Abstract
The wood-inhabiting fungi play an integral role in wood degradation and the cycle of matter in the ecological system. They are considered as the “key player” in wood decomposition, because of their ability to produce lignocellulosic enzymes that break down woody lignin, cellulose and hemicellulose. In the present study, four new wood-inhabiting fungal species, Adustochaetealbomarginata, Ad.punctata, Alloexidiopsisgrandinea and Al.xantha collected from southern China, are proposed, based on a combination of morphological features and molecular evidence. Adustochaetealbomarginata is characterised by resupinate basidiomata with cream to buff, a smooth, cracked, hymenial surface, a monomitic hyphal system with clamped generative hyphae and subcylindrical to allantoid basidiospores (12–17.5 × 6.5–9 µm). Adustochaetepunctata is characterised by resupinate basidiomata with cream, a smooth, punctate hymenial surface, a monomitic hyphal system with clamped generative hyphae and subcylindrical to allantoid basidiospores (13.5–18 × 6–8.2 µm). Alloexidiopsisgrandinea is characterised by resupinate basidiomata with buff to slightly yellowish, a grandinioid hymenial surface, a monomitic hyphal system with clamped generative hyphae and allantoid basidiospores (10–12.3 × 5–5.8 µm). Additionally, Alloexidiopsisxantha is characterised by resupinate basidiomata with cream to slightly buff, a smooth hymenial surface, a monomitic hyphal system with clamped generative hyphae and subcylindrical to allantoid basidiospores measuring 20–24 × 5–6.2 µm. Sequences of the internal transcribed spacers (ITS) and the large subunit (nrLSU) of the nuclear ribosomal DNA (rDNA) markers of the studied samples were generated. Phylogenetic analyses were performed with the Maximum Likelihood, Maximum Parsimony and Bayesian Inference methods. The phylogram, based on the ITS+nLSU rDNA gene regions, revealed that four new species were assigned to the genera Adustochaete and Alloexidiopsis within the order Auriculariales, individually. The phylogenetic tree inferred from the ITS sequences highlighted that Ad.albomarginata was retrieved as a sister to Ad.yunnanensis and the species Ad.punctata was sister to Ad.rava. The topology, based on the ITS sequences, showed that Al.grandinea was retrieved as a sister to Al.schistacea and the taxon Al.xantha formed a monophyletic lineage. Furthermore, two identification keys to Adustochaete and Alloexidiopsis worldwide are provided.
Key words: Biodiversity, molecular systematics, taxonomy, wood-inhabiting fungi, Yunnan Province
Introduction
In forest ecosystems, fungi play an essential ecological role to drive carbon cycling in forest soils, mediate mineral nutrition of plants and alleviate carbon limitations (Tedersoo et al. 2014). The fungal order Auriculariales is a group mainly composed of wood-inhabiting fungi in Agaricomycetes Doweld (Basidiomycota) (Hibbett et al. 2007). The type genus of this order is Auricularia Bull., in which several other gelatinous genera Exidia Fr., Guepinia Fr. and Pseudohydnum P. Karst., comprise important edible and medicinal fungi (Wu et al. 2019; Liu et al. 2022). Therefore, interest in species diversity in gelatinous genera has increased significantly in recent years (Chen et al. 2020; Shen and Fan 2020; Ye et al. 2020; Wang and Thorn 2021; Wu et al. 2021; Tohtirjap et al. 2023).
Contrary to the gelatinous genera, most species in the order Auriculariales are tough, include saprophytic species with resupinate, effused-reflexed, hydnoid, cerebriform, coralloid or pileate basidiomata (Wells and Bandoni 2001; Miettinen et al. 2012; Hibbett et al. 2014; Malysheva and Spirin 2017; Alvarenga et al. 2019; Spirin et al. 2019a, 2019b; Liu et al. 2022; Tohtirjap et al. 2023). Species with the stereoid basidiocarps are widely distributed in many orders of the Agaricomycetes, although they are certainly a minority in the order Auriculariales (Malysheva and Spirin 2017).
The genus Adustochaete Alvarenga & K.H. Larss. was erected by Alvarenga and Larsson and typed by the taxon Ad.rava Alvarenga & K.H. Larss. It is characterised by the resupinate basidiomata, spiny or tuberculate hymenophore, a monomitic hyphal structure with clamp connections on generative hyphae, present cystidia and hyphidia, ellipsoid-ovoid to obconical basidia, cylindrical to broadly cylindrical, straight or curved basidiospores (Alvarenga et al. 2019). The genus Alloexidiopsis L.W. Zhou & S.L. Liu is typified by Al.schistacea L.W. Zhou & S.L. Liu, which is c characterised by annual, resupinate basidiomata, smooth or with sterile spines hymenophore, a monomitic hyphal structure with clamp connections on generative hyphae, present cystidia and hyphidia, ellipsoid to ovoid, septate basidia, and cylindrical to broadly cylindrical, slightly curved (allantoid) basidiospores (Liu et al. 2022). Based on the MycoBank database (http://www.mycobank.org, accessed on 25 July 2024) and the Index Fungorum (http://www.indexfungorum.org, accessed on 25 July 2024), the genera Adustochaete and Alloexidiopsis have registered four and six species, respectively (Alvarenga et al. 2019; Guan et al. 2020; Hyde et al. 2020; Li et al. 2022a, 2022b; Li and Zhao 2022; Liu et al. 2022; Dong et al. 2024).
Classification of the kingdom of fungi has been updated continuously, based on the frequent inclusion of data from DNA sequences in many phylogenetic studies (Wijayawardene et al. 2020, 2022). Based on the early embrace of molecular systematics by mycologists, both the discovery and classification of fungi, amongst the more basal branches of the tree, are now coming to light from genomic analyses and environmental DNA surveys that have been conducted (James et al. 2020). Based on both the morphological and phylogenetic evidence, the generic concepts of Eichleriella Bres., Hirneolina (Pat.) Bres. and Tremellochaete Raitv. were revised, in which Malysheva and Spirin (2017) proposed that the genus Heteroradulum Lloyd ex Spirin and Malysheva was validated. The genus Eichleriella was accepted to be a monophyletic genus, while both genera Exidiopsis (Bref.) Möller and Heterochaete Pat. seemed to be synonymous, with priority given to the latter genus (Malysheva and Spirin 2017; Alvarenga et al. 2019; Alvarenga and Gibertoni 2021). However, certain species of Exidiopsis, even sequenced ones such as E.calcea (Pers.) K. Wells and E.grisea (Bres.) Bourdot & Maire, still have no appropriate placement at the generic level (Malysheva and Spirin 2017; Li et al. 2022a; Liu et al. 2022).
In recent years, the species diversity of the resupinate Auriculariales have been described or better defined using morphological and molecular analyses and the results showed the hidden diversity of this group and several corticioid genera, for example, Adustochaete, Alloexidiopsis, Amphistereum Spirin & Malysheva, Crystallodon Alvarenga, Heteroradulum, Metulochaete Alvarenga, Proterochaete Spirin & Malysheva and Sclerotrema Spirin & Malysheva, which have been established and described, based on the morphological and phylogenetic studies (Malysheva and Spirin 2017; Alvarenga et al. 2019; Spirin et al. 2019a, 2019b; Alvarenga and Gibertoni 2021; Liu et al. 2022).
During investigations on wood-inhabiting fungi in the Yunnan-Guizhou Plateau, China, many specimens were collected. To clarify the placement and relationships of these specimens, we carried out a phylogenetic and taxonomic study, based on the ITS+nLSU and ITS sequences. These specimens were assigned to the genera Adustochaete and Alloexidiopsis within the order Auriculariales. Therefore, four new species Ad.albomarginata, Ad.punctata, Al.grandinea and Al.xantha are proposed with description and illustrations, based on the morphological characteristics and phylogenetic analyses.
Materials and methods
Sample collection and herbarium specimen preparation
The fresh fruiting bodies were collected on the fallen angiosperm branches from Dali, Dehong, Diqing, Lincang and Zhaotong of Yunnan Province, China. The samples were photographed in situ and fresh macroscopic details were recorded. Photographs were recorded by a Nikon D7100 camera. All the photos were focus-stacked using Helicon Focus software. Macroscopic details were recorded and transported to a field station where the fruit body was dried on an electronic food dryer at 45 °C. Once dried, the specimens were sealed in an envelope and zip-lock plastic bags and labelled (Zhang et al. 2024). The dried specimens were deposited in the Herbarium of the Southwest Forestry University (SWFC), Kunming, Yunnan Province, China.
Morphology
The macromorphological descriptions were based on field notes and photos captured in the field and lab. The colour terminology follows Petersen (1996). The micromorphological data were obtained from the dried specimens after observation under a light microscope with a magnification of 10 × 100 oil (Zhao et al. 2023). Sections mounted in 5% potassium hydroxide (KOH) and 2% phloxine B dye (C20H2Br4Cl4Na2O5) and we also used other reagents, including Cotton Blue and Melzer’s reagent to observe micromorphology following Wu et al. (2022b). To show the variation in spore sizes, 5% of measurements were excluded from each end of the range and shown in parentheses. At least thirty basidiospores from each specimen were measured. Stalks were excluded from basidia measurements and the hilar appendage was excluded from basidiospores measurements. The following abbreviations are used: KOH = 5% potassium hydroxide water solution, CB– = acyanophilous, IKI– = both inamyloid and non-dextrinoid, L = mean spore length (arithmetic average for all spores), W = mean spore width (arithmetic average for all spores), Q = variation in the L/W ratios between the specimens studied, Qm represented the average Q of basidiospores measured ± standard deviation and n = a/b (number of spores (a) measured from given number (b) of specimens).
Molecular phylogeny
The CTAB rapid plant genome extraction kit-DN14 (Aidlab Biotechnologies Co., Ltd., Beijing, China) was used to obtain genomic DNA from the dried specimens according to the manufacturer’s instructions. The ITS region was amplified with ITS5 and ITS4 primers (White et al. 1990). The nLSU region was amplified with the LR0R and LR7 (Vilgalys and Hester 1990; Rehner and Samuels 1994). The PCR procedure for ITS was as follows: initial denaturation at 95 °C for 3 min, followed by 35 cycles at 94 °C for 40 s, 58 °C for 45 s and 72 °C for 1 min and a final extension of 72 °C for 10 min. The PCR procedure for nLSU was as follows: initial denaturation at 94 °C for 1 min, followed by 35 cycles at 94 °C for 30 s, 48 °C for 1 min and 72 °C for 1.5 min and a final extension of 72 °C for 10 min. The PCR products were purified and sequenced at Kunming Tsingke Biological Technology Limited Company (Yunnan Province, P.R. China). The newly-generated sequences were deposited in NCBI GenBank (Table 1).
Table 1.
List of species, specimens, and GenBank accession number of sequences used in this study.
| Species Name | Sample No. | GenBank Accession No. | Country | References | |
|---|---|---|---|---|---|
| ITS | nLSU | ||||
| Adustochaetealbomarginata | CLZhao 22774 * | PP852049 | PP849033 | China | Present study |
| Adustochaeteinterrupta | LR 23435 | MK391518 | MK391527 | Brazil | Alvarenga et al. (2019) |
| Adustochaetenivea | RLMA 531 | MN165954 | MN165989 | USA | Liu et al. (2022) |
| Adustochaetepunctata | CLZhao 29669 | PP852050 | — | China | Present study |
| Adustochaetepunctata | CLZhao 29671 | PP852051 | PP849034 | China | Present study |
| Adustochaetepunctata | CLZhao 29675 * | PP852052 | PP849035 | China | Present study |
| Adustochaetepunctata | CLZhao 29685 | PP852053 | PP849036 | China | Present study |
| Adustochaetepunctata | CLZhao 29686 | PP852054 | PP849037 | China | Present study |
| Adustochaetepunctata | CLZhao 29706 | PP852055 | — | China | Present study |
| Adustochaetepunctata | CLZhao 29710 | PP852056 | PP849038 | China | Present study |
| Adustochaetepunctata | CLZhao 29711 | PP852057 | PP849039 | China | Present study |
| Adustochaeterava | RC 841 | MK391516 | — | Brazil | Alvarenga et al. (2019) |
| Adustochaeterava | KHL 15526 | MK391517 | MK391526 | Brazil | Alvarenga et al. (2019) |
| Adustochaeteyunnanensis | CLZhao 8212 | MZ911964 | MZ950629 | China | Li and Zhao (2022) |
| Adustochaeteyunnanensis | CLZhao 4671 | MZ911965 | — | China | Li and Zhao (2022) |
| Adustochaeteyunnanensis | CLZhao 4401 | MZ911966 | MZ950630 | China | Li and Zhao (2022) |
| Alloexidiopsisaustraliensis | LWZ 20180514-18 | OM801934 | OM801919 | China | Liu et al. (2022) |
| Alloexidiopsisaustraliensis | LWZ 20180513-22 | OM801933 | OM801918 | China | Liu et al. (2022) |
| Alloexidiopsiscalcea | LWZ 20180904-14 | OM801935 | OM801920 | China | Liu et al. (2022) |
| Alloexidiopsiscalcea | MW 331 | AF291280 | AF291326 | Germany | Weiß and Oberwinkler (2001) |
| Alloexidiopsisgrandinea | CLZhao 33798 * | PP852058 | — | China | Present study |
| Alloexidiopsisgrandinea | CLZhao 34279 | PP852059 | — | China | Present study |
| Alloexidiopsisnivea | CLZhao 11204 | MZ352947 | MZ352938 | China | Li et al. (2022a) |
| Alloexidiopsisnivea | CLZhao 11210 | MZ352948 | MZ352939 | China | Li et al. (2022a) |
| Alloexidiopsisschistacea | LWZ 20200819-21a | OM801939 | OM801932 | China | Liu et al. (2022) |
| Alloexidiopsisxantha | CLZhao 25093 * | PP852060 | PP849040 | China | Present study |
| Alloexidiopsisyunnanensis | CLZhao 8106 | MT215569 | MT215565 | China | Guan et al. (2020) |
| Alloexidiopsisyunnanensis | CLZhao 4023 | MT215568 | MT215564 | China | Guan et al. (2020) |
| Amphistereumleveilleanum | FP-106715 | KX262119 | KX262168 | USA | Malysheva and Spirin (2017) |
| Amphistereumschrenkii | HHB 8476 | KX262130 | KX262178 | USA | Malysheva and Spirin (2017) |
| Aporpiumcaryae | Miettinen 14774 | JX044145 | — | Finland | Miettinen et al. (2012) |
| Aporpiumcaryae | WD 2207 | AB871751 | AB871730 | Japan | Sotome et al. (2014) |
| Auriculariaauricula-judae | JT 04 | KT152099 | KT152115 | UK | Tohtirjap et al. (2023) |
| Auriculariacornea | Dai 13621 | MZ618936 | MZ669905 | China | Tohtirjap et al. (2023) |
| Auriculariapolytricha | TUFC 12920 | AB871752 | AB871733 | Japan | Sotome et al. (2014) |
| Auriculariatibetica | Dai 13336 | MZ618943 | MZ669915 | China | Tohtirjap et al. (2023) |
| Bourdotiagalzinii | Otto MiettinenX3067 | MG757511 | MG757511 | Spain | Malysheva et al. (2018) |
| Crystallodonsubgelatinosum | RC 1609-URM93444 | MN475884 | MN475888 | Brazil | Alvarenga and Gibertoni (2021) |
| Crystallodonsubgelatinosum | TBG BF-18001-URM93445 | MN475885 | MN475889 | Brazil | Alvarenga and Gibertoni (2021) |
| Ductiferasucina | KW3886 | AY509551 | AY509551 | Canada | Liu et al. (2022) |
| Eichleriellabactriana | TAAM 55071 | KX262121 | KX262170 | Russia | Malysheva and Spirin (2017) |
| Eichleriellacrocata | TAAM 101077 | KX262100 | KX262147 | Russia | Malysheva and Spirin (2017) |
| Eichleriellaleucophaea | Barsukova LE 303261 | KX262111 | KX262161 | Russia | Malysheva and Spirin (2017) |
| Eichleriellatenuicula | ValCB 1 | MK391515 | MK391525 | Brazil | Alvarenga et al. (2019) |
| Elmerinacladophora | Miettinen 14314 | MG757509 | MG757509 | Indonesia | Malysheva et al. (2018) |
| Elmerinasclerodontia | Miettinen 16431 | MG757512 | MG757512 | Malaysia | Malysheva et al. (2018) |
| Exidiaglandulosa | YC Dai 21232 | MT663362 | MT664781 | China | Wu et al. (2020) |
| Exidiaglandulosa | YC Dai 21233 | MT663363 | MT664782 | China | Wu et al. (2020) |
| Exidiapithya | MW 313 | AF291275 | AF291321 | Germany | Weiß and Oberwinkler (2001) |
| Grammatuslabyrinthinus | Yuan 1600 | KM379139 | KM379140 | China | Alvarenga et al. (2019) |
| Grammatussemis | OM10618 | KX262146 | KX262194 | China | Malysheva and Spirin (2017) |
| Heteroradulumadnatum | LR 23453 | KX262116 | KX262165 | Mexico | Tohtirjap et al. (2023) |
| Heteroradulumkmetii | VS 6466 | KX262104 | KX262152 | Russia | Malysheva and Spirin (2017) |
| Hyalodonpiceicola | Spirin 2689 | MG735414 | MG735422 | Russia | Spirin et al. (2019a) |
| Hyalodonpiceicola | Spirin 11063 | MG735415 | MG735423 | Russia | Spirin et al. (2019a) |
| Mycostillavermiformis | Spirin 11330 | MG735417 | MG735425 | Russia | Spirin et al. (2019a) |
| Mycostillavermiformis | OF 188059 | MG735418 | — | Russia | Spirin et al. (2019a) |
| Myxariumcinnamomescens | OF160494 | KY801882 | KY801909 | Russia | Spirin et al. (2018) |
| Myxariumgrilletii | VS9016 | MK098896 | MK098944 | Russia | Spirin et al. (2019b) |
| Myxariumhyalinum | TL2012 443455 | KY801880 | KY801907 | Russia | Spirin et al. (2018) |
| Myxariumlegonii | VS 8986 | MK098899 | MK098947 | Russia | Spirin et al. (2019b) |
| Protodaedaleafoliacea | Miettinen 13 054 | MG757507 | MG757507 | Finland | Malysheva et al. (2018) |
| Protodaedaleahispida | Spirin 5139 | MG757510 | MG757510 | Finland | Malysheva et al. (2018) |
| Protodontiaafricana | AS 171126 1104 | MK098978 | MK098973 | Russia | Spirin et al. (2019b) |
| Protohydnumcartilagineum | SP 467240 | MG735419 | MG735426 | Russia | Malysheva et al. (2018) |
| Protomeruliusdubius | VS 3019 | MK484041 | MK480553 | Russia | Spirin et al. (2019a) |
| Protomeruliusminor | KHL 15937 | MK484060 | MK480569 | Russia | Spirin et al. (2019a) |
| Protomeruliussubstuppeus | O 19171 | JX134482 | JQ764649 | China | Spirin et al. (2019a) |
| Pseudohydnumgelatinosum | F14063 | AF384861 | AF384861 | Canada | Weiß and Oberwinkler (2001) |
| Pseudohydnumgelatinosum | AFTOL ID1875 | DQ520094 | DQ520094 | Germany | Lutzoni et al. (2004) |
| Stypellopsisfarlowii | Larsson 12337 | MG857095 | MG857099 | Russia | Spirin et al. (2018) |
| Stypellopsishyperborea | J Norden 9751 | MG857097 | MG857101 | Russia | Spirin et al. (2018) |
| Tremellochaeteatlantica | URM90199 | MG594381 | MG594383 | Brazil | Alvarenga et al. (2019) |
| Tremellochaetejaponica | TAA 42689 | AF291274 | AF291320 | Russia | Weiß and Oberwinkler (2001) |
| Tremiscushelvelloides | AFTOL ID1680 | DQ520100 | DQ520100 | Germany | Lutzoni et al. (2004) |
| Sistotremabrinkmannii | isolate 236 | JX535169 | JX535170 | Netherlands | Alvarenga and Gibertoni (2021) |
New species is shown in bold; * is shown type material, holotype.
The sequences were aligned in MAFFT v. 7 (Katoh et al. 2019) using the G-INS-i strategy. The alignment was adjusted manually using AliView v. 1.27 (Larsson 2014). The dataset was aligned first and then the sequences of ITS+nLSU were combined with Mesquite v. 3.51. The combined ITS+nLSU sequences and ITS datasets were used to infer the position of the new species and related species. The sequence of Sistotremabrinkmannii (Bres.) J. Erikss. obtained from GenBank was used as an outgroup to root trees in the ITS+nLSU analysis (Fig. 1) in the order Auriculariales (Tohtirjap et al. 2023). The sequence of Amphistereumleveilleanum (Berk. & M.A. Curtis) Spirin & Malysheva obtained from GenBank was used as an outgroup to root trees in the ITS analysis in the genus Adustochaete (Fig. 2). The sequence of Heteroradulumkmetii (Bres.) Spirin & Malysheva obtained from GenBank was used as an outgroup to root trees in the ITS analysis in the genus Alloexidiopsis (Fig. 3).
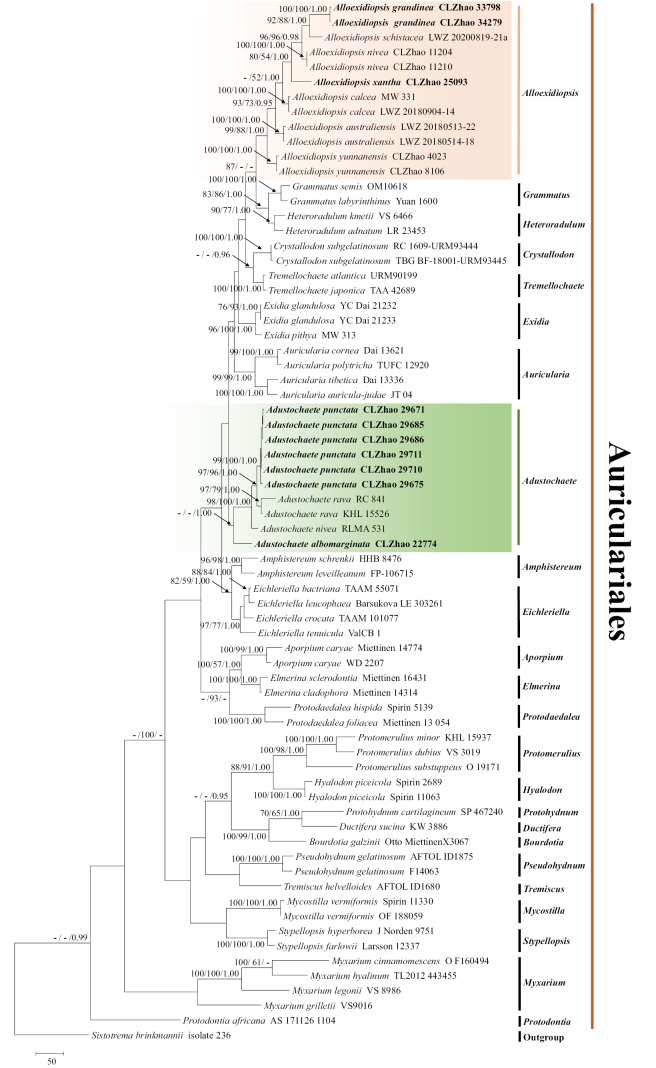
Figure 1.
Maximum parsimony strict consensus tree illustrating the phylogeny of Adustochaete and Alloexidiopsis and related genera in the order Auriculariales, based on ITS+nLSU sequences. Branches are labelled with Maximum Likelihood bootstrap value ≥ 70%, parsimony bootstrap value ≥ 50% and Bayesian posterior probabilities ≥ 0.95.
Figure 2.
Maximum parsimony strict consensus tree illustrating the phylogeny of the two new species and related genera in the genus Adustochaete, based on ITS sequences. Branches are labelled with Maximum Likelihood bootstrap value ≥ 70%, parsimony bootstrap value ≥ 50% and Bayesian posterior probabilities ≥ 0.95.
Figure 3.
Maximum parsimony strict consensus tree illustrating the phylogeny of the two new species and related genera in the genus Alloexidiopsis, based on ITS sequences. Branches are labelled with Maximum Likelihood bootstrap value ≥ 70%, parsimony bootstrap value ≥ 50% and Bayesian posterior probabilities ≥ 0.95.
Maximum Parsimony (MP), Maximum Likelihood (ML) and Bayesian Inference (BI) analyses were applied to the combined three datasets following a previous study (Zhao and Wu 2017) and the tree construction procedure was performed in PAUP* v. 4.0b10 (Swofford 2002). All of the characters were equally weighted and gaps were treated as missing data. Using the heuristic search option with TBR branch swapping and 1000 random sequence additions, trees were inferred. Maxtrees were set to 5000, branches of zero length were collapsed and all parsimonious trees were saved. Clade robustness was assessed using bootstrap (BT) analysis with 1000 replicates (Felsenstein 1985). Descriptive tree statistics, tree length (TL), the consistency index (CI), the retention index (RI), the rescaled consistency index (RC) and the homoplasy index (HI) were calculated for each maximum parsimonious tree generated. The multiple sequence alignment was also analysed using Maximum Likelihood (ML) in RAxML-HPC2 on XSEDE v. 8.2.8 with default parameters (Miller et al. 2012). Branch support (BS) for ML analysis was determined by 1000 bootstrap replicates.
jModelTest v. 2 (Darriba et al. 2012) was used to determine the best-fit evolution model for each dataset for the purposes of Bayesian Inference (BI), which was performed using MrBayes 3.2.7a with a GTR+I+G model of DNA substitution and a gamma distribution rate variation across sites (Ronquist et al. 2012). The first one-quarter of all the generations were discarded as burn-in. The majority-rule consensus tree of all the remaining trees was calculated. Branches were considered significantly supported if they received a Maximum Likelihood bootstrap value (BS) of > 70%, a Maximum Parsimony bootstrap value (BT) of > 70% or Bayesian Posterior Probabilities (BPP) of > 0.95.
Results
Sequence similarity search
The results of BLAST queries in NCBI, based on ITS and nLSU separately, showed the sequences producing significant alignment descriptions:
Adustochaetealbomarginata: in ITS BLAST results, Ad.rava, Exidiasaccharina Fr., Ea.qinghaiensis S.R. Wang & Thorn, Ad.nivea Alvarenga and Exidiopsismucedinea (Pat.) K. Wells were found as the top ten taxa (maximum record descriptions: Max score 830; Total score 830; Query cover 96%; E value 0.0; Ident 92.93%). In nLSU BLAST results, Alloexidiopsisyunnanensis (C.L. Zhao) L.W. Zhou & S.L. Liu, Auriculariaasiatica Bandara & K.D. Hyde, Au.brasiliana Y.C. Dai & F. Wu and Steccherinumnandinae (F. Wu, P. Du & X.M. Tian) Z.B. Liu, Y.C. Dai & Jing Si were found as the top ten taxa (maximum record descriptions: Max score 2398; Total score 2398; Query cover 98%; E value 0.0; Ident 98.60%).
Adustochaetepunctata: in ITS BLAST results, Ad.rava, Ad.nivea, Exidiopsismucedinea and Exidiacandida Lloyd were found as the top ten taxa (maximum record descriptions: Max score 959; Total score 959; Query cover 96%; E value 0.0; Ident 96.74%). In nLSU BLAST results, Ad.rava, Ad.yunnanensis Y.F. Li & C.L. Zhao., Auriculariathailandica Bandara & K.D. Hyde, Au.scissa Looney, Birkebak & Matheny, Au.nigricans (Sw.) Birkebak, Looney & Sánchez-García and Alloexidiopsisyunnanensis were found as the top ten taxa (maximum record descriptions: Max score 2464; Total score 2464; Query cover 98%; E value 0.0; Ident 99.34%).
Alloexidiopsisgrandinea: in ITS BLAST results, Ad.nivea and Al.schistacea were found as the top ten taxa (maximum record descriptions: Max score 861; Total score 861; Query cover 91%; E value 0.0; Ident 94.94%).
Alloexidiopsisxantha: in ITS BLAST results, Al.sinensis J.H. Dong & C.L. Zhao was found as the top ten taxa (maximum record descriptions: Max score 832; Total score 832; Query cover 98%; E value 0.0; Ident 92.42%). In nLSU BLAST results, Al.sinensis and Al.yunnanensis were found as the top ten taxa (maximum record descriptions: Max score 2457; Total score 2457; Query cover 99%; E value 0.0; Ident 99.05%).
The aligned dataset comprised 70 specimens representing 53 species. Four Markov chains were run for two runs from random starting trees, each for two million generations for the combine ITS+nLSU (Fig. 1) dataset with trees and parameters sampled every 1000 generations. The dataset had an aligned length of 2333 characters, of which 1301 characters are constant, 368 are variable and parsimony uninformative and 664 are parsimony informative. Maximum parsimony analysis yielded 120 equally parsimonious trees (TL = 4342, CI = 0.4000, HI = 0.6000, RI = 0.5288 and RC = 0.2115). The best model for the ITS+nLSU dataset, estimated and applied in the Bayesian analysis, was SYM+I+G. Both Bayesian analysis and ML analysis resulted in a similar topology to MP analysis with an average standard deviation of split frequencies = 0.008542 (BI) and the effective sample size (ESS) for Bayesian analysis across the two runs is double of the average ESS (avg. ESS) = 395.5.
The aligned dataset comprised 17 specimens representing seven species. Four Markov chains were run for two runs from random starting trees, each for 0.5 million generations for the ITS (Fig. 2) dataset with trees and parameters sampled every 1000 generations. The dataset had an aligned length of 522 characters, of which 413 characters are constant, 47 are variable and parsimony uninformative and 62 are parsimony informative. Maximum parsimony analysis yielded four equally parsimonious trees (TL = 161, CI = 0.8075, HI = 0.1925, RI = 0.8306 and RC = 0.6707). The best model for the ITS dataset, estimated and applied in the Bayesian analysis, was SYM+G. Both Bayesian analysis and ML analysis resulted in a similar topology to MP analysis with an average standard deviation of split frequencies = 0.006786 (BI) and the effective sample size (ESS) for Bayesian analysis across the two runs is double the average ESS (avg. ESS) = 617.
The aligned dataset comprised 13 specimens representing eight species. Four Markov chains were run for two runs from random starting trees, each for 0.3 million generations for the ITS (Fig. 3) dataset with trees and parameters sampled every 1000 generations. The dataset had an aligned length of 562 characters, of which 417 characters are constant, 64 are variable and parsimony uninformative and 81 are parsimony informative. Maximum parsimony analysis yielded two equally parsimonious trees (TL = 218, CI = 0.784, HI = 0.2156, RI = 0.7814 and RC = 0.6129). The best model for the ITS dataset, estimated and applied in the Bayesian analysis, was SYM+G. Both Bayesian analysis and ML analysis resulted in a similar topology to MP analysis with an average standard deviation of split frequencies = 0.007707 (BI) and the effective sample size (ESS) for Bayesian analysis across the two runs is double of the average ESS (avg. ESS) = 639.5.
The phylogram, based on the combined ITS+nLSU sequences (Fig. 1) analysis, showed that four new species Ad.albomarginata, Ad.punctata, Al.grandinea and Al.xantha were assigned to the genera Adustochaete and Alloexidiopsis within the order Auriculariales, individually. The phylogenetic tree, based on ITS sequences (Fig. 2), revealed that Ad.albomarginata was retrieved as a sister to Ad.yunnanensis. The taxon Ad.punctata was sister to Ad.rava. The topology, based on the ITS sequences (Fig. 3), revealed that Al.grandinea was retrieved as a sister to Al.schistacea and the species Al.xantha formed a monophyletic lineage.
Taxonomy
. Adustochaete albomarginata
J.H Dong & C.L. Zhao sp. nov.
FE1AB077-33E7-57BC-81AB-45982E09F628
854168
Figure 4.
Basidiomata of Adustochaetealbomarginata in general and detailed views (CLZhao 22774, holotype). Scale bars: 1 cm (A); 1 mm (B).
Figure 5.
Sections of hymenium of Adustochaetealbomarginata (holotype, CLZhao 22774) A basidiospores B basidia C cystidia D hyphidia. Scale bars: 20 µm (A–D); 10 × 100 Oil.
Figure 6.
Microscopic structures of Adustochaetealbomarginata (holotype, CLZhao 22774) A basidiospores B basidia C basidioles D cystidia E hyphidia F part of the vertical section of hymenium. Scale bars: 10 µm (A–F).
Diagnosis.
Differs from other Adustochaete species by its soft membranaceous basidiomata with cream to buff, smooth, cracked hymenial surface, a monomitic hyphal system with clamped generative hyphae and subcylindrical to allantoid basidiospores measuring 12–17.5 × 6.5–9 µm.
Holotype.
China • Yunnan Province, Dali, Weishan County, Leqiu Town, Zhongyao Village, 25°01′N, 100°19′E, altitude 1910 m, on the fallen branch of angiosperm, leg. C.L. Zhao, 19 July 2022, CLZhao 22774 (SWFC).
Etymology.
albomarginata (Latin or Greek origin): referring to the white margin of the basidiomata.
Basidiomata.
Annual, resupinate, closely adnate, soft membranaceous, very hard to separate from substrate, without odour or taste when fresh, becoming coriaceous upon drying, up to 5 cm long, 1.5 cm wide, 50–100 µm thick. Hymenial surface smooth, white to cream when fresh, turning to cream to buff upon drying, cracked. Sterile margin white, thinning out, up to 0.5 mm wide.
Hyphal system.
Monomitic, generative hyphae with clamp connections, colourless, thin-walled, unbranched, interwoven, 2.5–3.5 µm in diameter; IKI–, CB–, tissues unchanged in KOH. Hymenium. Cystidia numerous, thin-walled, subclavate to fusiform with an acute or obtuse apex, occasionally sinuous in the basal, 23.5–48.5 × 10–13.5 µm, with a clamp connection at base; cystidioles absent. Hyphidia arising from generative hyphae, nodulose, branched, colourless, thin-walled, 2.5–5 µm in diameter. Basidia ellipsoid to ovoid, longitudinally septate, two to four-celled, 17–24.5 × 11–16.5 µm; basidioles dominant, similar to basidia in shape, but slightly smaller. Basidiospores. Subcylindrical to allantoid, slightly curved, colourless, smooth, thin-walled, with 1–2 oil drops, IKI–, CB–, (11.5–)12–17.5(–18) × 6.5–9(–9.5) µm, L = 14.66 µm, W = 7.80 µm, Q = 1.72–1.99, Qm = 1.88 ± 0.08 (n = 30/1).
. Adustochaete punctata
J.H Dong & C.L. Zhao sp. nov.
7CFE9A66-C65B-593E-876E-BFC40F3BEA23
854170
Figure 7.
Basidiomata of Adustochaetepunctata in general and detailed views (CLZhao 29675, holotype). Scale bars: 1 cm (A); 1 mm (B).
Figure 8.
Sections of hymenium of Adustochaetepunctata (holotype, CLZhao 29675) A basidiospores B basidia C cystidia D hyphidia. Scale bars: 20 µm (A–D); 10 × 100 Oil.
Figure 9.
Microscopic structures of Adustochaetepunctata (holotype, CLZhao 29675) A basidiospores B basidia C basidioles D cystidia E hyphidia F part of the vertical section of hymenium. Scale bars: 10 µm (A–F).
Diagnosis.
Differs from other Adustochaete species by its membranaceous basidiomata with cream, smooth, punctate hymenial surface, a monomitic hyphal system with clamped generative hyphae and subcylindrical to allantoid basidiospores measuring 13.5–18 × 6–8.2 µm.
Holotype.
China • Yunnan Province, Dehong, Yingjiang County, Tongbiguan Provincial Nature Reserve, 23°48′N, 97°38′E, altitude 1500 m, on the fallen branch of angiosperm, leg. C.L. Zhao, 17 July 2023, CLZhao 29675 (SWFC).
Etymology.
punctata (Latin or Greek origin): referring to the punctate hymenial surface of the specimen.
Basidiomata.
Annual, resupinate, closely adnate, membranaceous, very hard to separate from substrate, without odour or taste when fresh, becoming coriaceous upon drying, up to 10 cm long, 1.5 cm wide, 100–250 µm thick. Hymenial surface smooth, punctate, white to cream when fresh, turning to cream upon drying. Sterile margin cream, thinning out, up to 1 mm wide.
Hyphal system.
Monomitic, generative hyphae with clamp connections, colourless, thin-walled, unbranched, interwoven, 1.5–3.5 µm in diameter; IKI–, CB–, tissues unchanged in KOH. Hymenium. Cystidia numerous, thin-walled, subcylindrical to clavate with an obtuse apex, occasionally sinuous in the basal, 15.5–23.5 × 5.5–7.5 µm, with a clamp connection at base; cystidioles absent. Hyphidia arising from generative hyphae, nodulose, branched, colourless, thin-walled, 1.5–5 μm in diameter. Basidia ellipsoid to ovoid, longitudinally septate, two to four-celled, 17–25 × 16.5–21 µm; basidioles dominant, similar to basidia in shape, but slightly smaller. Basidiospores. Subcylindrical to allantoid, slightly curved, colourless, smooth, thin-walled, with several oil drops, IKI–, CB–, (13–)13.5–18(–18.5) × (5.5–)6–8.2(–8.5) µm, L = 15.78 µm, W = 6.79 µm, Q = 2.15–2.40 Qm = 2.32 ± 0.08 (n = 90/3).
Additional specimens examined.
China • Yunnan Province, Dehong, Yingjiang County, Tongbiguan Provincial Nature Reserve, 23°48′N, 97°38′E, altitude 1500 m, on the fallen branch of angiosperm, leg. C.L. Zhao, 17 July 2023, CLZhao 29669; CLZhao 29671; CLZhao 29685; CLZhao 29686; CLZhao 29706; CLZhao 29710; CLZhao 29711 (SWFC).
. Alloexidiopsis grandinea
J.H Dong & C.L. Zhao sp. nov.
E5A16478-6DE6-54DD-92AC-32E3CFE78F86
854171
Figure 10.
Basidiomata of Alloexidiopsisgrandinea in general and detailed views (CLZhao 33798, holotype). Scale bars: 1 cm (A); 1 mm (B).
Figure 11.
Sections of hymenium of Alloexidiopsisgrandinea (holotype, CLZhao 33798) A basidiospores B basidia C cystidia D hyphidia. Scale bars: 10 µm (A); 20 µm (B–D); 10 × 100 Oil.
Figure 12.
Microscopic structures of Alloexidiopsisgrandinea (holotype, CLZhao 33798) A basidiospores B basidia C basidioles D cystidia E hyphidia F part of the vertical section of hymenium. Scale bars: 10 µm (A–F).
Diagnosis.
Differs from other Alloexidiopsis species by its membranaceous basidiomata with buff to slightly yellowish, grandinioid hymenial surface, a monomitic hyphal system with clamped generative hyphae and cylindrical to allantoid basidiospores measuring 10–12.3 × 5–5.8 µm.
Holotype.
China • Yunnan Province, Zhaotong, Wumengshan National Nature Reserve, 28°03′N, 104°20′E, altitude 1500 m, on the fallen branch of angiosperm, leg. C.L. Zhao, 21 September 2023, CLZhao 33798 (SWFC).
Etymology.
grandinea (Latin or Greek origin): referring to the grandinioid hymenial surface.
Basidiomata.
Annual, resupinate, closely adnate, membranaceous, very hard to separate from substrate, without odour or taste when fresh, becoming coriaceous upon drying, up to 20 cm long, 3 cm wide, 50–100 µm thick. Hymenial surface grandinioid, white to buff when fresh, turning to buff to slightly yellowish upon drying. Sterile margin cream to buff, thinning out, up to 1 mm wide.
Hyphal system.
Monomitic, generative hyphae with clamp connections, colourless, thin-walled, rarely branched, interwoven, 2–4 µm in diameter; IKI–, CB–, tissues unchanged in KOH. Hymenium. Cystidia numerous, thin-walled, fusiform with an acute apex, occasionally sinuous in the basal, 20–42.5 × 5.5–9.5 µm, with a clamp connection at base; cystidioles absent. Hyphidia arising from generative hyphae, nodulose, frequently branched, colourless, thin-walled, 2–5 µm in diameter. Basidia ellipsoid to ovoid, longitudinally septate, two to four-celled, 12.5–14.5 × 9–11.5 µm; basidioles dominant, similar to basidia in shape, but slightly smaller. Basidiospores. Cylindrical to allantoid, slightly curved, colourless, smooth, thin-walled, with 1–2 oil drops, IKI–, CB–, (9.5–)10–12.3(–12.5) × (4.8–)5–5.8(–6) µm, L = 11.08 µm, W = 5.38 µm, Q = 1.95–2.20, Qm = 2.06 ± 0.04 (n = 60/2).
Additional specimen examined.
China • Yunnan Province, Diqing, Weixi County, Weiden Town, Fuchuan Village, 27°06′N, 99°10′E, altitude 2900 m, on the fallen branch of angiosperm, leg. C.L. Zhao, 12 October 2023, CLZhao 34279 (SWFC).
. Alloexidiopsis xantha
J.H. Dong & C.L. Zhao sp. nov.
41994D3C-BA51-5BCA-9B54-998A6CDECB73
854172
Figure 13.
Basidiomata of Alloexidiopsisxantha in general and detailed views (CLZhao 25093, holotype). Scale bars: 1 cm (A); 1 mm (B).
Figure 14.
Sections of hymenium of Alloexidiopsisxantha (holotype, CLZhao 25093) A basidiospores B basidia C hyphidia D cystidia. Scale bars: 20 µm (A–D); 10 × 100 Oil.
Figure 15.
Microscopic structures of Alloexidiopsisxantha (holotype, CLZhao 25093) A basidiospores B basidioles C cystidia D basidia E hyphidia F part of the vertical section of hymenium. Scale bars: 10 µm (A–F).
Diagnosis.
Differs from other Alloexidiopsis species by its coriaceous basidiomata with cream to buff to yellow, smooth, slightly cracked hymenial surface, a monomitic hyphal system with clamped generative hyphae and allantoid to sickle-shaped basidiospores measuring 20–24 × 5–6.2 µm.
Holotype.
China • Yunnan Province, Lincang, Yun County, Dumu Village, 24°32′N, 100°23′E, altitude 2100 m, on the fallen branch of angiosperm, leg. C.L. Zhao, 20 October 2022, CLZhao 25093 (SWFC).
Etymology.
xantha (Latin or Greek origin): referring to the buff to yellow hymenial surface of the type specimen.
Basidiomata.
Annual, resupinate, closely adnate, coriaceous, very hard to separate from substrate, without odour or taste when fresh, becoming leathery upon drying, up to 10 cm long, 2 cm wide, 200–300 µm thick. Hymenial surface smooth, slightly cracked, cream when fresh, turning to cream to buff to yellow upon drying. Sterile margin cream, thinning out, up to 1 mm wide.
Hyphal system.
Monomitic, generative hyphae with clamp connections, colourless, thin- to thick walled, branched, interwoven, 2.5–3.5 µm in diameter; IKI–, CB–, tissues unchanged in KOH. Hymenium. Cystidia numerous, thin-walled, subcylindrical to subconiform with an obtuse apex, 12.5–17.5 × 3.5–6 µm, with a clamp connection at base; cystidioles absent. Hyphidia arising from generative hyphae, nodulose, frequently branched, colourless, thin-walled, 2.5–4 µm in diameter. Basidia ellipsoid to ovoid, obconical, longitudinally septate, two to four-celled, 18–20.5 × 12–15.5 µm; basidioles dominant, similar to basidia in shape, but slightly smaller. Basidiospores. Allantoid, curved, sickle-shaped, colourless, smooth, thin-walled, IKI–, CB–, (18.5–)20–24(–24.5) × 5–6.2(–6.5) µm, L = 21.66 µm, W = 5.63 µm, Q = 3.60–4.05, Qm = 3.85 ± 0.10 (n = 30/1).
Discussion
In the present study, four new species Ad.albomarginata, Ad.punctata, Al.grandinea and Al.xantha are described, based on the phylogenetic analyses and morphological characteristics.
The corticioid species of the order Auriculariales are traditionally placed in Eichleriella, Exidiopsis and Heterochaete according to the morphological characteristics (Liu et al. 2022). On the basis of the erection of six new genera as Adustochaete, Alloexidiopsis, Amphistereum, Crystallodon, Proterochaete and Sclerotrema, they were placed in the corticioid species and three previously known genera were reinstated, for example, Hirneolina, Heteroradulum and Tremellochaete (Malysheva and Spirin 2017; Alvarenga et al. 2019; Alvarenga and Gibertoni 2021; Liu et al. 2022). A multilocus-based phylogeny with a wider sampling of various morphological groups in Auriculariales is urgently needed to achieve a more natural classification of this order, as in other orders within Agaricomycetes (Wang et al. 2021).
Phylogenetically, based on the combined ITS+nLSU sequence data (Fig. 1), it demonstrated that the four new species were nested in the genera Adustochaete and Alloexidiopsis within the order Auriculariales. Based on ITS topology tree (Fig. 2), Ad.albomarginata was retrieved as a sister to Ad.yunnanensis and the species Ad.punctata was sister to Ad.rava. However, Ad.yunnanensis differs from Ad.albomarginata by its grandinioid hymenial surface, longer basidia (25–47.5 × 8.5–14 µm) and smaller cystidia (17.5–24.5 × 3.5–5.8 µm; Li and Zhao (2022)). Ad.rava can be distinguished from Ad.punctata by its spined, sharp-tipped hymenial surface, smaller basidia (14.9–16.2 × 9.7–10.1 μm) and basidiospores (10.2–13.6 × 4.6–5.9 µm; Hyde et al. (2020)). Based on ITS topology tree (Fig. 3), Al.grandinea was retrieved as a sister to Al.schistacea and Al.xantha formed a monophyletic lineage. However, Al.schistacea differs from Al.grandinea by its smooth hymenial surface and longer basidia (15–20 × 7–10 µm; Liu et al. (2022)).
Morphologically, two new species Adustochaetealbomarginata and Ad.punctata resemble four similar species in the genus Adustochaete, Ad.interrupta Spirin & Malysheva, Ad.nivea, Ad.rava and Ad.yunnanensis. A morphological comparison between two new Adustochaete species and four similar species are presented in Table 2. Two new species Al.grandinea and Al.xantha are similar to five species in the genus Alloexidiopsis, Al.australiensis S.L. Liu, Z.Q. Shen & L.W. Zhou, Al.calcea (Pers.) L.W. Zhou & S.L. Liu, Al.nivea (J.J. Li & C.L. Zhao) L.W. Zhou & S.L. Liu, Al.schistacea and Al.yunnanensis. A morphological comparison between two new Alloexidiopsis species and six similar species are presented in Table 3.
Table 2.
A morphological comparison between two new Adustochaete species and four similar species in the genus Adustochaete.
| Species name | Hymenial surface | Hyphae | Cystidia | Basidia | Basidiospores | References |
|---|---|---|---|---|---|---|
| Adustochaetealbomarginata | Smooth/ Cream to buff | Thin-walled, unbranched | Subclavate to fusiform; 23.5–48.5 × 10–13.5 µm | Ellipsoid to ovoid, two to four-celled; 17–24.5 × 11–16.5 µm | Subcylindrical to allantoid; 12–17.5 × 6.5–9 µm | Present study |
| Adustochaeteinterrupta | Smooth/ Light ochraceous-grey to brownish | Thin-walled | Clavate to fusiform; 45–96 × 6–13.5 µm | Narrowly ovoid to obconical, four-celled; 15.1–24 × 9.1–11.8 µm | Broadly cylindrical; 11.3–14.3 × 5.7–6.2 µm | Alvarenga et al. (2019) |
| Adustochaetenivea | Sharp-tipped spines/ White | Thin-walled | — | Narrowly ovoid to obconical, four-celled; 14.9–16.2 × 9.7–10.1 μm | Cylindrical; 10.2–13.6 × 4.6–5.9 µm | Hyde et al. (2020) |
| Adustochaetepunctata | Smooth/ Punctate, white to cream | Thin-walled, unbranched | Subcylindrical to clavate; 15.5–23.5 × 5.5–7.5 µm | Ellipsoid to ovoid, two to four-celled; 17–25 × 16.5–21 µm | Subcylindrical to allantoid; 13.5–18 × 6–8.2 µm | Present study |
| Adustochaeterava | Sharp-tipped spines/ Pale to dark grey | Thin-walled | Clavate to fusiform; 27–52 × 4–8 µm | Narrowly ovoid to obconical, four-celled; 10.8–15.2 × 7.3–10 µm | Cylindrical; 10.2–13.7 × 3.8–4.7 µm | Alvarenga et al. (2019) |
| Adustochaeteyunnanensis | Grandinioid/ Dark greyish to brownish | Thin-walled, branched | Clavate to fusiform; 17.5–24.5 × 3.5–5.8 µm | Narrowly ovoid to obconical, four-celled; 25–47.5 × 8.5–14 µm | Narrow cylindrical to allantoid; 12–20 × 5–7 µm | Li and Zhao (2022) |
Table 3.
A morphological comparison between two new Alloexidiopsis species and six similar species in the genus Alloexidiopsis.
| Species name | Hymenial surface | Hyphae | Cystidia | Basidia | Basidiospores | References |
|---|---|---|---|---|---|---|
| Alloexidiopsisaustraliensis | Smooth, covered by sterile spines/ Cream to pale orange | Thin-walled, branched | Cylindrical, ventricose; 21.5–24.5 × 9.5–12 µm | Ellipsoid to ovoid, four-celled; 18–21 × 13–18 µm | Cylindrical to broadly cylindrical; 13–25 × 7–11 µm | Li et al. (2022b) |
| Alloexidiopsiscalcea | Granulose to pruinose/ Greyish-white to light ochraceous | Thin-walled, branched | — | Obovate to clavate, two to four-celled; 14–25 × 9.5–15 µm | Allantoid to cylindrical, sometimes helicoid; 12–18 × 5–7 µm | Wells (1961) |
| Alloexidiopsisgrandinea | Grandinioid/ Buff to slightly yellowish | Thin-walled, branched | Fusiform; 20–42.5 × 5.5–9.5 µm | Ellipsoid to ovoid, two to four-celled; 12.5–14.5 × 9–11.5 µm | Cylindrical to allantoid; 10–12.3 × 5–5.8 µm | Present study |
| Alloexidiopsisnivea | Smooth/ White to slightly cream | Thin-walled, unbranched | Tubular; 15–34 × 2.5–7 µm | Narrowly ovoid to obconical, two to four-celled; 9–19 × 8–15 µm | Allantoid; 6.5–13.5 × 2.7–5.5 µm | Li et al. (2022a) |
| Alloexidiopsisschistacea | Smooth/ Greyish | Thin-walled, branched | Cylindrical; 25–50 × 4–6 µm | Ellipsoid to ovoid, four-celled; 15–20 × 7–10 µm | Cylindrical to broadly cylindrical; 9.5–11 × 4.5–5.5 µm | Liu et al. (2022) |
| Alloexidiopsissinensis | Grandinoid/ Yellowish-brown to rose to slightly purple | Thin- to thick-walled, branched | Cylindrical; 11.5–15.5 × 3–5.5 µm | Ellipsoid to ovoid, two to four-celled; 16–22 × 7.5–10 µm | Allantoid; 14.5–23 × 4.5–6.5 µm | Dong et al. (2024) |
| Alloexidiopsisxantha | Smooth/ Cream to slightly buff | Thin- to thick walled, branched | Subcylindrical to subconiform; 12.5–17.5 × 3.5–6 µm | Ellipsoid to ovoid, obconical; 18–20.5 × 12–15.5 µm | Allantoid, sickle-shaped; 20–24 × 5–6.2 µm | Present study |
| Alloexidiopsisyunnanensis | Odontoid/ White to smoke grey | Thin-walled, unbranched | Clavate to fusiform; 13–35 × 2–6 µm | Narrowly ovoid to obconical, two to three-celled; 28–41 × 9–14 µm | Cylindrical; 17–24 × 5–8 µm | Guan et al. (2020) |
In the ecological distribution, both genera species are not an extensively studied group, distributed worldwide and mainly found on hardwood (Alvarenga et al. 2019; Liu et al. 2022). The species of Adustochaeteinterrupta Spirin & Malysheva was found in Mexico, Ad.nivea was described in Brazil, Ad.rava was found in Brazil and Ad.yunnanensis was found in China. The species of Alloexidiopsisaustraliensis was found in Australia, Al.calcea was found in Germany and Al.nivea, Al.schistacea, Al.sinensis and Al.yunnanensis were found in China.
Fungi are one of the most diverse groups of organisms on Earth and play a crucial role in ecosystem processes and functions (Hyde 2022). New DNA sequencing techniques have revolutionised the studies of fungal taxonomy and diversity, in which about 150 k species of fungi have been described (Hyde 2022). In recent years, the wood-inhabiting fungi are an extensively studied group of Basidiomycota, which includes a number of poroid, smooth, grandinoid, odontioid and hydnoid basidiomata in China (Wu et al. 2022a, 2022b; Dong et al. 2023a, 2023b; Guan et al. 2023; Liu et al. 2023; Mao et al. 2023; Yang et al. 2023, 2024; Deng et al. 2024; Li et al. 2024; Luo et al. 2024; Zhang et al. 2024; Zhao et al. 2024; Zhou et al. 2024). In the past several years, many corticioid species have been reported and described in the order Auriculariales (Malysheva and Spirin 2017; Alvarenga et al. 2019; Spirin et al. 2019a, 2019a; Alvarenga and Gibertoni 2021; Li et al. 2022a, 2022b; Li and Zhao 2022; Liu et al. 2022), but many new taxa have not yet been discovered. Thus, the corticioid species diversity of the order Auriculariales is still not well known in China, especially in the subtropical and tropical areas. In the present study, four new species, Ad.albomarginata, Ad.punctata, Al.grandinea and Al.xantha were found and reported. This paper enriches our knowledge of fungal diversity in the order Auriculariales. We anticipate that more undescribed corticioid taxa will be discovered throughout China after extensive collection combined with morphological and molecular analyses.
Key to the known species of Adustochaete worldwide
| 1 | Hymenial surface smooth | 2 |
| – | Hymenial surface grandinioid | 4 |
| 2 | Basidia > 16.5 µm wide | Adustochaetepunctata |
| – | Basidia < 16.5 µm wide | 3 |
| 3 | Basidiospores > 6.5 µm wide | Adustochaetealbomarginata |
| – | Basidiospores < 6.5 µm wide | Adustochaeteinterrupta |
| 4 | Cystidia absent | Adustochaetenivea |
| – | Cystidia present | 5 |
| 5 | Basidiospores > 5 µm wide, basidia > 16 µm long | Adustochaeteyunnanensis |
| – | Basidiospores < 5 µm wide, basidia < 16 µm long | Adustochaeterava |
Key to the known species of Alloexidiopsis worldwide
| 1 | Basidiospores allantoid | 2 |
| – | Basidiospores cylindrical | 6 |
| 2 | Hymenial surface smooth | 3 |
| – | Hymenial surface grandinoid, granulose to pruinose | 4 |
| 3 | Basidiospores > 13.5 µm long, cystidia subcylindrical to subconiform | Alloexidiopsisxantha |
| – | Basidiospores < 13.5 µm long, cystidia tubular | Alloexidiopsisnivea |
| 4 | Cystidia absent | Alloexidiopsiscalcea |
| – | Cystidia present | 5 |
| 5 | Cystidia > 5.5 µm wide | Alloexidiopsisgrandinea |
| – | Cystidia < 5.5 µm wide | Alloexidiopsissinensis |
| 6 | Basidia > 28 µm long, cystidia clavate to fusiform | Alloexidiopsisyunnanensis |
| – | Basidia < 28 µm long, cystidia cylindrical | 7 |
| 7 | Basidiospores > 11 µm long, cystidia < 25 µm long | Alloexidiopsisaustraliensis |
| – | Basidiospores < 11 µm long, cystidia > 25 µm long | Alloexidiopsisschistacea |
Supplementary Material
Citation
Dong J, Zhu Y, Qian C, Zhao C (2024) Taxonomy and phylogeny of Auriculariales (Agaricomycetes, Basidiomycota) with descriptions of four new species from south-western China. MycoKeys 108: 115–146. https://doi.org/10.3897/mycokeys.108.128659
Funding Statement
The research was supported by the National Natural Science Foundation of China (Project Nos. 32170004, U2102220), Forestry Innovation Programs of Southwest Forestry University (Grant No: LXXK-2023Z07), the High-level Talents Program of Yunnan Province (YNQR-QNRC-2018-111), and the Research Project of Key Laboratory of Forest Disaster Warning and Control in Universities of Yunnan Province (ZKJS-S-202208)
Additional information
Conflict of interest
The authors have declared that no competing interests exist.
Ethical statement
No ethical statement was reported.
Funding
The research was supported by the National Natural Science Foundation of China (Project Nos. 32170004, U2102220), Forestry Innovation Programs of Southwest Forestry University (Grant No: LXXK-2023Z07), the High-level Talents Program of Yunnan Province (YNQR-QNRC-2018-111) and the Research Project of Key Laboratory of Forest Disaster Warning and Control in Universities of Yunnan Province (ZKJS-S-202208).
Author contributions
Conceptualisation, CZ and JD; methodology, CZ and JD; software, CZ, JD and YZ; validation, CZ and JD; formal analysis, CZ and J JD; investigation, CZ and JD; resources CZ; writing – original draft preparation, CZ, JD, YZ and CQ; writing – review and editing, CZ and JD; visualisation, CZ and JD; supervision, CZ and JD; project administration, CZ; funding acquisition, CZ. All authors have read and agreed to the published version of the manuscript.
Author ORCIDs
Junhong Dong https://orcid.org/0000-0001-8740-0805
Yonggao Zhu https://orcid.org/0009-0008-5341-3796
Chengbin Qian https://orcid.org/0009-0003-5329-1016
Changlin Zhao https://orcid.org/0000-0002-8668-1075
Data availability
All of the data that support the findings of this study are available in the main text.
References
- Alvarenga RLM, Gibertoni TB. (2021) Crystallodon Alvarenga gen. nov., a new genus of the Auriculariales from the Neotropics. Cryptogamie. Mycologie 42(2): 17–24. 10.5252/cryptogamie-mycologie2021v42a2 [DOI] [Google Scholar]
- Alvarenga RLM, Spirin V, Malysheva V, Gibertoni TB, Larsson KH. (2019) Two new genera and six other novelties in Heterochaete sensu lato (Auriculariales, Basidiomycota). Botany 97(8): 439–451. 10.1139/cjb-2019-0046 [DOI] [Google Scholar]
- Chen YL, Su MS, Zhang LP, Zou Q, Wu F, Zeng NK, Liu M. (2020) Pseudohydnumbrunneiceps (Auriculariales, Basidiomycota), a new species from Central China. Phytotaxa 441: 87–94. 10.11646/phytotaxa.441.1.8 [DOI] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. (2012) jModelTest 2: More models, new heuristics and parallel computing. Nature Methods 9(8): 772. 10.1038/nmeth.2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Jabeen S, Zhao C. (2024) Species diversity and taxonomy of Vararia (Russulales, Basidiomycota) with descriptions of six species from Southwestern China. MycoKeys 103: 97–128. 10.3897/mycokeys.103.118980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong JH, Gu JY, Zhao CL. (2023a) Diversity of wood-decaying fungi in Wenshan Area, Yunnan Province, China. Junwu Xuebao 42: 638–662. 10.13346/j.mycosystema.220205 [DOI] [Google Scholar]
- Dong JH, Zhang XC, Chen JJ, Zhu ZL, Zhao CL. (2023b) A phylogenetic and taxonomic study on Steccherinum (Polyporales, Basidiomycota): Focusing on three new Steccherinum species from Southern China. Frontiers in Cellular and Infection Microbiology 12: 1103579. 10.3389/fcimb.2022.1103579 [DOI] [PMC free article] [PubMed]
- Dong JH, Gu ZR, Wang YY, Shi YM, Zhao CL. (2024) A new corticioid fungus, Alloexidiopsissinensis (Auriculariales, Basidiomycota), in China, evidenced by morphological characteristics and phylogenetic analyses. Phytotaxa 658(3): 227–239. 10.11646/phytotaxa.658.3.1 [DOI] [Google Scholar]
- Felsenstein J. (1985) Confidence intervals on phylogenetics: An approach using bootstrap. Evolution; International Journal of Organic Evolution 39(4): 783–791. 10.2307/2408678 [DOI] [PubMed] [Google Scholar]
- Guan QX, Liu CM, Zhao TJ, Zhao CL. (2020) Heteroradulumyunnanensis sp. nov. (Auriculariales, Basidiomycota) evidenced by morphological characters and phylogenetic analyses in China. Phytotaxa 437(2): 51–59. 10.11646/phytotaxa.437.2.1 [DOI] [Google Scholar]
- Guan QX, Huang J, Huang J, Zhao CL. (2023) Five new species of Schizoporaceae (Basidiomycota, Hymenochaetales) from East Asia. MycoKeys 96: 25–56. 10.3897/mycokeys.96.99327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibbett DS, Binder M, Bischoff JF, Blackwell M, Cannon PF, Eriksson OE, Huhndorf S, James T, Kirk PM, Lücking R, Thorsten Lumbsch H, Lutzoni F, Matheny PB, McLaughlin DJ, Powell MJ, Redhead S, Schoch CL, Spatafora JW, Stalpers JA, Vilgalys R, Aime MC, Aptroot A, Bauer R, Begerow D, Benny GL, Castlebury LA, Crous PW, Dai YC, Gams W, Geiser DM, Griffith GW, Gueidan C, Hawksworth DL, Hestmark G, Hosaka K, Humber RA, Hyde KD, Ironside JE, Kõljalg U, Kurtzman CP, Larsson KH, Lichtwardt R, Longcore J, Miadlikowska J, Miller A, Moncalvo JM, Mozley-Standridge S, Oberwinkler F, Parmasto E, Reeb V, Rogers JD, Roux C, Ryvarden L, Sampaio JP, Schüssler A, Sugiyama J, Thorn RG, Tibell L, Untereiner WA, Walker C, Wang Z, Weir A, Weiss M, White MM, Winka K, Yao YJ, Zhang N. (2007) A higher-level phylogenetic classification of the Fungi. Mycological Research 111(5): 509–547. 10.1016/j.mycres.2007.03.004 [DOI] [PubMed] [Google Scholar]
- Hibbett DS, Bauer R, Binder M, Giachini AJ, Hosaka K, Justo A, Larsson E, Larsson KH, Lawrey JD, Miettinen O, Nagy LG, Nilsson RH, Weiss M, Thorn RG. (2014) 14 Agaricomycetes. In: McLaughlin D, Spatafora J. (Eds) Systematics and Evolution.The Mycota, vol 7A. Springer, Berlin, Heidelberg, 373–430. 10.1007/978-3-642-55318-9_14 [DOI]
- Hyde KD. (2022) The numbers of fungi. Fungal Diversity 114(1): 1. 10.1007/s13225-022-00507-y [DOI] [Google Scholar]
- Hyde KD, Dong Y, Phookamsak R, Jeewon R, Bhat DJ, Jones EBG, Liu NG, Abeywickrama PD, Mapook A, Wei DP, Perera RH, Manawasinghe IS, Pem D, Bundhun D, Karunarathna A, Ekanayaka AH, Bao DF, Li JF, Samarakoon MC, Chaiwan N, Lin CG, Phutthacharoen K, Zhang SN, Senanayake IC, Goonasekara ID, Thambugala KM, Phukhamsakda C, Tennakoon DS, Jiang HB, Yang J, Zeng M, Huanraluek N, Liu JK, Wijesinghe SN, Tian Q, Tibpromma S, Brahmanage RS, Boonmee S, Huang SK, Thiyagaraja V, Lu YZ, Jayawardena RS, Dong W, Yang EF, Singh SK, Singh SM, Rana S, Lad SS, Anand G, Devadatha B, Niranjan M, Sarma VV, Liimatainen K, Aguirre‐Hudson B, Niskanen T, Overall A, Alvarenga RLM, Gibertoni TB, Pfliegler WP, Horváth E, Imre A, Alves AL, Santos ACS, Tiago PV, Bulgakov TS, Wanasinghe DN, Bahkali AH, Doilom M, Elgorban AM, Maharachchikumbura SSN, Rajeshkumar KC, Haelewaters D, Mortimer PE, Zhao Q, Lumyong S, Xu JC, Sheng J. (2020) Fungal diversity notes 1151–1276: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Diversity 100(1): 5–277. 10.1007/s13225-020-00439-5 [DOI] [Google Scholar]
- James TY, Stajich JE, Hittinger CT, Rokas A. (2020) Toward a fully resolved fungal tree of life. Annual Review of Microbiology 74(1): 291–313. 10.1146/annurev-micro-022020-051835 [DOI] [PubMed] [Google Scholar]
- Katoh K, Rozewicki J, Yamada KD. (2019) MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics 20(4): 1160–1166. 10.1093/bib/bbx108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson A. (2014) AliView: A fast and lightweight alignment viewer and editor for large data sets. Bioinformatics (Oxford, England) 30(22): 3276–3278. 10.1093/bioinformatics/btu531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YF, Zhao CL. (2022) Adustochaeteyunnanensis sp. nov. from China. Mycotaxon 137(2): 261–270. 10.5248/137.261 [DOI] [Google Scholar]
- Li JJ, Zhao CL, Liu CM. (2022a) The morphological characteristics and phylogenetic analyses revealed an additional taxon in Heteroradulum (Auriculariales). Diversity 14(1): 40. 10.3390/d14010040 [DOI] [Google Scholar]
- Li QZ, Liu SL, Wang XW, May TW, Zhou LW. (2022b) Redelimitation of Heteroradulum (Auriculariales, Basidiomycota) with H.australiense sp. nov. MycoKeys 86: 87–101. 10.3897/mycokeys.86.76425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Luo YX, Zhao CL. (2024) Molecular systematics and taxonomy reveal three new wood-inhabiting fungal species (Basidiomycota) from Yunnan Province, southern China. Mycological Progress 23(1): 30. 10.1007/s11557-024-01968-y [DOI] [Google Scholar]
- Liu SL, Shen ZQ, Li QZ, Liu XY, Zhou LW. (2022) Alloexidiopsis gen. nov., a revision of generic delimitation in Auriculariales (Basidiomycota). Frontiers in Microbiology 13: 894641. 10.3389/fmicb.2022.894641 [DOI] [PMC free article] [PubMed]
- Liu S, Shen LL, Xu TM, Song CG, Gao N, Wu DM, Sun YF, Cui BK. (2023) Global diversity, molecular phylogeny and divergence times of the brown-rot fungi within the Polyporales. Mycosphere: Journal of Fungal Biology 14(1): 1564–1664. 10.5943/mycosphere/14/1/18 [DOI] [Google Scholar]
- Luo KY, Su JQ, Zhao CL. (2024) Morphological and molecular identification for four new wood-inhabiting species of Trechispora (Basidiomycota) from China. MycoKeys 105: 155–178. 10.3897/mycokeys.105.120438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutzoni F, Kauff F, Cox CJ, McLaughlin D, Celio G, Dentinger B, Padamsee M, Hibbett D, James TY, Baloch E, Grube M, Reeb V, Hofstetter V, Schoch C, Arnold AE, Miadlikowska J, Spatafora J, Johnson D, Hambleton S, Crockett M, Shoemaker R, Sung GH, Lücking R, Lumbsch T, O’Donnell K, Binder M, Diederich P, Ertz D, Gueidan C, Hansen K, Harris RC, Hosaka K, Lim YW, Matheny B, Nishida H, Pfister D, Rogers J, Rossman A, Schmitt I, Sipman H, Stone J, Sugiyama J, Yahr R, Vilgalys R. (2004) Assembling the fungal tree of life: Progress, classification, and evolution of subcellular traits. American Journal of Botany 91(10): 1446–1480. 10.3732/ajb.91.10.1446 [DOI] [PubMed] [Google Scholar]
- Malysheva V, Spirin V. (2017) Taxonomy and phylogeny of the Auriculariales (Agaricomycetes, Basidiomycota) with stereoid basidiocarps. Fungal Biology 121(8): 689–715. 10.1016/j.funbio.2017.05.001 [DOI] [PubMed] [Google Scholar]
- Malysheva V, Spirin V, Miettinen O, Motato-Vásquez V, Hernawati JSSS, Larsson KH. (2018) Revision of Protohydnum (Auriculariales, Basidiomycota). Mycological Progress 17(7): 805–814. 10.1007/s11557-018-1393-6 [DOI] [Google Scholar]
- Mao WL, Wu YD, Liu HG, Yuan Y, Dai YC. (2023) A contribution to Porogramme (Polyporaceae, Agaricomycetes) and related genera. IMA Fungus 14(1): 5. 10.1186/s43008-023-00110-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miettinen O, Spirin V, Niemelä T. (2012) Notes on the genus Aporpium (Auriculariales, Basidiomycota), with a new species from temperate Europe. Annales Botanici Fennici 49(5–6): 359–368. 10.5735/085.049.0607 [DOI] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. (2012) The CIPRES science gateway. In: Miller MA, Pfeiffer W, Schwartz T(Eds) Proceedings of the 1st Conference of the Extreme Science and Engineering Discovery Environment: Bridging from the Extreme to the Campus and Beyond, Chicago, IL, 39 pp. 10.1145/2335755.2335836 [DOI] [Google Scholar]
- Petersen JH. (1996) Farvekort. In: Petersen JH (Ed. ) The Danish Mycological Society’s Colour-Chart, Foreningen til Svampekundskabens Fremme: Greve, Germany, 6 pp. [Google Scholar]
- Rehner SA, Samuels GJ. (1994) Taxonomy and phylogeny of Gliocladium analysed fromnuclear large subunit ribosomal DNA sequences. Mycological Research 98(6): 625–634. 10.1016/S0953-7562(09)80409-7 [DOI] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. (2012) Mrbayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61(3): 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Fan L. (2020) Morphological and molecular analyses reveal two new species of Guepinia from China. Phytotaxa 475(2): 91–101. 10.11646/phytotaxa.475.2.3 [DOI] [Google Scholar]
- Sotome K, Maekawa N, Nakagiri A, Lee SS, Hattori T. (2014) Taxonomic study of Asian species of poroid Auriculariales. Mycological Progress 13(4): 987–997. 10.1007/s11557-014-0984-0 [DOI] [Google Scholar]
- Spirin V, Malysheva V, Larsson KH. (2018) On some forgotten species of Exidia and Myxarium (Auriculariales, Basidiomycota). Nordic Journal of Botany 2018(3): e01601. 10.1111/njb.01601 [DOI]
- Spirin V, Malysheva V, Miettinen O, Vlasák J, Alvarenga RLM, Gibertoni TB, Ryvarden L, Larsson KH. (2019a) On Protomerulius and Heterochaetella (Auriculariales, Basidiomycota). Mycological Progress 18(9): 1079–1099. 10.1007/s11557-019-01507-0 [DOI] [Google Scholar]
- Spirin V, Malysheva V, Roberts P, Trichies G, Savchenko A, Larsson KH. (2019b) A convolute diversity of the Auriculariales (Agaricomycetes, Basidiomycota) with sphaeropedunculate basidia. Nordic Journal of Botany 2019(7): e02394. 10.1111/njb.02394 [DOI]
- Swofford DL. (2002) PAUP*: Phylogenetic analysis using parsimony (*and Other Methods), Version 4.0b10, Sinauer Associates: Sunderland, MA, USA.
- Tedersoo L, Bahram M, Põlme S, Kõljalg U, Yorou NS, Wijesundera R, Villarreal Ruiz L, Vasco-Palacios AM, Thu PQ, Suija A, Smith ME, Sharp C, Saluveer E, Saitta A, Rosas M, Riit T, Ratkowsky D, Pritsch K, Põldmaa K, Piepenbring M, Phosri C, Peterson M, Parts K, Pärtel K, Otsing E, Nouhra E, Njouonkou AL, Nilsson RH, Morgado LN, Mayor J, May TW, Majuakim L, Lodge DJ, Lee SS, Larsson KH, Kohout P, Hosaka K, Hiiesalu I, Henkel TW, Harend H, Guo LD, Greslebin A, Grelet G, Geml J, Gates G, Dunstan W, Dunk C, Drenkhan R, Dearnaley J, De Kesel A, Dang T, Chen X, Buegger F, Brearley FQ, Bonito G, Anslan S, Abell S, Abarenkov K. (2014) Fungal biogeography. Global diversity and geography of soil fungi. Science 346(6213): 1256688. 10.1126/science.1256688 [DOI] [PubMed] [Google Scholar]
- Tohtirjap A, Hou SX, Rivoire B, Gates G, Wu F, Dai YC. (2023) Two new species of Exidia sensu lato (Auriculariales, Basidiomycota) based on morphology and DNA sequences. Frontiers in Microbiology 13: 1080290. 10.3389/fmicb.2022.1080290 [DOI] [PMC free article] [PubMed]
- Vilgalys R, Hester M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172(8): 4238–4246. 10.1128/jb.172.8.4238-4246.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Thorn RG. (2021) Exidiaqinghaiensis, a new species from China. Mycoscience 62(3): 212–216. 10.47371/mycosci.2021.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XW, May TW, Liu SL, Zhou LW. (2021) Towards a natural classification of Hyphodontia sensu lato and the trait evolution of basidiocarps within Hymenochaetales (Basidiomycota). Journal of Fungi 7: 478. 10.3390/jof7060478 [DOI] [PMC free article] [PubMed]
- Weiß M, Oberwinkler F. (2001) Phylogenetic relationships in Auriculariales and related groups – hypotheses derived from nuclear ribosomal DNA sequences. Mycological Research 105(4): 403–415. 10.1017/S095375620100363X [DOI] [Google Scholar]
- Wells K. (1961) Studies of some Tremellaceae. IV. Exidiopsis. Mycologia 53(4): 317–370. 10.1080/00275514.1961.12017967 [DOI] [Google Scholar]
- Wells K, Bandoni RJ. (2001) Heterobasidiomycetes, In: McLaughlin DJ, McLaughlin EG, Lemke PA. (Eds) The Mycota.(A Comprehensive Treatise on Fungi as Experimental Systems for Basic and Applied Research), vol 7B. Springer, Berlin, Heidelberg, 85–120. 10.1007/978-3-662-10189-6_4 [DOI]
- White TJ, Bruns T, Lee S, Taylor J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ. (Eds) PCR protocols: A guide to methods and applications.Academic Press, San Diego, 315–322. 10.1016/B978-0-12-372180-8.50042-1 [DOI]
- Wijayawardene NN, Hyde KD, Al-Ani LKT, Tedersoo L, Haelewaters D, Rajeshkumar KC, Zhao RL, Aptroot A, Leontyev DV, Saxena RK, Tokarev YS, Dai DQ, Letcher PM, Stephenson SL, Ertz D, Lumbsch HT, Kukwa M, Issi IV, Madrid H, Phillips AJL, Selbmann L, Pfliegler WP, Horváth E, Bensch K, Kirk PM, Kolaříková K, Raja HA, Radek R, Papp V, Dima B, Ma J, Malosso E, Takamatsu S, Rambold G, Gannibal PB, Triebel D, Gautam AK, Avasthi S, Suetrong S, Timdal E, Fryar SC, Delgado G, Réblová M, Doilom M, Dolatabadi S, Pawłowska J, Humber RA, Kodsueb R, Sánchez-Castro I, Goto BT, Silva DKA, de Souza FA, Oehl F, da Silva GA, Silva IR, Błaszkowski J, Jobim K, Maia LC, Barbosa FR, Fiuza PO, Divakar PK, Shenoy BD, Castañeda-Ruiz RF, Somrithipol S, Lateef AA, Karunarathna SC, Tibpromma S, Mortimer PE, Wanasinghe DN, Phookamsak R, Xu J, Wang Y, Tian F, Alvarado P, Li DW, Kušan I, Matočec N, Maharachchikumbura SSN, Papizadeh M, Heredia G, Wartchow F, Bakhshi M, Boehm E, Youssef N, Hustad VP, Lawrey JD, Santiago ALCMA, Bezerra JDP, Souza-Motta CM, Firmino AL, Tian Q, Houbraken J, Hongsanan S, Tanaka K, Dissanayake AJ, Monteiro JS, Grossart HP, Suija A, Weerakoon G, Etayo J, Tsurykau A, Vázquez V, Mungai P, Damm U, Li QR, Zhang H, Boonmee S, Lu YZ, Becerra AG, Kendrick B, Brearley FQ, Motiejūnaitė J, Sharma B, Khare R, Gaikwad S, Wijesundara DSA, Tang LZ, He MQ, Flakus A, Rodriguez-Flakus P, Zhurbenko MP, McKenzie EHC, Stadler M, Bhat DJ, Liu JK, Raza M, Jeewon R, Nassonova ES, Prieto M, Jayalal RGU, Erdoğdu M, Yurkov A, Schnittler M, Shchepin ON, Novozhilov YK. (2020) Outline of Fungi and fungus-like taxa. Mycosphere: Journal of Fungal Biology 11(1): 1060–1456. 10.5943/mycosphere/11/1/8 [DOI] [Google Scholar]
- Wijayawardene NN, Hyde KD, Dai DQ, Sánchez-García M, Goto BT, Saxena RK, Erdoğdu M, Selçuk F, Rajeshkumar KC, Aptroot A, Błaszkowski J, Boonyuen N, da Silva GA, de Souza FA, Dong W, Ertz D, Haelewaters D, Jones EBG, Karunarathna SC, Kirk PM, Kukwa M, Kumla J, Leontyev DV, Lumbsch HT, Maharachchikumbura SSN, Marguno F, Martínez-Rodríguez P, Mešić A, Monteiro JS, Oehl F, Pawłowska J, Pem D, Pfliegler WP, Phillips AJL, Pošta A, He MQ, Li JX, Raza M, Sruthi OP, Suetrong S, Suwannarach N, Tedersoo L, Thiyagaraja V, Tibpromma S, Tkalčec Z, Tokarev YS, Wanasinghe DN, Wijesundara DSA, Wimalaseana SDMK, Madrid H, Zhang GQ, Gao Y, Sánchez-Castro I, Tang LZ, Stadler M, Yurkov A, Thines M. (2022) Outline of Fungi and fungus-like taxa – 2021. Mycosphere: Journal of Fungal Biology 13(1): 53–453. 10.5943/mycosphere/13/1/2 [DOI] [Google Scholar]
- Wu F, Zhou LW, Yang ZL, Bau T, Li TH, Dai YC. (2019) Resource diversity of Chinese macrofungi: Edible, medicinal and poisonous species. Fungal Diversity 98(1): 1–76. 10.1007/s13225-019-00432-7 [DOI] [Google Scholar]
- Wu F, Zhao Q, Yang ZL, Ye SY, Rivoire B, Dai YC. (2020) Exidiayadongensis, a new edible species from East Asia. Junwu Xuebao 39: 1203–1214. 10.13346/j.mycosystema.200205 [DOI] [Google Scholar]
- Wu F, Tohtirjap A, Fan LF, Zhou LW, Alvarenga RLM, Gibertoni TB, Dai YC. (2021) Global diversity and updated phylogeny of Auricularia (Auriculariales, Basidiomycota). Journal of Fungi (Basel, Switzerland) 7(11): 933. 10.3390/jof7110933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Man XW, Tohtirjap A, Dai YC. (2022a) A comparison of polypore funga and species composition in forest ecosystems of China, North America, and Europe. Forest Ecosystems 9: 100051. 10.1016/j.fecs.2022.100051 [DOI]
- Wu F, Zhou LW, Vlasák J, Dai YC. (2022b) Global diversity and systematics of Hymenochaetaceae with poroid hymenophore. Fungal Diversity 113(1): 1–192. 10.1007/s13225-021-00496-4 [DOI] [Google Scholar]
- Yang Y, Li R, Liu CM, Zhao CL. (2023) Morphological and molecular identification for two new species of wood-inhabiting macrofungi (Basidiomycota) from Yunnan-Guizhou Plateau, China. Phytotaxa 591(1): 1–18. 10.11646/phytotaxa.591.1.1 [DOI] [Google Scholar]
- Yang Y, Li R, Jiang QQ, Zhou HM, Muhammad A, Wang HJ, Zhao CL. (2024) Phylogenetic and taxonomic analyses reveal three new wood-inhabiting fungi (Polyporales, Basidiomycota) in China. Journal of Fungi (Basel, Switzerland) 10(1): 55. 10.3390/jof10010055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye SY, Zhang YB, Wu F, Liu HX. (2020) Multi-locus phylogeny reveals two new species of Exidia (Auriculariales, Basidiomycota) from China. Mycological Progress 19(9): 859–868. 10.1007/s11557-020-01601-8 [DOI] [Google Scholar]
- Zhang XC, Li YC, Wang YY, Xu Z, Zhao CL, Zhou HM. (2024) Xylodonasiaticus (Hymenochaetales, Basidiomycota), a new species of corticioid fungus from southern China. Phytotaxa 634(1): 1–15. 10.11646/phytotaxa.634.1.1 [DOI] [Google Scholar]
- Zhao CL, Wu ZQ. (2017) Ceriporiopsiskunmingensis sp. nov. (Polyporales, Basidiomycota) evidenced by morphological characters and phylogenetic analysis. Mycological Progress 16(1): 93–100. 10.1007/s11557-016-1259-8 [DOI] [Google Scholar]
- Zhao CL, Qu MH, Huang RX, Karunarathna SC. (2023) Multi‐gene phylogeny and taxonomy of the wood‐rotting fungal genus Phlebia sensu lato (Polyporales, Basidiomycota). Journal of Fungi (Basel, Switzerland) 9(3): 320. 10.3390/jof9030320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Wu YD, Yang ZR, Liu HG, Wu F, Dai YC, Yuan Y. (2024) Polypore funga and species diversity in tropical forest ecosystems of Africa, America and Asia, and a comparison with temperate and boreal regions of the Northern Hemisphere. Forest Ecosystems 11: 100200. 10.1016/j.fecs.2024.100200 [DOI]
- Zhou HM, Gu ZR, Zhao CL. (2024) Molecular phylogeny and morphology reveal a new species of Asterostroma from Guizhou Province, China. Phytotaxa 634(1): 1–15. 10.11646/phytotaxa.640.1.1 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the data that support the findings of this study are available in the main text.