Abstract
One of the multifactorial worldwide health syndromes is diabetes mellitus which is increasing at a disturbing rate. The inhibition of α-glucosidase, an enzyme that catalyzes starch hydrolysis in the intestine, is one helpful therapeutic approach for controlling hyperglycemia related to type-2 diabetes. To discover α-glucosidase inhibitors, some 2-hydrazolyl-4-thiazolidinone hybrids (3a-e) were synthesized from new one-pot reaction procedures. Next, their chemical structures were confirmed by 1H NMR, 13C NMR, and FT-IR spectra, and elemental analysis technique. Then, the α-glucosidase inhibitory activity of the titled compounds was evaluated. Among them, derivatives 3b and 3c revealed the highest activity against α-glucosidase compared to acarbose as a drug. Enzyme kinetic studies of the most active derivative (3b) indicated a competitive inhibition. Finally, molecular modeling studies were accomplished to describe vital interactions of the most potent compounds (3b and 3c) with the α-glucosidase enzyme.
Ketwords: Diabetes, α-glucosidase, In silico study, Molecular dynamics simulations, Docking study
Graphical abstract

1. Introduction
One of the risk factors for Alzheimer’s, cognitive decline, and cardiovascular diseases is diabetes [1]. Type 2 diabetes mellitus (T2DM) is known as one of the most widespread health problems in the world involving about 462 million patients equivalent to 6.28 % of the world’s people and more than 1 million deaths per year [2,3].
Diabetes is commonly treated with insulin and oral antidiabetic drugs like 4-thiazolidinones, biguanides, sulphonylureas, α-glucosidase and β-glucosidase inhibitors [4]. α-Glucosidase (EC.3.2.1.20) is an enzyme found at the intestine's brush border that breaks down oligosaccharides, trisaccharides, and disaccharides into monosaccharides [5]. α-Glucosidase inhibitors were used in the 1970s and the first α-glucosidase inhibitors for diabetes therapy were approved in the 1990s [6]. An FDA-approved α-glucosidase inhibitor titled acarbose, is used orally to regulate blood sugar levels. Other FDA-approved α-glucosidase inhibitors are voglibose, nojirimycin, and miglitol [7,8]. These drugs show serious side effects for example gastrointestinal disturbances like diarrhea, abdominal distension, and flatulence. Furthermore, numerous small molecules of α-glucosidase inhibitors have been synthesized with imidazole, quinazolinone, isatin, pyrazoles, xanthone, and azole scaffolds [9].
4-Thiazolidinones are among the most extensively studied organic compounds and their derivatives have been shown diversity of biological activities such as anti-diabetics, anti-bacterial [10], anti-fungal [11], antioxidant [12], anti-inflammatory [13], and anti-tuberculosis [14]. In nature, penicillin derivatives have the thiazolidine ring which was the first identification of medicinal features of this scaffold [15,16]. Thiazolidinone analogues are used also as α-glucosidase inhibitors [17].
Thiazolidinone-based derivatives showed capacity as single- and multi-targeted agents for regulatory hyperglycemia in T2DM [18]. In specific, the 2,4-thiazolidindione pioglitazone is a famous oral drug applied for the administration of T2DM, and the only drug approved to treat diabetic neuropathy is epalrestat, a 2-thioxo-4-thiazolidinone analogue [19]. Moreover, thiazolidinones, such as ciglitazone, rosiglitazone, troglitazone, and pioglitazone, are insulin-sensitizing drugs beneficial in the diagnosis of T2DM. Unfortunately, all drugs due to having high side effects were withdrawn from the market [20].
There are some studies on thiazolidinone derivatives as α-glucosidase inhibitors. For example, Azam et al. assessed the various thiazolidinones against α-glucosidase. Compound 1 (Fig. 1) exhibited potent α-glucosidase inhibition compared to acarbose [21]. Khan et al. reported a series of 2-mercaptobenzimidazole-based 1,3-thiazolidin-4-ones as α-glucosidase inhibitors. Compound 2 (Fig. 1) displayed excellent activity [22]. Khan et al. evaluated thiazolidinone-based indole derivatives against α-glucosidase activity. Analogue 3 (Fig. 1) exhibited a few folds better inhibitory activity than acarbose [23]. Yakaiah Chintala et al. described the α-glucosidase inhibitory activity of triazole linked to thiazolidinedione. Compound 4 (Fig. 1) revealed potent α-glucosidase inhibition [24].
Fig. 1.
Drugs and reported synthetic α-glucosidase inhibitors.
According to the high activity of the thiazolidinone derivatives as α-glucosidase inhibitors, in the current study, a series of 2-hydrazolyl-4-thiazolidinone hybrids with a new synthesis route and more yield were provided and evaluated against α-glucosidase enzyme. Furthermore, in vitro kinetic analysis and in silico investigations were carried out to clarify the interactions between these derivatives and the α-glucosidase active site.
2. Materials and methods
2.1. General
Alkylidene or benzylidene hydrazines were prepared from the reaction between aldehydes and hydrazine hydrate, based on the reported procedure [25]. Other starting materials and solvents were obtained from Merck (Germany) and were used without further purification. Progress of the reaction was monitored by analytical thin-layer chromatography (TLC) on silica gel 60 F254 aluminum-backed silica plates (Merck). Melting points (uncorrected) were measured with a Stuart SMP-3 apparatus. Elemental analysis (C, H, N, and S) was performed using a CHNS-923 LECO analyzer (USA). FT-IR spectra of compounds were measured with an FT-IR PerkinElmer RXI. NMR spectra were reported on a Bruker 250 MHz (250.1 MHz for 1H and 62.9 MHz for 13C) with CDCl3 as the solvent. Chemical shifts are given in ppm (δ) relative to internal TMS, and coupling constants (J) are reported in Hertz (Hz).
2.2. Synthesis
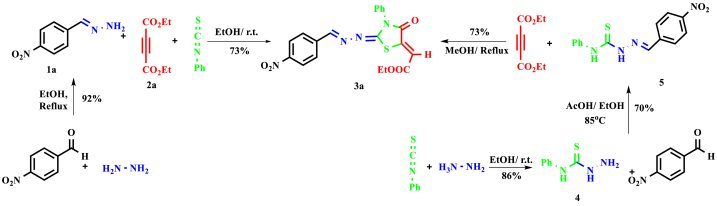
General procedure for the preparation of alkyl 2-(2-(arylidenehydrazino)-4-oxo-3-phenylthiazolidin-5-ylidene)acetates (3a-e), exemplified by 3a.
A mixture of 2-(4-nitrophenyl)ethen-1-amine 1a (2 mmol) and phenyl isothiocyanate (2 mmol) in absolute ethanol (5 mL) was stirred for 15 min at room temperature. After that, diethyl acetylenedicarboxylate 2a (2 mmol) was added to the reaction mixture, and the mixture was stirred for 14 h at room temperature. Then, the light-yellow precipitate was filtered and washed with a mixture of n-hexane/ethyl acetate to produce pure product 3a.
2.2.1. (Z)-Ethyl-2-(2-((4-nitrobenzylidene)hydrazono)-4-oxo-3-phenylthiazolidin-5-ylidene) acetate (3a)
Yellow powder, yield: 0.62 g (73 %), mp: 208–209 °C, FT-IR (KBr, cm−1): 1725 (C=O), 1692 (C=O), 1602 (C=N). 1H NMR (250.1 MHz, CDCl3): δ ppm 1.35 (3H, t, 3JHH = 7.0 Hz, CH3), 4.36 (2H, q, 3JHH = 7.0 Hz, OCH2), 7.00 (1H, s, CH), 7.40–7.58 (5H, m, Ar–H), 7.92 (2H, d, 3JHH = 8.8 Hz, Ar–H), 8.32 (2H, d, 3JHH = 8.8 Hz, Ar–H), 8.57 (1H, s, CH). 13C NMR (62.9 MHz, CDCl3): δ ppm 14.2 (CH3), 62.0 (OCH2), 117.5 (CH), 124.0 (2CH), 127.6 (2CH), 128.9 (2CH), 129.4 (3CH), 133.7 (C), 139.5 (C), 140.6 (C), 149.2 (C), 157.6 (CH), 163.3 (C), 164.6 (C), 166.0 (C). Anal. Calc. for C20H16N4O5S (424.08): C, 56.60; H, 3.80; N, 13.20; S: 7.55 found: C, 56.94; H, 3.92; N, 13.49; S: 7.76 %.
2.2.2. (Z)-Methyl-2-(2-((4-nitrobenzylidene)hydrazono)-4-oxo-3-phenylthiazolidin-5-ylidene) acetate (3b)
Yellow powder, yield: 0.56 g (68 %), mp: 199–200 °C, FT-IR (KBr, cm−1): 1723 (C=O), 1696 (C=O), 1602 (C=N). 1H NMR (250.1 MHz, CDCl3): δ ppm 3.92 (3H, s, OCH3), 7.03 (1H, s, CH), 7.40–7.58 (5H, m, Ar–H), 7.94 (2H, d, 3JHH = 8.8 Hz, Ar–H), 8.27 (2H, d, 3JHH = 8.8 Hz, Ar–H), 8.41 (1H, s, CH). 13C NMR (62.9 MHz, CDCl3): δ ppm 52.7 (OCH3), 117.1 (CH), 124.0 (2CH), 127.6 (2CH), 129.0 (2CH), 129.4 (3CH), 132.3 (C), 139.2 (C), 141.5 (C), 143.8 (C), 157.7 (CH), 161.0 (C), 161.2 (C), 166.5 (C). Anal. Calc. for C19H14N4O5S (410.40): C, 55.61; H, 3.44; N, 13.65; S: 7.81 found: C, 55.24; H, 3.65; N, 13.89; S: 7.66 %.
2.2.3. Ethyl (Z)-2-((E)-2-(((1E,2E)-3-(2-nitrophenyl)allylidene)hydrazono)-4-oxo-3-phenylthiazolidin-5-ylidene) acetate (3c)
Yellow powder, yield:0.58 g (64 %), mp: 200–201 °C, FT-IR (KBr, cm−1): 1727 (C=O), 1696 (C=O), 1601 (C=N). 1H NMR (250.1 MHz, CDCl3): δ ppm 1.37 (3H, t, 3JHH = 7.3 Hz, CH3), 4.34 (2H, q, 3JHH = 7.3 Hz, OCH2), 6.98 (1H, s, CH), 6.93–7.55 (8H, m, Ar–H and 2CH), 7.66 (1H, t, 3JHH = 8.0 Hz Ar–H), 7.74 (1H, m, Ar–H), 8.00 (1H, d, 3JHH = 8.3 Hz, Ar–H), 8.19 (1H, d, 3JHH = 9.7 Hz, CH). 13C NMR (62.9 MHz, CDCl3): δ ppm 14.2 (CH3), 61.7 (CH2), 117.2 (CH), 125.0 (CH), 127.6 (2CH), 129.1 (CH), 129.4 (CH), 129.9 (3CH), 131.3 (C), 133.4 (CH), 133.7 (CH), 136.6 (C), 136.8 (CH), 140.6 (C), 147.9 (C), 161.2 (C), 161.3 (C), 164.7 (C), 165.9 (C). Anal. Calc. for C22H18N4O5S (450.47): C, 58.66; H, 4.03; N, 12.44; S: 7.12 found: C, 58.35; H, 3.85; N, 12.75; S: 7.28 %.
2.2.4. (Z)-Methyl-2-(2-((4-cholorobenzylidene) hydrazono)-4-oxo-3-phenylthiazolidin-5-ylidene) acetate (3d)
Yellow powder, yield: 0.60 g (75 %), mp: 207–208 °C, FT-IR (KBr, cm−1): 1720 (C=O), 1704 (C=O), 1613 (C=N). 1H NMR (250.1 MHz, CDCl3): δ ppm 3.90 (3H, s, OCH3), 6.98 (1H, s, CH), 7.25–7.86 (9H, m, Ar–H), 8.30 (1H, s, CH). 13C NMR (62.9 MHz, CDCl3): δ ppm 52.6 (OCH3), 116.5 (CH), 127.6 (2CH), 129.1 (2CH), 129.3 (CH), 129.4 (2CH). 129.6 (2CH), 132.5 (C), 133.8 (C), 137.3 (C), 141.4 (C), 159.0 (CH), 161.1 (C), 164.6 (C), 166.5 (C). Anal. Calc. for C19H14ClN3O3S (399.85): C, 57.07; H, 3.53; N, 10.51; S: 8.02 found: C, 57.38; H, 3.36; N, 10.25; S: 7.73 %.
2.2.5. Ethyl (Z)-2-((E)-2-(((E)-benzylidene)hydrazono)-4-oxo-3-phenylthiazolidin-5-ylidene)acetate (3e)
Yellow powder, yield: 0.38 g (50 %), mp: 223–225 °C, FT-IR (KBr, cm−1): 1717 (C=O), 1688 (C=O), 1611 (C=N). 1H NMR (250.1 MHz, CDCl3): δ ppm 1.38 (3H, t, 3JHH = 7.0 Hz, CH3), 4.36 (2H, q, 3JHH = 7.0 Hz, OCH2), 6.99 (1H, s, CH), 7.41–7.55 (8H, m, Ar–H), 7.78 (2H, d, 3JHH = 7.3 Hz, Ar–H), 8.36 (1H, s, CH). 13C NMR (62.9 MHz, CDCl3): δ ppm 13.4 (CH3), 60.9 (OCH2), 116.0 (CH), 126.5 (C), 126.9 (2CH), 127.7 (2CH), 128.0 (2CH), 128.6 (3CH), 130.5 (CH), 133.0 (C), 140.5 (C), 159.5 (CH), 160.1 (C), 164.0 (C), 165.3 (C). Anal. Calc. for C20H17N3O3S (379.43): C, 63.31; H, 4.52; N, 11.07; S: 8.45 found: C, 63.65; H, 4.41; N, 11.34; S: 8.26 %.
2.3. α-Glucosidase inhibition assay
The inhibitory activity of 2-hydrazolyl-4-thiazolidinone hybrids was evaluated on the α-glucosidase enzyme according to the previous procedure [26]. The Saccharomyces cerevisiae form of the α-glucosidase enzyme (EC 3.2.1.20) (purchased from Sigma-Aldrich) (20 U/mg) and p-nitrophenyl glucopyranoside (pNPG) as substrate were ready in the buffer of potassium phosphate (pH 6.8, 50 mM). After that, derivatives were dissolved in DMSO (10 % final concentration). The reaction mixture was added to the 96-well plates (including enzyme buffer (20 μL, 20 U/mg), potassium phosphate buffer (135 μL), and derivatives (20 μL)). Then, the mixture was incubated at 37 °C for 10 min. In the next step, incubation continued after the addition of the substrate (25 μL, 4 mM) at 37 °C for 20 min. Finally, a yellow color was produced due to the presence of the privation of p-nitrophenol. After that, the absorbance of compounds was recorded at 405 nm by using a spectrophotometer (Gen5, Power wave xs2, BioTek, America). DMSO (10 % final concentration) as negative control and acarbose as positive control were used. The percentage of enzyme inhibition for each compound was calculated by using the following formula:
| % Inhibition = [-Abs control Abs sample)/Abs control] × 100 |
IC50 values were calculated from a non-linear regression curve using the Logit method. All investigations were performed three times.
2.4. Enzyme kinetic studies
Enzyme kinetic studies were carried out on derivative 3c, the most potent derivative recognized with the highest IC50, against α-glucosidase activity. It was performed in the presence and absence of derivative 3c at diverse concentrations (0, 43.5, 87.0, and 174.0 μM) with varied concentrations of p-nitrophenyl α-D-glucopyranoside (1–16 mM) as substrate. The enzyme kinetic methodology was similar to the procedure performed in the previous research [5].
2.5. Molecular docking study
The docking studies of the compounds into the isomaltase of Saccharomyces cerevisiae binding site were performed by software Autodock 4.2 [27]. The crystal structure of isomaltase of Saccharomyces cerevisiae (PDB code 3A4A) was reached from the protein data bank (PDB). The docking methodology was similar to the procedure of docking in the previous research [5].
2.6. Molecular dynamics simulations study
Molecular dynamics simulations (MD) was carried out via the GROMACS V4.5.5 computational package. MD methodology of derivatives was similar to the procedure of MD in the previous article [5].
2.7. Binding free energy analyses
Binding free energy was calculated for derivatives by the g_mmpbsa tool. This tool uses the MM/PBSA method to calculate binding free energy derivatives. MM/PBSA methodology of derivatives was similar to the procedure of MM/PBSA in the previous article [28].
3. Results and discussion
3.1. Chemistry
The synthesis of hydrazolyl-4-thiazolidinone hybrids with various biological activities has attracted the attention of researchers [29,30]. The described procedures for synthesizing these derivatives need several stages, and thiosemic carbazide derivatives are usually the key intermediates in these methods [29,31]. For example, highly functionalized thiazolidinone 3a was synthesized by Benmohammad et al. [31] in three stages, achieving an overall yield of 44 %. This approach provided thiosemicarbazide derivatives 4 and 5 as intermediates. We suggested a different pathway to synthesize 3a that could be a one-pot reaction involving benzylidene hydrazine 1a and phenylisothiocyanate, in the presence of diethyl acetylenedicarboxylate 2a (Scheme 1). Initially, we produced functionalized benzylidene hydrazine 1a from the reaction of 4-nitobenzaldehyde and hydrazinehydrate, following the optimal conditions cited in previous studies [25]. Subsequently, we synthesized compound 3a via a one-pot reaction between benzylidene hydrazine 1a and phenyl isothiocyanate, with diethylacetylenedicarboxylate 2a present. This method allowed us to reduce the synthesis steps to two steps and enhance the overall yield by approximately 67 % (Scheme 1).
Scheme 1.
Synthesis of 2-hydrazolyl-4-thiazolidinone 3a via two routes - Benmohammed et al.'s 2014 [31] method (right) and our method (left).
In the current work, we report a facile synthesis of functionalized hydrazolyl-4-thiazolidinone derivatives (3a-e) through a one-pot reaction between functionalized benzylidene/alkylidene hydrazines 1 and phenyl isothiocyanate in the presence of dialkylacetylenedicarboxylates 2.
Functionalized thiazolidine-4-one derivatives 3, including two critical pharmacophores thiazolidinedin-4-one and imino/hydrazine groups, were synthesized by the one-pot reaction between functionalized benzylidene/alkylidene hydrazines 1 and dialkyl acetylenedicarboxylates 2 in the presence of phenylisothiocyanate. The first step of the synthesis included the preparation of benzylidene hydrazines 1 from the condensation reaction between hydrazine hydrate with substituted aldehydes, according to the literature [25]. Next, a one-pot reaction between benzylidene/alkylidene hydrazines 1 and dialkyl acetylenedicarboxylates 2 in the presence of phenylisothiocyanat produced hydrazinyl-4-thiazolidinone derivatives (3a-e) in good yields (Scheme 2).
Scheme 2.
The one-pot reaction between functionalized benzylidene/alkylidene hydrazines and phenyl isothiocyanate in the presence of dialkyl acetylenedicarboxylates.
The synthetic methodology employed was straightforward, and the resulting compounds were efficiently purified by simple washing with a n-hexane/ethyl acetate solvent system. The high purity of the isolated compounds was confirmed by TLC, sharp melting points, and corroborative elemental analysis data (see Experimental). The structures of compounds 3a and 3c were assigned by comparison of their spectroscopic data with literature precedents [31]. The structures of the remaining new compounds were established through comprehensive spectroscopic analysis, including 1H NMR, 1³C NMR, and FT-IR spectroscopy, as well as elemental analysis. The results of elemental analysis for all products were in good agreement with the calculated values. FT-IR spectra exhibited characteristic peaks at approximately 3057–3070 cm⁻1 (aromatic C–H), 2854–2926 cm⁻1 (aliphatic C–H), 1717–1727 cm⁻1 (ester carbonyl), 1688–1704 cm⁻1 (thiazolidinone carbonyl), and 1601–1613 cm⁻1 (-C=N-). 1H NMR spectra revealed signals at δ 8.19–8.57 ppm (aldimine –HC=N- proton), 6.98–7.03 ppm (vinylic = CHCO2R proton), 4.34–4.36 ppm (ester CH₂ in 3a, 3c, and 3e), 3.90–3.92 ppm (ester CH₃ in 3b and 3d), 1.35–1.38 ppm (ester CH₃ in 3a, 3c, and 3e), and 6.93–8.32 ppm (aromatic protons). 1³C NMR spectra exhibited resonances at δ 13.4–62.0 ppm (ester aliphatic carbons), 116.0–117.5 ppm (vinylic = CHCO2R carbon), 157.6–161.2 ppm (aldimine –HC=N- carbon), 161.0–166.5 ppm (carbonyl and imino carbon of thiazolidinone ring and ester carbonyl), and 124.0–149.2 ppm (aromatic carbons and C-5 of thiazolidinone ring).
The reaction mechanism being proposed is depicted in Scheme 3. The process begins with the addition of benzylidene/alkylidene hydrazines to phenylisothiocyanate, resulting in the formation of an intermediate known as I. This intermediate then proceeds to react with dialkyl acetylenedicarboxylate, leading to the production of intermediates referred to as II. Following this, these intermediates undergo a process of intramolecular cyclization, culminating in the production of 2-hydrazolyl-4-thiazolidinone derivatives 3, as shown in Scheme 3.
Scheme 3.
The proposed mechanism for the synthesis of 2-hydrazolyl-4-thiazolidinone derivatives (3a-e).
3.2. α-Glucosidase inhibition assay
To evaluate the inhibitory activity of the derivatives (3a-e), the α-glucosidase enzyme (Saccharomyces cerevisiae form) was applied. Based on the findings, some of the evaluated compounds (IC50 = 174–488 μM) were more potent toward the acarbose (IC50 = 750 μM) (Table 1). Among compounds, the derivative 3c was the most potent compound with IC50 values equal to 174 μM.
Table 1.
Evaluation of α-glucosidase inhibitory activity of compounds (3a-e).
| Compound | R1 | R2 | IC50 (μM) |
|---|---|---|---|
| 3a | 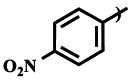 |
Et | >750 |
| 3b | 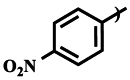 |
Me | 215.4 ± 0.9 |
| 3c |  |
Et | 174.0 ± 0.4 |
| 3d |  |
Me | 488.9 ± 1.6 |
| 3e | 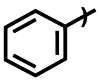 |
Et | >750 |
| Acarbose | – | - | 750 ± 2.0 |

The assay results showed that the position and the nature of substituents on the phenyl ring of hydrazolyl moiety play an important role against the α-glucosidase. Compound 3c, the most potent derivative, was bearing ortho-nitrophenyl-allylidene in hydrazolyl moiety. Shortening the length of the liker between the phenyl ring and hydrazolyl moiety and moving the nitro group from the ortho to para position decreased inhibitory activity. For example, compound 3c (IC50 = 174.0 μM) showed higher inhibitory activity than compounds 3b (IC50 = 215.4 μM), 3a (IC50 > 750 μM), and 3d (IC50 = 488.9 μM). Changing the nitro group on the benzyl ring with hydrogen or chlorine atoms 3e (IC50 > 750 μM) and 3d (IC50 = 488.9 μM) dramatically decreased activity. It seems that the presence of diverse groups at the para position of the phenyl ring of hydrazolyl moiety was effective against α-glucosidase.
Likewise, the presence of an electron-withdrawing and hydrophilic substituent on the phenyl ring increased inhibitory activity. Additionally, the introduction of ethyl ester in thiazolidinone moiety compared to methyl ester decreased inhibitory activity, which might be due to increased lipophilic features at this position [[32], [33], [34]].
3.3. Enzyme kinetic studies
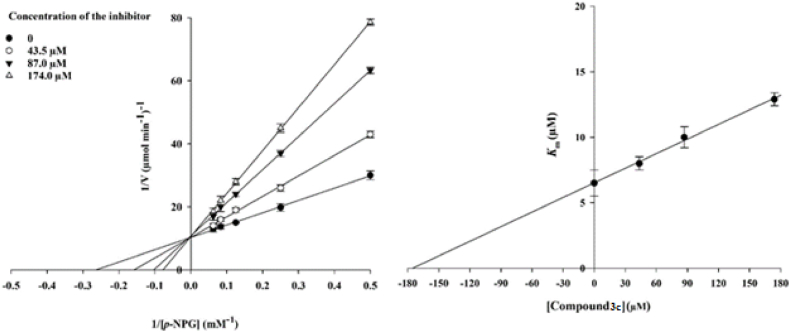
The Lineweaver-Burk plot exhibited that Vmax remained unchanged and the Km slowly increased with increasing inhibitor concentration representing a competitive inhibition (Fig. 2a). The findings exhibited that 3c competes with the substrate for binding to the active site. Moreover, the plot of the Km versus diverse concentrations of inhibitor provided a Ki of 174.0 μM (Fig. 2b).
Fig. 2.
Kinetics of α-glucosidase inhibition by derivative 3c. (a) The Lineweaver-Burk plot in the presence and absence of diverse concentrations of the derivative 3c; (b) The secondary plot between Km and various concentrations of the derivative 3c.
3.4. Molecular docking study
The screened derivatives based on the number and type of groups linked to the carboxylate moiety of the main scaffold, and positions of the functional group on the phenyl ring can inhibit α-glucosidase (Table 1). These derivatives were diverse in the kind and position of the substituted moieties (Table 1). Docking calculations of evaluated derivatives was done to achieve of estimated score docking and suitable interactions in active site enzyme. Validation of the docking methodology was carried out using a re-dock of native co-crystallized into the binding site of α-glucosidase. The docking protocol successfully restored the docked ligand alignment of the native co-crystallized with an RMSD equal to 1.32 Å.
Derivative 3c, the most potent compound, showed appropriate interactions with the active site residues. Fig. 1 shows the interactions of the best docked conformation of 3c with the active site residues. The oxygen atom of the nitro group of compound 3c made two hydrogen bonds with Asn415 and Tyr158 residues. The carbonyl group in the ethyl ester of the compound showed a hydrogen bond with Asp215. The carbonyl group of thiazolidinone moiety also exhibited another hydrogen bond with Glu277. This derivative displayed hydrophobic interactions with Tyr316, Phe314, Phe159, Ser157, Lys156, Val216, Glu411, Gln353, Arg315, Tyr72, Asp69, Phe178, Gln182, His112, Asp352, Arg442, Phe303 (Fig. 3).
Fig. 3.
Binding mode of (A) derivative 3b and (B) derivative 3c in α-glucosidase active site.
Compound 3b, second rank after compound 3c, showed hydrophobic interactions with Phe314, Glu411, Phe33, Tyr158, Arg315, Gln353, Phe178, Val216, Gln279, Asp352, Asp215, Glu277, Tyr72, Asp69 residues. This compound exhibited five hydrogen bonds with residues: the carbonyl group of the compound with Arg442, methyl ester of the ligand with His351, and three hydrogen bonds between the carbonyl group of the compound and Arg213, Asp215 and Glu277 (Fig. 3).
Increasing the length of the linker between the hydrazolyl moiety and the phenyl ring affected the activity. Docking results exhibited that increasing the length of the linker and moving the nitro group from para to ortho position might lead to a conformation change in the active site [[35], [36], [37]]. That could be effective in inhibitory activity (compound 3b (IC50 = 215.4 μM) (ΔGbinding = −10.23 kcal/mol) vs. compound 3c (IC50 = 174.0 μM) (ΔGbinding = −11.61 kcal/mol)). Based on previous reports all almost residues that interacted with these compounds play an important role in the binding and inhibitory activity of compounds [5,9,38].
Comparison of the binding mode of acarbose as the drug and positive control in the active site [5] with compounds 3b and 3c showed that these two compounds formed hydrogen bonds and hydrophobic interactions with Tyr72, Phe314, Tyr158, Phe303, Asp352, Arg442, His112, and Asp215 similar to acarbose. Moreover, they showed more interactions than acarbose with residues. Thus, extra interactions can be important for binding, which these compounds were more potent compared to acarbose. Further studies showed that compounds with lower inhibitory activity e.g. 3a have fewer interactions with the key residues of the active site than the most potent compounds (3b and 3c) and acarbose.
Replacing methyl ester (compound 3b) with the ethyl ester (compound 3a) dramatically reduced the inhibitory effect. The weak activity of 3a might be ascribed to a slight conformation change and dropping the establishment of hydrogen bonds and key hydrophobic interactions. It appears that extra hydrogen bonds hydrophobic interactions show a positive effect. Moreover, the ester of derivative 3a made hydrogen bonds with Asp69, Arg442 and Glu277 (Fig. 4), while the ester moiety of compounds 3b and 3c made hydrogen bonds with Asp215, Glu277, Arg213 and His351 residues.
Fig. 4.
Conformation of derivative 3a in α-glucosidase active site.
Replacement para-chlorophenyl ring of compound 3d with the para-nitrophenyl ring of compound 3a intensely decreased inhibitory activity. The differences between the groups along with the ΔGbind values for compounds 3b (ΔGbind = −10.23 kcal/mol) and 3d (ΔGbind = −9.55 kcal/mol) may account for the differences between the inhibitory potency of these compounds. Moreover, compound 3b showed lower hydrophobic features than compound 3d (CLogP = 3.45 for 3b vs. CLogP = 4.42 for 3d). This may be attributed to the poor penetration ability of this compound to the outer membrane of the cell.
Un-substituted phenyl ring in comparison to the substituted phenyl ring showed lower inhibitory activity and free binding energy. In comparison to 3a and 3e, the un-substituted phenyl ring is inappropriate for the activity and caused a reduction of interaction with key residues. That caused a decrease in the formation of hydrogen bonds at the ester moiety and eliminated hydrogen bonds at the phenyl ring. Therefore, these interactions might be important for the stability of the compound at the active site.
From the structural differences of these compounds, assay findings, and their molecular docking study, it was observed that the compounds that have attachment groups in the ortho position over the aromatic ring have good interaction networks as well as inhibitory activities as compared to the compounds having either para attachment or without group. Similarly, a good inhibition mode was observed for most electronegative groups than less electronegative groups attached over the aromatic phenyl ring. Moreover, increasing the length of the linker of the phenyl ring to the hydrazolyl moiety changes the binding mode and the binding to amino acids.
The comparison of compounds showed that: (1) extra hydrogen bonds have a positive effect on α-glucosidase inhibition, (2) Asp215 and Glu277 residues are important for the formation of hydrogen bonds, (3) Tyr158, Asp352, Phe303, His112, and Phe178 residues are main for the hydrophobic interactions, (4) presence nitro group at ortho position is better than para position due to more hydrogen bond formation and appropriate orientation of the compound in the active site, (5) the formation of hydrogen bond between carbonyl group of hydrazolyl moiety and Glu277 is important for activity, (6) increasing the length of the linker of the phenyl ring to the hydrazolyl moiety have a positive effect on α-glucosidase inhibition. These findings are aligned with acarbose and previous reports [5,[39], [40], [41]].
The active and moderately active compounds' binding modes were all oriented more or less in a similar fashion, which they were similar to the binding mode of acarbose, while in contrast, the least active compounds’ binding modes were all oriented differently from the active ones and acarbose.
3.5. Molecular dynamics simulation study
In the docking methodology, ligand-protein complex interactions are instant and potentially unstable. Therefore, molecular dynamics (MD) simulations is performed for the study of compound stability in the active site. It investigates the separate element motions and identifies the stability of the complex as a function of time more easily than experiments in the actual system [42].
3.6. Root mean square deviation (RMSD)
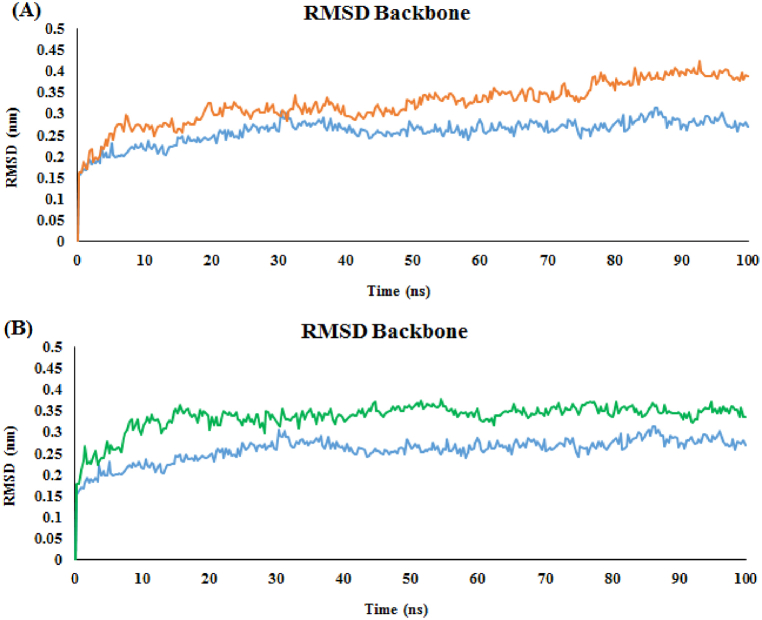
The dynamic stabilities of all systems can reflect via root mean square deviations (RMSDs) for all heavy atoms of each ligand, backbone atoms of amino acids in the active site within 5 Å around the ligand, and all backbone atoms of the protein. The RMSDs of the two complexes compared to the acarbose complex are shown in Fig. 5. The blue line in Fig. 5 displays the acarbose-α-glucosidase complex. The RMSD plot of acarbose became overall stable after 12 ns and the RMSD average value was at about 2.5 Å. The RMSD plot of derivative 3b also got an equilibration state after 12 ns with a higher RMSD value (>3 Å) (Fig. 5, brown line). However, The RMSD plot of derivative 3c touched an equilibration state after 9 ns with an RMSD value equal to 3.5 Å (Fig. 5, green line).
Fig. 5.
RMSD backbone plot of the α-glucosidase backbone in complexed with acarbose (in blue) and (A) compound 3b (in brown), (B) compound 3c (in green) 100 ns of the MD simulation time.
Compound 3c complexed α-glucosidase was stable after equilibrium. The compound 3b exhibited diverse dynamics performances in the α-glucosidase active site. According to Fig. 5, the fluctuations of the RMSD plot were about 3.0 Å at the first 50 ns. Then, a sharp increase was detected and kept stable with average RMSD values of 4.0 Å until the end of the MD simulation. This showed that the conformation of derivative 3b changes during simulation.
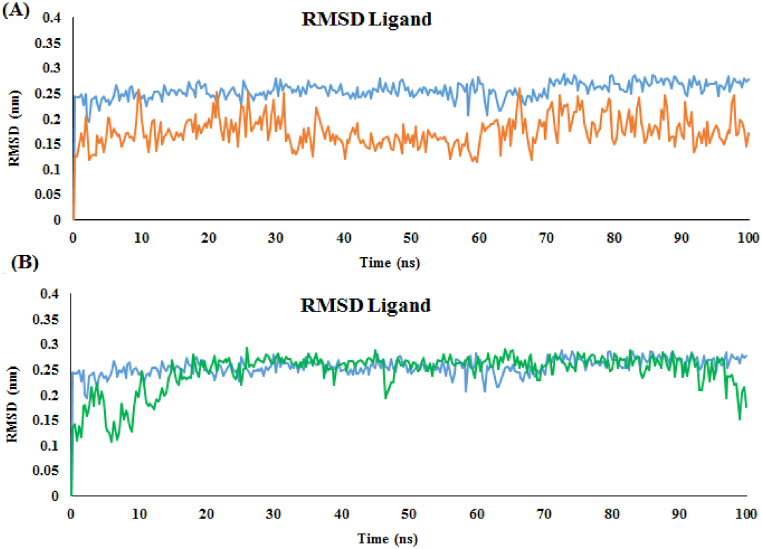
Likewise, the RMSD of molecules was also calculated separately (Fig. 6). The ligands 3c and 3b in the α-glucosidase enzyme exhibited a stable RMSD plot with small fluctuations of 2.5 and 2.0 Å, respectively. Based on the RMSD of ligand 3c, a large fluctuation was not observed and it was very stable during the simulation similar to acarbose (Fig. 6). However, compound 3b exhibited stability from 5ns to 100ns. Ligand 3b exhibited a lower RMSD value than acarbose with large fluctuation (Fig. 6).
Fig. 6.
RMSD ligand plot of the acarbose (in blue) and (A) compound 3b (in brown), (B) compound 3c (in green) 100 ns of the MD simulation time.
3.7. Root mean square fluctuation (RMSF)
The RMSF value of the enzyme’s amino acids was investigated to demonstrate the enzyme structure flexibility. Fig. 5 displays that the active site area was relatively rigid. The fluctuation of the active site for the 3b-α-glucosidase complex is more significant, and the 3c-α-glucosidase complex is smaller as compared to acarbose, which shows compound 3c has strong interactions with active site residues and is stable in the active site. Analysis of the results from the 3b-α-glucosidase revealed that this compound fluctuates to a greater degree. This proposes that the behavior of 3b is not an effect of the protein conformation and is characteristic of the compound itself. Furthermore, Fig. 7 displays that derivative 3c well fitted and firmly fastened the active site, which decreased the flexibility of the amino acids via interacting with main residues and resulted in the inhibition of α-glucosidase.
Fig. 7.
RMSF plot of the α-glucosidase residue in complexed with acarbose (in blue) and compound 3c (in green) and compound 3b (in brown).
For example, in acarbose Phe303, Glu411, Glu277, Asp215, Tyr158, His112, Phe178, Phe159, Asp352, His351, Arg442, and Asn415 showed maximum RMSFs of 0.767, 0.608, 0.725, 0.562, 0.568, 0.564, 0.532, 0.582, 0.637, 0.601, 0.565 and 0.838 Å. About in compound 3c Phe303, Glu411, Glu277, Asp215, Tyr158, His112, Phe178, Phe159, Asp352, His351, Arg442 and Asn415 showed maximum RMSFs of 0.709, 0.917, 0.699, 0.577, 0.566, 0.577, 0.518, 0.576, 0.579, 1.058 and 0.589 Å. In compound 3b Phe303, Glu411, Glu277, Asp215, Tyr158, His112, Phe178, Phe159, Asp352, His351, Arg442, Asn415 showed maximum RMSFs of 0.812, 0.631, 0.480, 0.50, 0.729, 0.545, 0.603, 0.720, 0.624, 0.644, 0.634 and 1.199 Å. A comparison of all complexes RMSF results exhibited that, Asp215, Tyr158, His112, Phe178, Phe159 and Arg442 residues displayed the smallest fluctuation with acarbose as the drug standard. While, Asp215, Tyr158, His112, Phe178, Phe159, Asp352, His351 and Asn415 exhibited the smallest fluctuation with compound 3c. Glu277, Asp215 and His112 displayed the smallest fluctuation with compound 3b. According to the results, all compounds showed the smallest fluctuation with Asp215 and His112. Acarbose and compound 3c showed strong interactions with Tyr158, Phe178, and Phe159, however, compound 3b showed higher fluctuation with these residues. These observations correlated with docking results and previous studies [5].
3.7.1. Radius of gyration (Rg)
To know the compact of the enzyme, the radius of gyration (Rg) was investigated. The number of residues present in the enzyme is important in the Rg value [43]. When the protein structure has proper folding and stability, the Rg value is lower and stable. However, improper folding and conformational flexibility of the enzyme has a very fluctuating Rg value [44].
The average Rg values of acarbose-α-glucosidase, 3c-α-glucosidase, and compound 3b-α-glucosidase complexes were 2.41 nm, 2.42 nm, and 2.40 nm, respectively. The Rg value of the compound 3b-α-glucosidase complex showed an increase after 70 ns however the Rg of acarbose-α-glucosidase dropped after 70ns. The 3c-α-glucosidase complex exhibited more a constant Rg value during MD simulation; a minor lowering was detected from 2 to 2.35 ns then increasing was observed from 2.35 to 2.43 ns. A slight decrease again was detected from 2.43 to 3.7 ns (Fig. 8). The Rg value shows that the 3b-α-glucosidase complex was fairly unstable toward the acarbose and compound 3c because they accepted a more controlled conformation for the period of the simulation. Thus, the compound has less stability in the flap, and interactions between residues, and hinge domains are mainly affected.
Fig. 8.
Radius of gyration (Rg) graphs of 3b (brown), 3c (green), and acarbose (blue) complexes.
3.7.2. Solvent accessible surface area (SASA)
The surface area of a compound-enzyme complex that interacts with solvent molecules is accounted for by SASA. An open or extended enzyme structure exposed more to solvent shows higher SASA. The average SASA for the acarbose was 232.04 nm2, while it was for compounds 3b and 3c 236.48 nm2 and 234.27 nm2, respectively. This funding shows solvent interaction with complexes was relatively similar and also confirms the residue flexibility and compactness analysis results. The SASA value slightly decreased for complexes and then it showed almost a constant SASA value throughout the simulation. Fluctuations were similar in very minor complexes. Comparatively, the compound 3b-α-glucosidase complex showed higher SASA suggesting less stability than the 3c-α-glucosidase complex (Fig. 9).
Fig. 9.
SASA plot for α-glucosidase complexes of acarbose, compound 3c and compound 3b (shown in blue, green and brown, respectively).
3.7.3. Hydrogen bonding analysis
One of the indicators of the binding stability of the compound-enzyme complex is the formation of hydrogen bonds between the compound and the enzyme. The MD findings exhibited intermolecular hydrogen bonds made significantly between compounds and amino acids (Fig. 10). The average hydrogen bonds for compounds 3c, 3b, and acarbose complexes are 1.86, 0.80, and 1.81, respectively. The hydrogen bonds of 3c-α-glucosidase were higher compared to 3b and acarbose complexes. However, the hydrogen bonds were constant throughout the simulation for compounds 3c and 3b complexes, indicating stronger binding between these compounds and α-glucosidase and leading to a highly stable conformation. The number of hydrogen bonds made throughout the simulations in compounds 3c and 3b was more than acarbose.
Fig. 10.
Hydrogen bond numbers between α-glucosidase and compound 3c (green), compound 3b (brown) and acarbose (blue) over MD production run.
3.7.4. Structural analysis during MD simulations
To understand the structural variations in 3c-, 3b-, and acarbose-α-glucosidase, the conformational snapshots at the distance of every 20 ns were taken overall 100 ns. In first 20 ns revealed more fluctuation for the 3c-α-glucosidase complex but in the following, because of more compactness in the active site enzyme (reflected by Rg and SASA findings), the complex showed less structural variations and more stability from 40 ns to 100 ns (Fig. 11).
Fig. 11.
Snapshots of compound 3c-α-glucosidase complex at interval of every 20 ns of MD simulations.
For compound 3b-α-glucosidase complex snapshots were provided. In the first 20 ns, this complex showed more fluctuation; then, it exhibited high stability and less structural variations from 40 ns to 80 ns. Afterward, due to extension in the active site (reflected by SASA and Rg studies), the complex showed less stability and more structural variations from 80 ns to 100 ns (Fig. 12).
Fig. 12.
Snapshots of compound 3b-α-glucosidase complex at interval of every 20 ns of MD simulations.
Moreover, the snapshots were achieved for acarbose-α-glucosidase. Acarbose demonstrated structural variations only for primary 40 ns and then kept almost constant until 100 ns (Fig. 13).
Fig. 13.
Snapshots of acarbose-α-glucosidase complex at interval of every 10 ns of MD simulations.
3.7.5. Binding free energy
The MD/MM-PBSA affords diverse separate components, such as ΔEvdW, ΔEelec, ΔGpol, ΔGnpol, and TΔS, to the total binding free energy (ΔGbinding) (Table 2). It is obvious from Table 2 that ΔEelec and ΔEvdW preferred the binding, whereas ΔEnpol and ΔEpol disfavored the complexation. For both cases, ΔEpol is positive, which means that the electrostatic and van der Waals interactions chiefly determine the binding.
Table 2.
Molecular energy terms for interactions of two compounds and acarbose with α-glucosidase.
| Terms | Energy (kcal/mol) |
||
|---|---|---|---|
| 3b | 3c | Acarbose | |
| ΔEbinding | −222.452 | −260.950 | −159.271 |
| ΔEelec | −308.575 | −355.092 | −226.624 |
| ΔEvdW | −48.521 | −57.730 | −42.915 |
| ΔEpol | 110.132 | 123.521 | 73.328 |
| ΔEnpol | −24.009 | −29.379 | −19.643 |
Moreover, Table 2 exposes that the ΔGbinding of 3c is higher (−260.950 kcal/mol) than 3b (−222.452 kcal/mol). This can be because ΔEvdW and ΔEelec are more favorable in 3c-α-glucosidase (ΔEvdW = −57.730 kcal/mol, ΔEelec = −355.092 kcal/mol) than 3b-α-glucosidase (ΔEvdW = −48.521 kcal/mol, ΔEelec = −308.575 kcal/mol). Generally, this finding proposes that 3c showed higher activity on α-glucosidase compared to 3b.
Next, the binding affinity of compounds 3b and 3c were estimated and compared with acarbose. Acarbose showed binding energy lower (−159.271 kcal/mol) than 3b and 3c (Table 2). It is also displayed that the van der Waals interactions (−42.915 kcal/mol) unfavored the complex formation less compared to the electrostatic interaction (−226.624 kcal/mol). This is in approving with what has been detected for compounds 3b and 3c. This result showed that the binding affinity reduces in the following order against α-glucosidase: 3c > 3b > acarbose (Table 2).
After that, the binding free energy of the crucial amino acids involved in the compound-enzyme binding was analyzed with MM-PBSA. The MM-PBSA investigates hotspot amino acids in interaction energy which are higher than −1.0 kcal/mol (Table 3). According to Table 3, compound 3c has interacted with a more significant number of crucial amino acids (Tyr72, Tyr158, Phe159, and Phe178) toward compound 3b and acarbose. This can be one of the causes for the higher affinity and inhibitory activity of compound 3c compared to compound 3b against α-glucosidase (see Table 4, Table 5).
Table 3.
Interaction energy of hotspot residues considered with 3c.
| Residues | ΔEbinding | ΔEMM | ΔEpol | ΔEnpol |
|---|---|---|---|---|
| Asp69 | −0.04 | −0.21 | 0.17 | −0.04 |
| Tyr72 | −8.03 | −11.54 | 4.33 | −0.85 |
| His112 | −0.90 | −1.88 | 1.03 | −0.04 |
| Lys156 | 6.41 | −5.64 | 12.52 | −0.49 |
| Tyr158 | −15.18 | −17.98 | 4.14 | −1.31 |
| Phe159 | −14.34 | −15.17 | 1.75 | −0.94 |
| Phe178 | −11.66 | −14.00 | 3.45 | −1.01 |
| Arg213 | −0.14 | 0.08 | −0.20 | 0 |
| Asp215 | 0.56 | 0.13 | 0.42 | 0 |
Table 4.
Interaction energy of hotspot residues considered with 3b.
| Residues | ΔEbinding | ΔEMM | ΔEpol | ΔEnpol |
|---|---|---|---|---|
| Asp69 | −0.43 | −0.31 | −0.12 | 0 |
| Tyr72 | −0.41 | −0.49 | 0.10 | −0.02 |
| His112 | −1.93 | −6.04 | 4.44 | −0.33 |
| Lys156 | 8.45 | −2.00 | 10.59 | −0.06 |
| Tyr158 | −10.06 | −12.61 | 3.24 | −0.69 |
| Phe159 | −15.31 | −16.52 | 1.98 | −0.79 |
| Phe178 | −5.42 | −7.51 | 2.62 | −0.55 |
| Arg213 | −0.76 | −2.07 | 1.30 | 0 |
| Asp215 | 3.02 | 5.13 | −2.15 | −0.01 |
Table 5.
Interaction energy of hotspot residues considered with acarbose.
| Residues | ΔEbinding | ΔEMM | ΔEpol | ΔEnpol |
|---|---|---|---|---|
| Asp69 | −0.38 | −0.22 | −0.15 | 0 |
| Tyr72 | 0.01 | −0.12 | 0.12 | 0 |
| His112 | 0.07 | 0.05 | 0.02 | 0 |
| Lys156 | 2.87 | −3.94 | 6.85 | −0.08 |
| Tyr158 | −7.61 | −12.26 | 5.46 | −0.88 |
| Phe159 | −4.07 | −4.61 | 0.92 | −0.41 |
| Phe178 | −0.98 | −1.06 | 0.19 | −0.12 |
| Arg213 | 1.39 | 1.62 | −0.21 | 0 |
| Asp215 | −1.40 | −1.06 | −0.35 | 0 |
4. Conclusion
In summary, some of the 2-hydrazolyl-4-thiazolidinone hybrids were synthesized using a new route with more yield and fewer steps. All titled derivatives showed weak to good activity against α-glucosidase relative to acarbose as a drug. Two compounds 3b and 3c showed the highest inhibitory activity. Compound 3c having a para-nitrophenyl ring and methyl ester moiety was the most potent one with an IC50 value of 174 μM. A kinetic finding exhibited that 3c inhibited via a competitive mechanism. Moreover, docking study and molecular dynamics simulations displayed that compound 3c as the most potent compound was stable and fit in the active site and showed key interactions with vital residues compared to compound 3b and acarbose.
CRediT authorship contribution statement
Saghi Sepehri: Writing – review & editing, Writing – original draft, Supervision, Software, Project administration, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Ghazaleh Farhadi: Writing – original draft, Methodology, Investigation, Data curation. Maryam Maghbul: Writing – original draft, Methodology, Investigation. Farough Nasiri: Writing – review & editing, Writing – original draft, Supervision, Methodology, Investigation, Data curation, Conceptualization. Mohammad Ali Faramarzi: Writing – original draft, Supervision, Methodology, Investigation. Karim Mahnam: Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Somayeh Mojtabavi: Methodology, Investigation, Formal analysis. Mohammad Mahdavi: Supervision, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Zhaleh Moharrami Oranj: Methodology, Formal analysis.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:Saghi Sepehri reports financial support was provided by Ardabil University of Medical Sciences. If there are other authors, they declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This research was supported by Ardabil University of Medical Sciences. Funding Number is IR.ARUMS.REC.1401.241.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e36408.
Contributor Information
Saghi Sepehri, Email: saghisepehridr@gmail.com, s.sepehri@arums.ac.ir.
Farough Nasiri, Email: nasiri@uma.ac.ir.
Mohammad Mahdavi, Email: momahdavi@tums.ac.ir.
Appendix A. Supplementary data
The following is/are the supplementary data to this article.
References
- 1.Chen L., Jiang Z., Yang L., Fang Y., Lu S., Akakuru O.U., Huang S., Li J., Ma S., Wu A. HPDA/Zn as a CREB inhibitor for ultrasound imaging and stabilization of atherosclerosis plaque. Chin. J. Chem. 2023;41:199–206. [Google Scholar]
- 2.Zhou Y., Chai X., Yang G., Sun X., Xing Z. Changes in body mass index and waist circumference and heart failure in type 2 diabetes mellitus. Front. Endocrinol. 2023;14 doi: 10.3389/fendo.2023.1305839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liang D., Cai X., Guan Q., Ou Y., Zheng X., Lin X. Burden of type 1 and type 2 diabetes and high fasting plasma glucose in Europe, 1990-2019: a comprehensive analysis from the global burden of disease study 2019. Front. Endocrinol. 2023;14 doi: 10.3389/fendo.2023.1307432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu Y., Wang L., Ni S., Li D., Liu J., Chu H.Y., Zhang N., Sun M., Li N., Ren Q., Zhuo Z., Zhong C., Xie D., Li Y., Zhang Z.-K., Zhang H., Li M., Zhang Z., Chen L., Pan X., Xia W., Zhang S., Lu A., Zhang B.-T., Zhang G. Targeting loop3 of sclerostin preserves its cardiovascular protective action and promotes bone formation. Nat. Commun. 2022;13:4241. doi: 10.1038/s41467-022-31997-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bakherad Z., Bakherad H., Sepehri S., Faramarzi M.A., Mahnam K., Mojtabavi S., Mahdavi M. In silico and in vitro studies of thiosemicarbazone-indole hybrid compounds as potent α-glycosidase inhibitors. Comput. Biol. Chem. 2022;97 doi: 10.1016/j.compbiolchem.2022.107642. [DOI] [PubMed] [Google Scholar]
- 6.Joshi S.R., Standl E., Tong N., Shah P., Kalra S., Rathod R. Therapeutic potential of α-glucosidase inhibitors in type 2 diabetes mellitus: an evidence-based review. Expet Opin. Pharmacother. 2015;16:1959–1981. doi: 10.1517/14656566.2015.1070827. [DOI] [PubMed] [Google Scholar]
- 7.Proença C., Freitas M., Ribeiro D., Oliveira E.F.T., Sousa J.L.C., Tomé S.M., Ramos M.J., Silva A.M.S., Fernandes P.A., Fernandes E. α-Glucosidase inhibition by flavonoids: an in vitro and in silico structure-activity relationship study. J. Enzym. Inhib. Med. Chem. 2017;32:1216–1228. doi: 10.1080/14756366.2017.1368503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mughal E.U., Hawsawi M.B., Naeem N., Hassan A., Alluhaibi M.S., Ali Shah S.W., Nazir Y., Sadiq A., Alrafai H.A., Ahmed S.A. Exploring fluorine-substituted piperidines as potential therapeutics for diabetes mellitus and Alzheimer's diseases. Eur. J. Med. Chem. 2024;273 doi: 10.1016/j.ejmech.2024.116523. [DOI] [PubMed] [Google Scholar]
- 9.Karami M., Hasaninejad A., Mahdavi H., Iraji A., Mojtabavi S., Faramarzi M.A., Mahdavi M. One-pot multi-component synthesis of novel chromeno[4,3-b]pyrrol-3-yl derivatives as alpha-glucosidase inhibitors. Mol. Divers. 2022;26:2393–2405. doi: 10.1007/s11030-021-10337-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ren J., Dai J., Chen Y., Wang Z., Sha R., Mao J. Physiochemical characterization and ameliorative effect of rice resistant starch modified by heat-stable α-amylase and glucoamylase on the gut microbial community in T2DM mice. Food Funct. 2024;15:5596–5612. doi: 10.1039/d3fo05456j. [DOI] [PubMed] [Google Scholar]
- 11.Patel N.B., Shaikh F.M. Synthesis and antimicrobial activity of new 4-thiazolidinone derivatives containing 2-amino-6-methoxybenzothiazole. Saudi Pharmaceut. J. : SPJ : the official publication of the Saudi Pharmaceutical Society. 2010;18:129–136. doi: 10.1016/j.jsps.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prieto P., Pineda M., Aguilar M. Spectrophotometric quantitation of antioxidant capacity through the formation of a phosphomolybdenum complex: specific application to the determination of vitamin E. Anal. Biochem. 1999;269:337–341. doi: 10.1006/abio.1999.4019. [DOI] [PubMed] [Google Scholar]
- 13.Vigorita M.G., Ottanà R., Monforte F., Maccari R., Trovato A., Monforte M.T., Taviano M.F. Synthesis and antiinflammatory, analgesic activity of 3,3′-(1,2-Ethanediyl)-bis[2-aryl-4-thiazolidinone] chiral compounds. Part 10. Bioorg. Med. Chem. Lett. 2001;11:2791–2794. doi: 10.1016/s0960-894x(01)00476-0. [DOI] [PubMed] [Google Scholar]
- 14.Bouzroura S., Bentarzi Y., Kaoua R., Poulain-Martini B.N.-K.S., Dunach E. A convenient one pot preparation of 4-thiazolidinones from enaminolactones. Org. Commun. 2010;3:8. [Google Scholar]
- 15.Crowfoot D., Bunn C.W., Rogers-Low B.W., Turner-Jones A. The X-ray crystallographic investigation of the structure of penicillin. Chemistry of penicillin. 1949:310–367. [Google Scholar]
- 16.Hussain Z., Mahmood A., Shah Q., Imran A., Mughal E.U., Khan W., Baig A., Iqbal J., Mumtaz A. Synthesis and evaluation of amide and thiourea derivatives as carbonic anhydrase (CA) inhibitors. ACS Omega. 2022;7:47251–47264. doi: 10.1021/acsomega.2c06513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ullah H., Uddin I., Rahim F., Khan F., Sobia, Taha M., Khan M.U., Hayat S., Ullah M., Gul Z., Ullah S., Zada H., Hussain J. In vitro α-glucosidase and α-amylase inhibitory potential and molecular docking studies of benzohydrazide based imines and thiazolidine-4-one derivatives. J. Mol. Struct. 2022;1251 [Google Scholar]
- 18.Khan S.A., Ali M., Latif A., Ahmad M., Khan A., Al-Harrasi A. Mercaptobenzimidazole-based 1, 3-Thaizolidin-4-ones as antidiabetic agents: synthesis, in vitro α-glucosidase inhibition activity, and molecular docking studies. ACS Omega. 2022;7:28041–28051. doi: 10.1021/acsomega.2c01969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szabó K., Maccari R., Ottanà R., Gyémánt G. Extending the investigation of 4-thiazolidinone derivatives as potential multi-target ligands of enzymes involved in diabetes mellitus and its long-term complications: a study with pancreatic α-amylase. Carbohydr. Res. 2021;499 doi: 10.1016/j.carres.2020.108220. [DOI] [PubMed] [Google Scholar]
- 20.Nanjan M.J., Mohammed M., Prashantha Kumar B.R., Chandrasekar M.J.N. Thiazolidinediones as antidiabetic agents: a critical review. Bioorg. Chem. 2018;77:548–567. doi: 10.1016/j.bioorg.2018.02.009. [DOI] [PubMed] [Google Scholar]
- 21.Aisha, Raza M.A., Sumrra S.H., Javed K., Saqib Z., Maurin J.K., Budzianowski A. Synthesis, characterization and molecular modeling of amino derived thiazolidinones as esterase and glucosidase inhibitors. J. Mol. Struct. 2020;1219 [Google Scholar]
- 22.Khan S.A., Ali M., Latif A., Ahmad M., Khan A., Al-Harrasi A. Mercaptobenzimidazole-based 1,3-Thaizolidin-4-ones as antidiabetic agents: synthesis, in vitro α-glucosidase inhibition activity, and molecular docking studies. ACS Omega. 2022;7:28041–28051. doi: 10.1021/acsomega.2c01969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khan S., Iqbal S., Rahim F., Shah M., Hussain R., Alrbyawi H., Rehman W., Dera A.A., Rasheed L., Somaily H.H., Pashameah R.A., Alzahrani E., Farouk A.E. New biologically hybrid pharmacophore thiazolidinone-based indole derivatives: synthesis, in vitro αlpha-amylase and αlpha-glucosidase along with molecular docking investigations. Molecules. 2022;27 doi: 10.3390/molecules27196564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chinthala Y., Kumar Domatti A., Sarfaraz A., Singh S.P., Kumar Arigari N., Gupta N., Satya S.K., Kotesh Kumar J., Khan F., Tiwari A.K., Paramjit G. Synthesis, biological evaluation and molecular modeling studies of some novel thiazolidinediones with triazole ring. Eur. J. Med. Chem. 2013;70:308–314. doi: 10.1016/j.ejmech.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 25.Shastin A.V., Korotchenko V.N., Nenajdenko V.G., Balenkova E.S. A novel synthetic approach to dichlorostyrenes. Tetrahedron. 2000;56:6557–6563. [Google Scholar]
- 26.Adib M., Peytam F., Rahmanian-Jazi M., Mohammadi-Khanaposhtani M., Mahernia S., Bijanzadeh H.R., Jahani M., Imanparast S., Faramarzi M.A., Mahdavi M. Design, synthesis and in vitro α-glucosidase inhibition of novel coumarin-pyridines as potent antidiabetic agents. New J. Chem. 2018;42:17268–17278. [Google Scholar]
- 27.Keivanloo A., Sepehri S., Bakherad M., Eskandari M. Click synthesis of 1,2,3-triazoles-linked 1,2,4-Triazino[5,6-b]indole, antibacterial activities and molecular docking studies. ChemistrySelect. 2020;5:4091–4098. [Google Scholar]
- 28.Razzaghi-Asl N., Mirzayi S., Mahnam K., Adhami V., Sepehri S. In silico screening and molecular dynamics simulations toward new human papillomavirus 16 type inhibitors. Res Pharm Sci. 2022;17:189–208. doi: 10.4103/1735-5362.335177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saeed A., Al-Masoudi N.A., Latif M. Synthesis and antiviral activity of new substituted methyl [2-(arylmethylene-hydrazino)-4-oxo-thiazolidin-5-ylidene]acetates. Arch. Pharmazie. 2013;346:618–625. doi: 10.1002/ardp.201300057. [DOI] [PubMed] [Google Scholar]
- 30.Pizzo C., Saiz C., Talevi A., Gavernet L., Palestro P., Bellera C., Blanch L.B., Benítez D., Cazzulo J.J., Chidichimo A., Wipf P., Mahler S.G. Synthesis of 2-hydrazolyl-4-thiazolidinones based on multicomponent reactions and biological evaluation against Trypanosoma Cruzi. Chem. Biol. Drug Des. 2011;77:166–172. doi: 10.1111/j.1747-0285.2010.01071.x. [DOI] [PubMed] [Google Scholar]
- 31.Benmohammed A., Khoumeri O., Djafri A., Terme T., Vanelle P. Synthesis of novel highly functionalized 4-thiazolidinone derivatives from 4-phenyl-3-thiosemicarbazones. Molecules. 2014;19:3068–3083. doi: 10.3390/molecules19033068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang Y.-Y., Chen Z., Yang X.-D., Deng R.-R., Shi L.-X., Yao L.-Y., Xiang D.-X. Piperazine ferulate prevents high-glucose-induced filtration barrier injury of glomerular endothelial cells. Exp. Ther. Med. 2021;22:1175. doi: 10.3892/etm.2021.10607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li J.M., Li X., Chan L.W.C., Hu R., Zheng T., Li H., Yang S. Lipotoxicity-polarised macrophage-derived exosomes regulate mitochondrial fitness through Miro1-mediated mitophagy inhibition and contribute to type 2 diabetes development in mice. Diabetologia. 2023;66:2368–2386. doi: 10.1007/s00125-023-05992-7. [DOI] [PubMed] [Google Scholar]
- 34.Xu L., Wang T., Shan Y., Wang R., Yi C. Soybean protein isolate inhibiting the retrogradation of fresh rice noodles: combined experimental analysis and molecular dynamics simulation. Food Hydrocolloids. 2024;151 [Google Scholar]
- 35.Li X., Liang J., Hu J., Ma L., Yang J., Zhang A., Jing Y., Song Y., Yang Y., Feng Z. Screening for primary aldosteronism on and off interfering medications. Endocrine. 2024;83:178–187. doi: 10.1007/s12020-023-03520-6. [DOI] [PubMed] [Google Scholar]
- 36.Cheng S., Huang M., Liu S., Yang M. Bisphenol F and bisphenol S induce metabolic perturbations in human ovarian granulosa cells. Arab. J. Chem. 2024 [Google Scholar]
- 37.Wang Q., Guo Q., Niu W., Wu L., Gong W., Yan S., Nishinari K., Zhao M. The pH-responsive phase separation of type-A gelatin and dextran characterized with static multiple light scattering (S-MLS) Food Hydrocolloids. 2022;127 [Google Scholar]
- 38.Shayegan N., Haghipour S., Tanideh N., Moazzam A., Mojtabavi S., Faramarzi M.A., Irajie C., Parizad S., Ansari S., Larijani B., Hosseini S., Iraji A., Mahdavi M. Synthesis, in vitro α-glucosidase inhibitory activities, and molecular dynamic simulations of novel 4-hydroxyquinolinone-hydrazones as potential antidiabetic agents. Sci. Rep. 2023;13:6304. doi: 10.1038/s41598-023-32889-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mohammadi-Khanaposhtani M., Rezaei S., Khalifeh R., Imanparast S., Faramarzi M.A., Bahadorikhalili S., Safavi M., Bandarian F., Nasli Esfahani E., Mahdavi M., Larijani B. Design, synthesis, docking study, α-glucosidase inhibition, and cytotoxic activities of acridine linked to thioacetamides as novel agents in treatment of type 2 diabetes. Bioorg. Chem. 2018;80:288–295. doi: 10.1016/j.bioorg.2018.06.035. [DOI] [PubMed] [Google Scholar]
- 40.Ernawati T. In silico evaluation of molecular interactions between known α-glucosidase inhibitors and homologous α-glucosidase enzymes from Saccharomyces cerevisiae, Rattus norvegicus, and GANC-human. Thai Journal of Pharmaceutical Sciences (TJPS) 2018:42. [Google Scholar]
- 41.Sun H., Ding W., Song X., Wang D., Chen M., Wang K., Zhang Y., Yuan P., Ma Y., Wang R., Dodd R.H., Zhang Y., Lu K., Yu P. Synthesis of 6-hydroxyaurone analogues and evaluation of their α-glucosidase inhibitory and glucose consumption-promoting activity: development of highly active 5,6-disubstituted derivatives. Bioorg. Med. Chem. Lett. 2017;27:3226–3230. doi: 10.1016/j.bmcl.2017.06.040. [DOI] [PubMed] [Google Scholar]
- 42.Ahangarzadeh N., Shakour N., Rezvanpoor S., Bakherad H., Pakdel M.H., Farhadi G., Sepehri S. Design, synthesis, and in silico studies of tetrahydropyrimidine analogs as urease enzyme inhibitors. Arch. Pharmazie. 2022 doi: 10.1002/ardp.202200158. [DOI] [PubMed] [Google Scholar]
- 43.Sivakumar M., Saravanan K., Saravanan V., Sugarthi S., Kumar S.M., Alhaji Isa M., Rajakumar P., Aravindhan S. Discovery of new potential triplet acting inhibitor for Alzheimer's disease via X-ray crystallography, molecular docking and molecular dynamics. J. Biomol. Struct. Dynam. 2020;38:1903–1917. doi: 10.1080/07391102.2019.1620128. [DOI] [PubMed] [Google Scholar]
- 44.Biswas R., Chowdhury N., Mukherjee R., Bagchi A. Identification and analyses of natural compounds as potential inhibitors of TRAF6-Basigin interactions in melanoma using structure-based virtual screening and molecular dynamics simulations. J. Mol. Graph. Model. 2018;85:281–293. doi: 10.1016/j.jmgm.2018.09.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.