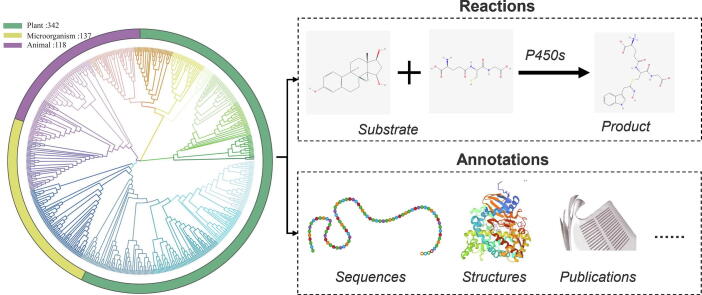
Graphical abstract
Keywords: Cytochrome P450 enzymes, Reactions, Substrates, Products, Database
Highlights
-
•
P450Rdb compiles a comprehensive catalog of over 1,600 reactions catalyzed by P450s.
-
•
P450Rdb has collected a diverse collection of more than 590 P450s from over 200 species.
-
•
P450Rdb systematically organizes all reactions based on their chemical reaction type and site.
-
•
P450Rdb provides a user-friendly interface on P450s and their associated reactions.
-
•
P450Rdb is beneficial for research in synthetic biology, pharmacology, and the chemical industry.
Abstract
Introduction
Cytochrome P450 enzymes (P450s) are recognized as the most versatile catalysts worldwide, playing vital roles in numerous biological metabolism and biosynthesis processes across all kingdoms of life. Despite the vast number of P450 genes available in databases (over 300,000), only a small fraction of them (less than 0.2 %) have undergone functional characterization.
Objectives
To provide a convenient platform with abundant information on P450s and their corresponding reactions, we introduce the P450Rdb database, a manually curated resource compiles literature-supported reactions catalyzed by P450s.
Methods
All the P450s and Reactions were manually curated from the literature and known databases. Subsequently, the P450 reactions organized and categorized according to their chemical reaction type and site. The website was developed using HTML and PHP languages, with the MySQL server utilized for data storage.
Results
The current version of P450Rdb catalogs over 1,600 reactions, involving more than 590 P450s across a diverse range of over 200 species. Additionally, it offers a user-friendly interface with comprehensive information, enabling easy querying, browsing, and analysis of P450s and their corresponding reactions. P450Rdb is free available at http://www.cellknowledge.com.cn/p450rdb/.
Conclusions
We believe that this database will significantly promote structural and functional research on P450s, thereby fostering advancements in the fields of natural product synthesis, pharmaceutical engineering, biotechnological applications, agricultural and crop improvement, and the chemical industry.
Introduction
Cytochrome P450 (CYP, P450s) is a superfamily of enzymes that utilize heme as a cofactor to catalyze monooxygenase reactions. They are found across all kingdoms of life, including plants, animals, fungi, protists, bacteria, archaea, and even viruses [1], [2]. Among these, plant P450s have undergone significant diversification compared to animals and microbes [3], [4]. They exhibit a wide range of abundance and functional diversity within the plant kingdom, playing a critical role in the biosynthetic pathways of natural products and providing a molecular foundation for plant plasticity [3], [5].
Accumulated studies have demonstrated that P450s harbor remarkable catalytic abilities, enabling them to facilitate over 20 types of oxido-reduction reactions, including hydroxylation, sulfoxidation, oxidation, epoxidation, decarboxylation, and cyclization [6]. These reactions involve various substrates and products, including terpenoids, alkaloids, fatty acids, steroids, antibiotics, and xenobiotics, making P450s nature's most versatile biological catalysts [7], [8], [9]. In vivo, P450s play vital roles in cellular metabolism, natural product biosynthesis and degradation, drug and environmental pollutant metabolism, as well as chemical defense in plants [3], [10], [11], [12], [13]. Due to their diverse catalytic capacities, P450s have garnered great interest in synthetic biology, biotechnological and agricultural applications, pharmaceuticals, bioremediation and environmental monitoring and the chemical industry [14], [15], [16], [17], [18], [19], [20], [21].
Advancements in “omics” technologies and synthetic biology in recent years have made significant contributions to the identification and functional interpretation of P450s [22], [23], [24]. However, certain challenges still persist, such as identification of crystal structures, enhancing the heterologous activities, and enzyme engineering [25], [26], [27], [28]. Additionally, although numerous databases have been developed to record, manage, and analyze the vast amount of P450s data (Table S1), such as the Cytochrome P450 Homepage [29], the Homepage of the Human Cytochrome P450 (CYP) Allele Nomenclature Committee [30], the Directory of P450-containing Systems [31], the Arabidopsis P450 database [32], SuperCYP database [33], the Insect P450 Site (http://p450.antibes.inra.fr/), the P450s in PROMISE (http://metallo.scripps.edu/PROMISE/P450.html), Fungal cytochrome P450 database [34], The Cytochrome P450 Engineering Database [35], PCPD database [36] and The Plant Cytochrome P450 Database [3], Some review articles have also summarized knowledge related to P450, including its protein structure, function, associated catalytic reactions, and interactions with drugs (Table S2) [37], [38], [39], [40]. these resources have identified and collected over 300,000 P450 genes, yet less than 0.2 % of them have been functionally characterized [41], [42], [43].
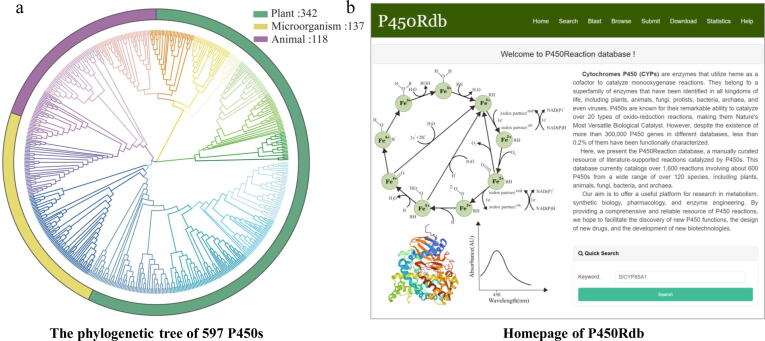
Recently, databases focusing on the catalytic function of plant P450s have emerged, The PCPD database [36], for [44] instance, has collected 181 plant P450s along with their sequences, structures, and reaction information from literature and other databases. The Plant Cytochrome P450 database provides an extensive compilation of CYPs known to metabolize one or more substrates, based on literature sources [3]. These resources serve as important reference data for functional interpretation and synthetic biology related to plant P450s. However, to the best of our knowledge, there is still a lack of a dedicated resource that stores and integrates the catalytic functions and reactions of P450s across all organisms. Herein, we present the P450Rdb database, a manually curated resource that focuses on literature-supported reactions catalyzed by P450s. The current version of P450Rdb documents over 590 P450s, with more than 1,600 reactions (Fig. 1a). Our aim with this database is to provide a convenient interface with abundant information on P450s and their corresponding reactions (Fig. 1b), thereby accelerating research in the synthesis of natural products and pharmacology.
Fig. 1.
The data and website of the P450Rdb. a. The phylogenetic tree of 597 P450s in the database. b. the homepage of the P450db website.
Materials and methods
Data collection
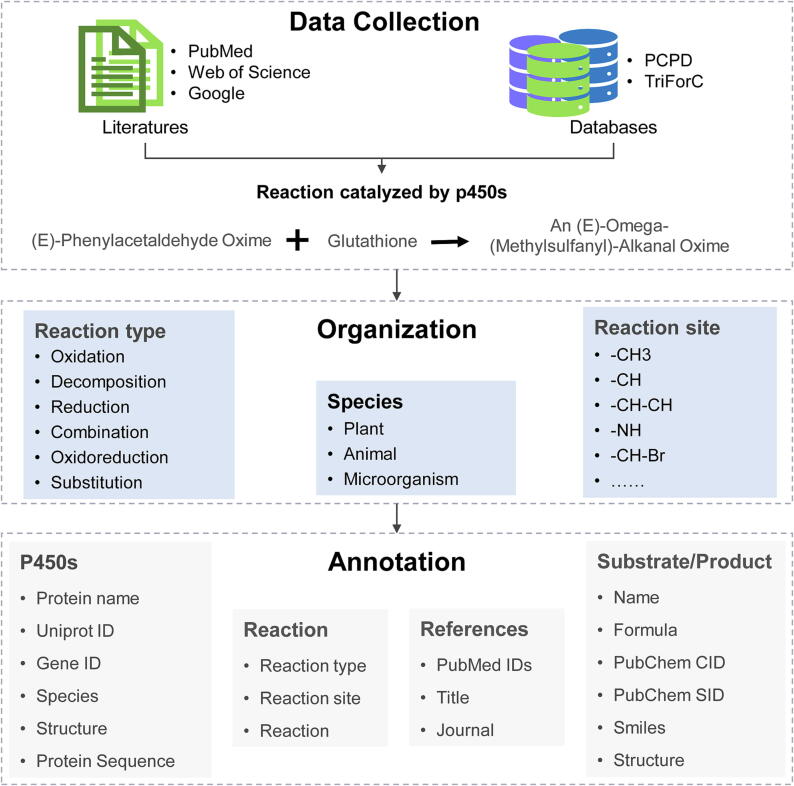
All the P450s and Reactions were manually curated from the literature (before april 2023) and two other known databases, including PCPD database and TriForC database (Fig. 2). Initially, a comprehensive search was performed on PubMed, bioRxiv, and Google Scholar using keywords such as “P450,” “CYPs,” “Cytochrome P450,” “monooxygenases,” “Reaction,” and “p450 enzyme.” The retrieved publications were then subjected to a preliminary check by expert curators to eliminate false-positive papers. Only experimentally supported reactions catalyzed by P450s were included in the P450Reaction database. The collected entries underwent a rigorous evaluation and double-checking process by at least two separate expert curators. Any discrepancies that arose were resolved through discussions with a third expert curator to ensure a consensus was reached. Additionally, the P450Reaction database incorporated 181 reactions from the PCPD database [36] and 228 reactions from the TriForC database [45].
Fig. 2.
Data collection, organization and annotation of P450Rdb.
Organization
At first, we categorized all the reactions into six types based on their chemical reaction type: oxidation reaction, reduction reaction, oxidoreduction reaction, combination reaction, decomposition reaction, and substitution reaction (Fig. 2). An oxidation reaction involves the loss of electrons by a molecule, atom, or ion during a chemical reaction. Conversely, a reduction reaction occurs when a reactant gains electron. In an oxidoreduction reaction, one part of the reaction decreases its oxidation number, typically by gaining electrons, while the other part undergoes oxidation, resulting in electron loss. A combination reaction takes place when two substrates combine to form a single product. On the other hand, a decomposition reaction occurs when a single reactant breaks down into two or more products. Lastly, a substitution reaction involves the replacement of an atom, ion, or group of atoms within a molecule by another atom, ion, or group. Furthermore, in addition to considering the chemical reaction type, we differentiated the reactions based on the specific reaction sites involved (Fig. 2). These sites include functional groups such as —CH3, —CH, —NH and chemical bonds like —CH—CH, —CH—OH, —CH-Br. In total, we identified 41 distinct reaction sites to comprehensively classify all the reactions.
Annotation
To establish consistency among the P450s and compounds (substrates/products) mentioned in various literature sources, we employed authoritative reference databases for mapping (Fig. 2). All the P450s were mapped to the UniProt database (Uniprot ID) [46] and NCBI gene database (Entrez ID) [47], and the substrate/product were mapped to PubChem database (PubChem CID and SID) [48]. In terms of the P450s details, we obtained the protein names, species information and sequences from the Uniprot database. And considering that many P450s lack experimentally validated crystal structures, we have included hyperlinks to access predicted protein structures from the AlphaFold database. In terms of the substrate/product details, we collected the respective formulas, Smiles notation, and structures from the PubChem database. These comprehensive mappings and data acquisitions from authoritative resources ensure standardized and reliable information in the database.
Architecture
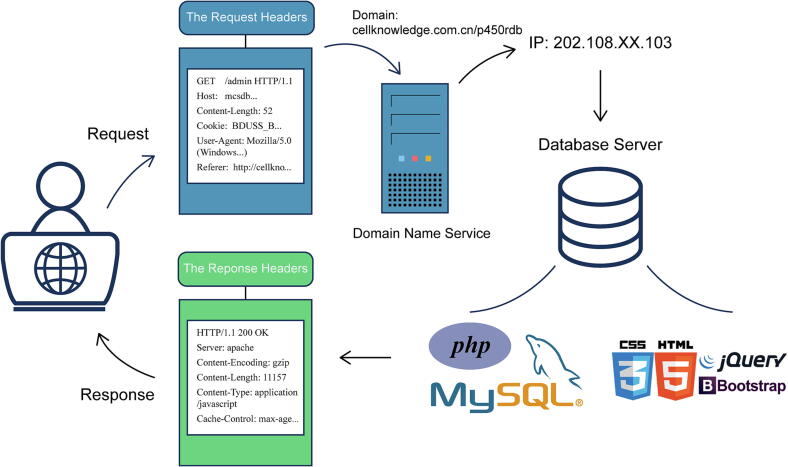
First, we adopt the framework of HTML + CSS + JavaScript for the front-end web development (Fig. 3). The three programming languages cooperate with each other to provide users with clear contents, clean interfaces and rich interactive applications. And the self-responsive layout adopted by the website is also compatible with different scenarios on the PC and mobile. Meanwhile, we adopt the PHP + MySQL architecture for the back-end, which can run different scripts based on different front-end access requests, and dynamically generates the corresponding returned data. This process has high data security and fast query speed. In addition, we adopt the Smarty template engine to separate the front-end and back-end, which separates the logical program and the external content to facilitate later management and maintenance. The network service of the platform is provided by EngineX (Nginx).
Fig. 3.
Architecture of P450Rdb.
Results
Data statistics
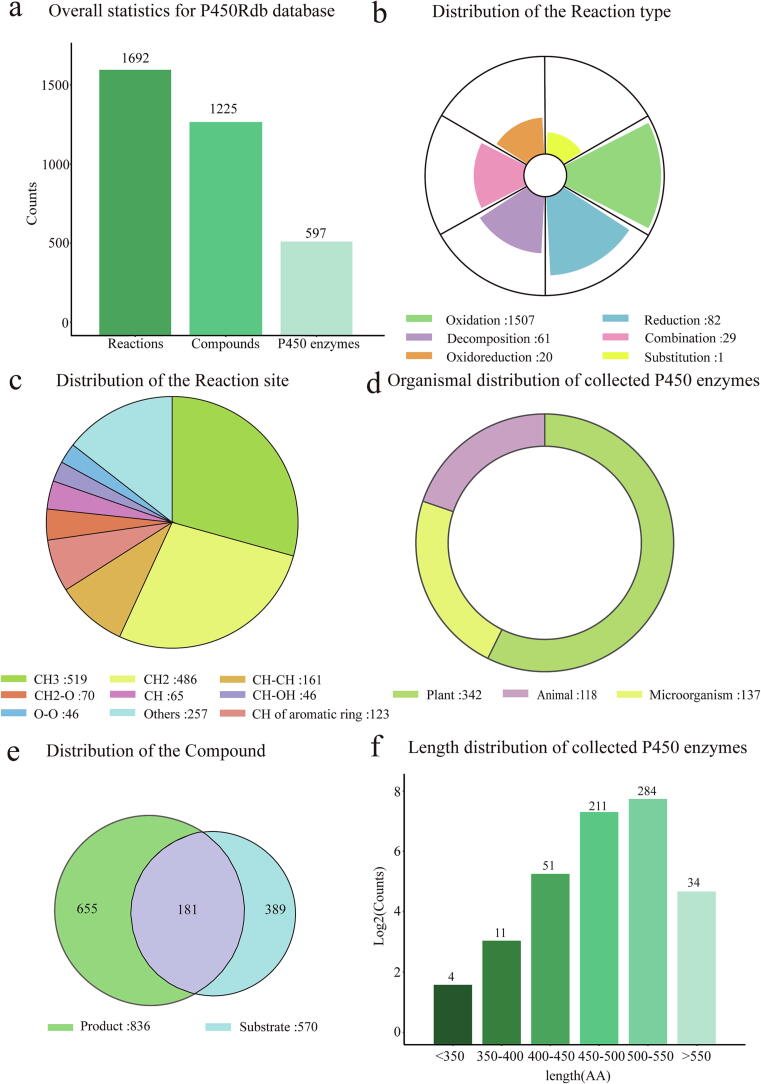
This current version of P450Rdb documents 1692 manually curated reactions supported by peer reviewed literatures, refers to 890 compounds (substrates/products), 597 P450s across a range of over 200 species (Fig. 4a). These reactions involve 1225 compounds serving as substrates or products and are catalyzed by P450s across more than 200 species (Fig. 4a). The distribution of reaction types is displayed in Fig. 4b, with 1,507 oxidation reactions, 82 reduction reactions, 61 decomposition reactions, 29 combination reactions, and one substitution reaction. Fig. 4c illustrates the distribution of reaction sites, highlighting that the majority of reactions occur on CH3 (519 entries) and CH2 (486 entries) functional groups. Furthermore, over 100 reactions take place on CH-CH bonds and CH of aromatic rings groups. The organismal distribution of P450s is depicted in Fig. 4d, with plants accounting for more than half of the P450s (342/597), followed by microorganisms (137 P450s) and animals (118 P450s). Fig. 4e presents the distribution of compounds involved in reactions catalyzed by P450s, indicating 836 products and 570 substrates. Notably, 181 compounds serve as both products and substrates. Fig. 4f showcases the length distribution of P450s, with over 80 % of the P450s having sequence lengths ranging between 450 and 550 amino acids.
Fig. 4.
Data statistics of P450Rdb. a Overall statistics for P450Rdb database. b Distribution of reaction type. c Distribution of reaction site. d Organismal distribution of P450s. e Distribution of the compounds. f Sequence length distribution of P450s.
Data querying and result presentation
P450Rdb offers a user-friendly web interface that allows users to easily query P450s and their catalyzed reactions. The navigation bar provides quick access to various pages, including 'Search', 'Blast', 'Browse', 'Download', and 'Statistics'. The 'Search' page in P450Rdb offers two search options: 'Protein search', where users can input the P450 symbol/Entrez ID/UniProt ID, and 'Compound search', where users can input the substrate or product name/PubChem CID/formula. The search results are summarized in a table on the 'Result' page (Fig. 5). By clicking 'more', users can access detailed information about specific protein entries on the 'Detail' page. The 'Detail' page provides comprehensive information related to the reactions and P450s, including details about the P450 protein involved in the reactions (such as 'P450 symbol', 'P450 name', 'Gene ID', 'UniProt IDs', 'Species', 'Txid', 'P450 protein structure', and 'protein sequence'), reaction information (including 'Reaction type', 'Reaction site', and equation), substrate/product information (such as 'Substrates/Products name', 'Substrates/Products Chemical Formula', 'Substrates/Products PubChem CID', 'substrates/products PubChem SID', 'Substrates/Products Smiles', and 'Substrates/Products Structure'), and references associated with the entry (including 'PMIDs', 'Title', 'Journal', and the year of publication). Additionally, P450Rdb provides a Blast webserver for sequence similarity searches. Users can input the protein (Blastp) or nucleotide (Blastx) sequence of the queried P450 protein in the query window. The 'Result' table displays three indexes (identity, E-value, Bit-score) of the Blast software to assist users in assessing the similarity of sequences. Overall, P450Rdb offers a comprehensive and user-friendly platform for querying P450s and their reactions, facilitating efficient access to relevant information for users.
Fig. 5.
The Search page, Result page and Detail page of P450Rdb.
‘Browse’, ‘Download’, ‘Statistics’, ‘Help’, and ‘Submit’ page
P450Rdb provides the ‘Browse’ page to help quickly browse certain category of reactions and P450s by selecting certain species, reaction type and reaction site. Then users could query the detail information by clicking each entry in the ‘Result’ table. The ‘Download’ page enables users to easily download the P450s, reaction, compound lists and the sequence data (fasta file) for nonprofit purposes. The ‘Statistics’ page presents the detail information the data in the most up to date version of P450Rdb using various statistical graphics. The ‘Help’ page provides step-by-step tutorial for users to manipulation, querying and browsing the P450Rdb database. Moreover, it is inevitable that the collections of P450Rdb not cover all reactions catalyzed by P450s. So, we provide a ‘Submit’ interface to make sure that researchers can submit new reactions catalyzed by P450s that are not documented in this database.
Discussion
P450Rdb provides a valuable platform to store, integrate and analyze P450s and their corresponding reactions for accelerating P450s identification and application studies. However, it still has some limitations. Firstly, P450Rdb does not include kinetic constants such as Km, Ks, and Kcat for P450 enzymes due to the lack of explicit enzyme kinetic experiments for the majority of documented P450s. Secondly, the database does not provide information on the specific conditions related to the reactions catalyzed by P450s. This is because most studies primarily focus on the identification and biological functional characterization of P450s without delving into the detailed information about the catalyzed reactions.
Nevertheless, we are committed to addressing these limitations by continuously accumulating new functional evidence and outcomes of P450s and their reactions in the future. We will periodically update P450Rdb by collecting P450s and their corresponding reactions from publications. Additionally, we will strive to gather information about kinetic constants and relevant reaction conditions to expand the coverage of the database. Furthermore, we aim to broaden the scope of P450Rdb by collecting and integrating biosynthetic pathways involving P450s, thus facilitating functional interpretation and synthetic biology investigations related to P450s.
Conclusions
In summary, P450Rdb currently catalogs over 1,600 reactions involving about 600 P450s from a wide range of over 200 species, including plants, animals, fungi, bacteria, and archaea. Meanwhile, it provides a convenient interface with multitudinous information to help in querying, browsing and analyzing P450s and corresponding reactions. We believe this database could greatly promote p450s structural and functional research, and contribute to the development of synthetic biology, bio-pharmaceutics, environmental science and enzyme engineering.
CRediT authorship contribution statement
Yang Zhang: Conceptualization, Methodology, Validation, Funding acquisition, Writing–original draft. Xianrun Pan: Methodology, Validation. Tianyu Shi: Investigation, Formal analysis. Zhifeng Gu: Data curation. Zhaochang Yang: Data curation. Minghao Liu: Data curation. Yi Xu: Data curation. Yu Yang: Software. Liping Ren: Validation, Funding acquisition. Xiaoming Song: Supervision, Project administration, Funding acquisition. Hao Lin: Writing –review & editing, Supervision, Project administration. Kejun Deng: Conceptualization, Writing–review & editing, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (82130112, 62202069), the Natural Science Foundation of Sichuan Province (2022NSFSC1610, 2023NSFSC0678, 2023NSFSC0569, 2022NCFSC00647), the Natural Science Foundation for Distinguished Young Scholar of Hebei Province (C2022209010), the China Postdoctoral Science Foundation (2023M730507) and the Sichuan Province Postdoctoral Research Project Special Support Foundation (TB2023012).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2023.10.012.
Contributor Information
Xiaoming Song, Email: songxm@ncst.edu.cn.
Hao Lin, Email: hlin@uestc.edu.cn.
Kejun Deng, Email: dengkj@uestc.edu.cn.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Urlacher V.B., Girhard M. Cytochrome P450 monooxygenases in biotechnology and synthetic biology. Trends Biotechnol. 2019;37(8):882–897. doi: 10.1016/j.tibtech.2019.01.001. [DOI] [PubMed] [Google Scholar]
- 2.Lamb D.C., Follmer A.H., Goldstone J.V., Nelson D.R., Warrilow A.G., Price C.L., et al. On the occurrence of cytochrome P450 in viruses. Proc Natl Acad Sci USA. 2019;116(25):12343–12352. doi: 10.1073/pnas.1901080116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hansen C.C., Nelson D.R., Møller B.L., Werck-Reichhart D. Plant cytochrome P450 plasticity and evolution. Mol Plant. 2021;14(8):1244–1265. doi: 10.1016/j.molp.2021.06.028. [DOI] [PubMed] [Google Scholar]
- 4.Werck-Reichhart D. Promiscuity, a driver of plant cytochrome P450 Evolution? Biomolecules. 2023;13(2) doi: 10.3390/biom13020394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Werck-Reichhart D., Feyereisen R. Cytochromes P450: a success story. Genome Biol. 2000;1(6) doi: 10.1186/gb-2000-1-6-reviews3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Durairaj P., Li S. Functional expression and regulation of eukaryotic cytochrome P450 enzymes in surrogate microbial cell factories. Eng Microbiol. 2022;2(1) doi: 10.1016/j.engmic.2022.100011. [DOI] [Google Scholar]
- 7.Rittle J., Green M.T. Cytochrome P450 Compound I: capture, characterization, and C-H bond activation kinetics. Science. 2010;330(6006):933–937. doi: 10.1126/science.1193478. [DOI] [PubMed] [Google Scholar]
- 8.Jensen S.B., Thodberg S., Parween S., Moses M.E., Hansen C.C., Thomsen J., et al. Biased cytochrome P450-mediated metabolism via small-molecule ligands binding P450 oxidoreductase. Nat Commun. 2021;12(1):2260. doi: 10.1038/s41467-021-22562-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coon M.J. CYTOCHROME P450: nature's most versatile biological catalyst. Annu Rev Pharmacol Toxicol. 2004;45(1):1–25. doi: 10.1146/annurev.pharmtox.45.120403.100030. [DOI] [PubMed] [Google Scholar]
- 10.Zhou Y., Lauschke V.M. The genetic landscape of major drug metabolizing cytochrome P450 genes—an updated analysis of population-scale sequencing data. Pharmacogenomics J. 2022;22(5):284–293. doi: 10.1038/s41397-022-00288-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guengerich F.P. Cytochrome p450 enzymes in the generation of commercial products. Nat Rev Drug Discov. 2002;1(5):359–366. doi: 10.1038/nrd792. [DOI] [PubMed] [Google Scholar]
- 12.Rudolf J.D., Chang C.Y., Ma M., Shen B. Cytochromes P450 for natural product biosynthesis in Streptomyces: sequence, structure, and function. Nat Product Rep. 2017;34(9):1141–1172. doi: 10.1039/c7np00034k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang H., Wang X., Song R., Ding W., Li F., Ji L. Emerging metabolic profiles of sulfonamide antibiotics by cytochromes P450: a computational-experimental synergy study on emerging pollutants. Environ Sci Tech. 2023;57(13):5368–5379. doi: 10.1021/acs.est.3c00071. [DOI] [PubMed] [Google Scholar]
- 14.Girvan H.M., Munro A.W. Applications of microbial cytochrome P450 enzymes in biotechnology and synthetic biology. Curr Opinion Chem Biol. 2016;31:136–145. doi: 10.1016/j.cbpa.2016.02.018. [DOI] [PubMed] [Google Scholar]
- 15.Thomson R.E.S., D'Cunha S.A., Hayes M.A., Gillam E.M.J. Use of engineered cytochromes P450 for accelerating drug discovery and development. Adv Pharmacol (San Diego, Calif) 2022;95:195–252. doi: 10.1016/bs.apha.2022.06.001. [DOI] [PubMed] [Google Scholar]
- 16.Luo Y., Jiang Y., Chen L., Li C., Wang Y. Applications of protein engineering in the microbial synthesis of plant triterpenoids. Synthetic Syst Biotechnol. 2023;8(1):20–32. doi: 10.1016/j.synbio.2022.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu X., Cheng J., Zhang G., Ding W., Duan L., Yang J., et al. Engineering yeast for the production of breviscapine by genomic analysis and synthetic biology approaches. Nat Commun. 2018;9(1):448. doi: 10.1038/s41467-018-02883-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sue Masters B., Marohnic C.C. Cytochromes P450—A family of proteins and scientists-understanding their relationships. Drug Metab Rev. 2006;38(1–2):209–225. doi: 10.1080/03602530600570065. [DOI] [PubMed] [Google Scholar]
- 19.Pandian B.A., Sathishraj R., Djanaguiraman M., Prasad P.V.V., Jugulam M. Role of Cytochrome P450 Enzymes in Plant Stress Response. Antioxidants (Basel, Switzerland) 2020;9 doi: 10.3390/antiox9050454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y., Liu T., Wang J., Zou B., Li L., Yao L., et al. Cellinker: a platform of ligand–receptor interactions for intercellular communication analysis. Bioinformatics. 2021;37(14):2025–2032. doi: 10.1093/bioinformatics/btab036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chao W., Quan Z.O.U. A machine learning method for differentiating and predicting human-infective coronavirus based on physicochemical features and composition of the spike protein. Chin J Electron. 2021;30(5):815–823. doi: 10.1049/cje.2021.06.003. [DOI] [Google Scholar]
- 22.Liu X., Cheng J., Zhang G., Ding W., Duan L., Yang J., et al. Engineering yeast for the production of breviscapine by genomic analysis and synthetic biology approaches. Nat Commun. 2018;9(1):448. doi: 10.1038/s41467-018-02883-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Christ B., Xu C., Xu M., Li F.S., Wada N., Mitchell A.J., et al. Repeated evolution of cytochrome P450-mediated spiroketal steroid biosynthesis in plants. Nat Commun. 2019;10(1):3206. doi: 10.1038/s41467-019-11286-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ning L., Liu M., Gou Y., Yang Y., He B., Huang J. Development and application of ribonucleic acid therapy strategies against COVID-19. Int J Biol Sci. 2022;18(13):5070–5085. doi: 10.7150/ijbs.72706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu B., Zhao X., Wang E., Zhou J., Li J., Chen J., et al. Efficient heterologous expression of cytochrome P450 enzymes in microorganisms for the biosynthesis of natural products. Crit Rev Biotechnol. 2023;43(2):227–241. doi: 10.1080/07388551.2022.2029344. [DOI] [PubMed] [Google Scholar]
- 26.Kim Y.H., Kwon T., Yang H.J., Kim W., Youn H., Lee J.Y., et al. Gene engineering, purification, crystallization and preliminary X-ray diffraction of cytochrome P450 p-coumarate-3-hydroxylase (C3H), the Arabidopsis membrane protein. Protein Expr Purif. 2011;79(1):149–155. doi: 10.1016/j.pep.2011.04.013. [DOI] [PubMed] [Google Scholar]
- 27.Li Z., Jiang Y., Guengerich F.P., Ma L., Li S., Zhang W. Engineering cytochrome P450 enzyme systems for biomedical and biotechnological applications. J Biol Chem. 2020;295(3):833–849. doi: 10.1016/S0021-9258(17)49939-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y., Liu T., Hu X., Wang M., Wang J., Zou B., et al. Cell Call: integrating paired ligand–receptor and transcription factor activities for cell–cell communication. Nucleic Acids Res. 2021;49(15):8520–8534. doi: 10.1093/nar/gkab638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nelson D.R. The Cytochrome P450 Homepage. Hum Genomics. 2009;4(1):59. doi: 10.1186/1479-7364-4-1-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sim S.C., Ingelman-Sundberg M. The Human Cytochrome P450 (CYP) Allele Nomenclature website: a peer-reviewed database of CYP variants and their associated effects. Hum Genomics. 2010;4(4):278. doi: 10.1186/1479-7364-4-4-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fábién P., Degtyarenko K.N. The Directory of P450-containing Systems in 1996. Nucleic Acids Res. 1997;25(1):274–277. doi: 10.1093/nar/25.1.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paquette S.M., Bak S., Feyereisen R. Intron-exon organization and phylogeny in a large superfamily, the paralogous cytochrome P450 genes of Arabidopsis thaliana. DNA Cell Biol. 2000;19(5):307–317. doi: 10.1089/10445490050021221. [DOI] [PubMed] [Google Scholar]
- 33.Preissner S., Kroll K., Dunkel M., Senger C., Goldsobel G., Kuzman D., et al. SuperCYP: a comprehensive database on Cytochrome P450 enzymes including a tool for analysis of CYP-drug interactions. Nucl Acids Res. 2009;38(suppl_1):D237–D243. doi: 10.1093/nar/gkp970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park J., Lee S., Choi J., Ahn K., Park B., Park J., et al. Fungal cytochrome P450 database. BMC Genomics. 2008;9(1):402. doi: 10.1186/1471-2164-9-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fischer M., Knoll M., Sirim D., Wagner F., Funke S., Pleiss J. The Cytochrome P450 Engineering Database: a navigation and prediction tool for the cytochrome P450 protein family. Bioinformatics. 2007;23(15):2015–2017. doi: 10.1093/bioinformatics/btm268. [DOI] [PubMed] [Google Scholar]
- 36.Wang H., Wang Q., Liu Y., Liao X., Chu H., Chang H., et al. PCPD: Plant cytochrome P450 database and web-based tools for structural construction and ligand docking. Synthetic Syst Biotechnol. 2021;6(2):102–109. doi: 10.1016/j.synbio.2021.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rendic S.P., Guengerich F.P. Human Family 1–4 cytochrome P450 enzymes involved in the metabolic activation of xenobiotic and physiological chemicals: an update. Arch Toxicol. 2021;95(2):395–472. doi: 10.1007/s00204-020-02971-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rendic S.P., Peter G.F. Human cytochrome P450 enzymes 5–51 as targets of drugs and natural and environmental compounds: mechanisms, induction, and inhibition - toxic effects and benefits. Drug Metab Rev. 2018;50(3):256–342. doi: 10.1080/03602532.2018.1483401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rendic S.P. Metabolism and interactions of Ivermectin with human cytochrome P450 enzymes and drug transporters, possible adverse and toxic effects. Arch Toxicol. 2021;95(5):1535–1546. doi: 10.1007/s00204-021-03025-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen C., Wang D.W. Cytochrome P450-CYP2 family-epoxygenase role in inflammation and cancer. Adv Pharmacol (San Diego, Calif) 2015;74:193–221. doi: 10.1016/bs.apha.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 41.Liu X., Zhu X., Wang H., Liu T., Cheng J., Jiang H. Discovery and modification of cytochrome P450 for plant natural products biosynthesis. Synthetic Syst Biotechnol. 2020;5(3):187–199. doi: 10.1016/j.synbio.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nelson D.R. Cytochrome P450 diversity in the tree of life. Biochimica et biophysica acta Proteins and proteomics. 2018;1866(1):141–154. doi: 10.1016/j.bbapap.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang Y., Wang J., Zhao Y., Wang H., Liu T., Li Y., et al. cncRNAdb: a manually curated resource of experimentally supported RNAs with both protein-coding and noncoding function. Nucleic Acids Res. 2020;49(D1):D65–D70. doi: 10.1093/nar/gkaa791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ning L., Abagna H.B., Jiang Q., Liu S., Huang J. Development and application of therapeutic antibodies against COVID-19. Int J Biol Sci. 2021;17(6):1486–1496. doi: 10.7150/ijbs.59149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miettinen K., Iñigo S., Kreft L., Pollier J., De Bo C., Botzki A., et al. The TriForC database: a comprehensive up-to-date resource of plant triterpene biosynthesis. Nucl Acids Res. 2018;46(D1):D586–D594. doi: 10.1093/nar/gkx925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.UniProt: the Universal Protein Knowledgebase in 2023. Nucleic Acids Res.51(D1)(2023):D523-d31. 10.1093/nar/gkac1052. [DOI] [PMC free article] [PubMed]
- 47.Sayers E.W., Bolton E.E., Brister J.R., Canese K., Chan J., Comeau D.C., et al. Database resources of the National Center for Biotechnology Information in 2023. Nucleic Acids Res. 2023;51(D1):D29–D38. doi: 10.1093/nar/gkac1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim S., Chen J., Cheng T., Gindulyte A., He J., He S., et al. PubChem 2023 update. Nucleic Acids Res. 2023;51(D1):D1373–D1380. doi: 10.1093/nar/gkac956. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.