VMRGs. Out of the 109 differentially expressed genes, 44 VMRGs showed significant prognostic value (Fig. 1C). The network of protein-protein interactions (PPI) (Fig. 1D) created with STRING showed connections between these VMRGs, which were enriched in phosphatidylinositol 3-kinase binding,promoter-specific chromatin binding, protease binding, and growth factor activity(strength>1). Furthermore, somatic mutations were detected in VMRGs, with 55 out of 411 samples exhibiting mutant VMRGs, resulting in a mutation rate of 13.38 %. Notably, KDR, FLT1, ARHGAP25, and EGFR exhibited the highest mutation rates (Fig. 1E). Furthermore, our study substantiated the occurrence of CNV in the VMRGs of patients with ccRCC. Substantial upregulation was observed in FLT4, EFNA1, and PECAM1, whereas WNT5A, TLP3, and FLT1 were notably downregulated (Fig. 1F). Fig. 1G shows the CNV loci of the 44 VMRGs on 23 chromosomes. This revealed significant disparities in the expression and mutation of VMRGs between normal and ccRCC samples, implying the potentially crucial involvement of VMRGs in ccRCC.
Fig. 1.
Expression and mutation of vasculogenic mimicry-related genes (VMRGs). (A) The differential expression of VMRGs between tumor tissue and normal tissue. (B) Heatmap showing the 109-differential expression VMRGs between normal and tumor tissues based on the expression. (C) The Cox survival analyses of differential expression VMRGs in the TCGA-KIRCC cohort and GSE29609 cohort. (D) The protein-protein interaction (PPI) network of the 44 VMRGs. (E) The incidence of somatic mutations of 44 VMRGs in ccRCC patients. (F) The CNV frequency of 44VMRGs. (G) The locus of CNV alterations of 44 VMRGs on 23 chromosomes.
3.2. Formation of VMRGs clusters in ccRCC
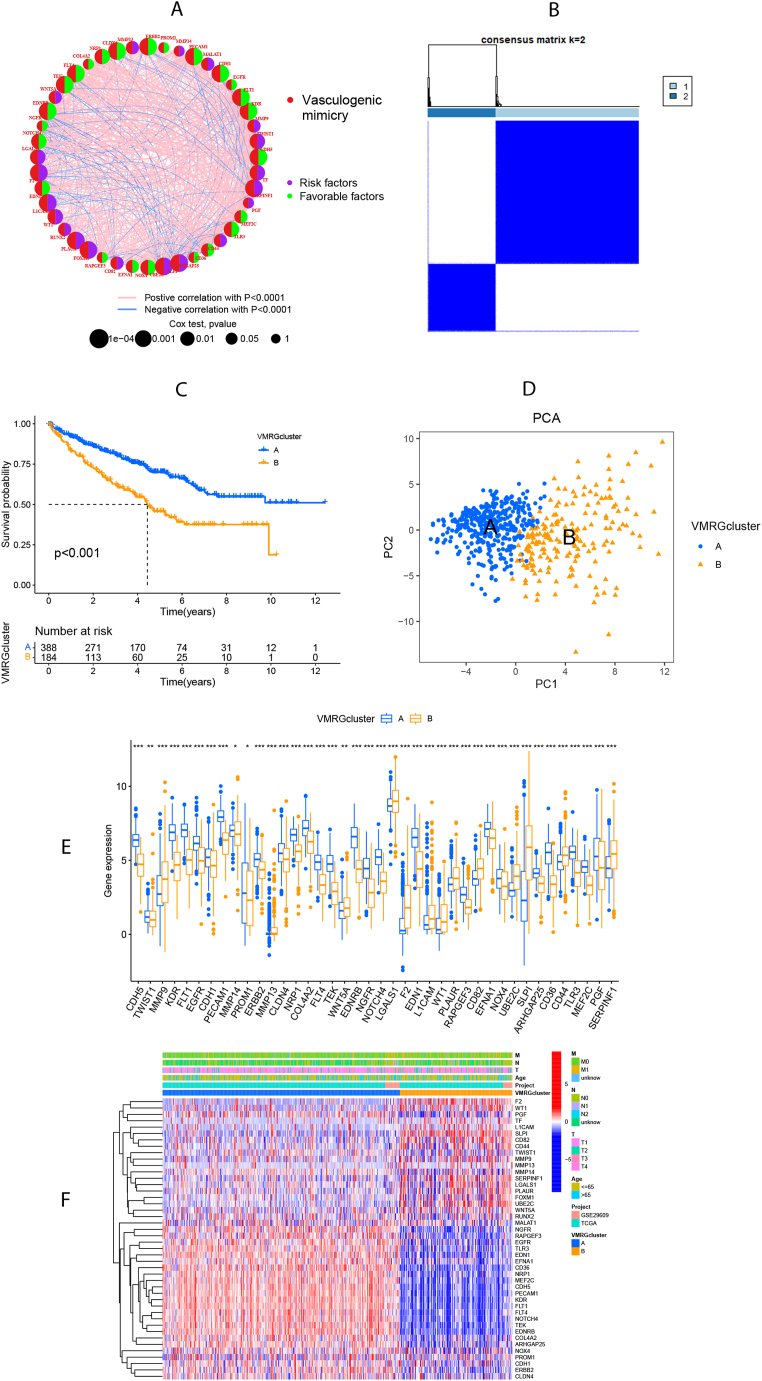
Fig. 2A illustrates the interactions and risk/favorable factors associated with VMRGs in ccRCC, demonstrating the intricate crosstalk among these prognosis-related VMRGs. To further explore the expression patterns of VMRGs in ccRCC, we performed a consensus clustering analysis, grouping patients from k = 2 to k = 9 (Supplementary Fig. 1). The findings indicated that k = 2 was the most suitable clustering variable (Fig. 2B). Survival analysis unveiled that VMRGs cluster A had more favorable survival outcomes (P < 0.001; Fig. 2C). Additionally, PCA confirmed a notable difference in the distributions of the two VMRGs clusters (Fig. 2D). Forty genes exhibited differential expression between the two VMRGs clusters (Fig. 2E and F).
Fig. 2.
Formation of vasculogenic mimicry-related genes clusters (VMRGs clusters). (A) The network of interactions between 44 VMRGs in the TCGA-KIRCC cohort and GSE29609 cohort, where the line thickness indicates the correlation strength. (B) All samples from the TCGA-KIRC cohort and GSE29609 cohort were divided into 2 clusters using the consensus clustering algorithm (k = 2). (C) Kaplan-Meier curves show the different overall survival (OS) between the two VMRGs clusters. (D) Principal component analysis (PCA) showed significant differences between the two VMRGs clusters. (E) VMRGs expression levels in two VMRGs clusters. (F)The heatmap showed the differences between the clusters in VMRGs expression.
3.3. Difference in biological characteristics and tumor immune infiltration between two VMRGs clusters
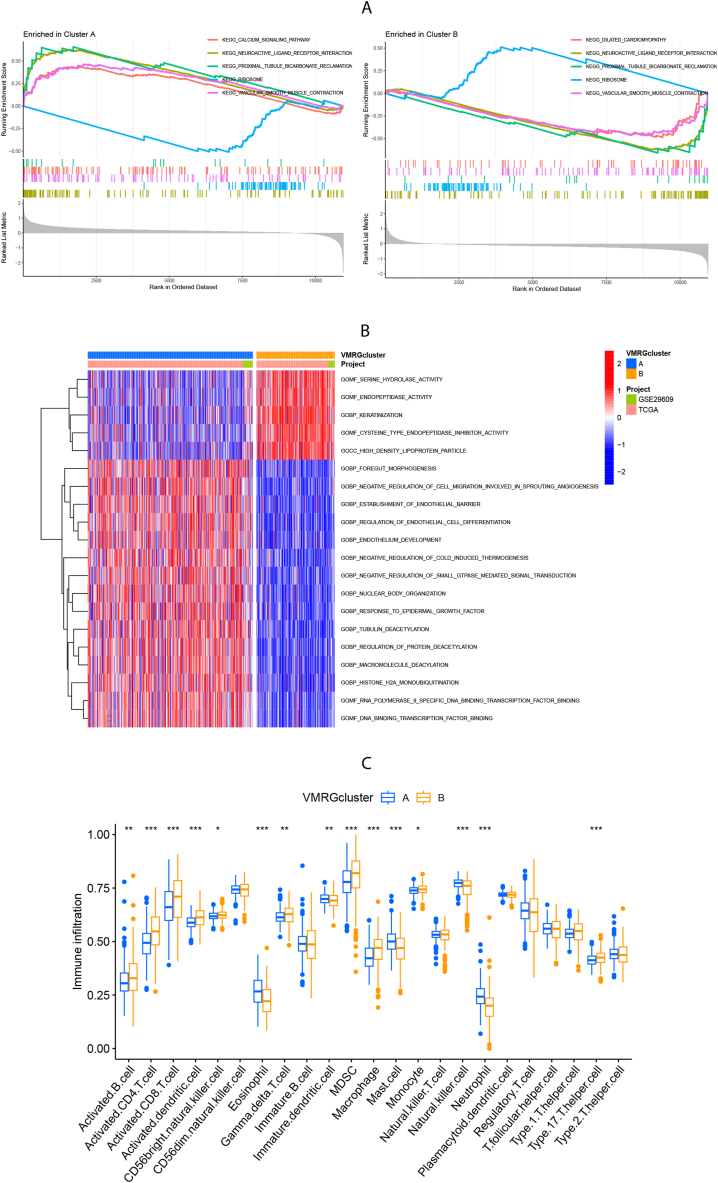
In VMRGs cluster A, the KEGG-based GSEA analysis indicated significant enrichment of the calcium signaling pathway, neuroactive ligand-receptor interaction, proximal tubule bicarbonate reclamation, and vascular smooth muscle contraction. In contrast, VMRGs cluster B showed enrichment of the ribosome (Fig. 3A). Furthermore, the results of Go-related GSVA indicated that catalytic activity (serine hydrolase, endopeptidase, and cysteine-type endopeptidase inhibitor activity), keratinization, and lipoprotein particles were more abundant in cluster B. Cluster A was enriched in endothelial cell development and migration, negative regulation of signal transduction, protein diacylation, protein ubiquitination, and transcription factor binding (Fig. 3B).
Fig. 3.
Analysis of biological features and tumor immune infiltration in two VMRGs clusters. (A) KEGG-related GSEA analysis showing the biological pathways of two VMRGs clusters. (B) GO-related GSVA analysis showing the biological pathways of two VMRGs clusters. (C) ssGSEA analysis showing the infiltration of 23 types of immune cells in two VMRGs clusters. Adjusted p-values were shown as insignificant, **P<0.01, ***P<0.001.
ssGSEA was employed to assess immune cell infiltration levels in the two VMRGs clusters, revealing notable differences in enrichment. The correlation between these clusters and the 23 tumor-infiltrating immune cells (TIICs) was analyzed using CIBERSORT (Fig. 3C). VMRGs cluster B is characterized by robust immune infiltration involving activated B cells, activated CD4/CD8 T cells, activated dendritic cells (DCs), CD56bright natural killer (NK) cells, γ.δ. T cells, myeloid-derived suppressor cells (MDSCs), mono/macro cells, and type 17 T helper cells suggest potential immunotherapy benefits. Conversely, VMRGs cluster A exhibits enrichment in eosinophils, immature DCs, mast cells, neutrophils, and NK cells.
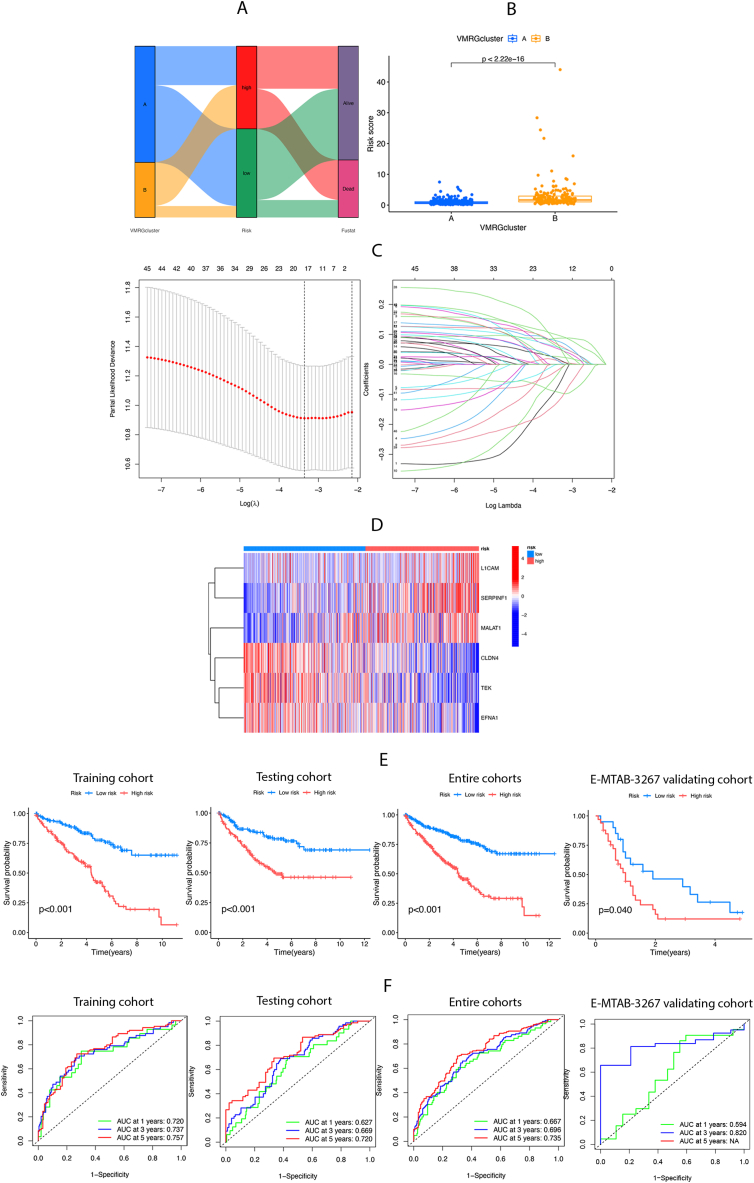
3.4. Construction and validation of the prognostic VMRGs risk score model
A VMRGs scoring system was devised based on distinct VMRGs expressions to assess the prognosis of individual patients with ccRCC. Fig. 4A depicts the patient distribution across two clusters and risk score groups. Notably, VMRGs cluster B exhibited a heightened risk score (Fig. 4B). All patients with ccRCC were randomly divided into training (n = 286) and testing (n = 286) cohorts. Subsequently, LASSO and multiCox analyses were conducted on 40 prognosis-related VMRGs within the training cohort to establish the prognostic model (Fig. 4C). Ultimately, six genes (L1CAM, SERPINF1, MALAT1, CLDN4, TEK, and EFNA1) were linked to ccRCC survival based on minimum partial likelihood deviation and multivariate Cox regression analysis. The VMRG risk score was calculated using the formula: Risk score = (0.144 × L1CAM expression) + (0.264 × SERPINF1 expression) + (0.302 × MALAT1 expression) + (−0.288 × CLDN4 expression) + (−0.174 × TEK expression) + (−0.226 × EFNA1 expression). In the high-risk group, levels of L1CAM, SERPINF1, and MALAT1 were elevated, while those of CLDN4, TEK, and EFNA1 were reduced (Fig. 4D). As shown in Fig. 4E, patients at low risk survived significantly longer than those at high risk across the training, testing, entire cohorts, and the E-MTAB-3267 validating cohort. (P < 0.05). In the training group, the AUC values for the ROC curves of survival rates at 1, 3, and 5 years were 0.720, 0.737, and 0.757. Similar analyses were performed on the testing, and the entire cohorts validated the risk-score model's accuracy. In the E-MTAB-3267 validating cohort, the AUC values for the ROC curves of survival rates at 1 and 3 years were 0.616 and 0.831 (Fig. 4F). Collectively, our results indicated a good performance of the 6-gene signature for survival prediction.
Fig. 4.
Construction and validation of the prognostic VMRGs score model. (A) Alluvial plot shows the distribution of patients in two VMRGs clusters, two risk groups, and their survival status. (B) The differences in risk score of two VMRGs clusters. (C) Significant differences in and expression of 6 prognosis-related genes between high-risk and low-risk groups. (D) Prognostic value of VMRGs in the training set. Multivariate Cox regression via LASSO is presented, and six candidate VMRGs were selected in the training cohort. (E) Survival analysis of the overall survival (OS) for high-risk and low-risk patients in the training, testing, entire cohorts, and the E-MTAB-3267 validating cohort. (F) The ROC curves for 1-,3-, and 5-year survival of ccRCC patients in the training, testing, entire cohorts, and the E-MTAB-3267 validating cohort.
3.5. Construction and validation of a nomogram to predict survival
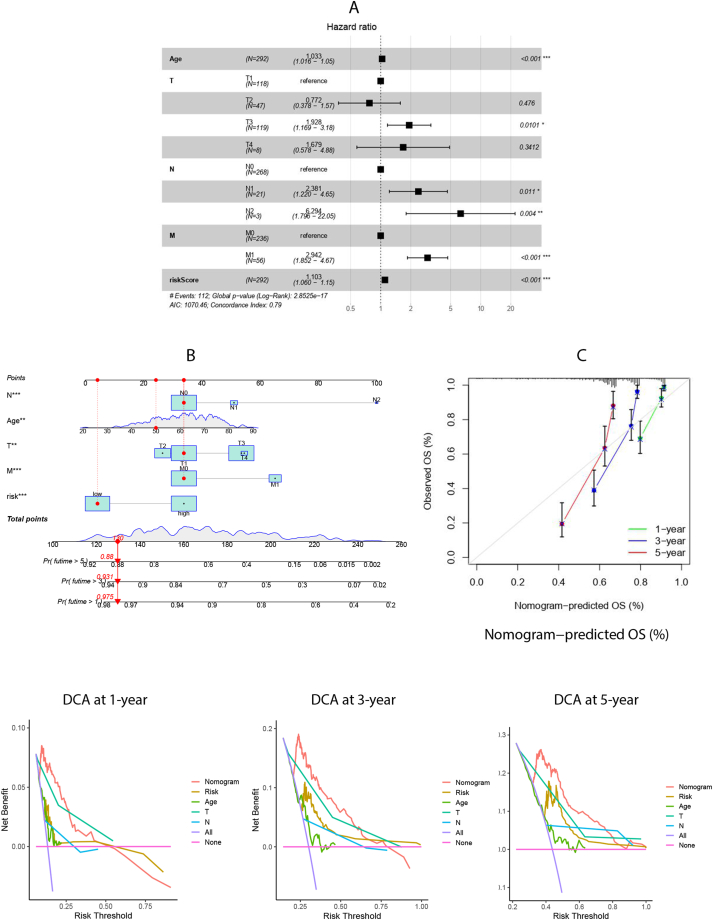
Cox regression analysis of the risk score alongside clinical characteristics such as age and stage demonstrated the VMRGs risk score's independence as a prognostic factor among patients with ccRCC (Fig. 5A). Recognizing the VMRGs risk score's pivotal role in the prognostic model, it was integrated with age and stage to develop a nomogram for predicting clinical outcomes at 1, 3, and 5 years (Fig. 5B). Patients with high VMRGs risk exhibited significantly lower survival rates than those with low VMRGs risk (P < 0.001; Supplementary Fig. 2). The calibration curve verified the nomogram's prediction accuracy (Fig. 5C). Furthermore, the decision curve analysis (DCA) values were assessed for predicting outcomes at 1, 3, and 5 years, highlighting the superior predictive performance of the nomogram combining VMRGs risk score, age, and stage (Fig. 5D).
Fig. 5.
Construction and validation of a nomogram. (A) Multivariate Cox regression analysis of risk scores and clinicopathological factors. (B) Nomogram construction for predicting the 1-,3-, and 5-year OS of ccRCC patients. (C) Calibration curve analysis for predicting patients’ survival at 1-,3-,5-year. (D) Decision curve analysis (DCA) for predicting the clinical utility of the nomogram at 1-,3-,5-year.
3.6. VMRGs risk was correlated with immune infiltration and TME
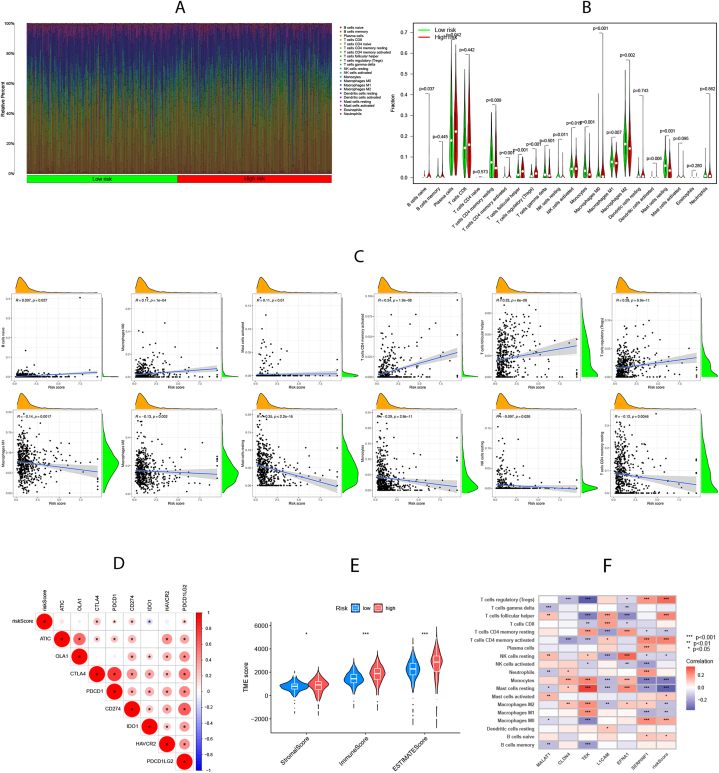
A notable discrepancy in the abundance of innate and adaptive immune cells was evident between the distinct risk groups (Fig. 6A and B). As depicted in Fig. 6C, the VMRGs risk score positively correlated with naïve B, Macro M0, activated mast cells, activated CD4 memory T cells, follicular helper T cells, and regulatory T cells. However, the opposite relationship was observed with Macro M1 and M2, resting mast, resting mono, resting NK, and resting CD4 memory cells. Additionally, we assessed the expression of ICPs across different risk groups, revealing a correlation between CTLA4, PDCD1, CD274, and PDCD1LG2 ICPs with the high-risk group, while IDO1 correlated with the low-risk group (Fig. 6D). Upon comparing the TME scores of these groups, we noted higher estimated and immune scores in the high-risk group, indicating elevated immunogenicity (Fig. 6E). Furthermore, we investigated the association between selected model genes in the prognostic signature and immune cell abundance (Fig. 6F). Based on these insights, the high-risk group can be characterized by increased immune cell infiltration and heightened responsiveness to immunotherapy.
Fig. 6.
Assessment of immune infiltration and tumor microenvironment (TME) characteristics between risk groups. (A-B) Differences in immune cell abundance. (C)Correlations between risk scores and immune cell abundance. (D) Expression levels of immune checkpoints (ICPs) in high-risk and low-risk groups. (E) Differences in Stromal score, Immune score, and ESTIMATE score between the two risk groups. (F) Correlations between 6 model genes in prognostic model and immune cell abundance. Adjusted p-values were shown as *P<0.05; **P<0.01; ***P<0.001.
3.7. Model gene expression and cells in ccRCC were examined by single-cell analysis, TCGA, IHC staining, and qRT-PCR
Using the scRNA-seq data of human ccRCC from GSE159115, 27,669 cells from eight patients were divided into 32 clusters [36] (Fig. 7A). The mRNA expression of CLDN4 was highest in malignant cells, with epithelial cells showing the next highest level. EDNA1 and TEK mRNA were mainly expressed in endothelial cell clusters. The mRNA expression of SERPINF14 predominated in mono- and macrocellular clusters. MALATI mRNA was predominantly expressed in CD8 + T cells, endothelial cells, epithelial cells, erythroblasts, malignant cells, monocyte/macrophages, pericytes, and plasma cells. (Fig. 7B and C).
Fig. 7.
Model genes mRNA expression of the single-cell-type clusters.
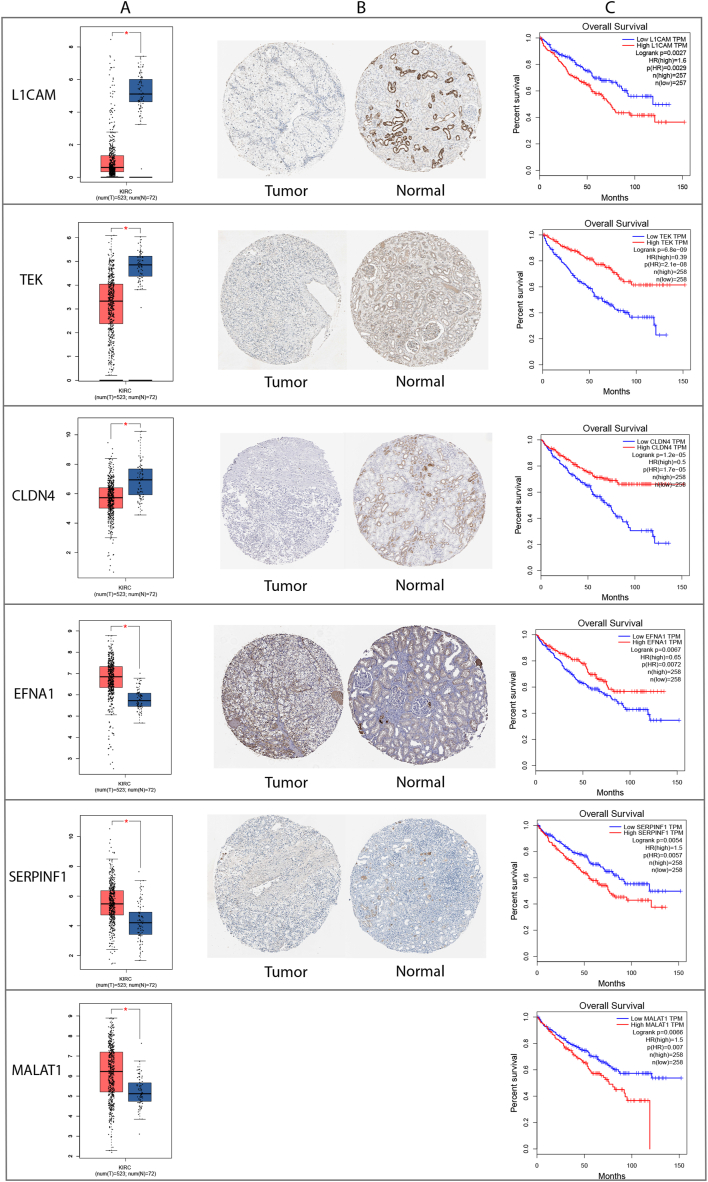
Differential expression analysis of six key genes showed higher expressions of EFNA1, SERPINF1, and MALAT1 and lower expressions of L1CAM, TEK, and CLDN4 in ccRCC than adjacent normal tissues. (Fig. 8A). Meanwhile, IHC staining from HPA showed higher expressions of EFNA1 and SERPINF1 and lower expressions of L1CAM, TEK, and CLDN4 relative to RCC tumor tissues than normal kidney tissues (Fig. 8B). Since MALAT1 is a long non-coding RNA (lncRNA) and does not encode a protein, it is likely not included in the HPA database. Analysis of TCGA data using GEPIA revealed that high expression of L1CAM, SERPINF1, and MALAT1 were significantly associated with poor survival prognosis in patients with RCC. Conversely, low expression of CLDN4, TEK, and EFNA1 were significantly associated with poor survival prognosis (Fig. 8C).
Fig. 8.
Validation of model gene expression identified in ccRCC. (A) Differential expression of six key genes between the kidney normal tissue and ccRCC using GEPIA online software. (B) Relative protein expressions of six key genes in the kidney normal tissue and ccRCC were measured by IHC staining from the HPA database. (C) Survival analysis of TCGA data using GEPIA online software.
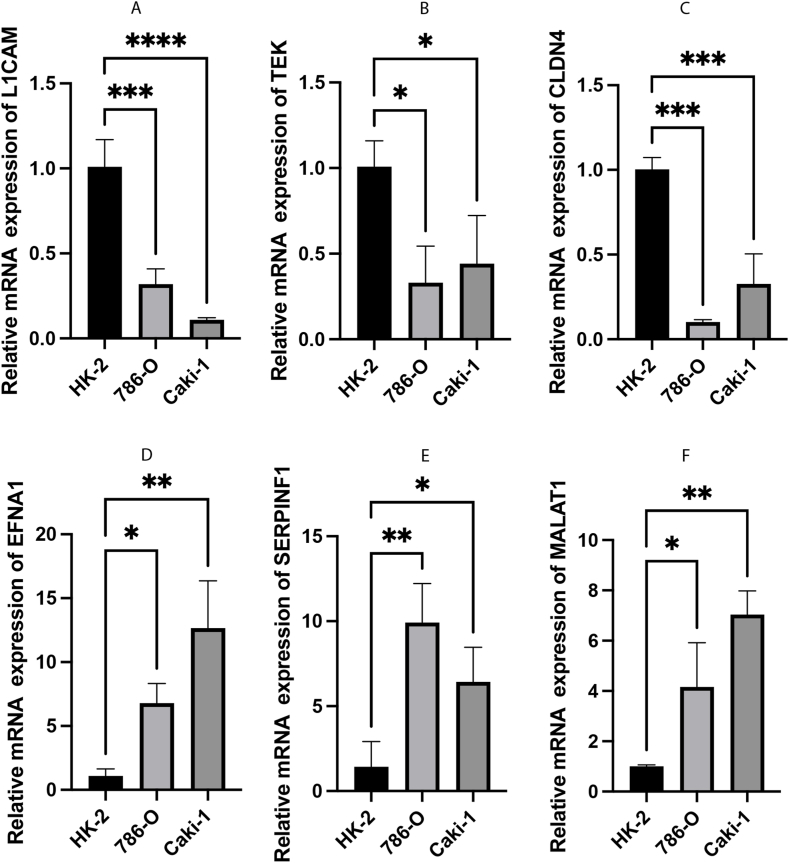
Furthermore, the expression of six VMRGs risk-associated genes was assessed using qRT-PCR in normal renal tubular epithelial cells (HK-2) and ccRCC cell lines, namely 786-O and Caki-1 cells. Compared to HK-2 cells, 786-O and Caki-1 cells showed significantly increased expression of EFNA1, SERPINF1, and MALAT1. Conversely, the expression of L1CAM, TEK, and CLDN4 was notably reduced in 786-O and Caki-1 cells (Fig. 9). The expression trend of the six key genes was consistent with the findings in this study.
Fig. 9.
The mRNA expression levels of 6 model genes in ccRCC cells (786-O and Caki-1) and the normal renal tubular epithelial cells HK-2. (A–F) The mRNA relative expression levels of L1CAM, TEK, CLDN4, EFNA1, SERPINF1, and MALAT1.
4. Discussion
ccRCC is a solid tumor that is characterized by significant vascularization [37]. Despite the current recommendations for surgery as the primary treatment for ccRCC, there is persistent recurrence and metastasis in patients who have undergone resection, posing a significant challenge. In situations where ccRCC is inoperable, at an advanced stage, recurrent, or has metastasized, the conventional therapeutic approach typically includes anti-angiogenic TKIs and immunotherapies [2,38]. Nonetheless, the efficacy of anti-angiogenic therapy is limited due to the emergence of drug resistance. Therefore, there is an urgent need to identify new therapeutic targets and prognostic factors for patients with ccRCC. VM serves as a mechanism for tumor vasculature replenishment, providing an alternative pathway for resistance to anti-angiogenic therapy [39]. Recent research has highlighted VM as an independent and significant predictor of disease-free survival [40,41]. However, the precise mechanisms and the correlation between VM and immune cell infiltration remain unclear. In this investigation, multiple subgroups of the VM were successfully identified, and a prognostic scoring model specifically for ccRCC was constructed, thereby emphasizing the crosstalk between the VM and TME.
In this study, a panel of six-gene signatures comprising L1CAM, TEK, CLDN4, EFNA1, SERPINF1, and MALAT1 was finally selected to generate a risk score model that can be exploited for predicting survival in ccRCC. The predictive performance of the signature was mutually verified in internal TCGA-KIRC, GSE29609, and external E-MTAB-3267 datasets. Patients with higher risk scores exhibited poorer survival rates at 1, 3, and 5 years than those with lower scores. A nomogram integrated with both the 6-gene-based signature and clinicopathological risk factors demonstrated that the model can accurately predict patients’ overall survival (OS). Data from TCGA, HPA, and qRT-PCR showed higher expressions of EFNA1, SERPINF1, and MALAT1 and lower expressions of L1CAM, TEK, and CLDN4 in ccRCC than adjacent normal tissues. Combining traditional anti-angiogenic therapies with the identified anti-VM targets, including L1CAM, TEK, CLDN4, EFNA1, SERPINF1, and MALAT1, could enhance outcomes for patients with ccRCC.
Interestingly, these six genes, which were used to establish our risk signature model, have been shown to be involved in tumor development and thus may be promising therapeutic targets for ccRCC. EFNA1 expression is elevated in ccRCC and could be stimulated under hypoxic conditions via a HIF-dependent pathway, thereby enhancing tumor angiogenesis [42]. SERPINH1 was the strongest association with poor prognosis and regulation on the expression levels of epithelial-to-mesenchymal transition markers in ccRCC in a previous study [43]. The secreted protein encoded by SERPINF1 strongly inhibits effects on angiogenesis [44]. Growing evidence shows that high MALAT1 expression in RCC tissues is inversely correlated with overall survival, and downregulation of MALAT1 inhibits ccRCC tumor growth by miR-182-5p regulation [45]. LncRNA MALAT1 was first recognized as a marker for metastasis development in the early stages of lung adenocarcinoma [46]. L1CAM, an axonal glycoprotein classified within the immunoglobulin supergene family, facilitates VM by upregulating hexokinase 2 expression in glioma [39]. It has been reported that LICAM plays a crucial role in the invasive ability of RCC [47]. TEK encodes a receptor belonging to the Tie2 family of protein tyrosine kinases that cooperate with VEGFs as critical regulators of vascular development [48]. In ccRCC, our data implied that TEK downregulation was associated with a poor prognosis, which was also demonstrated in previous studies [49]. Since Tie2 signaling influences vascular permeability, low expression of Tie2 may potentiate inflammatory cell migration into the tumor microenvironment. It has been reported that CLDN4, as a tight junction protein, might have implications in preserving the tumor microenvironment and could be involved in the aggressive characteristics of RCC [50]. Therefore, these model genes might be potential therapeutic targets for ccRCC.
There is mounting evidence that cell cycle processes are not only linked to tumor development but also play a role in immune escape and immunotherapy [51,52]. VM, an alternative mechanism of vasculature, is involved in the resistance to anti-angiogenic therapies. The concurrent implementation of anti-angiogenesis and immunotherapy has shown potential to improve clinical effectiveness [53,54]. The correlation between the VMRGs risk score and various immune cell subtypes highlights its significant role in modulating the tumor immune microenvironment. Its positive correlation with activated immune cells, such as activated mast cells and CD4 memory T cells, suggests that this signature may promote immune activation, supporting the immune system's response to tumors [55]. The upregulation of the VMRGs risk score in ccRCC was also concomitant with increased immune infiltration, as indicated by the elevated stromal, immune, and ESTIMATE scores. In addition, TLA4, PDCD1, CD274, and PDCD1LG2 ICPs were correlated with a high-risk group for VM. We imputed that once these immune checkpoint molecules were activated, they may promote tumor cells and other “bad cells” to escape the immune system. These results showed the great effectiveness of our six-gene-based risk signature in distributing patients with ccRCC into subtypes, which may benefit from different ICIs-based treatments. It might serve as a biomarker in tailoring individualized immunotherapy.
Considering the intra- and inter-tumor heterogeneity of ccRCC tumors, we sought to find variables in transcriptional levels that are more suitable to explain the complex interplay between tumors and the immune system compared to models using only clinical parameters. By integrating datasets from GEO, TCGA, and EBI ArrayExpress through rank aggregation analysis, a robust gene signature was identified. Recognizing that ccRCC comprises diverse molecular subtypes with varying prognoses and therapeutic responses, our six-gene signature, combined with clinicopathological parameters, may improve the predictive power and the possibility of clinical use. Our findings also suggest a link between VM risk and immune cells, indicating that VM may influence ccRCC pathogenesis and metastasis via immune modulation. However, the accuracy and effectiveness of the risk model need to be further confirmed in substantial clinical trials. Also, in vivo studies investigating the role of the six genes are needed to verify our results in the future.
5. Conclusions
In summary, our study identified a novel signature that would be applied as a prognosticator and a promising biomarker in individualized immunotherapy for patients with ccRCC. This study underscores the functional importance of VMRGs signatures and offers novel perspectives for potential anti-angiogenesis and therapeutic targets.
Data availability statement
Additional Supporting Information can be found online in the Supporting Information section. The publicly available datasets analyzed in this study can be found in TCGA (http://portal.gdc.cancer.gov/), EBI ArrayExpress (https://www.ebi.ac.uk/arrayexpress/), GEO (https://www.ncbi.nlm.nih.gov/geo) under accession numbers GSE29609, and TISCH under accession number GSE159115, respectively.
Ethics declarations
Not available.
Funding
This work was supported by the National Natural Science Foundation of China [grant number: 82273389].
CRediT authorship contribution statement
Junyong Ou: Conceptualization, Data curation, Formal analysis, Funding acquisition, Project administration, Supervision, Validation, Visualization, Writing – original draft. Haoming Yin: Data curation, Methodology, Project administration, Software, Writing – original draft. Fan Shu: Conceptualization, Funding acquisition, Methodology, Resources. Zonglong Wu: Resources, Software, Supervision. Shuai Liu: Resources, Supervision, Validation. Jianfei Ye: Funding acquisition, Investigation, Supervision, Validation, Visualization, Writing – review & editing. Shudong Zhang: Funding acquisition, Investigation, Project administration, Software, Supervision, Validation, Writing – review & editing.
Declaration of competing interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.
Acknowledgments
We deeply grateful to Ms. Shanshan Zhu (Carnegie Mellon University, PA, USA) for her invaluable assistance in the data processing for this thesis. Her expertise and dedication were crucial in ensuring the accuracy and reliability of our findings. The authors acknowledge the efforts of the researchers and study patients for their contributions.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e36235.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- 1.Barata P.C., Rini B.I. Treatment of renal cell carcinoma: current status and future directions. CA Cancer J Clin. 2017;67(6):507–524. doi: 10.3322/caac.21411. [DOI] [PubMed] [Google Scholar]
- 2.Motzer R.J., Hutson T.E., Cella D., et al. Pazopanib versus sunitinib in metastatic renal-cell carcinoma. N. Engl. J. Med. 2013;369(8):722–731. doi: 10.1056/NEJMoa1303989. [DOI] [PubMed] [Google Scholar]
- 3.Qu L., Ding J., Chen C., et al. Exosome-transmitted lncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell. 2016;29(5):653–668. doi: 10.1016/j.ccell.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 4.Maniotis A.J., Folberg R., Hess A., et al. Vascular channel formation by human melanoma cells in vivo and in vitro: vasculogenic mimicry. Am. J. Pathol. 1999;155(3):739–752. doi: 10.1016/S0002-9440(10)65173-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiang T., Lin Y.X., Ma W., et al. Vasculogenic mimicry formation in EBV-associated epithelial malignancies. Nat. Commun. 2018;9(1):5009. doi: 10.1038/s41467-018-07308-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williamson S.C., Metcalf R.L., Trapani F., et al. Vasculogenic mimicry in small cell lung cancer. Nat. Commun. 2016;7 doi: 10.1038/ncomms13322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delgado-Bellido D., Serrano-Saenz S., Fernández-Cortés M., Oliver F.J. Vasculogenic mimicry signaling revisited: focus on non-vascular VE-cadherin. Mol. Cancer. 2017;16(1):65. doi: 10.1186/s12943-017-0631-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun T., Sun B cun, lan Zhao X., et al. Promotion of tumor cell metastasis and vasculogenic mimicry by way of transcription coactivation by Bcl-2 and Twist1: a study of hepatocellular carcinoma. Hepatology. 2011;54(5):1690–1706. doi: 10.1002/hep.24543. [DOI] [PubMed] [Google Scholar]
- 9.Yang J.P., Liao Y.D., Mai D.M., et al. Tumor vasculogenic mimicry predicts poor prognosis in cancer patients: a meta-analysis. Angiogenesis. 2016;19(2):191–200. doi: 10.1007/s10456-016-9500-2. [DOI] [PubMed] [Google Scholar]
- 10.Heidegger I., Pircher A., Pichler R. Targeting the tumor microenvironment in renal cell cancer biology and therapy. Front. Oncol. 2019;9:490. doi: 10.3389/fonc.2019.00490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruf W., Seftor E.A., Petrovan R.J., et al. Differential role of tissue factor pathway inhibitors 1 and 2 in melanoma vasculogenic mimicry. Cancer Res. 2003;63(17):5381–5389. [PubMed] [Google Scholar]
- 12.Sun D., Sun B., Liu T., et al. Slug promoted vasculogenic mimicry in hepatocellular carcinoma. J. Cell Mol. Med. 2013;17(8):1038–1047. doi: 10.1111/jcmm.12087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee M., Cho H.J., Park K.S., Jung H.Y. ELK3 controls gastric cancer cell migration and invasion by regulating ECM remodeling-related genes. Int. J. Mol. Sci. 2022;23(7):3709. doi: 10.3390/ijms23073709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu Y., Liu X., Zhao P., Zhao H., Gao W., Wang L. Celastrol suppresses glioma vasculogenic mimicry formation and angiogenesis by blocking the PI3K/Akt/mTOR signaling pathway. Front. Pharmacol. 2020;11:25. doi: 10.3389/fphar.2020.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu J., May L., Milsom C., et al. Contribution of host-derived tissue factor to tumor neovascularization. Arterioscler. Thromb. Vasc. Biol. 2008;28(11):1975–1981. doi: 10.1161/ATVBAHA.108.175083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu X mei, Zhang Q ping, gao Mu Y., et al. Clinical significance of vasculogenic mimicry in human gliomas. J. Neuro Oncol. 2011;105(2):173–179. doi: 10.1007/s11060-011-0578-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morales-Guadarrama G., García-Becerra R., Méndez-Pérez E.A., García-Quiroz J., Avila E., Díaz L. Vasculogenic mimicry in breast cancer: clinical relevance and drivers. Cells. 2021;10(7):1758. doi: 10.3390/cells10071758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim H.S., Won Y.J., Shim J.H., et al. Morphological characteristics of vasculogenic mimicry and its correlation with EphA2 expression in gastric adenocarcinoma. Sci. Rep. 2019;9(1):3414. doi: 10.1038/s41598-019-40265-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mao X.G., Xue X.Y., Wang L., et al. CDH5 is specifically activated in glioblastoma stemlike cells and contributes to vasculogenic mimicry induced by hypoxia. Neuro Oncol. 2013;15(7):865–879. doi: 10.1093/neuonc/not029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Basu G.D., Liang W.S., Stephan D.A., et al. A novel role for cyclooxygenase-2 in regulating vascular channel formation by human breast cancer cells. Breast Cancer Res. 2006;8(6):R69. doi: 10.1186/bcr1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wechman S.L., Emdad L., Sarkar D., Das S.K., Fisher P.B. Vascular mimicry: triggers, molecular interactions and in vivo models. Adv. Cancer Res. 2020;148:27–67. doi: 10.1016/bs.acr.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herrera-Vargas A.K., García-Rodríguez E., Olea-Flores M., Mendoza-Catalán M.A., Flores-Alfaro E., Navarro-Tito N. Pro-angiogenic activity and vasculogenic mimicry in the tumor microenvironment by leptin in cancer. Cytokine Growth Factor Rev. 2021;62:23–41. doi: 10.1016/j.cytogfr.2021.10.006. [DOI] [PubMed] [Google Scholar]
- 23.Lu X.S., Sun W., Ge C.Y., Zhang W.Z., Fan Y.Z. Contribution of the PI3K/MMPs/Ln-5γ2 and EphA2/FAK/Paxillin signaling pathways to tumor growth and vasculogenic mimicry of gallbladder carcinomas. Int. J. Oncol. 2013;42(6):2103–2115. doi: 10.3892/ijo.2013.1897. [DOI] [PubMed] [Google Scholar]
- 24.Mo J., Zhao X., Wang W., et al. TFPI2 promotes perivascular migration in an angiotropism model of melanoma. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.662434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu K., Lin C.C.J., Hatcher A., et al. PIK3CA variants selectively initiate brain hyperactivity during gliomagenesis. Nature. 2020;578(7793):166–171. doi: 10.1038/s41586-020-1952-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Waddell N., Pajic M., Patch A.M., et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518(7540):495–501. doi: 10.1038/nature14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seiler M., Huang C.C., Szalma S., Bhanot G. ConsensusCluster: a software tool for unsupervised cluster discovery in numerical data. OMICS A J. Integr. Biol. 2010;14(1):109–113. doi: 10.1089/omi.2009.0083. [DOI] [PubMed] [Google Scholar]
- 28.Sabah A., Tiun S., Sani N.S., Ayob M., Taha A.Y. Enhancing web search result clustering model based on multiview multirepresentation consensus cluster ensemble (mmcc) approach. PLoS One. 2021;16(1) doi: 10.1371/journal.pone.0245264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hänzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-Seq data. BMC Bioinf. 2013;14(1):7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bloniarz A., Liu H., Zhang C.H., Sekhon J.S., Yu B. Lasso adjustments of treatment effect estimates in randomized experiments. Proc. Natl. Acad. Sci. USA. 2016;113(27):7383–7390. doi: 10.1073/pnas.1510506113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aran D., Hu Z., Butte A.J. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18(1):220. doi: 10.1186/s13059-017-1349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen B., Khodadoust M.S., Liu C.L., Newman A.M., Alizadeh A.A. In: Cancer Systems Biology: Methods And Protocols. Methods in Molecular Biology. von Stechow L., editor. Springer; 2018. Profiling tumor infiltrating immune cells with CIBERSORT; pp. 243–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoshihara K., Shahmoradgoli M., Martínez E., et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun D., Wang J., Han Y., et al. TISCH: a comprehensive web resource enabling interactive single-cell transcriptome visualization of tumor microenvironment. Nucleic Acids Res. 2021;49(D1):D1420–D1430. doi: 10.1093/nar/gkaa1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uhlen M., Zhang C., Lee S., et al. A pathology atlas of the human cancer transcriptome. Science. 2017;357(6352) doi: 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y., Narayanan S.P., Mannan R., et al. Single-cell analyses of renal cell cancers reveal insights into tumor microenvironment, cell of origin, and therapy response. Proc Natl Acad Sci U S A. 2021;118(24) doi: 10.1073/pnas.2103240118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bedke J., Albiges L., Capitanio U., et al. The 2021 updated European association of urology guidelines on renal cell carcinoma: immune checkpoint inhibitor–based combination therapies for treatment-naive metastatic clear-cell renal cell carcinoma are standard of care. Eur. Urol. 2021;80(4):393–397. doi: 10.1016/j.eururo.2021.04.042. [DOI] [PubMed] [Google Scholar]
- 38.Méjean A., Ravaud A., Thezenas S., et al. Sunitinib alone or after nephrectomy in metastatic renal-cell carcinoma. N. Engl. J. Med. 2018;379(5):417–427. doi: 10.1056/NEJMoa1803675. [DOI] [PubMed] [Google Scholar]
- 39.Angara K., Borin T.F., Arbab A.S. Vascular mimicry: a novel neovascularization mechanism driving anti-angiogenic therapy (aat) resistance in glioblastoma. Transl Oncol. 2017;10(4):650–660. doi: 10.1016/j.tranon.2017.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vartanian A.A., Stepanova E.V., Gutorov S.L., et al. Prognostic significance of periodic acid-Schiff-positive patterns in clear cell renal cell carcinoma. Can. J. Urol. 2009;16(4):4726–4732. [PubMed] [Google Scholar]
- 41.Zhang Y., Sun B., Zhao X., et al. Clinical significances and prognostic value of cancer stem-like cells markers and vasculogenic mimicry in renal cell carcinoma. J. Surg. Oncol. 2013;108(6):414–419. doi: 10.1002/jso.23402. [DOI] [PubMed] [Google Scholar]
- 42.Hao Y., Li G. Role of EFNA1 in tumorigenesis and prospects for cancer therapy. Biomed. Pharmacother. 2020;130 doi: 10.1016/j.biopha.2020.110567. [DOI] [PubMed] [Google Scholar]
- 43.Qi Y., Zhang Y., Peng Z., et al. SERPINH1 overexpression in clear cell renal cell carcinoma: association with poor clinical outcome and its potential as a novel prognostic marker. J. Cell Mol. Med. 2018;22(2):1224–1235. doi: 10.1111/jcmm.13495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ren J.G., Jie C., Talbot C. How PEDF prevents angiogenesis: a hypothesized pathway. Med. Hypotheses. 2005;64(1):74–78. doi: 10.1016/j.mehy.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 45.Abstract 2478: miR-182-5p suppresses progression of renal cancer through cell cycle arrest by targeting lncRNA MALAT-1 | Cancer Research | American Association for Cancer Research. Accessed July 1, 2024. https://aacrjournals.org/cancerres/article/78/13_Supplement/2478/626782/Abstract-2478-miR-182-5p-suppresses-progression-of.
- 46.Li J., Wang J., Chen Y., et al. LncRNA MALAT1 exerts oncogenic functions in lung adenocarcinoma by targeting miR-204. Am. J. Cancer Res. 2016;6(5):1099–1107. [PMC free article] [PubMed] [Google Scholar]
- 47.Wu Z., Wu Y., Liu Z., et al. L1CAM deployed perivascular tumor niche promotes vessel wall invasion of tumor thrombus and metastasis of renal cell carcinoma. Cell Death Dis. 2023;9(1):112. doi: 10.1038/s41420-023-01410-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Du S., Qian J., Tan S., et al. Tumor cell-derived exosomes deliver TIE2 protein to macrophages to promote angiogenesis in cervical cancer. Cancer Lett. 2022;529:168–179. doi: 10.1016/j.canlet.2022.01.005. [DOI] [PubMed] [Google Scholar]
- 49.Ha M., Son Y.R., Kim J., et al. TEK is a novel prognostic marker for clear cell renal cell carcinoma. Eur. Rev. Med. Pharmacol. Sci. 2019;23(4):1451–1458. doi: 10.26355/eurrev_201902_17102. [DOI] [PubMed] [Google Scholar]
- 50.Owari T., Sasaki T., Fujii K., et al. Role of nuclear claudin-4 in renal cell carcinoma. Int. J. Mol. Sci. 2020;21(21):8340. doi: 10.3390/ijms21218340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu J., Peng Y., Wei W. Cell cycle on the crossroad of tumorigenesis and cancer therapy. Trends Cell Biol. 2022;32(1):30–44. doi: 10.1016/j.tcb.2021.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bednarski J.J., Sleckman B.P. At the intersection of DNA damage and immune responses. Nat. Rev. Immunol. 2019;19(4):231–242. doi: 10.1038/s41577-019-0135-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Argentiero A., Solimando A.G., Krebs M., et al. Anti-angiogenesis and immunotherapy: novel paradigms to envision tailored approaches in renal cell-carcinoma. J. Clin. Med. 2020;9(5):1594. doi: 10.3390/jcm9051594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yi M., Jiao D., Qin S., Chu Q., Wu K., Li A. Synergistic effect of immune checkpoint blockade and anti-angiogenesis in cancer treatment. Mol. Cancer. 2019;18(1):60. doi: 10.1186/s12943-019-0974-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Monjaras-Avila C.U., Lorenzo-Leal A.C., Luque-Badillo A.C., D'Costa N., Chavez-Muñoz C., Bach H. The tumor immune microenvironment in clear cell renal cell carcinoma. Int. J. Mol. Sci. 2023;24(9):7946. doi: 10.3390/ijms24097946. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Additional Supporting Information can be found online in the Supporting Information section. The publicly available datasets analyzed in this study can be found in TCGA (http://portal.gdc.cancer.gov/), EBI ArrayExpress (https://www.ebi.ac.uk/arrayexpress/), GEO (https://www.ncbi.nlm.nih.gov/geo) under accession numbers GSE29609, and TISCH under accession number GSE159115, respectively.