Abstract
Background
Calcium-dependent protein kinase (CDPK) plays a key role in cotton tolerance to abiotic stress. However, its role in cotton heat stress tolerance is not well understood. Here, we characterize the GhCDPK gene family and their expression profiles with the aim of identifying CDPK genes associated with heat stress tolerance.
Results
This study revealed 48 GhCDPK members in the cotton genome, distributed on 18 chromosomes. Tree phylogenetic analysis showed three main clustering groups of the GhCDPKs. Cis-elements revealed many abiotic stress and phytohormone pathways conserved promoter regions. Similarly, analysis of the transcription factor binding sites (TFBDS) in the GhCDPK genes showed many stress and hormone related sites. The expression analysis based on qRT-PCR showed that GhCDPK16 was highly responsive to high-temperature stress. Subsequent protein-protein interactions of GhCDPK16 revealed predictable interaction with ROS generating, calcium binding, and ABA signaling proteins. Overexpression of GhCDPK16 in cotton and Arabidopsis improved thermotolerance by lowering ROS compound buildup. Under heat stress, GhCDPK16 transgenic lines upregulated heat-inducible genes GhHSP70, GHSP17.3, and GhGR1, as demonstrated by qRT-PCR analysis. Contrarily, GhCDPK16 knockout lines in cotton exhibited an increase in ROS accumulation. Furthermore, antioxidant enzyme activity was dramatically boosted in the GhCDPK16-ox transgenic lines.
Conclusions
The collective findings demonstrated that GhCDPK16 could be a viable gene to enhance thermotolerance in cotton and, therefore, a potential candidate gene for improving heat tolerance in cotton.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-024-05563-x.
Keywords: Cotton, CDPK gene family, Heat stress, Tolerance
Background
Increasing high temperatures is an all-encompassing issue worldwide. In contrast to humans, plants are sessile, which limits their capacity to withstand the negative impacts of unfavorable environmental circumstances that lead to improper growth and development. High temperature is an unfavorable factor that not only impairs photosynthetic activity and negatively affects cell division and growth but also reduces the yield of crops, threatens food security, and endangers human life [1]. Plants undergo a series of morpho-anatomical and physiochemical processes when exposed to extreme high temperatures, resulting in high levels of reactive oxygen species (ROS) in cells and the accumulation of oxides such as H2O2 and malondialdehyde (MDA), exposing plants to oxidative damage during oxidative stress [2].
To re-stand the continual consequences of stress, plants engage in natural defensive cellular and molecular processes. The molecular mechanism of heat stress responses in plants is well understood. An existing library of genes linked to heat stress have been found and classified, including RNA helicases, heat shock proteins (Hsps), and ring finger proteins, which are involved in heat stress tolerance [3–5]. Non-coding RNAs, including as microRNAs (miRNAs) and long non-coding RNAs (lncRNAs), are also thought to influence heat stress responses [2, 6]. Furthermore, regulatory proteins, including CDPKs [7], wall associated receptor-like protein kinases (RLKs) [8], and mitogen-activated protein kinases are crucial for controlling signal transduction in heat-stressed conditions.
Enhancing plant adaptability to environmental stress, the mechanisms regulated by stimulation and subsequent signal transduction often trigger diverse chemical, molecular, biochemical, and physiological changes. Moreover, during stress encounters, plant cells employ the well-known calcium-dependent protein kinase (CDPK) sensor protein family to launch quick signal transduction pathways by inducing phosphorylation cascades [9]. A number of significant families of calcium ion binding proteins, such as calmodulin (CaM), CAM-like proteins (CML), calcineurin B-like proteins (CBL), and Ca2+-dependent protein kinases (CDPK), have also been identified in plants [9, 10]. However, the CDPK family is the largest calcium-sensing family exclusive to plants, protozoa, oomycetes, and green algae [11]. In addition, they are encoded by a wide-ranging gene family that is important for the growth and development of plants, as well as their ability to resist biotic and abiotic stressors [9, 12]. For instance, CDPKs are highly responsive to cold stress in rice seedling stem tissues, involved in light signaling, positively regulate drought, and salt tolerance [13, 14]. Concerning high temperature tolerance, reports have shown that Arabidopsis AtCPK3, which is Ca2+/CaM-dependent, activates HSF to trigger the expression of downstream HSPs that enhance heat tolerance in response to high-temperature stress [15]. ZmCDPK7 is a heat-stress protein kinase involved in ABA signaling and heat-stress tolerance in maize [7], VaCPK29 is involved in grapevine responses to heat stress [16], while GhCDPK60 positively regulates drought stress tolerance in both transgenic Arabidopsis and cotton by controlling proline and ROS levels [17].
As a cash crop of global importance, cotton (Gossypium hirsutum L.) is crucial for national economic development [18]. Cotton thrives in warm temperatures and grows best at high temperatures ranging from 28 to 35 °C. However, temperatures over 35 °C harm cotton growth causing anther dehiscence, reduced pollen activity, and hampered reproductive process [4]. China, India, Pakistan, and Uzbekistan are the top cotton-producing nations, accounting for 80% of the world’s cotton output [19, 20]. Nevertheless, regions with heavy cotton farming often experience temperatures exceeding 40 °C, leading to significant production losses per unit [19]. As a result, screening and breeding plant species for heat tolerance is crucial.
Previous research has identified CDPK in the cotton genome and its impact on fiber development and abiotic stress tolerance [17, 21]. Although the role of the CDPK family in regulating abiotic stress tolerance is well understood, its functional significance and characterization under heat stress are still unclear. To address this gap, we used various bioinformatics techniques, such as phylogenetic analysis, protein-protein interaction analysis, chromosomal distribution, exon-intron structures, collinearity analysis, and identification of cis-regulatory elements and binding sites for transcription factors (TF), to characterize the cotton CDPK protein in the cotton genome. Out of the 48 GhCDPK genes that were identified, GhCDPK16 was found to be associated with heat stress tolerance and was selected for further investigation. Overexpression of GhCDPK16 in Arabidopsis facilitated additional functional roles. The study revealed new information about cotton’s ability to withstand heat stress and indicated that GhCDPK16 could be a promising candidate for genetic improvement.
Methods
Databases and identification of CDPK gene families
The upland cotton genome-wide database was obtained from the website (http://mascotton.njau.edu.cn) [22]. The Arabidopsis genomic database source was obtained at the website (http://www.arabidopsis.org/). Using DNATOOLS software, a local database of the complete genome sequences of Arabidopsis thaliana, Gossypium hirsutum, Oryza sativa, and Zea mays was established. CDPK family genes were screened in the local database using TblastN (E-value = 0.001) for comparison against the structural domain of CDPK’s kinase (PF00069. 16) in A. thaliana. Using the Pfam (http://pfam.xfam.org/) and NCBI (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) databases, all downloaded sequences were subjected to domain search to screen the CDPK sequence gene signature domains (Pkinase, EF-hand). The confirmed CDPK sequences were subjected to multiple sequence alignment using the ClustalW tool in MEGA 6.0 [23].
Physicochemical properties and molecular structure
The ExPAsy (http://www.expasy.org/) online tool was utilized to examine the amino acid sequence, isoelectric point (PI), molecular weight (MW) of the protein, and number of exons. Prediction of N-terminal myristoylation was carried out using the Myristoylator tool (https://web.expasy.org/myristoylator/) [24]. Subcellular localization prediction of proteins was performed using Cell-PLoc 2.0 software (http://www.Csbio.sjtu.Edu.cn/bioinf/Cell-PLoc-2/).
Cis-acting elements of CDPK promoter
For the prediction of cis-acting elements in the promoter region of CDPK genes in G. hirsutum, a sequence located 2000 bp upstream of the coding sequence start site was analyzed using the PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html) online website.
Phylogenetic, gene structure, motif pattern and gene duplication of CDPKs
To conduct tree phylogeny, 152 CDPK protein sequences were used from four plant species including G. hirsutum (48), A. thaliana (34), O. sativa (30), and Z. mays (40). The MEGA6.0 software was used to perform multiple sequence alignment by utilizing the ClustalW 1.8.1 tool. The phylogenetic tree was constructed using the neighboring grouping method, and the p-distance, Jones-Taylor-Tornton (JTT) model, and Poisson model were employed for this purpose. Nonparametric bootstrapping values of 1000 replicates were used to ensure the reliability of the constructed tree [9]. The structure of exons and introns in the CDPK family of G. hirsutum was obtained by using the GSDS online tool (http://gss.cbi.pku.edu.cn/). Based on the protein sequence obtained, the MEME online analysis tool (http://meme.sdsc.edu/) was used to analyze the motif pattern of the CDPK family protein in G. hirsutum [25]. The Plant Genome Duplication Database (PGDD: http://chibba.agtec.uga.edu/duplication/) was used to extract synonymous (Ks) and nonsynonymous (Ka) substitution rates, followed by KaKs computation using the KaKs_Calculator2.0 of Tbtools as previously described [24].
Collinearity, synteny and chromosome localization
The duplication and intraspecific covariance of GhCDPK family members were analyzed using the one-step MCSanX-Super Fast function in TBtools [26]. The evolutionary relationship between pairs of CDPK genes was analyzed using TBtools advanced circos function. Furthermore, to map the chromosomal localization information of G. hirsutum and three other species (A. thaliana, O. sativa and Z. mays), the collinearity connection was established using MapInspect software (https://mapinspect.software.informer.com).
Transcription factor binding site analysis of the GhCDPK family
Putative TFBDS of the GhCDPK family were determined using the G. hirsutum genome database. The predicted TF binding sites in the target genes were analyzed using the Plant Transcriptional Regulatory Map website (http://plantregmap.gao-lab.org/) [27].
Functional network interaction
The online STRING 10 (https://string-db.org/) tool was used to predict the interaction network between GhCDPK16 and interactive protein gene in A. thaliana and G. hirsutum based on text mining, databases, co-expression, neighborhood, gene fusion, co-occurrence, and experimental evidence [28].
Plant materials and heat treatment
The heat-sensitive upland cotton cultivar “FuZi mian” (18YZ589) was employed in this investigation. Seedlings were cultivated in a greenhouse, under optimal growth conditions of 24 °C with 16 h of light and 8 h of darkness. At the two true-leaf stages, seedlings of similar growth were selected and subjected to high-temperature stress treatment at 42 °C. Different samples were collected at different time intervals during a heat stress treatment. The collection times included 0 h of no stress, as well as 1, 2, 4, 8, and 12 h of heat stress. A. thaliana (Columbia) was grown in a conditioned greenhouse with a 16-hour/8-hour photoperiod at 22 °C and a relative humidity of 60%. Seeds of both wild-type and transgenic GhCDPK16-ox Arabidopsis lines were grown on MS plates to observe their response to heat stress treatments. The seeds were treated at 45 °C for 1 h and were then allowed to recover for 5 days for further observation. Next, 7-day-old seedlings of GhCDPK16-ox Arabidopsis lines and wild-type grown on MS plates were treated at 45 °C for 2 h and were then allowed to recover for 7 days for observation and survival rate was further analyzed. Afterwards, 4-week-old GhCDPK16-ox Arabidopsis lines and wild-type were grown in a soil medium and then subjected to 45 °C heat treatment for 24 h and the phonological changes were observed to study the function of genes in response to stress.
SgRNA design, pRGEB32-GhCDPKR16-Cas9 construction and Agrobacterium-mediated transient expression in cotton
The nucleic acid sequence of GhCDPK16 and the corresponding NGG structure were discovered in the CDS. The sgRNA primers for GhCDPK16 were designed using (http://cbi.hzau.edu.cn/crispr) with the forward primer as 5’-TTCTATGGAGAAACTGAGCA-3’ and the reverse primer as 5’-CTAAAGGTCATTTTTCAGGAG-3’. By one-step cloning method, the CRISPR-Cas9 knockout vector pRGEB32 [29] was enzymatically sliced with the restricted intracytoplasmic enzyme BsaI, and the long fragment containing tRNA + target + gDNA was linked to the linear vector. The cloned vector was introduced into the DH5a competent cell; positive clones were selected, sequenced, and gene-editing expression vector pRGEB32-GhCDPK16-Cas9 was obtained.
The vectors (pCAMBIA1305-GhXTH16 and pRGEB32-GhCDPK16-Cas9) were transformed into Agrobacterium GV3101, cultured at 28 °C in 200 µl Luria Bertani (LB) liquid medium with an appropriate amount of rifampicin and kanamycin until the OD600 = 1.5 and the bacterium was collected at 10,000 r/min. The mixtures were then resuspended with a transformation mixture containing 100 µmol/L acetylcholine (AS) and 10 mmol / LMgSO4 to obtain the conversion liquid at an OD600 value of 0.6. Cultivated cotton seedlings were selected and sterilized with 75% ethanol and washed with distilled water to clear the residual alcohol. Subsequently, syringes without the needle were used to inject the vacuum side of the leaf blades, and then the injection sides were properly cleaned with a cotton swab dipped in distilled water. Finally, the injected cotton plants were cultured at 25 °C with a light and dark period of 16 h/ 8 h for 2–3 days. Each treatment consisted of at least 5 seedlings. Successfully transformed plants were validated through qRT-PCR and RT-PCR analysis.
Vector construction and arabidopsis genetic transformation
Vector and genetic transformation was carried out as previously described by [30] with few modifications. Briefly, the coding sequence of GhCDPK16 was amplified using KOD One TM PCR Master Mix (Toyobo, Tokyo, Japan) with gene-specific forward and reverse primers ligated into the pCAMBIA1305 vector (Table S3). Next, the expression vector pCambia1305-GhCDPK16 was introduced into Agrobacterium tumefaciens strain EHA105. The ecotype Col-0 Arabidopsis was used in genetic transformation by the floral dip method. Then the harvested T0 generation seeds were selected on 1/2 Murashige and Skoog (MS) medium with 50 mg/L kanamycin, and the resistant plants were further validated using PCR. Single-copy lines with a segregation ratio of 3:1 were selected and planted until the T3 generation. The transgenic GhCDPK16-ox Arabidopsis lines were grown in a growth chamber with a 16-h light/8-h dark scheme, and the growth temperature was set at around 22 °C.
RNA extraction and real-time quantitative PCR
Total RNA was isolated from fresh 0.1 g samples using the RNAprep Pure Plant Plus Kit (polysaccharides and polyphenolics-rich) according to the manufacturer’s instructions. First-strand cDNAs were synthesized from 1 µg of total RNA using the StarScript III All-in-one RT Mix with gDNA Remover. Then, qRT-PCR analysis was performed from three biological replicates as previously described by [20]. The GhUBQ gene was used as a reference and the relative expression levels were calculated using the 2−ΔΔCt method. The primer sequences used for the experiments are shown in Table S2.
H2O2 and MDA determination and histochemical staining
H2O2 content was measured based on the kit instruction method described previously by [31]. For MDA content determination, 0.5 g of plant leaves was weighed and ground together with 2 mL of phosphoric acid medium solution (PBS) on ice. Afterwards, the mixture was centrifuged at 8000 g for 10 min at a temperature of 4 °C. Next, the supernatant was extracted, and 3 mL of 5% thiobarbital acid was added, then placed in a water bath to boil for 10 min, cooled under 4 °C for 2 to 5 min, then centrifuged again to extract the supernatant. With water as the control, the absorbance was measured at 532 nm and 600 nm. For histochemical staining, leaves were chosen at random and submerged in diaminobenzidine (DAB) and nitro-blue tetrazolium (NBT) solutions, respectively. Chlorophyll was removed by using 95% ethanol, followed by soaking in 70% glycerin for microscopic observation and photography. This entire process was repeated three times and the intensity was quantified using ImageJ software [32].
Determination antioxidant enzyme parameters
To measure the levels of antioxidant enzyme activity, a 0.1 g sample of leaf was ground with 1 mL of ice cooled enzyme buffer. The resulting mixture was then subjected to centrifugation at 12,000 g for 10 min at 4 °C. The supernatant obtained after centrifugation was used for determining the enzyme activity. The enzyme activities of APX, CAT, SOD, and POD were analyzed using different methods. APX was analyzed by measuring the decrease in optical density at A290. CAT was analyzed using the ultraviolet absorption method, while nitroblue tetrazolium NBT and guaiacol methods were used to analyze SOD and POD, respectively [33, 34]. To conduct ascorbate and glutathione content assays, a leaf sample of about 0.1 g was ground into a fine powder using liquid nitrogen and 1 mL of 0.2 M HCl was used for extraction. After centrifugation at 12,000 g for 10 min at a temperature of 4 °C, 0.5 mL of supernatant was mixed with 100 µL of 0.2 M phosphate buffer (pH 5.6). The mixture was then neutralized with 0.2 M NaOH to achieve a pH of 4–5. The extracts were neutralized and spectrophotometric assays were used to measure various compounds, including ascorbic acid (ASA), dehydroascorbic acid (DHA), glutathione (GSH), ascorbic acid/dehydroascorbate (AsA/DHA), glutathione disulfide (GSSG), and glutathione/oxidized glutathione (GSH/GSSG), following previously established methods [35].
Statistical analyses
Statistical analysis was performed using (Excel 2010; Microsoft Office 2010, Microsoft Corp., USA. Graphs were drawn using GraphPad Prism 5.0 (GraphPad Software, Inc., USA). Tukey’s post-hoc tests was used to determine which means differed significantly with P-values (*P < 0.05; to ***P < 0.01). RT-qPCR data were normalized to a normal distribution as previously described by [36].
Results
Identification, characteristics and structural analysis of GhCDPK gene family
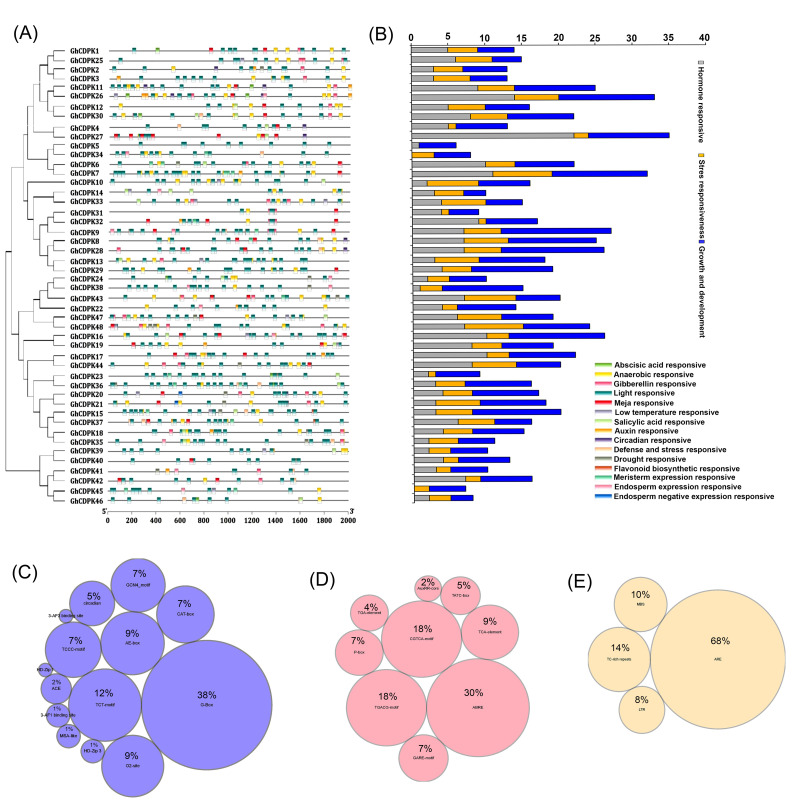
A total of 48 GhCDPKs were identified and classified as GhCDPK1 to GhCDPK48, grouped into three main clusters (Fig. S1A). All 48 putative CDPK genes were characterized using ExPASy software. The analysis included determination of the corresponding amino acid numbers, pI, molecular weight, number of exons, subcellular location, and respective EF-hands (Table 1). Based on the predicted gene biochemical properties, the amino acid lengths of GhCDPK proteins ranged from 487 to 655 amino acids. The molecular weight (MW) of these proteins ranged from 54.63 kD to 73.42 kD, and their isoelectric point (PI) varied from 5.06 to 6.96 (Table 1). Most of the GhCDPK genes exhibited 3–4 EF-hand motifs, whereas GhCDPK33, GhCDPK40, GhCDPK41, and GhCDPK42 contain only 2 EF-hand motifs (Table 1). Subsequent analysis revealed that 14 members possess potential N-myristoylation sites, including GhCDPK16, 17, 18, 20, 21, 22, 24, 35, 37, 39, 43, 44, and 48, while the remaining GhCDPK proteins were predicted to be non-myristoylated (Table 1). Based on subcellular localization prediction, all 48 GhCDPK genes are located in the nucleus (Table 1). Further results from the MEME software revealed 15 conserved motifs according to the ascending E-value of the alignment (Fig. 1B). In addition, motif 1–12 was present in all target GhCDPK genes, with the exception of GhCDPK44. Motif 13 was distributed in both subfamilies II and III and partially in subfamily I. The motif 14 was unique to subfamily I, whereas the motif 15 was present in subfamilies I and II but not in subfamily III (Fig. 2B). These findings show that the GhCDPK genes were both specific and conserved throughout evolution. Additionally, the differences in gene structure and conserved motifs indicate the relative conservation of the GhCDPK gene family during evolution and the diversity needed to adapt to the environment. The GhCDPK exon-intron structures showed that the intron numbers ranged from 6 to 7, while the exon numbers ranged from 7 to 9 (Fig. 1C).
Table 1.
Characteristics of calcium-dependent protein kinases (CDPKs) in cotton
| Gene name | Gene ID | Chr | Amino acid | MW (kDa) | PI | EF hands | N-terminal aa | N-myristoylation | Subcellular localization |
|---|---|---|---|---|---|---|---|---|---|
| GhCDPK1 | CotAD_21360 | At11 | 587 | 65.54 | 5.41 | 4 | MGNTCVG | No | Nucleus |
| GhCDPK2 | CotAD_33841 | sca | 573 | 64.03 | 5.87 | 3 | MGNTCVG | No | Nucleus |
| GhCDPK3 | CotAD_45432 | Dt1 | 579 | 64.64 | 5.77 | 4 | MGNTCVG | No | Nucleus |
| GhCDPK4 | CotAD_53506 | At12 | 605 | 67.32 | 5.66 | 4 | MGNACAG | No | Nucleus |
| GhCDPK5 | CotAD_34786 | At9 | 540 | 60.67 | 6.10 | 4 | MNKKIAG | No | Nucleus |
| GhCDPK6 | CotAD_05939 | Dt5 | 508 | 56.84 | 5.48 | 4 | MNNQSSS | No | Nucleus |
| GhCDPK7 | CotAD_36560 | At3 | 511 | 57.14 | 5.48 | 4 | MNNQSSS | No | Nucleus |
| GhCDPK8 | CotAD_66485 | sca | 568 | 63.62 | 5.78 | 4 | MGNTCRG | No | Nucleus |
| GhCDPK9 | CotAD_13019 | At8 | 607 | 67.88 | 5.76 | 3 | MGNTCRG | No | Nucleus |
| GhCDPK10 | CotAD_28390 | Dt12 | 552 | 61.94 | 5.26 | 4 | MGNTCRG | No | Nucleus |
| GhCDPK11 | CotAD_50659 | Dt2 | 648 | 71.66 | 5.32 | 4 | MGNVCAT | No | Nucleus |
| GhCDPK12 | CotAD_17127 | At6 | 610 | 68.28 | 5.16 | 4 | MGNNCFK | No | Nucleus |
| GhCDPK13 | CotAD_12120 | Dt13 | 487 | 54.63 | 5.06 | 4 | MRRAIDH | No | Nucleus |
| GhCDPK14 | CotAD_33393 | At13 | 523 | 58.56 | 6.51 | 3 | MGNTCLR | No | Nucleus |
| GhCDPK15 | CotAD_12430 | At9 | 532 | 59.52 | 5.38 | 4 | MGNLCSR | No | Nucleus |
| GhCDPK16 | CotAD_47701 | Dt8 | 534 | 60.25 | 5.98 | 3 | MGNCNSQ | Yes | Nucleus |
| GhCDPK17 | CotAD_70650 | sca | 538 | 60.51 | 5.28 | 4 | MGNCCSR | Yes | Nucleus |
| GhCDPK18 | CotAD_16448 | Dt1 | 513 | 57.31 | 5.27 | 4 | MGNCCSR | Yes | Nucleus |
| GhCDPK19 | CotAD_65954 | At3 | 519 | 58.60 | 5.62 | 3 | MGNCNGL | No | Nucleus |
| GhCDPK20 | CotAD_49677 | At6 | 523 | 58.55 | 5.69 | 4 | MGNCCSC | Yes | Nucleus |
| GhCDPK21 | CotAD_13649 | Dt6 | 524 | 58.57 | 5.44 | 4 | MGNCCSC | Yes | Nucleus |
| GhCDPK22 | CotAD_41076 | Dt9 | 524 | 58.82 | 5.98 | 4 | MGGCLTK | Yes | Nucleus |
| GhCDPK23 | CotAD_54795 | sca | 527 | 58.76 | 5.84 | 4 | MGNCCTR | No | Nucleus |
| GhCDPK24 | CotAD_73088 | sca | 537 | 60.46 | 6.14 | 4 | MGSCLTK | Yes | Nucleus |
| GhCDPK25 | CotAD_26791 | Dt11 | 583 | 64.87 | 5.40 | 4 | MGNTCVG | No | Nucleus |
| GhCDPK26 | CotAD_48046 | Dt12 | 600 | 66.47 | 5.48 | 4 | MGNVCAT | No | Nucleus |
| GhCDPK27 | CotAD_42895 | Dt12 | 593 | 65.99 | 5.44 | 4 | MGNACAG | No | Nucleus |
| GhCDPK28 | CotAD_56181 | Dt5 | 568 | 63.54 | 5.72 | 4 | MGNTCRG | No | Nucleus |
| GhCDPK29 | CotAD_33390 | At13 | 487 | 54.62 | 5.06 | 4 | MRRAIDH | No | Nucleus |
| GhCDPK30 | CotAD_31563 | Dt6 | 610 | 68.19 | 5.13 | 4 | MGNNCFK | No | Nucleus |
| GhCDPK31 | CotAD_34973 | At8 | 655 | 73.42 | 6.04 | 3 | MGNTCRG | No | Nucleus |
| GhCDPK32 | CotAD_34966 | At8 | 655 | 73.42 | 6.04 | 3 | MGNTCRG | No | Nucleus |
| GhCDPK33 | CotAD_12121 | Dt13 | 570 | 63.63 | 5.93 | 2 | MGNTCLG | No | Nucleus |
| GhCDPK34 | CotAD_16744 | Dt9 | 502 | 56.38 | 5.46 | 4 | MNKKIA | No | Nucleus |
| GhCDPK35 | CotAD_67913 | Dt1 | 513 | 57.26 | 5.27 | 4 | MGNCCTR | Yes | Nucleus |
| GhCDPK36 | CotAD_04751 | At4 | 527 | 58.72 | 6.09 | 4 | MGNCCTR | No | Nucleus |
| GhCDPK37 | CotAD_54317 | Dt9 | 532 | 59.51 | 5.38 | 4 | MGNLCSR | Yes | Nucleus |
| GhCDPK38 | CotAD_07025 | At2 | 537 | 60.54 | 6.06 | 4 | MGTCLTK | Yes | Nucleus |
| GhCDPK39 | CotAD_44026 | Dt8 | 550 | 62.29 | 6.60 | 3 | MGNCCAT | Yes | Nucleus |
| GhCDPK40 | CotAD_20538 | Dt6 | 531 | 60.23 | 6.32 | 2 | MGNCCVT | No | Nucleus |
| GhCDPK41 | CotAD_34534 | Dt1 | 506 | 56.85 | 6.06 | 2 | MGNCCAT | No | Nucleus |
| GhCDPK42 | CotAD_16484 | Dt1 | 529 | 59.39 | 6.96 | 2 | MGNCCAT | No | Nucleus |
| GhCDPK43 | CotAD_72552 | Dt13 | 536 | 60.42 | 5.98 | 4 | MGGCLTK | Yes | Nucleus |
| GhCDPK44 | CotAD_48692 | sca | 531 | 59.69 | 5.37 | 4 | MGNCCSR | Yes | Nucleus |
| GhCDPK45 | CotAD_13610 | Dt6 | 531 | 59.85 | 6.03 | 4 | MGNCCAT | No | Nucleus |
| GhCDPK46 | CotAD_20042 | At6 | 531 | 59.86 | 6.17 | 4 | MGNCCAT | No | Nucleus |
| GhCDPK47 | CotAD_13386 | Dt9 | 517 | 58.35 | 5.69 | 4 | MGICQSL | No | Nucleus |
| GhCDPK48 | CotAD_39468 | sca | 526 | 59.24 | 5.98 | 4 | MGLCQSL | Yes | Nucleus |
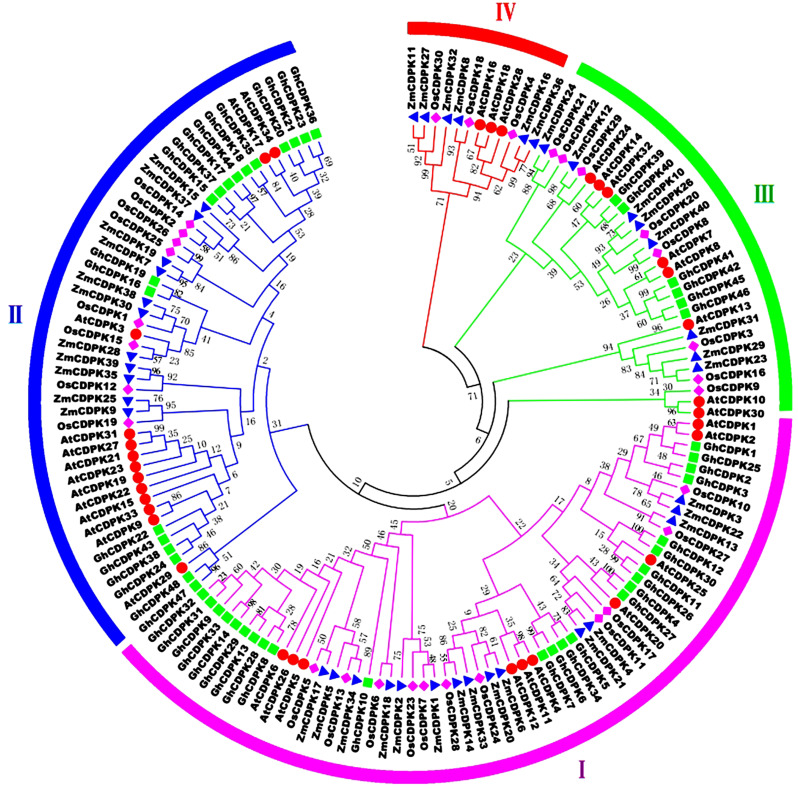
Fig. 1.
Phylogenetic tree of CDPK proteins in G.hirsutum, A.thaliana, Z.mays and O.sativa. The phylogenetic tree was derived using the MEGA6.0 software, neighboring grouping method with a bootstrapping value of 1000 replicates. Species abbreviations are listed as follows: Gh: cotton; At: Arabidopsis; Os: rice; Zm: maize
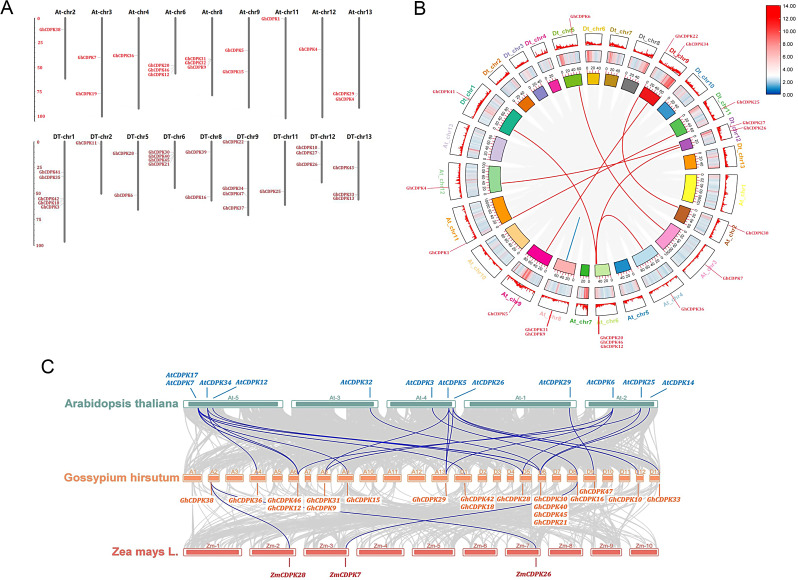
Fig. 2.
Chromosomal localization, gene duplication, and collinearity analysis of the GhCDPKs. A designated GhCDPK gene locations on the cotton genome. B Fragment duplication analysis of the CDPK gene family in G. hirsutum. The red lines indicate duplicated CDPK gene pairs. C Collinearity analysis of the CDPK gene family in cotton, Arabidopsis, and Zea mays
Phylogenetic relationships and divergence of cotton CDPK proteins
The conserved protein sequences of the GhCDPK and AtCDPK, ZmCDPK, and OsCDPK genes were used to build the phylogenetic tree (Fig. 1). Based on the similar classified results from Arabidopsis CDPKs classification, four distinct groups were identified. Among them, the GhCDPK gene family was primarily distributed in groups I, II, and III, with 24, 18, and 6 representative genes, respectively, whereas group IV contained no GhCDPK gene (Fig. 1). Moreover, group I had 10, 15, and 11 CDPKs of A. thaliana, Z. mays, and O. sativa, respectively. Group II showed 13, 11, and 8 CDPKs; group III revealed a similar number of CDPK representative genes; and group IV showed 3, 6, and 3 CDPK genes of AtCDPK, ZmCDPK, and OsCDPK, respectively (Fig. 1).
Chromosomal localization and gene-duplication analysis of GhCDPKs
To understand gene evolution of the GhCDPK family, the chromosome localization of CDPK genes was analyzed using MapInspect software. Based on the results from Fig. 2A, GhCDPK genes were distributed on eighteen chromosomes, namely, At2, At3, At4, At6, At8, At9, At11, At12, At13, Dt13, Dt1, Dt2, Dt5, Dt6, Dt8, Dt9, Dt11, and Dt12 (Fig. 2A). In addition, most GhCDPK genes were distributed on the chromosome arms, with a few genes, including GhCDPK4, GhCDPK7, GhCDPK5, and GhCDPK43, located in the heterochromatin regions around the centromeric repeats. The chromosome Dt1 contained the largest number of CDPK genes, followed by chromosomes (At6, Dt6, and Dt9) and chromosomes (At8, Dt12, and Dt13) having 5, 4, and 3 chromosomes, respectively. Two chromosomes were located on At3, At9, At13, Dt8, and Dt5, while a single chromosome was found on At2, At4, At11, Dt2, and Dt11 chromosomes (Fig. 2A). Moreover, for the evolutionary relationship among GhCDPK members, the (Ks), (Ka), and Ka/Ks ratio values for each duplication event were calculated (Table S1). The Ka values ranged from 0.002 to 3.366, while the Ks values ranged from 0.026 to 3.104 (Table S1). The Ka/Ks ratio of paralogous pairs including GhCDPK4/27, GhCDPK5/34, GhCDPK31/32, GhCDPK47/48, GhCDPK16/19, GhCDPK17/44, and GhCDPK41/42 segmental duplication was more than one, suggesting that this gene pair underwent positive selection while the other pairs of segmental duplications had Ka/Ks < 1, indicating a negative selection during evolution (Fig. 2B; Table S1). Further analysis was necessary to determine the GhCDPK family’s evolution and development. To ascertain the degree of collinearity among CDPK genes, we examined the genomes of maize, cotton, and Arabidopsis (Fig. 2C). Findings showed that there were 25 homologous gene pairs between the AtCDPK and GhCDPK genes, and 3 gene pairs between ZmCDPK and GhCDPK (Fig. 2C). The cotton CDPK gene shared more homology with dicotyledons than monocots, implying that the cotton and Arabidopsis CDPK families descended from a common ancestor.
Analysis of cis-acting elements of CDPK promoter
The cisregulatory elements of GhCDPK in the upstream sequences (~ 2000 bp) of their promoters linked to phytohormone signaling, plant growth and development, and response to biotic and abiotic stress were examined using the PlantCARE service online tool (Fig. 3; Additional file 1). Plant growth regulatory elements, including light responsiveness, endosperm expression responsiveness, circadian responsiveness, and flavonoid biosynthetic responsiveness, were predicted. The highest number of cisregulatory elements involved in stress, phytohormone signaling, and plant growth and development were found in group I GhCDPK genes (GhCDPK11, 26, 30, 27, 6, 7, 32, 9, 8, and 28) and group II GhCDPK genes (GhCDPK47, 48, 16, 19, 17, and 44) (Fig. 3B). Recorded plant growth and development elements were G-Box (38%), TCT-motif (12%), AE-Box (9%), O2-Site (7%), and CAT-Box (7%) (Fig. 3C). In addition, ABREs (30%), CGTCA-motif (18%), TGACG motifs (18%), and TGA elements (9%), and other phytohormone signaling elements, were also widely distributed in the GhCDPK gene promoter (Fig. 3D). Furthermore, stress response regulation elements for anaerobic responsiveness, defense and stress responsiveness, drought responsiveness, and low temperature responsiveness were found in the GhCDPK genes. Elements such as ARE (58%), TC-rich repeats (14%), LTR (8%), and MBS (10%) were the most abundant (Fig. 3E).
Fig. 3.
Cis-element prediction in the 2000 bp region upstream of GhCDPKs. A Different types of predicted cis regulatory elements in GhCDPK promoters. B The number of cis-regulatory elements involved in phytohormone signaling, plant growth and development, and abiotic and biotic stress. C-E The type and quantity of the GhCDPK cis elements responsive to plant growth and development, phytohormone signaling, and biotic and abiotic stress, respectively
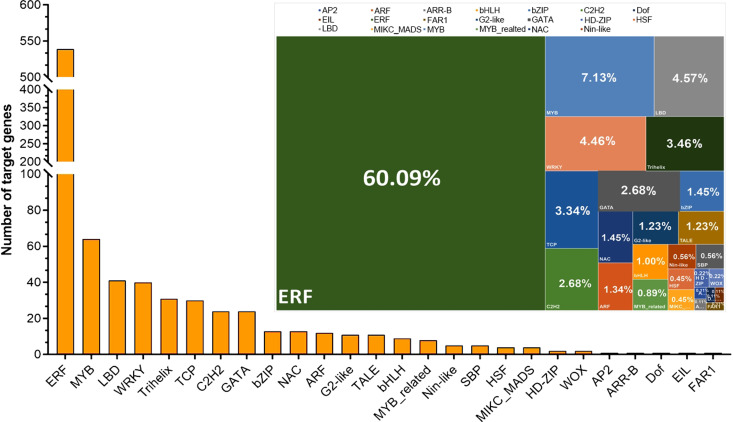
Predicted TF binding site analysis
TFBDS in CDPK genes were predicted. As displayed in Fig. 4, a total of 26 different TFs were observed in the GhCDPK family, with their length ranging from 7 to 29 bp. (Fig. 4; Additional file 2). Among them, the ethylene responsive transcription factor (ERFs) biding site constituted 60.09% of target sites in the GhCDPK family. Moreover, TFBDS, including LBD, C2H2, and MYB, were found to have a high interaction score with the GhCDPK gene family, indicating that a strong interaction might exist between TFs and the GhCDPK family proteins (Fig. 4). Although stress-responsive sites including ERF, MYB, NAC, TCP, Bzip, and WRKY were found in high quantity on target genes, hormone-related binding sites such as AP2, ARF, bHLH, and reproductive growth sites including ARR, MIKC_MADS, G2-like, and TALE were also recorded, suggesting that the GhCDPK family not only plays vital roles in the response to stress but may also perform some important functions in the hormone transduction and reproductive growth processes (Fig. 4).
Fig. 4.
Predicted transcription factor binding sites of GhCDPK members in the cotton genome. The tree map chart showed the percentage of the various transcription factors in each GhCDPK family gene
Heat stress responsive expression pattern of the GhCDPK family
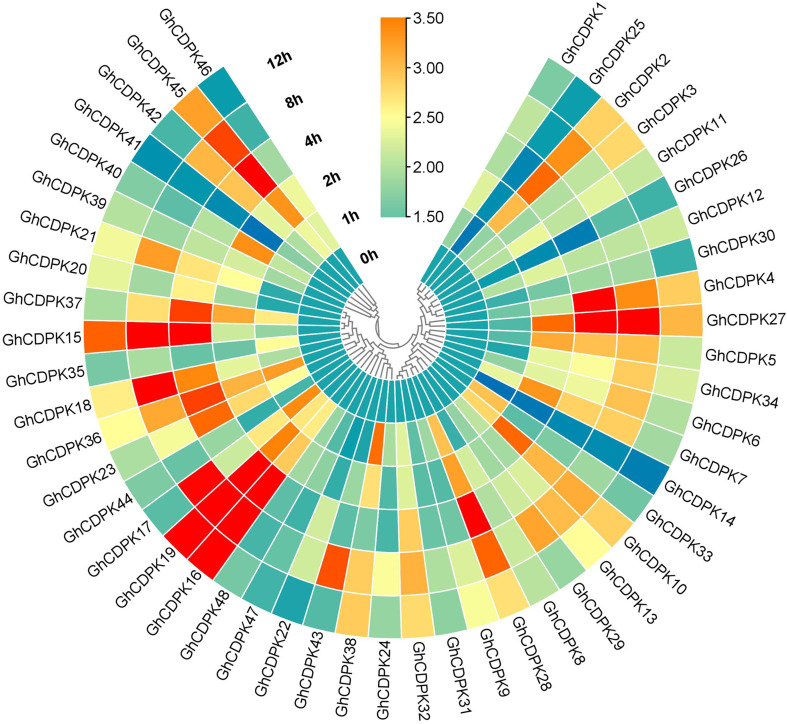
To deeply verify the roles of the GhCDPK family in response to heat stress, the expression levels of all 48 target genes were examined under 0, 1, 2, 4, 8, and 12 h of stress treatment (Fig. 5). Numerous GhCDPK genes exhibit increased expression in response to increased stress treatment, as seen in Fig. 5. For instance, following one and two hours of heat stress treatment, the expression patterns of GhCDPK45, GhCDPK40, GhCDPK37, GhCDPK38, and GhCDPK10 were responsively high; however, this response decreased as treatment levels increased (Fig. 5). After 4 h of heat stress treatment, target genes including GhCDPK45, GhCDPK37, GhCDPK15, GhCDPK18, GhCDPK36, GhCDPK23, GhCDPK19, GhCDPK16, GhCDPK28, GhCDPK27, GhCDPK4, and GhCDPK2 were highly upregulated, whereas GhCDPK45, GhCDPK15, GhCDPK18, GhCDPK17, GhCDPK19, GhCDPK16, and GhCDPK27 were significantly expressed after 8 h of stress treatment (Fig. 5). After 12 h of stress treatment, GhCDPK15, GhCDPK16, and GhCDPK19 were highly expressed. However, significant expressions were observed in GhCDPK16 and GhCDPK19 (Fig. 5). Contrarily, the expression levels of GhCDPK46, GhCDPK14, GhCDPK46, GhCDPK1, GhCDPK25, GhCDPK26, GhCDPK30, GhCDPK48 and GhCDPK47 showed no significant responses to heat stress treatments.
Fig. 5.
The expression profiles and functional network analysis of GhCDPK proteins. Circular heatmap of GhCDPK family genes at different heat stress time points
Predicted functional interaction network of GhCPK16 proteins
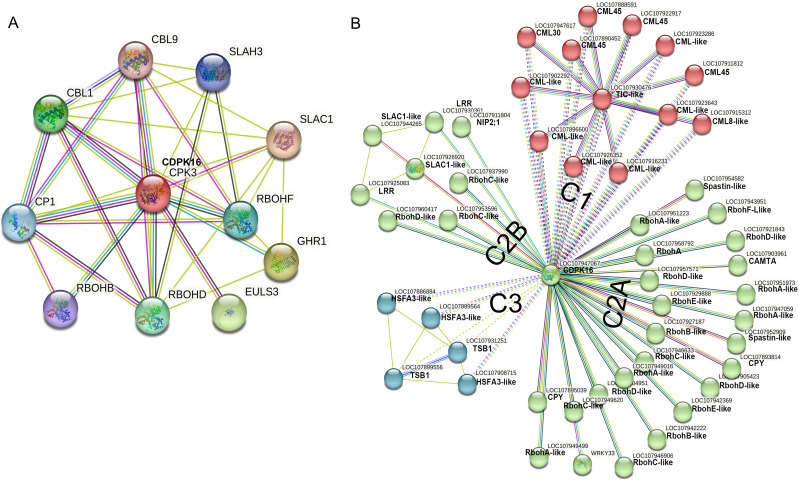
The expression pattern of the GhCDPK16 and GhCDPK19 genes validates a possible role in heat stress responses. However, the interacting role with other proteins remains unclear. Thus, an interaction analysis was carried out based on GhCDPK16 to investigate the interaction patterns in A. thaliana and G. hirsutum genome proteins, was required (Fig. 6). The results showed that various ROS generating protein genes in the A. thaliana genome interacted with GhCDPK16 (Fig. 6A) such as RbohD, RbohB and RbohF. In addition, the guard cell S-type anion channel (SLAC1), leucine-rich repeat receptor-like protein kinases (LRR), Ricin B-like lectin (EULS3) and calcineurin B-like (CBL1 and 9) proteins were found to interact with the putative GhCDPK protein in reference to A. thaliana genome (Fig. 6A). In relation to G. hirsutum genome proteins, predicted results showed that GhCDPK16 similarly associated with several respiratory oxidase homolog proteins (RBOH), which may promote ROS content during stress (Fig. 6B). The analysis showed that GhCDPK16 genes interact with three different groups of proteins. The cluster I proteins comprises calmodulin-like (CML-like), calcium-binding protein (CML45 and CML30), and protein time for coffee-like (Fig. 6B). Furthermore, various RBOH-generating proteins were found to interact with the target gene, grouped as cluster IIAB. In this group, several respiratory oxidase homolog proteins, including RbohA-like, RbohB-like, RbohC-like, RbohC-like, RbohD-like, RbohE-like, and RbohF-like, were found as predicted interactors (Fig. 6B). Other proteins including WRKY33, LRR, SLAC1-like, aquaporin (NIP2-1-like), and spastin-like also interacted with the target protein. Furthermore, the cluster gene group III included heat stress transcription factor A-3-like (HSFA-like) and N-(5-phosphoribosyl) anthranilate isomerase-1 chloroplastic-like (TSB1) protein genes (Fig. 6B). This indicated that the GhCDPK16 gene may have a role in heat stress tolerance by regulating ROS generation protein genes.
Fig. 6.
Predicted interaction networks based on the orthologs in A. thaliana and G. hirsutum. AGhCDPK16 protein interaction in A. thaliana; the colored nodes denote query proteins and the first shell of interactors; the white nodes are the second shell of interactors. Light blue, pink, green, red, blue, black, and purple edges represent known interactions; experimentally known interactions; predicted gene neighborhoods; predicted gene fusions; predicted gene co-occurrences; coexpression; and protein homology, respectively. BGhCDPK16 protein interaction in G. hirsutum: black, green, and blue lines indicate predicted co-expression, textmining and database interaction evidence. Different colored nodes indicate K-means clustering based on their centroids. The edges indicate both functional and physical protein associations
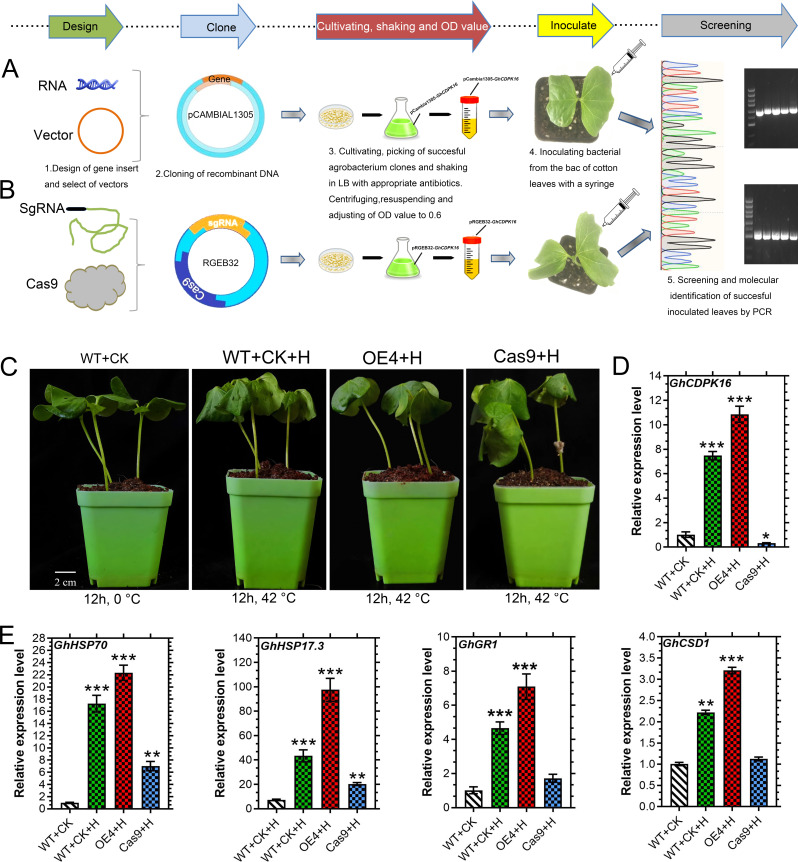
GhCDPK16 expression improves thermotolerance in transgenic plants
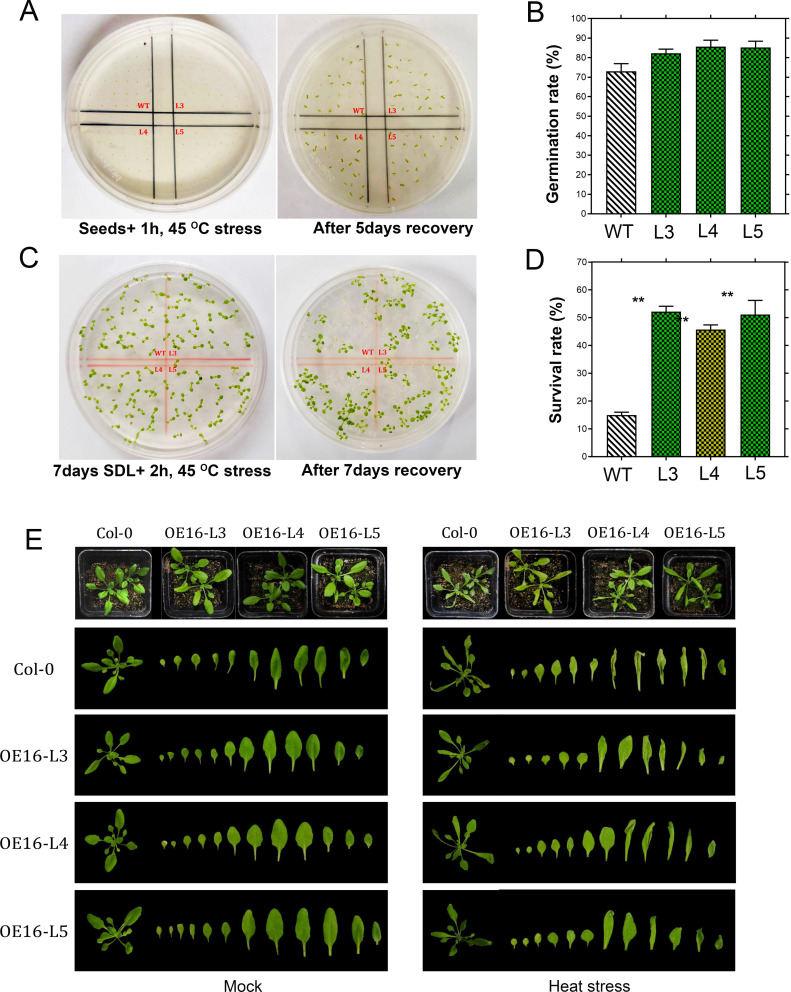
Transgenic cotton and Arabidopsis lines were subjected to high temperature stress to examine the function of GhCDPK16. As seen in Fig. 7A and B, transient overexpression and cas9 knockout techniques were employed in this study. The observed phenotype was leaf curling; however, the heat stress effect intensity was significantly higher in wild-type and Cas9 knockout plants than in GhCDPK16-OE lines (Fig. 7C). Furthermore, in comparison to the wild-type and the cas9 lines, the overexpressed lines had higher levels of GhCDPK16 expression (Fig. 7D). For further validation, the expression levels of heat-inducible genes GhHSP70, GhHSP17.3, GhGR1 were dramatically increased in overexpressed cotton lines (Fig. 7E), possibly contributing to cotton’s thermotolerance. In Arabidopsis, thermotoleance validation was conducted at different growth stages. For seed germination responses after an hour of stress treatment 5 days of recovery, results showed an increase in germination rate in GhCDPK16-ox transgenic lines compared to the wild type (Fig. 8A and B). In addition, line 4 and line 5 showed increased germination rates compared to line 3 (Fig. 8B). At the seedling stage of 7 days and 2 h of heat stress, the Arabidopsis line exhibits more significant signs of tolerance to heat stress than the wild type. Moreover, transgenic GhCDPK16 seedlings line 3 and line 5 showed a significant survival rate in comparison to the wild-type (Fig. 8C and D). After 4 weeks of plant growth and high temperature treatments at 45 °C for 24 h, the wild-type leaves showed severe wrinkling and drying, while the leaves of the overexpression lines showed fewer wrinkling compared to the WT (Fig. 8E).
Fig. 7.
Schematic views of transient expressions and CRISPR-Cas9 technology and thermotolerance assay in cotton transgenic lines. A Agrobacterium-mediated transient transfromation. B Agrobacterium-mediated CRISPR-Cas9 knockout transformation. C phenotype expression of cotton transgenic plants under heat stress. D The expression of GhCDPK16 in transient overexpression in CRISPR-cas9 knockout cotton plants. E Expresion analysis of heat-inducible genes in the transgenic plants. Asterisks * to *** represent significant differences (P < 0.05 to P < 0.01 two-way ANOVA with Tukey’s HSD post hoc test), while no asterisks denote no significant differences
Fig. 8.
Different growth stages of the GhCDPK16 transgenic line and wild-type responses under heat stress treatments. A Seed germination response to heat stress and 5-day recovery after treatment. B Germination rate analysis. C Seven-day-old seedling responses to heat stress and 7 days of recovery after treatment. D Survival rate analysis. E Phenologic changes of 4-week-old plants under heat stress treatments. SDL denotes seedlings. Each data point represents the mean (with SD bar) of three replicates. Asterisks * to *** represent significant differences (P < 0.05 to P < 0.01 two-way ANOVA with Tukey’s HSD post hoc test), while no asterisks denote no significant differences
Fig. 9.
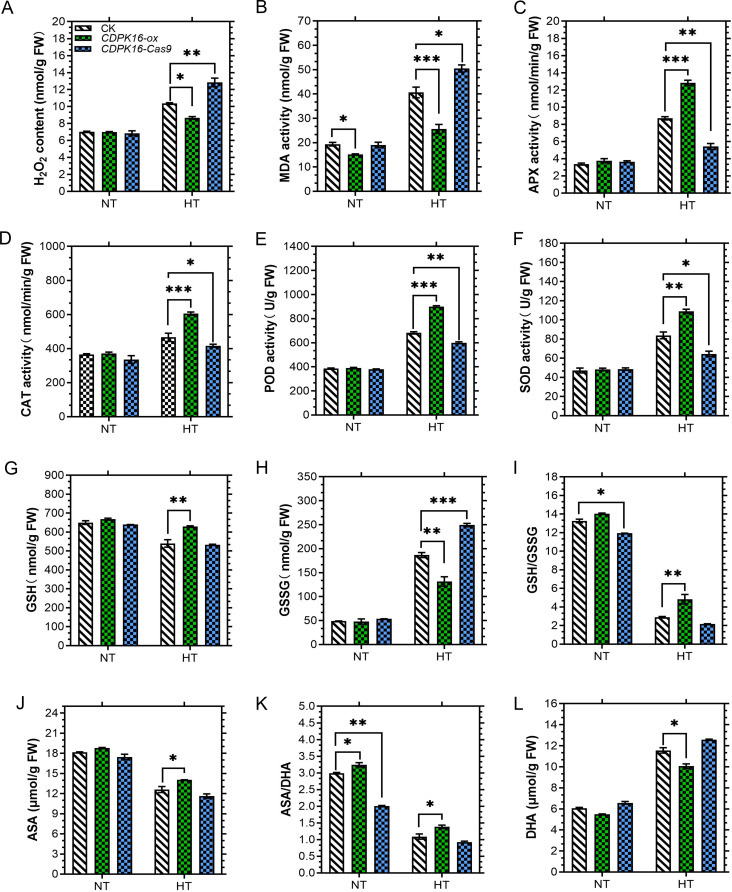
Overexpression of GhCDPK16 in cotton reduces ROS accumulation. A H2O2 content. B MDA content. C Ascorbate peroxidase (APX) activity. D Catalase (CAT) activity. E Peroxidase (POD) activity. F Superoxide dismutase (SOD) activity. G Glutathione (GSH) content. H Glutathione disulfide (GSSG) content. I Glutathione/oxidized glutathione (GSH/GSSG) content. J Ascorbate (ASA) content. K Ascorbic acid/dehydroascorbate (AsA/DHA) content. L Dehydroascorbate (DHA) content. Asterisks * to *** represent significant differences (P < 0.05 to P < 0.01 two-way ANOVA with Tukey’s HSD post hoc test), while no asterisks denote no significant differences
GhCDPK16 thermotolerance hinders ROS accumulation
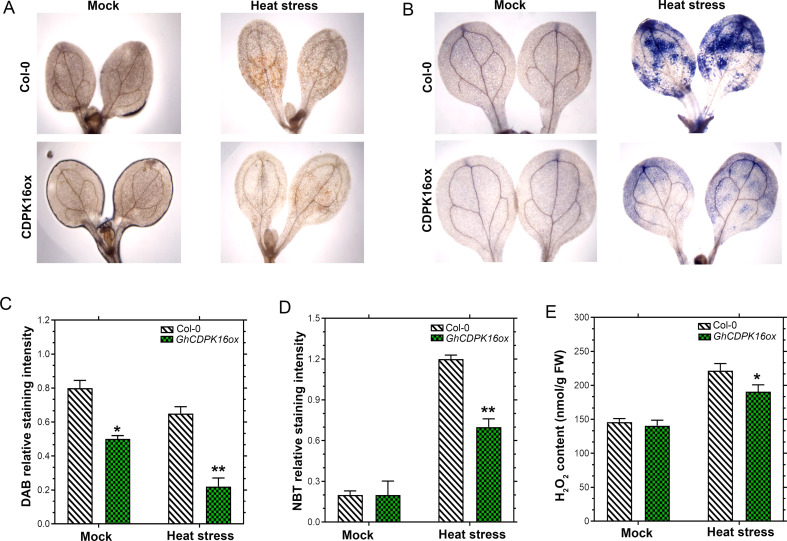
To investigate the expectations of ROS accumulation in transgenic and wild-type plants, the concentrations of H2O2 and MDA were evaluated in transgenic cotton following a 42 °C stress. To be precise, H2O2 content in overexpressed cotton leaves decreased by 0.16 folds compared to the control, whereas GhCDPK16 knockout lines showed a significant increase of 0.28 folds in comparison with the control under heat stress conditions. Contrarily, H2O2 levels under no stress were negligible (P > 0.05) (Fig. 9A). Likewise, MDA content in GhCDPK16-OE cotton leaves greatly decreased by 0.36 folds, contrary to an enhancement of 0.2 folds in knockout lines, respectively (Fig. 9B). Similarly, the accumulation of H2O2 and O2− were evaluated in Arabidopsis leaves using the DAB and NBT staining techniques. Under normal growth conditions, DAB and NBT results showed no significant difference between WT and transgenic Arabidopsis, whereas brown and blue staining increased significantly in WT leaves compared to transgenic plants after 2 h of heat stress (Fig. 10A and B), signifying the presence of a high ROS concentration. Staining intensity levels confirmed the concentration levels in plant leaves (Fig. 10C and D). Furthermore, the levels of H2O2 were quantified in leaf samples. As depicted in Fig. 10E, the H2O2 concentration under normal conditions showed no significant difference between the transgenic plants and the wild-type. However, H2O2 levels were lower in transgenic plants than in wild-type after high-temperature treatment (Fig. 10E). This indicates that ROS may have played a significant role in heat stress treatment.
Fig. 10.
Histochemical and H2O2 levels in GhCDPK16 transgenic lines. A H2O2 quantity visualized by 3,3-diaminobenzidine (DAB) staining. B Superoxide anion radicals (O2-) quantity visualized by nitroblue tetrazolium (NBT). C and D staining intensity as determined with ImageJ software. E Quantification of H2O2 content. Asterisks * to *** represent significant differences (P < 0.05 to P < 0.01 two-way ANOVA with Tukey’s HSD post hoc test), while no asterisks denote no significant differences
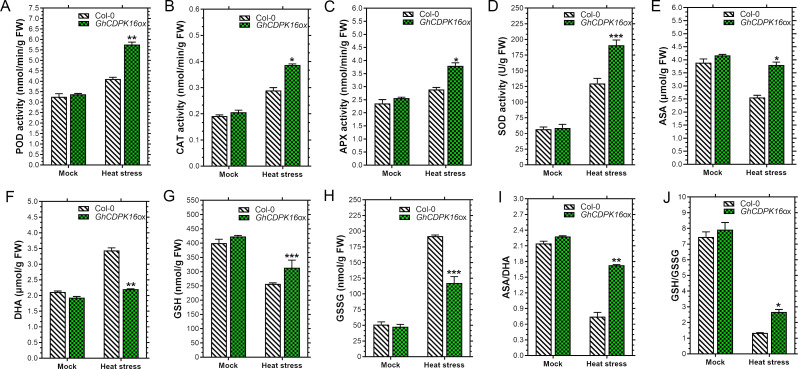
Overexpression of GhCDPK16 increases antioxidants protection under heat stress
Understanding the endogenous amounts of antioxidant compounds under GhCDPK16 thermotolerance will be beneficial in this study in terms of lowered ROS component concentrations. To do so, major antioxidant enzyme levels, including APX, POD, SOD, CAT, ascorbate, and glutathione, were determined in both cotton and Arabidopsis wild-type and transgenic plants. The results showed no significant differences in antioxidant enzyme activities between transgenic plants and WT plants under normal growth conditions. Nonetheless, POD, CAT, APX, and SOD were significantly higher in GhCDPK16-OE transgenic cotton and Arabidopsis lines compared to the wild-type (Figs. 9C-F and 11A-D). Similar increasing trends were observed in ASA, GSH, GSH/GSSG, and ASA/DHA antioxidant concentration levels in the transgenic leaves, but ASA, GSH, GSH/GSSG, and ASA/DHA content under normal temperatures were higher than heat stress (Figs. 9G, I, J and K and 11E, G, I and J). Whereas antioxidant levels in GhCDPK16-OE leaves increased dramatically, GhCDPK16-knockout cotton leaves decreased significantly when compared to the control. Contrary to the above mentioned antioxidants, only DHA and GSSG showed a decreased in antioxidant levels in overexpressed lines when compared to the wild-type (Figs. 9H and L and 11F and H). Furthermore, in cotton genotypes, GhCDPK16-Cas9 levels rose and GhCDPK16-ox antioxidant levels declined.
Fig. 11.
Endogenous antioxidant enzyme activities in GhCDPK16 transgenic and wild-type plants under heat stress. A Peroxidase (POD) activity. B Catalase (CAT) activity. C Ascorbate peroxidase (APX) activity. D Superoxide dismutase (SOD) activity. E Ascorbate (ASA) content. F Dehydroascorbate (DHA) content. G Glutathione (GSH) content. H Glutathione disulfide (GSSG) content. I Ascorbic acid/dehydroascorbate (AsA/DHA) content. J Glutathione/oxidized glutathione (GSH/GSSG) content. Each data point represents the mean (with SD bar) of three replicates. Asterisks * to *** represent significant differences (P < 0.05 to P < 0.01 two-way ANOVA with Tukey’s HSD post hoc test), while no asterisks denote no significant differences
Discussion
The CDPK gene was first reported to be physiologically activated by calcium ions in pea shoot membranes [37]. After a considerable years of research, the CDPK gene family was found to comprise of a sizable multigene family of CPK proteins that are present in all plants species with additional functional role in stress tolerance [9, 11]. Over the past few years, the characterization and evolution of the CDPK genes in the cotton genome has been extensively explored. For example, 41 putative cotton CDPKs were identified in a diploid Gossypium raimondi [21, 38], eighty-four CDPK genes were identified in G. barbadense, and 96, 44, and 57 CDPKs were identified in Gossypium hirsutum, Gossypium raimondii, and Gossypium arboretum [39] respectively. In addition, the cotton CDPK genes have been reported to be functionally involved in both biotic and abiotic roles such as verticillium wilt resistance, drought stress, and positive regulators of salt stress [17, 39, 40]. However, to date, the abiotic functional role of the plant CDPK gene family in heat stress tolerance has not been well understood. The CDPK biochemical structure of the Gossypium barbadense cultivar showed encoded amino acid lengths ranging from 648 to 155 with molecular weights ranging from 17.99 kDa to 71.854 kDa and predicted isoelectric point ranges from 4.313 to 9.48 [24]. In G. raimondii, G. arboretum, and G. hirsutum CDPK proteins, amino acid length varied from 64 to 907 amino acids, the molecular weight (MW) ranged from 6.726 kDa to 101.033 kDa, and the isoelectric point (IP) ranged from 4.128 to 10.72 [39]. In the present study, the amino acid lengths of GhCDPK proteins ranged from 487 to 655, the molecular weight (MW) ranged from 54.63 kDa to 73.42 kDa, and the PI varied from 5.06 to 6.96 (Table 1).
The tree phylogeny studied among GhCDPK members clustered into 3 major groups, with reference to A. thaliana, Z. mays, and O. sativa (Fig. 1 and S1A). However, contrary to previous reports, the evolutionary relationships among AtCDPKs, OsCDPKs, FaCDPKs, and GrCDPKs revealed four classified groups, indicating a possible divergence during evolution [9, 38]. According to existing reports, AtCDPK23 responds to drought and salt stresses, while GhCDPK60 positively regulates drought stress [17, 41]. In addition, AtCDPK7 and AtCDPK8 cluster in the same subgroup with GhCDPK60 [17]. In the present study, both AtCDPK7 and AtCDPK8 were present in a similar subgroup with GhCDPK41, 42, 46, and 46, signifying a possible involvement in drought stress response. The expression profile of GbCDPK24 revealed high responses after 24 h of heat stress [24]. Moreover, AtCDPK17 and AtCDPK34 were present in a similar group with GbCDPK24 deducing a similar role under abiotic stress responses. In the present work, AtCDPK17 and AtCDPK34 were similarly present in Group II. Additionally, the ZmCDPK7 gene, known to function in heat stress tolerance [7] was present in a similar subgroup with the GhCDPK gene in group II. A genome-wide study in Brachypodium distachyon showed the expression patterns of heat-tolerant BdCDPK genes, including BdCDPK24 and BdCDPK10. Moreover, in references to AtCDPKs, BdCDPK24 was present in the same cluster as AtCDPK34, 17, and 3 [42]. In the study, AtCDPK34, 17, and 3 clustered together in GhCDPK genes in group II, but AtCDPK3 was present in the same subgroup as GhCDPK16/19 (Fig. 1). Thus, we speculated that group II GhCDPK genes may have a role in heat stress tolerance. Orthologous genes are thought to have similar biological functions. Therefore, the function of GhCDPK genes can be inferred from orthologous genes to provide a scientific theoretical basis for subsequent functional studies [24, 43].
A typical characteristic of a CDPK structure is thought to have an N-terminal domain, a junction domain, a calmodulin-like Ca2+ binding domain, a protein kinase domain, and an EF-hand domain [9]. The EF hand allows for calcium binding, and N-myristoylation terminal domains function as part of a primary signaling process that directs proteins to the cell membrane, cytoplasm, or nucleus [9, 44]. We found all CDPKs possessing an EF-hand and a kinase domain; also, 14 putative CDPK genes were observed to have the N-terminal domain, indicating the possibility of Ca2+ binding ability and signaling functions (Table 1). The subcellular locations of CDPKs are widely distributed throughout plant cells. The plasma membrane, cytoplasm, endoplasmic reticulum, nucleus, mitochondria, oily bodies, peroxisomes, chloroplasts, and the Golgi complex are already reported locations of CDPKs [45]. In the present study, all GhCDPK genes were localized in the nucleus (Table 1). The binding sites of cis-regulatory in the gene promoter sequences play a major role in regulating the expression of the genes and controlling the growth, development, and stress responses in plants [27, 46]. As a matter of fact, the structure of the promoter could suggest the possible functions and regulatory mechanisms of the genes [47]. The MYC promoter element was reported to be involved in drought, salt, and ABA stress responses during the plant’s life cycle [48]. In addition, ABRE responded to drought and ABA through ABRE-binding proteins (AREBs) [49]. MYB is known to respond to drought, low temperatures, salt, ABA, and GA stress in plants [50]. Moreover, in Morus atropurpurea Roxb, an ABA response element (MBS, ABRE, and GARE-motif) was reported to positively respond to drought and salt stresses in Morus atropurpurea by interaction with MaCDPK1 [48]. Consistent with the present study, these mentioned promoters were among predicted cisregulatory analysis revealed to associate with GhCDPKs (Fig. 3). Stress-responsive promoter elements such as MBS, ARE, TC-rich repeats, LTR, and cis-regulatory elements involved in phytohormone pathways (ABA, GA, MeJA, and SA) were recorded, suggesting that GhCDPK could stress-responsively interact with plant hormones to defend against plant stress. Furthermore, the highest number of cis-elements was observed in A. thaliana, Z. mays, and P. sativum, implying multiple stress resistance pathways.
Gene expressions are usually controlled by sequence-specific DNA-binding proteins known as transcription factors. These factors recognize specific DNA sequences TFBSs and are thus targeted to specific genomic regions where they can recruit transcriptional co-factors and chromatin regulators to fine-tune spatiotemporal gene regulation [51]. Thus, the identification of TFBSs in genomic sequences is essential for understanding and predicting gene expression. In the present study, we examined the TFB sites of the GhCDPK genome sequence, revealing TFBS sites including ERF, MYB, LBD, WRKY, Trihelix, TCP, NAC, etc. (Fig. 4). The ethylene responsive transcription factor (ERF) was reported to regulate primary and secondary metabolism, growth, and developmental programs, as well as the tolerance of plants to various stresses, including heat stress [52, 53]. The MYB TF responds to various environmental stresses and plays a role in tolerance to high temperatures [54]. The WRKY TF family is known to be involved in high-temperature responses and can bind to the W-box cis-acting elements of target gene promoters, thereby regulating the expression of multiple types of target genes [55]. Moreover, the high number of trihelix, TCP and NAC biding sites indicates plant growth, development regulation, and stress response signaling in the GhCDPK genomic sequence [56–58].
The responsive gene expression patterns of all GhCDPKs were clearly presented in Fig. 5A. Among them, GhCDPK16/19 genes were significantly responsive to increasing heat stress as stress time increased. These results suggest the involvement of GhCDPK16/19 in tolerance to abiotic stress, especially heat stress tolerance. Moreover, the underlying biochemical mechanisms are still unknown. Therefore, using protein-protein interactions analysis will be essential towards understanding the mechanisms involved in GhCDPK16 towards heat stress tolerance. The interaction network based on the co-expression analysis revealed that GhCDPK16 is significantly associated with RBOHs, followed by WRKY, LRRs, CBLs, SLAC1-like, HSFs, and CML-related gene families (Fig. 5B). RBOHs are integral membrane proteins that convert superoxide anions into H2O2. The structure of RBOHs are made up of two calcium-binding EF hand motifs and multiple phosphorylation sites at their N-termini, enabling them to participate in the regulation of enzyme activity and regulate responses to abiotic stresses [9]. For instance, RBOHA and RBOHD are required for the accumulation of ROS during the plant defense response against cold stress in strawberries [59]. Moreover, the expression levels of VvRBOHA and VvRBOHB were significantly increased in grapes upon salt and drought stress treatments [9]. Further existing studies confirmed that, H2O2 enhances thermotolerance; therefore mutations in the ROS-generating RBOHs may cause defects in plant thermotolerance [7]. Similarly, in our investigation, transient overexpression of GhCDPK16 in cotton decreased MDA and H2O2 were found to be increased significantly under heat stress, whereas Cas9:GhCDPK16 plants sowed enhanced concentrations of ROS compounds. In addition, tested antioxidant activities were higher in overexpressed lines than in Cas9:GhCDPK16 plants, signifying that GhCDPK16 may have played a role in ROS scavenging and protection of leave membrane damages (Figs. 7 and 9). Thus, we hypothesized that H2O2 and its regulatory gene may have role in cotton tolerance to heat stress. In addition to RBOH proteins, Other heat stress-related proteins interacted with the GhCDPK gene, including LRR, HsfA3, and CML-related calcium ion-binding protein, known to control heat sensors [60] and regulate the heat stress response in Arabidopsis [61].
Thermotolerance is significantly influenced by the abscisic acid (ABA) hormone. Also, the WRKY TFs play a triggering role in activating ABA hormone signals [62]. In Arabidopsis, calcineurin B-like protein 9 (CBL9) was reported to be a calcium sensor involved in abscisic acid (ABA) signaling and stress-induced ABA biosynthesis pathways [63]. Similarly, a loss of WRKY33 function resulted in elevated ABA levels [62]. Moreover, Arabidopsis AtWRKY25, AtWRKY26, and AtWRKY33 promote the expression of ethylene insensitive protein 2 (EIN2) and the ethylene-mediated signal transduction pathway, thereby improving the high-temperature tolerance of plants [55]. Here, CBL9 and WRKY33 proteins interacted with GhCDPK16. We speculate that GhCDPK16 may have interacted with WRKY and CBL proteins to induce hormonal tolerance by regulating ABA and ethylene levels, during heat stress. Crizel et al. [9] further reported that FaCDPK4 and FaCDPK11 utilize ABA as the signaling molecule to increase the content of phenolics, ascorbic acid, and sugar compounds involved in drought stress tolerance. The ABA-mediated AsA-GSH promotes regeneration and improves the antioxidant capacity, alleviates the damage of ROS, reduces growth inhibition, and enhances the tolerance of desiccation stress [64]. Moreover, ABA acts as a defensive stress hormone against plant heat stress by inducing endogenous ABA content, which promotes water balance and strengthens heat tolerance [65]. Under extreme temperatures of 42 °C, 48 h in Vicia faba L and 38/28°C day/night, 7 days in L. esculentum Mill, the AsA, GSH, GSSG, AsA + DHA, and DHA content significantly increased [66]. Consistent with the current study, antioxidant concentrations in the leaves of the transgenic line significantly increased compared to the wild-type, indicating the activation of antioxidant protection (Fig. 8). Furthermore, we speculated that ABA might have played a role in the control of endogenous antioxidant levels.
Conclusions
In this study, a total of 48 predicted CDPKs were identified from upland cotton (Gossypium hirsutum L.), and phylogenetic analyses classified them into three groups. All CDPK proteins are localized in the nucleus and contain EF-hand motifs and kinase structural domains. The predicted ERF, MYB, and WRKY transcription binding sites showed the possibility of abiotic stress tolerance. Significant expression profiles of GhCDPK16/19 during 24 h of heat stress indicated a connection with heat stress responses. Moreover, the protein-protein interaction network analysis highlighted proteins involved in ROS regulation, heat stress TFs, and ABA-dependent Ca2+ signaling. Transient overexpression of GhCDPK16 in cotton and permanent overexpression in Arabidopsis substantially reduced H2O2 content and augmented tolerance by increasing protective antioxidant enzymes through ROS scavenging. This study provides a basis for subsequent studies on the functions of the GhCDPK16 gene family members in cotton and provides a theoretical basis for breeding new heat-tolerant cotton varieties. We propose future functional genomic studies on the interactive role of GhCDPK16 with heat stress-responsive proteins, which will allow deeper elucidation of the GhCDPK16 role in plant heat stress tolerance.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Not applicable.
Abbreviations
- CDPK
Calcium-dependent protein kinase
- BLAST
Basic Local Alignment Search tool
- Aa
Amino acids
- KDa
Kilo Dalton
- pI
Protein Isoelectric Point
- ABA
Abscisic Acid
- IAA
Auxin
- KDa
Kilo
- GA
Gibberellin
- MeJA
Methyl Jasmonate
- SA
Salicylic Acid
- ERF
Ethylene Responsive Transcription Factor
- ABRE
Abscisic Acid Responsiveness Element
- LRT
Low Temperature Response
- MBS
MYB Binding Site
- RY-element
Seed specifc regulation
- O2-site
Zein metabolism regulation elements
- Ox
Overexpression
- qRT-PCR
Quantitative Real-time Polymerase Chain Reaction
- CAT
Catalase
- POD
Peroxidase
- SOD
Superoxide Dismutase
- ROD
Reactive Oxygen Species
- WT
Wild Type
- TFs
Transcription Factors
- TFBDS
Transcription Factor Binding Site
Appendix A. Supporting information
Additional supporting information may be found online in the Supporting Information section at the end of the article.
Author contributions
WBL and JSG conceived and designed the experiments. WBL and MCS finished the main experiments. CHD, YTC, ML and AGO analyzed the data. WBL and CCM wrote the manuscript. JSG and AGO revised the manuscript. NG and DHL guided the experiments.
Funding
This work was supported by State Key Laboratory of Cotton Bio-breeding and Integrated Utilization and Sponsored by State Key Laboratory of Cotton Bio-breeding and Integrated Utilization Open Fund (CB2024A12).
Data availability
The data provided within the manuscript or supplementary information of this article will be made available by the corresponding authors, without undue reservation.
Declarations
Competing interests
The authors declare no competing interests.
Consent of publication
Not applicable.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Arshad MS, Farooq M, Asch F, Krishna JSV, Prasad PVV, Siddique KHM, et al. Thermal stress impacts Reproductive Development and Grain Yield in Rice. Plant Physiol Biochem. 2017;115(3):57–72. 10.1016/j.plaphy.2017.03.011 [DOI] [PubMed] [Google Scholar]
- 2.Huang LZ, Zhou M, Ding YF, Zhu C, et al. Gene Networks Involved in Plant Heat Stress Response and tolerance. Int J Mol Sci. 2022;23(19):1–19. 10.3390/ijms231911970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim JH, Lim SD, Jang CS, et al. Oryza Sativa Heat-Induced RING finger protein 1 (OsHIRP1) positively regulates Plant Response to heat stress. Plant Mol Biol. 2019;99(6):545–59. 10.1007/s11103-019-00835-9 [DOI] [PubMed] [Google Scholar]
- 4.Guo LM, Li J, He J, Liu H, Zhang HM, et al. A class I cytosolic HSP20 of Rice enhances Heat and Salt Tolerance in different organisms. Sci Rep. 2020;10(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feng ZJ, Liu N, Zhang GW, Niu FG, Xu SC, Gong YM, et al. Investigation of the AQP family in soybean and the promoter activity of TIP2;6 in heat stress and hormone responses. Int J Mol Sci. 2019;20(2):1–19. 10.3390/ijms20020262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song Y, Chen P, Liu P, Bu C, Zhang D, et al. High-temperature-responsive Poplar LncRNAs modulate target gene expression via RNA interference and act as RNA scaffolds to Enhance Heat Tolerance. Int J Mol Sci. 2020;21(18):1–22. 10.3390/ijms21186808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao Y, Du H, Wang Y, Wang H, Yang S, Li C, Chen N, Yang H, Zhang Y, Zhu Y, Yang L, Hu X, et al. The calcium-dependent protein kinase ZmCDPK7 functions in heat-stress tolerance in Maize. J Integr Plant Biol. 2021;63(3):510–27. 10.1111/jipb.13056 [DOI] [PubMed] [Google Scholar]
- 8.Wang H, Niu H, Liang M, Zhai Y, Huang W, Ding Q, Du Y, Lu M, et al. A Wall-Associated kinase gene Cawakl20 from Pepper negatively modulates Plant Thermotolerance by reducing the expression of ABA-Responsive genes. Front Plant Sci. 2019;10(5):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crizel RL, Perin EC, Vighi IL, Woloski R, Seixas A, da Silva Pinto L, Rombaldi CV, Galli V, et al. Genome-wide identification, and characterization of the CDPK Gene Family Reveal their involvement in Abiotic Stress Response in Fragaria x Ananassa. Sci Rep. 2020;10(1):1–17. 10.1038/s41598-020-67957-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edel KH, Marchadier E, Brownlee C, Kudla J, Hetherington AM, et al. The evolution of calcium-based signalling in plants. Curr Biol. 2017;27(13):R667–79. 10.1016/j.cub.2017.05.020 [DOI] [PubMed] [Google Scholar]
- 11.Valmonte GR, Arthur K, Higgins CM, Macdiarmid RM, et al. Calcium-dependent protein kinases in plants: evolution, expression and function. Plant Cell Physiol. 2014;55(3):551–69. 10.1093/pcp/pct200 [DOI] [PubMed] [Google Scholar]
- 12.Klimecka M, Muszyńska G, et al. Structure and functions of Plant Calcium-Dependent protein kinases. Acta Biochim Pol. 2007;54(2):219–33. 10.18388/abp.2007_3242 [DOI] [PubMed] [Google Scholar]
- 13.Giammaria V, Grandellis C, Bachmann S, Gargantini PR, Feingold SE, Bryan G, Ulloa RM, et al. StCDPK2 expression and activity reveal a highly responsive potato calcium-dependent protein kinase involved in light signalling. Planta. 2011;233(3):593–609. 10.1007/s00425-010-1319-2 [DOI] [PubMed] [Google Scholar]
- 14.Xu J, Tian YS, Peng RH, Xiong AS, Zhu B, Jin XF, Gao F, Fu XY, Hou XL, Yao QH, et al. AtCPK6, a functionally redundant and positive Regulator involved in Salt/Drought stress tolerance in Arabidopsis. Planta. 2010;231(6):1251–60. 10.1007/s00425-010-1122-0 [DOI] [PubMed] [Google Scholar]
- 15.Liu HT, Gao F, Li GL, Han JL, Liu DL, Sun DY, Zhou RG, et al. The calmodulin-binding protein kinase 3 is part of Heat-Shock Signal Transduction in Arabidopsis Thaliana. Plant J. 2008;55(5):760–73. 10.1111/j.1365-313X.2008.03544.x [DOI] [PubMed] [Google Scholar]
- 16.Dubrovina AS, Kiselev KV, Khristenko VS, Aleynova OA, et al. The calcium-dependent protein kinase gene VaCPK29 is involved in grapevine responses to heat and osmotic stresses. Plant Growth Regul. 2017;82(1):79–89. 10.1007/s10725-016-0240-5 [DOI] [Google Scholar]
- 17.Yan M, Yu X, Zhou G, Sun D, Hu Y, Huang C, Zheng Q, Sun N, Wu J, Fu Z, Li L, Feng Z, Yu S, et al. GhCDPK60 positively regulates Drought stress tolerance in both transgenic Arabidopsis and Cotton by regulating Proline Content and ROS Level. Front. Plant Sci. 2022;13(12):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan R, Liang C, Meng Z, Malik W, Zhu T, Zong X, Guo S, Zhang R, et al. Progress in genome sequencing will accelerate molecular breeding in cotton (Gossypium Spp). 3 Biotech. 2016;6(2):1–9. 10.1007/s13205-016-0534-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zahid KR, Ali F, Shah F, Younas M, Shah T, Shahwar D, Hassan W, Ahmad Z, Qi C, Lu Y, Iqbal A, Wu W, et al. Response and tolerance mechanism of Cotton Gossypium Hirsutum L. to elevated temperature stress: a review. Front. Plant Sci. 2016;7(6):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Owusu AG, Lv Y-P, Liu M, Wu Y, Li C-L, Guo N, Li D-H, Gao J-S, et al. Transcriptomic and metabolomic analyses reveal the potential mechanism of Waterlogging Resistance in Cotton (Gossypium Hirsutum L.). Front. Plant Sci. 2023;14(6):1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu W, Li W, He Q, Daud MK, Chen J, Zhu S, et al. Genome-wide survey and expression analysis of calcium-dependent protein kinase in Gossypium Raimondii. PLoS ONE. 2014;9(6):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang T, Hu Y, Jiang W, Fang L, Guan X, Chen J, Zhang J, Saski CA, Scheffler BE, Stelly DM, Hulse-Kemp AM, Wan Q, Liu B, Liu C, Wang S, Pan M, Wang Y, Wang D, Ye W, Chang L, Zhang W, Song Q, Kirkbride RC, Chen X, Dennis E, Llewellyn DJ, Peterson DG, Thaxton P, Jones DC, Wang Q, Xu X, Zhang H, Wu H, Zhou L, Mei G, Chen S, Tian Y, Xiang D, Li X, Ding J, Zuo Q, Tao L, Liu Y, Li J, Lin Y, Hui Y, Cao Z, Cai C, Zhu X, Jiang Z, Zhou B, Guo W, Li R, Chen ZJ, et al. Sequencing of Allotetraploid Cotton (Gossypium Hirsutum L. Acc. TM-1) provides a resource for Fiber improvement. Nat Biotechnol. 2015;33(5):531–7. 10.1038/nbt.3207 [DOI] [PubMed] [Google Scholar]
- 23.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S, et al. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol. 2013;30(12):2725–9. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi G, Zhu X, et al. Genome-wide identification and functional characterization of CDPK Gene Family Reveal their involvement in response to Drought stress in Gossypium Barbadense. PeerJ. 2022;10(2):1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao C, Gao K, Yang H, Ju T, Zhu J, Tang Z, Zhao L, Chen Q, et al. Genome-wide analysis of Metallothionein Gene Family in Maize to Reveal its role in development and stress resistance to Heavy Metal. Biol Res. 2022;55(1):1–13. 10.1186/s40659-021-00368-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang Z, Zhang H, Gao S, Zhai H, He S, Zhao N, Liu Q, et al. Genome-wide identification and expression analysis of the sucrose synthase Gene Family in Sweet Potato and its two diploid relatives. Int J Mol Sci. 2023;24(15):1–18. 10.3390/ijms241512493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hui W, Zheng H, Fan J, Wang J, Saba T, Wang K, Wu J, Wu H, Zhong Y, Chen G, Gong W, et al. Genome-wide characterization of the MBF1 Gene Family and its expression pattern in different tissues and stresses in Zanthoxylum Armatum. BMC Genomics. 2022;23(1):1–21. 10.1186/s12864-022-08863-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang Z, Zhang R, Zhou Z, et al. The XTH Gene Family in Schima Superba: genome-wide identification, expression profiles, and Functional Interaction Network Analysis. Front. Plant Sci. 2022;13(6):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alok A, Chauhan H, Upadhyay SK, Pandey A, Kumar J, Singh K, et al. Compendium of Plant-Specific CRISPR vectors and their technical advantages. Life. 2021;11(10):1–19. 10.3390/life11101021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun S, Xiong X, Zhang X, Feng H, Zhu Q, Sun J, et al. Characterization of the Gh4CL Gene Family reveals a role of Gh4CL7 in Drought Tolerance. BMC Plant Biol. 2021;20(2020):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wen Z, Chen Z, Liu X, Sun J, Zhang F, Zhang M, Dong C, et al. 24-Epibrassinolide facilitates adventitious Root formation by coordinating cell-wall polyamine oxidase- and plasma membrane respiratory burst oxidase homologue-derived reactive oxygen species in Capsicum Annuum L. Antioxidants. 2023;12(7):1–22. 10.3390/antiox12071451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li P, Cai Q, Wang H, Li S, Cheng J, Li H, Yu Q, Wu S, et al. Hydrogen Peroxide Homeostasis provides beneficial Micro-environment for SHR-Mediated Periclinal Division in Arabidopsis Root. New Phytol. 2020;228(6):1926–38. 10.1111/nph.16824 [DOI] [PubMed] [Google Scholar]
- 33.Sarker U, Oba S, et al. Catalase, Superoxide dismutase and ascorbate-glutathione cycle enzymes Confer Drought Tolerance of Amaranthus Tricolor. Sci Rep. 2018;8(1):1–12. 10.1038/s41598-018-34944-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou H, Zhou KH, Zhao G, Wang PP, Yang DG, Ma XF, Gao JS, et al. Physiological and biochemical properties of Cotton seedlings in response to Cu2 + stress. Curr Issues Mol Biol. 2023;45(5):4050–62. 10.3390/cimb45050258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Noctor G, Mhamdi A, Foyer CH, et al. Oxidative stress and Antioxidative systems: recipes for successful data Collection and Interpretation. Plant Cell Environ. 2016;39(5):1140–60. 10.1111/pce.12726 [DOI] [PubMed] [Google Scholar]
- 36.Niu M-X, Feng C-H, Liu M, Liu X, Liu S, Liu C, Yin W, Xia X, et al. Genome-wide identification of Poplar GSTU Gene Family and its PtrGSTU23 and PtrGSTU40 to Improve Salt Tolerance in Poplar. Ind Crops Prod. 2024;209(10):1–14. [Google Scholar]
- 37.Hetherington A, Trewavas A, et al. Calcium-dependent protein kinase in pea shoot membranes. FEBS Lett. 1982;145(1):67–71. 10.1016/0014-5793(82)81208-8 [DOI] [Google Scholar]
- 38.Li Lbei, Yu D, wei, Zhao F, li, Pang C you, Song M, zhen, Wei H ling, Fan S li, Yu S et al. xun,. Genome-Wide Analysis of the Calcium-Dependent Protein Kinase Gene Family in Gossypium Raimondii. J. Integr. Agric. 2015; 14(1): 29–41.
- 39.Wu Y, Zhang L, Zhou J, Zhang X, Feng Z, Wei F, Zhao L, Zhang Y, Feng H, Zhu H, et al. Calcium-dependent protein kinase GhCDPK28 was dentified and involved in Verticillium Wilt Resistance in Cotton. Front Plant Sci. 2021;12(11):1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao W, Xu FC, Guo DD, Zhao JR, Liu J, Guo YW, Singh PK, Ma XN, Long L, Botella JR, Song CP, et al. Calcium-dependent protein kinases in cotton: insights into early plant responses to salt stress. BMC Plant Biol. 2018;18(1):1–15. 10.1186/s12870-018-1230-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma SY, Wu WH, et al. AtCPK23 functions in Arabidopsis responses to Drought and Salt stresses. Plant Mol Biol. 2007;65(4):511–8. 10.1007/s11103-007-9187-2 [DOI] [PubMed] [Google Scholar]
- 42.Wen F, Ye F, Xiao Z, Liao L, Li T, Jia M, Liu X, Wu X, et al. Genome-wide survey and expression analysis of calcium-dependent protein kinase (CDPK) in Grass Brachypodium Distachyon. BMC Genomics. 2020;21(1):1–17. 10.1186/s12864-020-6475-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thornton JW, Desalle R, et al. GENE FAMILY EVOLUTION AND HOMOLOGY: Genomics meets phylogenetics. New York. 2000;1(48):41–73. [DOI] [PubMed] [Google Scholar]
- 44.Wang CT, Shao JM, et al. Characterization of the ZmCK1 gene encoding a calcium-dependent protein kinase responsive to multiple abiotic stresses in Maize. Plant Mol Biol Rep. 2013;31(1):222–30. 10.1007/s11105-012-0496-5 [DOI] [Google Scholar]
- 45.Asai S, Ichikawa T, Nomura H, Kobayashi M, Kamiyoshihara Y, Mori H, Kadota Y, Zipfel C, Jones JDG, Yoshioka H, et al. The variable domain of a plant calcium-dependent protein kinase (CDPK) confers subcellular localization and substrate recognition for NADPH oxidase. J Biol Chem. 2013;288(20):14332–40. 10.1074/jbc.M112.448910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang H, Zhou Y, Zhang Y, Wang J, Shi H, et al. Identification of Transcription Factors of Nitrate Reductase Gene Promoters and NRE2 Cis-element through yeast one-hybrid screening in Nicotiana Tabacum. BMC Plant Biol. 2019;19(1):1–8. 10.1186/s12870-019-1724-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li J, Chen X, Zhou X, Huang H, Wu D, Shao J, Zhan R, Chen L, et al. Identification of Trihelix Transcription Factors in Pogostemon Cablin reveals PatGT-1 negatively regulates Patchoulol Biosynthesis. Ind Crops Prod. 2021;161(7):113182. 10.1016/j.indcrop.2020.113182 [DOI] [Google Scholar]
- 48.Cao Z, He Q, Wang P, Yan J, Awais MM, Liu Z, Yan H, Sun J, et al. Functional characteristics of a calcium-dependent protein kinase (MaCDPK1) Enduring stress tolerance from Morus Atropurpurea Roxb. Plant Cell Tissue Organ Cult. 2020;141(1):131–43. 10.1007/s11240-020-01773-x [DOI] [Google Scholar]
- 49.Li W, Cui X, Meng Z, Huang X, Xie Q, Wu H, Jin H, Zhang D, Liang W et al. Transcriptional regulation of Arabidopsis MIR168a and ARGONAUTE1 Homeostasis in Abscisic Acid and Abiotic stress responses, vol. 158. 2012. [DOI] [PMC free article] [PubMed]
- 50.Zhu C, Schraut D, Hartung W, Schäffner AR, et al. Differential responses of Maize MIP genes to salt stress and ABA. J Exp Bot. 2005;56(421):2971–81. 10.1093/jxb/eri294 [DOI] [PubMed] [Google Scholar]
- 51.Lai X, Stigliani A, Vachon G, Carles C, Smaczniak C, Zubieta C, Kaufmann K, Parcy F, et al. Building transcription factor binding site models to understand gene regulation in plants. Mol Plant. 2019;12(6):743–63. 10.1016/j.molp.2018.10.010 [DOI] [PubMed] [Google Scholar]
- 52.Licausi F, Ohme-Takagi M, Perata P, et al. APETALA2/Ethylene Responsive factor (AP2/ERF) transcription factors: mediators of stress responses and Developmental Programs. New Phytol. 2013;199(3):639–49. 10.1111/nph.12291 [DOI] [PubMed] [Google Scholar]
- 53.Huang J, Zhao X, Bürger M, Wang Y, Chory J, et al. Two interacting Ethylene response factors regulate heat stress response. Plant Cell. 2021;33(2):338–57. 10.1093/plcell/koaa026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.El-kereamy A, Bi YM, Ranathunge K, Beatty PH, Good AG, Rothstein SJ, et al. The Rice R2R3-MYB transcription factor OsMYB55 is involved in the tolerance to high temperature and modulates amino acid metabolism. PLoS ONE. 2012;7(12):1–16. 10.1371/journal.pone.0052030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cheng Z, Luan Y, Meng J, Sun J, Tao J, Zhao D, et al. WRKY Transcription Factor Response to high-temperature stress. Plants. 2021;10(10):1–13. 10.3390/plants10102211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tong Y, Huang H, Wang Y, et al. Genome-wide analysis of the Trihelix Gene Family and their response to cold stress in Dendrobium Officinale. Sustain. 2021;13(5):1–15. [Google Scholar]
- 57.Alshareef NO, Otterbach SL, Allu AD, Woo YH, de Werk T, Kamranfar I, Mueller-Roeber B, Tester M, Balazadeh S, Schmöckel SM, et al. NAC Transcription Factors ATAF1 and ANAC055 affect the heat stress response in Arabidopsis. Sci Rep. 2022;12(1):1–15. 10.1038/s41598-022-14429-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang M, He G, Hou Q, Fan Y, Duan L, Li K, Wei X, Qiu Z, Chen E, He T, et al. Systematic analysis and expression profiles of TCP Gene Family in Tartary Buckwheat (Fagopyrum Tataricum (L.) Gaertn.) Revealed the potential function of FtTCP15 and FtTCP18 in response to Abiotic Stress. BMC Genomics. 2022;23(1):1–19. 10.1186/s12864-022-08618-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Y, Li Y, He Y, Hu W, Zhang Y, Wang X, Tang H, et al. Identification of NADPH oxidase Family Members Associated with Cold stress in Strawberry. FEBS Open Bio. 2018;8(4):593–605. 10.1002/2211-5463.12393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marchetti F, Cainzos M, Cascallares M, Distéfano AM, Setzes N, López GA, Zabaleta E, Pagnussat GC, et al. Heat stress in Marchantia Polymorpha: sensing and mechanisms underlying a dynamic response. Plant Cell Environ. 2021;44(7):2134–49. 10.1111/pce.13914 [DOI] [PubMed] [Google Scholar]
- 61.Schramm F, Larkindale J, Kiehlmann E, Ganguli A, Englich G, Vierling E, Von Koskull-Döring P, et al. A Cascade of transcription factor DREB2A and heat stress transcription factor HsfA3 regulates the heat stress response of Arabidopsis. Plant J. 2008;53(2):264–74. 10.1111/j.1365-313X.2007.03334.x [DOI] [PubMed] [Google Scholar]
- 62.Liu S, Kracher B, Ziegler J, Birkenbihl RP, Somssich IE, et al. Negative regulation of ABA signaling by WRKY33 is critical for Arabidopsis Immunity towards Botrytis Cinerea 2100. Elife. 2015;4(6):1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pandey GK, Yong HC, Kim KN, Grant JJ, Li L, Hung W, D’Angelo C, Weinl S, Kudla J, Luan S, et al. The Calcium Sensor Calcineurin B-like 9 modulates abscisic acid sensitivity and biosynthesis in Arabidopsis. Plant Cell. 2004;16(7):1912–24. 10.1105/tpc.021311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jiang Z, Zhu H, Zhu H, Tao Y, Liu C, Liu J, Yang F, Li M, et al. Exogenous ABA enhances the antioxidant Defense System of Maize by regulating the AsA-GSH cycle under Drought stress. Sustain. 2022;14(5):1–15. [Google Scholar]
- 65.Sezgin Muslu A, Kadıoğlu A, et al. Role of Abscisic Acid, Osmolytes and Heat Shock factors in high temperature thermotolerance of Heliotropium Thermophilum. Physiol Mol Biol Plants. 2021;27(4):861–71. 10.1007/s12298-021-00975-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hasanuzzaman M, Borhannuddin Bhuyan MHM, Anee TI, Parvin K, Nahar K, Al Mahmud J, Fujita M, et al. Regulation of ascorbate-glutathione pathway in mitigating oxidative damage in plants under abiotic stress. Antioxidants. 2019;8(9):1–50. 10.3390/antiox8090384 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data provided within the manuscript or supplementary information of this article will be made available by the corresponding authors, without undue reservation.