Abstract
Peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α) is an inducible transcriptional coactivator protein involved in mitochondrial biogenesis and metabolism. PGC-1α exhibits a short half-life and low abundance, rendering its detection challenging by immunoblotting. This study compared the specificity and sensitivity of seven commercially available PGC-1α antibodies towards the full-length mouse PGC-1α1 isoform, using overexpression and knockdown in primary mouse hepatocytes. While all antibodies identified overexpressed PGC-1α1, only one detected endogenous PGC-1α1. This study demonstrates wide variability in sensitivity and specificity of PGC-1α antibodies and suggests that controls should be used to differentiate PGC-1α protein from non-specific bands.
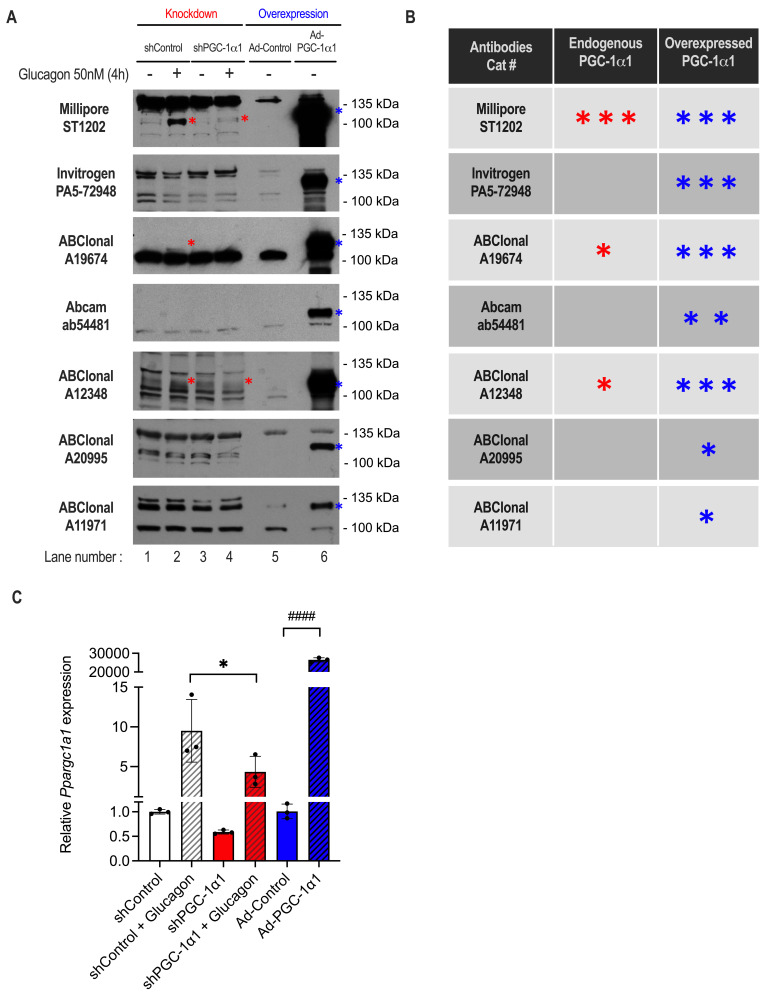
Figure 1. Comparison of PGC-1α antibody sensitivity and specificity for endogenous and overexpressed mouse PGC-1α1 protein by Western blot .
(A) Seven antibodies were tested on protein lysates from primary mouse hepatocytes following PGC-1α1 knockdown using shRNA (shPGC-1α1) or overexpression (Ad-PGC-1α1) using adenoviral vectors. Cells expressing shPGC-1α1 or a control shRNA (shControl) were treated with or without 50 nM glucagon for 4 hours to increase endogenous expression. As a positive control, mouse PGC-1α1 was overexpressed (Ad-PGC-1α1) and compared to vector alone (Ad-Control). For each blot, a red star is placed beside the band representing endogenous PGC-1α1 and a blue star beside overexpressed PGC-1α1. Unlabeled bands are either non-specific or other PGC-1α isoforms. (B) Comparison table of PGC-1α antibodies. Red and blue stars indicate whether the antibody recognizes endogenous and/or overexpressed PGC-1α1, respectively. Based on qualitative analysis of similarly exposed Western blot films, more stars mean greater sensitivity. (C) mRNA transcript levels of mouse Ppargc1a1 in a separate experiment where primary hepatocytes were treated as in (A). Data are mean±SD of biological replicates from one representative experiment of three independent trials. Statistical analysis was performed using a one-way ANOVA between shControl and shPGC-1α1 or shControl + glucagon and shPGC-1α1 + glucagon [p ≤ 0.05 (*)]. An unpaired t-test was performed between Ad-Control and Ad-PGC-1α1 [p ≤ 0.0001 (####)].
Description
Peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α) regulates genes involved in mitochondrial biogenesis, gluconeogenesis and thermogenesis in response to extracellular stimuli, including fasting, exercise, and cold (Goto et al., 2000; Pilegaard et al., 2003; Puigserver et al., 1998; Rhee et al., 2003; Yoon et al., 2001) . Full-length PGC-1α (also referred to as PGC-1α1 (Ruas et al., 2012) ) is the most well-studied since it was the first to be cloned and functionally described (Puigserver et al., 1998; Vega et al., 2000; Wu et al., 1999) . Multiple antibodies against PGC-1α have been developed and sold commercially for studies of expression, localization, and function in various tissues, cellular contexts and protein applications. However, the specificity and sensitivity of commercially available antibodies targeting PGC-1α are difficult to determine, in part due to multiple non-specific bands at a similar molecular weight that are difficult to identify in the absence of appropriate controls (overexpression and/or knockdown). PGC-1α also has a very short half-life and low abundance in most cell types, largely attributed to its rapid degradation through the ubiquitin-proteasome system (Olson et al., 2008; Sano et al., 2007; Trausch-Azar et al., 2010) , so high sensitivity techniques are needed to detect endogenous levels. Post-translational modifications of PGC-1α can also affect its protein stability, intracellular localization, and molecular weight (Housley et al., 2009; Jäger et al., 2007; Lerin et al., 2006; Puigserver et al., 2001; Rodgers et al., 2005; Rytinki & Palvimo, 2009; Teyssier et al., 2005) . Despite a predicted size of 90 kDa, multiple groups have shown that canonical PGC-1α (PGC-1α1) protein migrates between 110 and 120 kDa on SDS-PAGE gels (Gettys & Chang, 2019; Martínez-Redondo et al., 2016; Ruas et al., 2012; Zhang et al., 2009) . Since PGC-1α1 is often used as a general biomarker of mitochondrial biogenesis, it is important to identify which commercially available antibodies accurately and sensitively identify changing levels of PGC-1α1.
The purpose of this study was to evaluate the performance of commercially available PGC-1α antibodies. Primary hepatocytes were chosen as a model because they express canonical PGC-1α1 protein (Puigserver et al., 2003; Yoon et al., 2001) , yet have a moderate abundance compared to other cell types (i.e. oxidative muscle). To identify endogenous PGC-1α1 protein, PGC-1α1 was knocked down or overexpressed using adenoviral vectors expressing an shRNA (shPGC-1α1) or PGC-1α1 cDNA (Ad-PGC-1α1) respectively, alongside their appropriate control empty vectors (shControl or Ad-Control). As basal hepatic PGC-1α1 protein levels are low, shRNA-transduced hepatocytes were stimulated with glucagon as a fasting signal to induce PGC-1α1 expression (Lin et al., 2002, 2003; Yoon et al., 2001) . We then performed Western blot analysis for mouse PGC-1α1 using seven distinct antibodies and compared their relative abilities to detect both endogenous and overexpressed PGC-1α1 ( Figure 1A,B ). qPCR detecting canonical exons 1 and 2 of PGC-1α was also performed, illustrating the relative extent of knockdown and overexpression of Ppargc1a1 transcript levels in our model system ( Figure 1C ).
Our findings revealed that all the tested antibodies could detect overexpressed mouse PGC-1α1 (Ad-PGC-1α1), as shown by an intense band between 100 and 135 kDa, compared to vector alone lysates ( Figure 1A, lane 5 vs 6), albeit with varying sensitivity levels. Based on similar exposure times and antibody dilutions, the Millipore ST1202, Invitrogen PA5-72948, ABClonal A19674 and A12348 antibodies exhibited the highest sensitivity towards overexpressed PGC-1α1 ( Figure 1A,B ), while the least sensitive antibodies were ABClonal A20995 and A11971.
Using the positive control to identify the PGC-1α1 migration profile, we evaluated which antibodies were able to detect endogenous PGC-1α1 protein ( Figure 1A, lanes 1-4). All PGC-1α1 antibodies produced bands at various heights. However, when comparing the intensity of these bands across vehicle, glucagon-treated or knocked down conditions, only three antibodies showed the diminished intensity of a band around 110-120 kDa expected following knockdown by shPGC-1α1. Millipore ST1202, ABClonal A19674 and A12348 antibodies detected the glucagon-induced increases in endogenous PGC-1α1 ( Figure 1A, lane 1 vs 2, marked by a red star) which were also reduced by shPGC-1α1 ( Figure 1A, lane 3 vs 4). This suggests that the remaining signals on the Western blots were either non-specific bands or representative of other PGC-1α isoforms. Thus, in our model system, Invitrogen PA5-72948, Abcam ab54481, ABClonal A20995, and ABClonal A11971 antibodies failed to detect endogenous PGC-1α1, summarized in Figure 1B . It is noteworthy that for ABclonal A19674, the faint endogenous PGC-1α1 band was greatly obscured by a closely associated non-specific band ( Figure 1A, lane 2, marked by a red star). Overall, inclusion of appropriate controls was crucial to differentiate endogenous PGC-1α1 from non-specific bands with all commercial antibodies.
For accurate detection of the endogenous mouse PGC-1α1 isoform, we recommend inducing PGC-1α expression using stimuli (such as glucagon in liver) or an expression vector as a positive control for estimation of antibody sensitivity, relative expression levels and migration pattern in all biological contexts. The use of a negative control (knockdown or knockout) is extremely useful to determine specificity of the chosen PGC-1α1 antibody. Our findings suggest that the Millipore ST1202 antibody has both high specificity and sensitivity for PGC-1α1 protein.
In conclusion, this study underscores the importance of validating PGC-1α antibodies. Four of the seven commercially available PGC-1α antibodies tested lacked the sensitivity to detect endogenous PGC-1α1 in primary mouse hepatocytes, whereas all could detect vastly overexpressed levels, but to highly varying degrees. Given the potential for cell-type and species-specific variability in results, it is essential to validate PGC-1α antibodies using appropriate controls.
Methods
Hepatocyte cell isolation and culture
Primary hepatocytes from 2–3 chow-fed (Teklad Global 18% Protein Rodent Diet, TD.01432) C57/B6N male mice (aged 10–12 weeks) were isolated using collagenase (Liberase, Roche) perfusion and Percoll gradient purification. Cells were plated at a density of 450,000/well in 6-well plates in DMEM (Wisent, #319-005-CL) Plating Medium containing 10% FBS (Wisent, #090-150), 2 mM sodium pyruvate (Wisent, #600-110-EL), 1 µM dexamethasone (Sigma D4902), 1% penicillin/streptomycin (Wisent, #450-201-EL), and 0.1 µM insulin (Sigma I-6634). After 2 hours, medium was exchanged with DMEM Maintenance Medium supplemented with 0.2% fat-free BSA (GenDEPOT, #A0100-010), 2 mM sodium pyruvate, 1% penicillin/streptomycin, 100 nM dexamethasone, and 1 nM insulin. The next day, primary hepatocytes were infected overnight with different adenoviral constructs in Maintenance Medium. Media was renewed 24 hours later.
Hepatocyte cell transduction
To knockdown mouse PGC-1α1, primary hepatocytes were transduced overnight with a silencing adenovirus pAdTrack-U6-shPGC-1α1 or shControl vector expressing a hairpin that does not recognize any known mouse or human mRNA. The vector expresses a GFP reporter transcribed by a separate CMV promoter. 48 hours post-transduction, primary hepatocytes were incubated overnight in DMEM medium containing 0.2% fat-free BSA, 2 mM sodium pyruvate, and 1% penicillin/streptomycin and stimulated with 50 nM of glucagon or vehicle (PBS) for 4 hours prior to cell lysis.
To overexpress mouse PGC-1α1, primary hepatocytes were transduced overnight in Maintenance Medium with an adenoviral vector pAdTrack-CMV expressing mouse PGC-1α1 or empty vector (expressing a GFP reporter transcribed by a separate CMV promoter). After 16 hours, the media was renewed and cells were incubated up to 48 hours.
Western blotting
Primary hepatocytes were lysed on ice in 100 µL RIPA buffer (150 mM sodium chloride, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris, pH 8.0) with protease and phosphatase inhibitors (ThermoFisher, Pierce Protease (A32963) and Phosphatase (A32957) Inhibitor Tablets). Total protein was quantified by DC protein assay (Bio-Rad, #5000113-5000114) as per the manufacturer’s instructions. Protein was denatured in 5x Laemmli sample buffer (10% sodium dodecyl sulfate, 25% 2-mercaptoethanol, 30% glycerol, 0.05% bromophenol blue, 292 mM Tris HCl pH 6.8) at 95°C for ten minutes before loading onto a 7.5% - 10% gradient acrylamide gel and electrophoresing at 200V in 1x running buffer (25 mM Tris, 190 mM glycine, 0,1% SDS). 40 µg of protein was loaded to visualize endogenous PGC-1α1 and 20 µg of protein was used for samples with overexpressed PGC-1α1. Protein was transferred onto a nitrocellulose membrane at 25 volts for 20 minutes in 1x transfer buffer (Biorad, #10026938) using the Trans-Blot® Turbo™ Transfer System (Biorad, #1704150EDU). Membranes were blocked for one hour at room temperature in TBS with 5% skim milk powder (20 mM Tris, 130 mM NaCl, 2.5mM EDTA pH 7.6), and incubated overnight in primary antibody (anti-PGC-1α, see table 1) diluted 1:1000 as per manufacturer’s instructions in TBS-0.1% Tween, 5% BSA (or TBS-0.1% Tween, 2% milk for Millipore ST1202 antibody) and incubated at 4°C with continuous rocking. Following 3 x 10 minutes washes in TBS-0.1% Tween at room temperature, membranes were incubated in secondary antibody conjugated with HRP in TBS-0.1% Tween, 5% milk for one hour at room temperature, washed 3 x 10 minutes in TBS-0.1% Tween and visualized using Clarity Western ECL Prime (Biorad, #1705061) reagents. Protein signals were revealed after 1 hour of Blu-Lite UHC™ film exposition (Ultident, #39-20810).
RNA extraction and qRT-PCR
Total RNA was extracted from primary hepatocytes using TRIzol reagent (Invitrogen, #15596018) as indicated by the manufacturer’s protocol. RNA concentration and purity were determined by Nanodrop 2000 spectrophotometer (Thermo Scientific). DNAse-treated RNA (1 µg) was reversed transcribed (High-Capacity cDNA Reverse Transcription Kit; Applied Biosystems) and RT-qPCR performed (G892, BlasTaq qPCR MasterMix; Applied Biological Materials) in a 20 µL reaction according to the manufacturer's protocol. The primer sequences used were as follows: Ppargc1a1 (forward primer, 5'-GGA CAT GTG CAG CCA AGA CTC T-3'; reverse, 5’-CAC TTC AAT CCA CCC AGA AAG CT -3’) and Hprt (forward primer, 5'-GGC CAG ACT TTG TTG GAT TTG-3'; reverse, 5’- TGC GCT CAT CTT AGG CTT TGT-3’). Relative gene expression levels were calculated using the ΔΔC t method and normalized to Hprt . All reactions were performed in triplicate, and data were analyzed using GraphPad Prism.
Reagents
We are happy to provide any vectors upon request.
|
Adenoviral plasmids and species |
shRNA sequences |
|
pAdTrack-U6-shControl |
CAACAGCCACAACGTCTATA |
|
pAdTrack-U6-shPGC-1α1 ( mus musculus ) |
ACTCTGTATGGAGTGACATA |
|
Adenoviral plasmids and species |
Company & catalog number |
|
pAdTrack-CMV-Control vector |
Addgene # 16405 |
|
pAdTrack-CMV-PGC-1α1 ( mus musculus ) |
Addgene # 14426 |
|
Source & catalog number |
Dilution |
Animal and clonality |
Immunogen |
Species reactivity |
Tested WB applications by the company |
|
Millipore, ST1202 |
1:1000, TBS-0.1% Tween, 2% milk |
Mouse Monoclonal |
A recombinant protein corresponding to amino acids 1-120 of mouse PGC-1α |
Mouse, human and rat |
Tested in mouse, human and rat |
|
Invitrogen, PA5-72948 |
1:1000 in TBS- 0.1% Tween, 5% BSA |
Rabbit Polyclonal |
A synthetic peptide corresponding to amino acids 400-550 of human PGC-1α protein (#Q9UBK2) |
Hamster, human, and rat, but published in human, mouse, and rat |
Tested in human and mouse cell lines: A-431, HeLa, HepG2, NIH/3T3, as well as in human adipose and skeletal muscle |
|
Abcam, ab54481 |
1:1000 in TBS- 0.1% Tween, 5% BSA |
Rabbit Polyclonal |
A synthetic peptide corresponding to amino acids 750-798 of human PGC-1α (#Q9UBK2) |
Human and mouse |
Tested in human and mouse |
|
ABClonal, A19674 |
1:1000 in TBS- 0.1% Tween, 5% BSA |
Rabbit Monoclonal |
A synthetic peptide corresponding to amino acids 699-798 of human PGC-1α/β (NP_037393.1) |
Mouse and rat |
Tested in mouse liver and rat heart |
|
ABClonal, A12348 |
1:1000 in TBS- 0.1% Tween, 5% BSA |
Rabbit Polyclonal |
A recombinant fusion protein corresponding to amino acids 610-798 of human PGC-1α (NP_037393.1, #Q9UBK2) |
Human, mouse and rat |
Tested in 293T cells, mouse skeletal muscle, mouse kidney and rat kidney |
|
ABClonal, A20995 |
1:1000 in TBS- 0.1% Tween, 5% BSA |
Rabbit Monoclonal |
A recombinant fusion protein corresponding to amino acids 340-480 of human PGC-1α (NP_037393.1, #Q9UBK2) |
Human, mouse and rat |
Tested in HeLa cells, mouse kidney, rat kidney and rat stomach |
|
ABClonal, A11971 |
1:1000 in TBS- 0.1% Tween, 5% BSA |
Rabbit Polyclonal |
A recombinant fusion protein corresponding to amino acids 610-710 of human PGC-1α (NP_037393.1, #Q9UBK2) |
Human, mouse and rat |
Tested in mouse kidney, mouse heart and rat heart |
Acknowledgments
Acknowledgments
We thank Dr. Nathalie Jouvet (Institut de Recherches Cliniques de Montréal, Qc, Canada) for her scientific input and review of this paper. We also extend our gratitude to the animal facility staff for their diligent care of the mice used in our study.
Funding Statement
This work was supported by a CIHR grant (PJT-168853) to J.L.E.
References
- Gettys Thomas W., Chang Ji Suk. 2019. An Optimized Immunoblotting Protocol for Accurate Detection of Endogenous PGC-1α Isoforms in Various Rodent Tissues. Nuclear Receptors: Methods and Experimental Protocols: 7. [DOI] [PubMed]
- Goto Masahide, Terada Shin, Kato Miyuki, Katoh Masao, Yokozeki Toshiko, Tabata Izumi, Shimokawa Teruhiko. cDNA Cloning and mRNA Analysis of PGC-1 in Epitrochlearis Muscle in Swimming-Exercised Rats. Biochemical and Biophysical Research Communications. 2000 Aug 1;274(2):350–354. doi: 10.1006/bbrc.2000.3134. [DOI] [PubMed] [Google Scholar]
- Housley Michael P., Udeshi Namrata D., Rodgers Joseph T., Shabanowitz Jeffrey, Puigserver Pere, Hunt Donald F., Hart Gerald W. A PGC-1α-O-GlcNAc Transferase Complex Regulates FoxO Transcription Factor Activity in Response to Glucose. Journal of Biological Chemistry. 2009 Feb 1;284(8):5148–5157. doi: 10.1074/jbc.m808890200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäger Sibylle, Handschin Christoph, St.-Pierre Julie, Spiegelman Bruce M. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1α. Proceedings of the National Academy of Sciences. 2007 Jul 17;104(29):12017–12022. doi: 10.1073/pnas.0705070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerin Carles, Rodgers Joseph T., Kalume Dario E., Kim Seung-hee, Pandey Akhilesh, Puigserver Pere. GCN5 acetyltransferase complex controls glucose metabolism through transcriptional repression of PGC-1α. Cell Metabolism. 2006 Jun 1;3(6):429–438. doi: 10.1016/j.cmet.2006.04.013. [DOI] [PubMed] [Google Scholar]
- Lin Jiandie, Puigserver Pere, Donovan Jerry, Tarr Paul, Spiegelman Bruce M. Peroxisome Proliferator-activated Receptor γ Coactivator 1β (PGC-1β), A Novel PGC-1-related Transcription Coactivator Associated with Host Cell Factor. Journal of Biological Chemistry. 2002 Jan 1;277(3):1645–1648. doi: 10.1074/jbc.c100631200. [DOI] [PubMed] [Google Scholar]
- Lin Jiandie, Tarr Paul T., Yang Ruojing, Rhee James, Puigserver Pere, Newgard Christopher B., Spiegelman Bruce M. PGC-1β in the Regulation of Hepatic Glucose and Energy Metabolism. Journal of Biological Chemistry. 2003 Aug 1;278(33):30843–30848. doi: 10.1074/jbc.m303643200. [DOI] [PubMed] [Google Scholar]
- Martínez-Redondo Vicente, Jannig Paulo R., Correia Jorge C., Ferreira Duarte M.S., Cervenka Igor, Lindvall Jessica M., Sinha Indranil, Izadi Manizheh, Pettersson-Klein Amanda T., Agudelo Leandro Z., Gimenez-Cassina Alfredo, Brum Patricia C., Dahlman-Wright Karin, Ruas Jorge L. Peroxisome Proliferator-activated Receptor γ Coactivator-1 α Isoforms Selectively Regulate Multiple Splicing Events on Target Genes. Journal of Biological Chemistry. 2016 Jul 1;291(29):15169–15184. doi: 10.1074/jbc.m115.705822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson Brian L., Hock M. Benjamin, Ekholm-Reed Susanna, Wohlschlegel James A., Dev Kumlesh K., Kralli Anastasia, Reed Steven I. SCF Cdc4 acts antagonistically to the PGC-1α transcriptional coactivator by targeting it for ubiquitin-mediated proteolysis . Genes & Development. 2008 Jan 15;22(2):252–264. doi: 10.1101/gad.1624208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilegaard Henriette, Saltin Bengt, Neufer P. Darrell. Exercise induces transient transcriptional activation of the PGC‐1α gene in human skeletal muscle. The Journal of Physiology. 2003 Feb 1;546(3):851–858. doi: 10.1113/jphysiol.2002.034850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigserver Pere, Rhee James, Donovan Jerry, Walkey Christopher J., Yoon J. Cliff, Oriente Francesco, Kitamura Yukari, Altomonte Jennifer, Dong Hengjiang, Accili Domenico, Spiegelman Bruce M. Insulin-regulated hepatic gluconeogenesis through FOXO1–PGC-1α interaction. Nature. 2003 May 18;423(6939):550–555. doi: 10.1038/nature01667. [DOI] [PubMed] [Google Scholar]
- Puigserver Pere, Rhee James, Lin Jiandie, Wu Zhidan, Yoon J.Cliff, Zhang Chen-Yu, Krauss Stefan, Mootha Vamsi K, Lowell Bradford B, Spiegelman Bruce M. Cytokine Stimulation of Energy Expenditure through p38 MAP Kinase Activation of PPARγ Coactivator-1. Molecular Cell. 2001 Nov 1;8(5):971–982. doi: 10.1016/s1097-2765(01)00390-2. [DOI] [PubMed] [Google Scholar]
- Puigserver Pere, Wu Zhidan, Park Cheol Won, Graves Reed, Wright Margaret, Spiegelman Bruce M. A Cold-Inducible Coactivator of Nuclear Receptors Linked to Adaptive Thermogenesis. Cell. 1998 Mar 1;92(6):829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- Rhee James, Inoue Yusuke, Yoon J. Cliff, Puigserver Pere, Fan Melina, Gonzalez Frank J., Spiegelman Bruce M. Regulation of hepatic fasting response by PPARγ coactivator-1α (PGC-1): Requirement for hepatocyte nuclear factor 4α in gluconeogenesis. Proceedings of the National Academy of Sciences. 2003 Mar 21;100(7):4012–4017. doi: 10.1073/pnas.0730870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers Joseph T., Lerin Carlos, Haas Wilhelm, Gygi Steven P., Spiegelman Bruce M., Puigserver Pere. Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature. 2005 Mar 1;434(7029):113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- Ruas Jorge L., White James P., Rao Rajesh R., Kleiner Sandra, Brannan Kevin T., Harrison Brooke C., Greene Nicholas P., Wu Jun, Estall Jennifer L., Irving Brian A., Lanza Ian R., Rasbach Kyle A., Okutsu Mitsuharu, Nair K. Sreekumaran, Yan Zhen, Leinwand Leslie A., Spiegelman Bruce M. A PGC-1α Isoform Induced by Resistance Training Regulates Skeletal Muscle Hypertrophy. Cell. 2012 Dec 1;151(6):1319–1331. doi: 10.1016/j.cell.2012.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rytinki Miia M., Palvimo Jorma J. SUMOylation Attenuates the Function of PGC-1α. Journal of Biological Chemistry. 2009 Sep 1;284(38):26184–26193. doi: 10.1074/jbc.m109.038943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano Motoaki, Tokudome Satori, Shimizu Noriaki, Yoshikawa Noritada, Ogawa Chie, Shirakawa Kousuke, Endo Jin, Katayama Takaharu, Yuasa Shinsuke, Ieda Masaki, Makino Shinji, Hattori Fumiyuki, Tanaka Hirotoshi, Fukuda Keiichi. Intramolecular Control of Protein Stability, Subnuclear Compartmentalization, and Coactivator Function of Peroxisome Proliferator-activated Receptor γ Coactivator 1α. Journal of Biological Chemistry. 2007 Aug 1;282(35):25970–25980. doi: 10.1074/jbc.m703634200. [DOI] [PubMed] [Google Scholar]
- Teyssier Catherine, Ma Han, Emter Roger, Kralli Anastasia, Stallcup Michael R. Activation of nuclear receptor coactivator PGC-1α by arginine methylation. Genes & Development. 2005 Jun 15;19(12):1466–1473. doi: 10.1101/gad.1295005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trausch-Azar Julie, Leone Teresa C., Kelly Daniel P., Schwartz Alan L. Ubiquitin Proteasome-dependent Degradation of the Transcriptional Coactivator PGC-1α via the N-terminal Pathway. Journal of Biological Chemistry. 2010 Dec 1;285(51):40192–40200. doi: 10.1074/jbc.m110.131615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega Rick B., Huss Janice M., Kelly Daniel P.. 2000. The Coactivator PGC-1 Cooperates with Peroxisome Proliferator-Activated Receptor α in Transcriptional Control of Nuclear Genes Encoding Mitochondrial Fatty Acid Oxidation Enzymes. Molecular and Cellular Biology. 20: 1868. [DOI] [PMC free article] [PubMed]
- Wu Zhidan, Puigserver Pere, Andersson Ulf, Zhang Chenyu, Adelmant Guillaume, Mootha Vamsi, Troy Amy, Cinti Saverio, Lowell Bradford, Scarpulla Richard C., Spiegelman Bruce M. Mechanisms Controlling Mitochondrial Biogenesis and Respiration through the Thermogenic Coactivator PGC-1. Cell. 1999 Jul 1;98(1):115–124. doi: 10.1016/s0092-8674(00)80611-x. [DOI] [PubMed] [Google Scholar]
- Yoon J. Cliff, Puigserver Pere, Chen Guoxun, Donovan Jerry, Wu Zhidan, Rhee James, Adelmant Guillaume, Stafford John, Kahn C. Ronald, Granner Daryl K., Newgard Christopher B., Spiegelman Bruce M. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature. 2001 Sep 1;413(6852):131–138. doi: 10.1038/35093050. [DOI] [PubMed] [Google Scholar]
- Zhang Yubin, Huypens Peter, Adamson Aaron W., Chang Ji Suk, Henagan Tara M., Boudreau Anik, Lenard Natalie R., Burk David, Klein Johannes, Perwitz Nina, Shin Jeho, Fasshauer Mathias, Kralli Anastasia, Gettys Thomas W. Alternative mRNA Splicing Produces a Novel Biologically Active Short Isoform of PGC-1α. Journal of Biological Chemistry. 2009 Nov 1;284(47):32813–32826. doi: 10.1074/jbc.m109.037556. [DOI] [PMC free article] [PubMed] [Google Scholar]