Abstract
Background and Objectives
Amyotrophic lateral sclerosis (ALS) is an age-associated, fatal neurodegenerative disorder causing progressive paralysis and respiratory failure. The genetic architecture of ALS is still largely unknown.
Methods
We performed a genome-wide association study (GWAS) and transcriptome-wide association study (TWAS) to understand genetic risk factors for ALS using a population-based case-control study of 435 ALS cases and 279 controls from Northern New England and Ohio. Single nucleotide polymorphism (SNP) genotyping was conducted using the Illumina NeuroChip array. Odds ratios were estimated using covariate-adjusted logistic regression. We also performed a genome-wide SNP-smoking interaction screening. TWAS analyses used PrediXcan to estimate associations between predicted gene expression levels across 15 tissues (13 brain tissues, skeletal muscle, and whole blood) and ALS risk.
Results
GWAS analyses identified the p.A382T missense variant (rs367543041, p = 3.95E-6) in the TARDBP gene, which has previously been reported in association with increased ALS risk and was found to share a close affinity with the Sardinian haplotype. Both GWAS and TWAS analyses suggested that ZNF235 is associated with decreased ALS risk.
Discussion
Our results support the need for future evaluation to clarify the role of these potential genetic risk factors for ALS and to understand genetic susceptibility to environmental risk factors.
Introduction
Amyotrophic lateral sclerosis (ALS) is an age-associated, fatal neurodegenerative disorder causing progressive paralysis and respiratory failure.1,2 With approximately 400,000 individuals worldwide estimated to be afflicted with ALS by 2040,2 ALS is the third most prevalent neurodegenerative disorder after Alzheimer disease (AD) and Parkinson disease (PD).3,4 Family history of ALS is identifiable in approximately 10% of ALS cases, with the remaining 90% being of sporadic origin.2,5 In addition, a recent study found that ALS has an elevated prevalence in Northern New England compared with other regions in the United States.6 Therefore, it is important to understand genetic and environmental risk factors specific to this region.
Since 1993, researchers have made substantial progress in unraveling genetic mechanisms involved in ALS development and progression, leading to the identification of over 2 dozen genes associated with the disorder.2,7 Recent progress in genotyping and sequencing technologies has improved our understanding of ALS pathology.3 Multiple studies have revealed overlapping susceptibility variants of ALS with other neurodegenerative diseases, including frontotemporal dementia (FTD), AD, and PD.1,5,8 Exploring genetic risk factors for ALS can help uncover etiopathogenic mechanisms across the spectrum of neurodegenerative diseases and potentially unveil fundamental processes involved in neuronal degeneration.1
Genome-wide association studies (GWAS) have played an essential role in identifying common ALS-susceptibility variants, but many of these variants have small effect sizes and are located within noncoding genomic regions.8-10 Transcriptome-wide association studies (TWAS) can clarify the association between genetically regulated gene expression and ALS risk and potentially identify novel genes related to ALS risk while reducing the multiple testing burden.11 Previous TWAS have successfully identified several ALS-associated genes expressed in various brain-related tissues and blood.11,12
Despite multiple studies indicating that ALS has a moderately high heritability (40%–60%),12,13 previously identified loci only account for a small proportion of the overall genetic predisposition to ALS.3,5,7 Smoking is a known risk factor for ALS.14 It may interact with genetic factors to influence the risk of developing ALS. For instance, smoking has been shown to induce oxidative stress, which is associated with higher ALS risk.14 Therefore, smoking may interact with the ALS risk gene SOD1, which plays a critical role in regulating oxidative stress.15 There are very few previous studies examining genome-wide smoking-gene interactions associated with ALS risk and our research aims to bridge this gap. This study, seeking more insight into ALS's genetic architecture, integrates GWAS and TWAS methods to detect the genetic risk factors and assesses gene-smoking interactions using sporadic ALS cases and controls based in Northern New England and Ohio that have been collected in part from previous studies.
Methods
Study Population
The enrollment procedure for ALS cases and controls is outlined by Andrew et al.16,17 In summary, we recruited ALS cases and controls from Northern New England and Ohio, with their signed consent to provide blood or saliva samples, demographic and clinical information, and complete the environmental questionnaire. Cases were newly diagnosed patients with ALS from medical centers in these regions. Controls consist of both population controls and clinic controls. Population controls were recruited randomly by mail using the US Postal Service Delivery Sequence File (USPS DSF2). Clinic controls were patients diagnosed with non-neurodegenerative diseases. All participants were at least aged 18 years.
Between 2020 and 2023, the Laboratory of Neurogenetics at the National Institute on Aging genotyped DNA on 435 ALS cases and 279 controls from Northern New England and Ohio participants. Genotyping was conducted using the Illumina NeuroChip according to the manufacturer's instructions, a platform designed to target curated variants in neurologic diseases.18 We measured genotypes for 487,374 single nucleotide polymorphisms (SNPs) from the arrays prior to quality control filtering, including 305,670 SNPs from a GWAS backbone and 179,467 custom SNPs selected throughout the genome.
Quality Control and Genotype Imputation
We used PLINK19,20 software to perform standard quality control procedures for genotype data, and we implemented the following steps outlined by Chia et al.21 Briefly, we excluded samples with over 5% missing genotypes based on the sample call rate and removed samples with heterozygosity values beyond a threshold (F > 0.15 or F < −0.15). We removed non-European individuals from the principal component analysis because of the low numbers in the New Hampshire population, using the HapMap 3 Genome Reference Panel22 as the reference for ancestral information. Given that most instances of ALS are sporadic, we excluded all familial ALS cases; however, we cannot entirely exclude the rare occurrence of a monogenic gene variant because when this study was performed, sporadic cases were not undergoing clinical genetic testing. In addition, we removed variants (1) containing over 5% missing genotypes, (2) with less than 5% minor allele frequency, (3) showing deviation from Hardy-Weinberg equilibrium (p < 1.0E-3), and (4) with a p-value below 1E-4 in the case/control nonrandom missingness test.
After quality control, 613 individuals, including 378 sporadic ALS cases and 235 controls, were included in analyses, and 242,090 SNPs were available for imputation. We conducted genotype imputation by Michigan Imputation Server23 in GRCh37/hg19, using the European population data of the 1000 Genomes Project24,25 (phase 3, version 5, available at reference 26) as the imputation reference. Only SNPs with an imputation accuracy R2 ≥ 0.3 were included in analyses. The quantile-quantile (QQ) plots show no genomic inflation after quality control (eFigure 1).
Genome-Wide Association Analysis
For the GWAS analysis, we performed covariate-adjusted logistic regression using PLINK, adjusting for sex, age at symptom onset, and the first 10 principal components of genetic ancestry. The Manhattan plot was generated using the “CMplot”27 package in R version 4.0.2. We validated the significant SNP identified in previous GWAS results using publicly available data with a larger sample size, conducting logistic regression and adjusting for the same covariates. For this validation analysis, genotype data were obtained from 10,067 ALS cases and 2,251 controls from the database of Genotypes and Phenotypes (dbGaP)28 with the study accession number phs000101.v5.p1. To balance the case-control ratio, we included an additional 11,887 controls from 2 other dbGaP data sets (phs000187 and phs000428). After quality control, 22,419 individuals and 335,021 variants were available for imputation in the validation study. The genotype imputation was also conducted on this validation data using the Michigan Imputation Server. A threshold p-value of 5E-8 was set for genome-wide significance after Bonferroni correction for multiple testing in the GWAS.
In addition to examining the main effect of SNPs, we also evaluated SNP-smoking interactions associated with ALS susceptibility. We performed interaction analysis by including cigarette smoking status (ever-smoker vs never-smoker) and a multiplicative SNP-smoking interaction term adjusting in covariate-adjusted models. Participants without smoking status were removed from this analysis. Interaction analyses could not be pursued using the validation data set because smoking status was unavailable.
Transcriptome-Wide Association Analysis
We employed the widely used TWAS approach PrediXcan29 to predict the expression levels of participants from the Northern New England and Ohio ALS cohort. PrediXcan trains predictive models using reference data sets consisting of transcriptome and genotype information. Prediction weights were obtained from PredictDB, which derived these weights through the elastic net method using the Genotype-Tissue Expression version 7 as the reference panel.29,30 We examined associations between ALS risk and predicted gene expression levels across 15 tissues related to ALS, which included 13 brain and spinal cord regions (amygdala, anterior portion, caudate, cerebellar hemisphere, cerebellum, nucleus accumbens, cortex, frontal cortex BA9, hippocampus, hypothalamus, putamen, substantia nigra, C1 spinal cord), skeletal muscle, and whole blood tissues. We standardized predicted gene expression levels and tested associations using logistic regression with adjustment for sex, age at symptom onset, and the first 3 principal components of genetic ancestry. The false discovery rate (FDR) of 0.30 was used as the threshold for suggestively significant gene expression levels.
Standard Protocol Approvals, Registrations, and Patient Consents
All participants involved were consented. All study procedures have been approved by the Committee for the Protection of Human Subjects at Dartmouth Health.
Data Availability
We are in the process of uploading the genotype data used in this study to dbGaP with the accession number phs000101. Once processed, the data will be available through application on dbGaP.
Results
Genome-Wide Association Study Between ALS Risk and SNPs
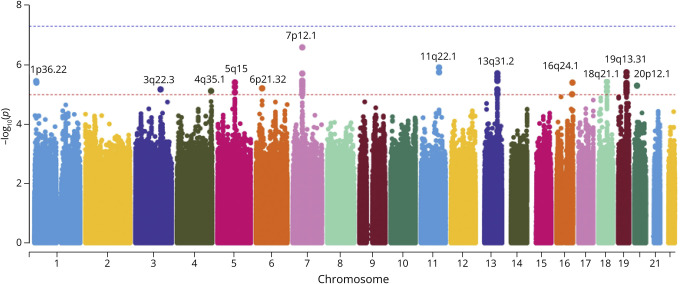
A total of 378 ALS cases and 235 controls of European ancestry passed the quality control. These participants' demographics information was provided in eTable 1. We calculated 14,125,267 association statistics for imputed genotypes. None of the SNPs passed the genome-wide significance threshold of 5E-8, but there were 150 SNPs with p-values less than 1E-5 (eTable 2). These 150 SNPs were localized to 12 cytogenetic locations (Figure 1).
Figure 1. Manhattan Plot for SNPs in Northern New England and Ohio ALS Cohort.
The dashed line in blue indicates the significance threshold of 5E-8, and the line in red indicates the threshold of 1E-5. Twelve loci passed the suggestive significance threshold of 1E-5.
The variants of suggestive statistical significance were all located within 15 genes. Table 1 lists the most statistically significant variant for each of the 15 genes. Among them, rs367543041, also known as c.1144G>A (p.A382T), is a missense variant of TARDBP (TAR DNA binding protein), which has been identified to be associated with ALS risk in multiple previous studies.31-33 This SNP was not available among imputed variants in the validation data set from dbGaP. Instead, we tested 2 nearby variants upstream and downstream, rs3835416 and rs148414479, as proxies. The association p-values for the 2 variants were 0.008 and 0.202, respectively.
Table 1.
Fifteen Genes With Suggestive Significant SNPs in Northern New England and Ohio ALS Cohort
| CHR | Gene | Cytogenetic band | SNP IDa | SNP positionb | Allelesc | MAFd | Odds ratio | Pe |
| 1 | KIF1B | 1p36.22 | rs12131785 | 1:10341516 | C/T | 0.19 | 0.48 | 3.57E-6 |
| 1 | TARDBP | 1p36.22 | rs367543041 | 1:11082610 | G/A | 0.17 | 2.95 | 3.95E-6 |
| 3 | IL20RB | 3q22.3 | rs79105994 | 3:136721632 | A/T | 0.44 | 0.56 | 6.67E-6 |
| 5 | MCTP1 | 5q15 | rs73133908 | 5:94085726 | G/A | 0.10 | 0.40 | 3.94E-6 |
| 6 | HLA-DMA | 6p21.32 | rs129654 | 6:32916699 | C/T | 0.23 | 2.05 | 6.18E-6 |
| 11 | CNTN5 | 11q22.1 | rs7949592 | 11:99035893 | G/A | 0.12 | 0.40 | 1.23E-6 |
| 13 | LINC01047 | 13q31.2 | rs7490607 | 13:89868217 | A/G | 0.25 | 0.51 | 3.20E-6 |
| 13 | LINC00440 | 13q31.2 | rs12877053 | 13:89901248 | T/C | 0.25 | 0.50 | 1.94E-6 |
| 18 | MIR4527HG | 18q21.1 | rs28505643 | 18:45081583 | A/T | 0.14 | 0.44 | 3.70E-6 |
| 19 | ZNF226 | 19q13.31 | rs35526214 | 19:44688732 | TA/T | 0.47 | 0.57 | 4.08E-6 |
| 19 | ZNF227 | 19q13.31 | rs2051059 | 19:44724723 | T/C | 0.45 | 0.57 | 4.41E-6 |
| 19 | ZNF233 | 19q13.31 | rs8106766 | 19:44769596 | T/C | 0.45 | 0.57 | 5.52E-6 |
| 19 | ZNF235 | 19q13.31 | rs2125579 | 19:44792701 | G/T | 0.45 | 0.57 | 5.25E-6 |
| 19 | ZNF112 | 19q13.31 | rs2722733 | 19:44845759 | G/T | 0.23 | 0.49 | 2.39E-6 |
| 20 | MACROD2 | 20p12.1 | rs67253970 | 20:14387190 | G/C | 0.23 | 2.07 | 5.03E-6 |
This table lists the most significant SNP of each gene, and the other SNPs can be found in eTable 2.
Positions are encoded in GRCh37/hg19.
Major allele/minor allele (effect allele).
Minor allele frequency (MAF).
p- Values of SNPs were calculated from logistic regression adjusting for sex, age at symptom onset, and the first 10 principal components.
Association Study Between ALS Risk and SNP-Smoking Interactions
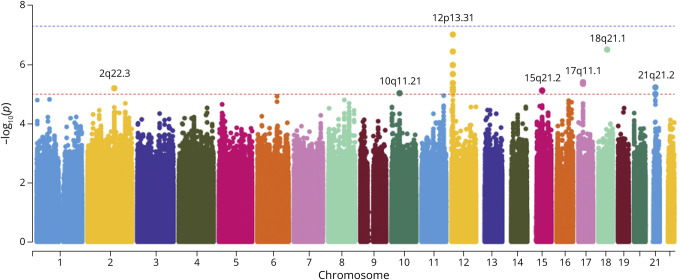
For SNP-smoking interaction analysis, 276 ALS cases and 230 controls had available data on smoking status. We found 19 SNPs from 7 cytogenetic locations with evidence of interaction with smoking based on the P < 1E-5 threshold (Figure 2). None of the p values for interaction reached P < 5E-8.
Figure 2. Manhattan Plot for SNP-Smoking Interactions in Northern New England and Ohio ALS Cohort.
The dashed line in blue indicates the significance threshold of 5E-8, and the line in red indicates the threshold of 1E-5. Seven loci passed the suggestive significance threshold of 1E-5.
Among these 19 SNPs, rs3815479 and rs201995562 are intronic variants located at the GDF3 gene (growth differentiation factor 3) and MYO5B gene (myosin VB), as displayed in Table 2. The rest of the SNPs were not located within any genes. The entire list of the 19 SNPs with evidence of interaction with smoking is provided in eTable 3.
Table 2.
Two Genes With Suggestive Smoking-Associated SNPs in Northern New England and Ohio ALS Cohort
| CHR | Gene | Cytogenetic band | SNP ID | SNP positiona | Allelesb | MAFc | Odds ratio | Pd |
| 12 | GFD3 | 12p13.31 | rs3815479 | 12:7843373 | C/T | 0.49 | 0.53 | 4.23E-6 |
| 18 | MYO5B | 18q21.1 | rs201995562 | 18:47698658 | C/A | 0.33 | 2.19 | 3.13E-7 |
Positions are encoded in GRCh37/hg19.
Major allele/minor allele (effect allele).
Minor allele frequency (MAF).
p- Values of SNP-smoking interaction were calculated from logistic regression adjusting for sex, age at symptom onset, smoking main effect, and the first 10 principal components.
Transcriptome-Wide Association Study of Tissue-Specific Predicted Gene Expression Levels and ALS Risk
We identified 8 genes from 5 tissues suggestively associated with ALS risk with FDR-adjusted P < 0.30, as displayed in Table 3. Higher predicted expression levels of ZNF235 showed marginal significant associations with lower ALS risk in the brain caudate tissue (p = 6.76E-5, FDR = 0.14) and skeletal muscle tissue (p = 3.84E-5, FDR = 0.29). SNPs within ZNF235 were also found to be associated with ALS risk in our GWAS analysis. The Miami plots, including both GWAS and TWAS results in brain caudate and skeletal muscle within the 1 Mb region of ZNF235, can be found in eFigure 2.
Table 3.
Eight Gene Expressions Identified by TWAS to be Suggestively Associated With ALS Risk With FDR <0.3
| Tissue | Gene | CHR | Cytogenetic band | Odds ratio | p Value | FDRa |
| Brain caudate | CEP43 b | 6 | 6q27 | 0.68 | 8.10E-6 | 0.03 |
| Brain caudate | ZNF235 | 19 | 19q13.31 | 0.71 | 6.76E-5 | 0.14 |
| Brain hippocampus | RPL7P18 | 5 | 5q15 | 0.71 | 5.05E-5 | 0.14 |
| Brain hippocampus | ANO5 | 11 | 11p14.3 | 0.72 | 1.35E-4 | 0.19 |
| Brain hippocampus | RP11-381K20.5 | 5 | 5q31.2 | 1.38 | 2.23E-4 | 0.21 |
| Brain substantia | ZBTB14 | 18 | 18p11.31 | 0.71 | 9.72E-5 | 0.20 |
| Whole blood | STIL | 1 | 1p33 | 0.64 | 3.36E-5 | 0.21 |
| Skeletal muscle | ZNF235 | 19 | 19q13.31 | 0.70 | 3.84E-5 | 0.29 |
FDR values were calculated in each tissue separately.
The CEP43 gene is also known as FGFR1OP.
Discussion
In this study, we conducted GWAS and TWAS on ALS cases and controls from Northern New England and Ohio to explore the underlying genetic architecture of ALS. Our GWAS identified 15 genes associated with ALS risk, characterized by SNPs with suggestive significance (P < 1E-5), while TWAS identified 8 gene expression levels across 5 tissues suggestively associated with ALS risk (FDR <0.3).
For GWAS findings, we identified a suggestively significant variant, rs367543041, located within the ALS-associated gene TARDBP. This result aligns with previous studies reporting rs367543041 as an ALS risk variant.31-33 The TARDBP variation, particularly the p.A382T missense variant (rs367543041), has been linked to approximately 30% of ALS cases within the genetically conserved Sardinian population.32,33 Although we could not directly replicate this variant in the larger dbGaP data set because this SNP was not included in the imputed genotype, we observed an association with a nearby proxy SNP, supporting the involvement of TARDBP variation in ALS susceptibility. The GWAS also identified KIF1B, a member of the kinesin family, associated with ALS risk.34 While there is no direct evidence linking KIF1B variants to ALS,35 a study observed a differential regulation of KIF1B in sciatic nerve cells and the spinal cord, suggesting its potential significance in ALS.36 In addition, another member of the kinesin family, KIF5A, has been identified to be associated with ALS in previous studies.10,37 Another gene identified as suggestively significant in the GWAS is MACROD2, a mono-ADP ribosylhydrolase that responds to DNA damage by nuclear export to the cytoplasm.38 In previous studies, MACROD2 has been reported as a neurodevelopmental-related gene,39,40 recognized as a susceptibility gene for autism spectrum disorders and schizophrenia.39,41
We additionally explored SNP-smoking interactions, identifying variants in GDF3 and MYO5B as suggestively interacting with cigarette smoking to influence ALS risk. GDF3 is a member of growth differentiation factors, which constitutes a subfamily of the transforming growth factor-β (TGF-β) superfamily.42,43 GDF3 has been identified as an inhibitor of bone morphogenetic proteins (BMPs).44 BMPs play a key role in inducing the formation of cartilage, bone, and skeletal muscle.45,46 The participation of GDF3 in the development of bone and cartilage, acting as an inhibitor of BMPs, may provide insights into its potential association with ALS. MYO5B is a member of the class V myosins participating in intracellular transport.47,48 Another member of the class V myosins, MYO5C, showed an association with late-onset AD based on its gene expression level.49 Despite SNP-smoking interactions being identified for GDF3 and MYO5B, our validation cohort's lack of smoking data precluded replicating these findings. In future analyses, we plan to leverage geographic information system technology and pollution databases from government agencies to explore more potential gene-environment interactions within our cohort, including lead, mercury, pesticides, and air pollution.
For TWAS findings, our results indicate ZNF235 as a potential ALS risk gene. ZNF235 encodes a zinc finger protein that acts as a transcriptional repressor, potentially participating in neuronal differentiation.50,51 Both GWAS and TWAS analyses suggest associations between ZNF235 and ALS, with predicted ZNF235 expression levels in the caudate and skeletal muscle tissues showing reduced expression levels among ALS cases. Despite limited knowledge regarding its role in ALS, our results suggest ZNF235 as a candidate gene warranting further functional investigation. Another TWAS-identified gene, CEP43 (also known as FGFR1OP), is a fusion partner for FGFR1.52 A previous study indicates that FGFR1 can mediate motor neuron apoptosis in ALS.53
Our study had several limitations. The modest sample size likely constrained our power to detect genome-wide significant associations. As a result, our study failed to identify significant associations with other known ALS genetic risk factors, such as SOD1, NUP50, and ERBB4, possibly because of differences in population structure or sample size. To mitigate this limitation, we sought to supplement our cohort with a larger dbGaP data set, but this introduced challenges with genotyping platform differences and unavailable smoking data. Future studies with expanded sample sizes and harmonized genotyping arrays are needed. This study was restricted to individuals of European ancestry. Future research should also include more populations to validate or extend our findings across different ancestral groups and geographical regions. In addition, the significant variant in TARDBP was not directly validated in the dbGaP database, so we can only conclude it as a tentative association. Despite these constraints, our integrated GWAS and TWAS of ALS provided useful insights into genetic susceptibility. Moreover, TWAS enabled us to explore the gene expression levels across 15 potential ALS-related tissues, including 13 brain-related tissues, skeletal muscle tissue, and whole blood tissue, to understand better the underlying genetic mechanisms of ALS in different tissues.
In summary, this study identified variants and genes associated with ALS risk through GWAS and TWAS analyses. We validated the TARDBP association and identified ZNF235 as a potential novel ALS risk gene. Our findings also reinforce the likely complex interplay of genetic and environmental factors in ALS etiology. Follow-up genetic research is important to uncover how identified variants and genes influence motor neuron degeneration in ALS.
Acknowledgment
This research was supported by the Centers for Disease Control (CDC) (R01TS000288) and the NIH Intramural Research Programs, National Institute on Aging (Z01-AG000933). This research used the high-performance computational resources of the Biowulf Linux cluster located at the NIH, Bethesda, MD, USA (biowulf.nih.gov). The authors express our gratitude to the personnel at the Laboratory of Neurogenetics (NIH) for their support. The data sets used for the analyses detailed in this manuscript were acquired from dbGaP through accession numbers phs000101.v5.p1, phs000187, and phs000428 (ncbi.nlm.nih.gov/sites/entrez?db=gap). Data collection and application development for the dbGaP data phs000187 were supported by grants 3P50CA093459, 5P50CA097007, 5R01ES011740, and 5R01CA133996.
Glossary
- AD
Alzheimer disease
- ALS
amyotrophic lateral sclerosis
- BMP
bone morphogenetic protein
- FDR
false discovery rate
- FTD
frontotemporal dementia
- GWAS
genome-wide association study
- PD
Parkinson disease
- SNP
single nucleotide polymorphism
- TWAS
transcriptome-wide association study
Appendix. Authors
| Name | Location | Contribution |
| Siting Li, PhD | Departments of Biomedical Data Science and Epidemiology, Dartmouth College, Hanover, NH | Drafting/revision of the manuscript for content, including medical writing for content; study concept or design; analysis or interpretation of data |
| Jiang Gui, PhD | Department of Biomedical Data Science, Dartmouth College, Hanover, NH | Drafting/revision of the manuscript for content, including medical writing for content; study concept or design; analysis or interpretation of data |
| Michael N. Passarelli, PhD | Department of Epidemiology, Dartmouth College, Hanover, NH | Drafting/revision of the manuscript for content, including medical writing for content; study concept or design |
| Angeline S. Andrew, PhD | Dartmouth Health, Lebanon, NH | Drafting/revision of the manuscript for content, including medical writing for content; study concept or design; analysis or interpretation of data |
| Kathleen M. Sullivan, MBA | Dartmouth Health, Lebanon, NH | Drafting/revision of the manuscript for content, including medical writing for content; major role in the acquisition of data |
| Kevin A. Cornell, MS | Dartmouth Health, Lebanon, NH | Drafting/revision of the manuscript for content, including medical writing for content; major role in the acquisition of data |
| Bryan J. Traynor, MD, PhD | Neuromuscular Diseases Research Section, National Institute on Aging; National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda; RNA Therapeutics Laboratory, National Center for Advancing Translational Sciences, National Institutes of Health, Rockville, MD | Drafting/revision of the manuscript for content, including medical writing for content; major role in the acquisition of data; study concept or design; analysis or interpretation of data |
| Ali Stark, BS | Neuromuscular Diseases Research Section, National Institute on Aging, National Institutes of Health, Bethesda, MD | Drafting/revision of the manuscript for content, including medical writing for content; major role in the acquisition of data |
| Ruth Chia, PhD | Neuromuscular Diseases Research Section, National Institute on Aging, National Institutes of Health, Bethesda, MD | Drafting/revision of the manuscript for content, including medical writing for content |
| Rebecca M. Kuenzler, MD | Cleveland Clinic, OH | Drafting/revision of the manuscript for content, including medical writing for content |
| Erik P. Pioro, MD, PhD | Department of Medicine, University of British Columbia, Vancouver, BC, Canada | Drafting/revision of the manuscript for content, including medical writing for content |
| Walter G. Bradley, DM, FRCP | University of Miami Miller School of Medicine, Miami, FL | Drafting/revision of the manuscript for content, including medical writing for content; study concept or design |
| Elijah W. Stommel, MD, PhD | Dartmouth Health, Lebanon, NH; Department of Neurology, Geisel School of Medicine at Dartmouth, Hanover, NH | Drafting/revision of the manuscript for content, including medical writing for content; major role in the acquisition of data; study concept or design |
Study Funding
This research was supported by the Centers for Disease Control (CDC) (R01TS000288) and the NIH Intramural Research Programs, National Institute on Aging (Z01-AG000933).
Disclosure
B. J. Traynor holds patents on the clinical testing and therapeutic intervention for the hexanucleotide repeat expansion of C9orf72. Go to Neurology.org/NG for full disclosures.
References
- 1.Renton AE, Majounie E, Waite A, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72(2):257-268. doi: 10.1016/j.neuron.2011.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chia R, Chiò A, Traynor BJ. Novel genes associated with amyotrophic lateral sclerosis: diagnostic and clinical implications. Lancet Neurol. 2018;17(1):94-102. doi: 10.1016/S1474-4422(17)30401-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Renton AE, Chiò A, Traynor BJ. State of play in amyotrophic lateral sclerosis genetics. Nat Neurosci. 2014;17(1):17-23. doi: 10.1038/nn.3584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mitchell JD, Borasio GD. Amyotrophic lateral sclerosis. Lancet. 2007;369(9578):2031-2041. doi: 10.1016/S0140-6736(07)60944-1 [DOI] [PubMed] [Google Scholar]
- 5.Van Rheenen W, van der Spek RA, Bakker MK, et al. Common and rare variant association analyses in amyotrophic lateral sclerosis identify 15 risk loci with distinct genetic architectures and neuron-specific biology. Nat Genet. 2021;53(12):1636-1648. doi: 10.1038/s41588-021-00973-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mehta P, Raymond J, Nair T, et al. Prevalence of ALS in all 50 states in the United States, data from the National ALS Registry, 2011-2018. Amyotroph Lateral Scler Front Degener. 2024;1-7. doi: 10.1080/21678421.2024.2358786 [DOI] [PubMed] [Google Scholar]
- 7.Masrori P, Van Damme P. Amyotrophic lateral sclerosis: a clinical review. Eur J Neurol. 2020;27(10):1918-1929. doi: 10.1111/ene.14393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diekstra FP, Van Deerlin VM, Van Swieten JC, et al. C9orf72 and UNC13A are shared risk loci for amyotrophic lateral sclerosis and frontotemporal dementia: a genome-wide meta-analysis. Ann Neurol. 2014;76(1):120-133. doi: 10.1002/ana.24198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Rheenen W, Shatunov A, Dekker AM, et al. Genome-wide association analyses identify new risk variants and the genetic architecture of amyotrophic lateral sclerosis. Nat Genet. 2016;48(9):1043-1048. doi: 10.1038/ng.3622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nicolas A, Kenna KP, Renton AE, et al. Genome-wide analyses identify KIF5A as a novel ALS gene. Neuron. 2018;97(6):1267-1288. doi: 10.1016/j.neuron.2018.02.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park S, Kim D, Song J, Joo JWJ. An integrative transcriptome-wide analysis of amyotrophic lateral sclerosis for the identification of potential genetic markers and drug candidates. Int J Mol Sci. 2021;22(6):3216. doi: 10.3390/ijms22063216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Megat S, Mora N, Sanogo J, et al. Integrative genetic analysis illuminates ALS heritability and identifies risk genes. Nat Commun. 2023;14(1):342. doi: 10.1038/s41467-022-35724-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ryan M, Heverin M, McLaughlin RL, Hardiman O. Lifetime risk and heritability of amyotrophic lateral sclerosis. JAMA Neurol. 2019;76(11):1367-1374. doi: 10.1001/jamaneurol.2019.2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D'Amico E, Factor-Litvak P, Santella RM, Mitsumoto H. Clinical perspective on oxidative stress in sporadic amyotrophic lateral sclerosis. Free Radic Biol Med. 2013;65:509-527. doi: 10.1016/j.freeradbiomed.2013.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsang CK, Liu Y, Thomas J, Zhang Y, Zheng XFS. Superoxide dismutase 1 acts as a nuclear transcription factor to regulate oxidative stress resistance. Nat Commun. 2014;5:3446. doi: 10.1038/ncomms4446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andrew AS, Bradley WG, Peipert D, et al. Risk factors for amyotrophic lateral sclerosis: a regional United States case-control study. Muscle Nerve. 2021;63(1):52-59. doi: 10.1002/mus.27085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andrew AS, Pioro EP, Li M, et al. The incidence of amyotrophic lateral sclerosis in Ohio 2016-2018: the Ohio population-based ALS Registry. Neuroepidemiology. 2021;55(3):196-205. doi: 10.1159/000515103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blauwendraat C, Faghri F, Pihlstrom L, et al. NeuroChip, an updated version of the NeuroX genotyping platform to rapidly screen for variants associated with neurological diseases. Neurobiol Aging. 2017;57:247.e9-e247.e13. doi: 10.1016/j.neurobiolaging.2017.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559-575. doi: 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4(1):7-015. doi: 10.1186/s13742-015-0047-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chia R, Saez-Atienzar S, Murphy N, et al. ; International Myasthenia Gravis Genomics Consortium. Identification of genetic risk loci and prioritization of genes and pathways for myasthenia gravis: a genome-wide association study. Proc Natl Acad Sci U S A. 2022;119(5):e2108672119. doi: 10.1073/pnas.2108672119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.International HapMap 3 Consortium, Altshuler DM, Gibbs RA, et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467(7311):52-58. doi: 10.1038/nature09298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Das S, Forer L, Schönherr S, et al. Next-generation genotype imputation service and methods. Nat Genet. 2016;48(10):1284-1287. doi: 10.1038/ng.3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.1000 Genomes Project Consortium, Auton A, Brooks LD, et al. A global reference for human genetic variation. Nature. 2015;526(7571):68-74. doi: 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sudmant PH, Rausch T, Gardner EJ, et al. An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526(7571):75-81. doi: 10.1038/nature15394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.internationalgenome.org/ The International Genome Sample Resource. Accessed August 22, 2022.
- 27.Yin L, Zhang H, Tang Z, et al. rMVP: a memory-efficient, visualization-enhanced, and parallel-accelerated tool for genome-wide association study. Genomics Proteomics Bioinformatics. 2021;19(4):619-628. doi: 10.1016/j.gpb.2020.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mailman MD, Feolo M, Jin Y, et al. The NCBI dbGaP database of genotypes and phenotypes. Nat Genet. 2007;39(10):1181-1186. doi: 10.1038/ng1007-1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gamazon ER, Wheeler HE, Shah KP, et al. ; GTEx Consortium. A gene-based association method for mapping traits using reference transcriptome data. Nat Genet. 2015;47(9):1091-1098. doi: 10.1038/ng.3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.GTEx Consortium, Laboratory, Data Analysis &Coordinating Center LDACC—Analysis Working Group, Statistical Methods groups—Analysis Working Group, et al. Genetic effects on gene expression across human tissues. Nature. 2017;550(7675):204-213. doi: 10.1038/nature24277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Corrado L, Ratti A, Gellera C, et al. High frequency of TARDBP gene mutations in Italian patients with amyotrophic lateral sclerosis. Hum Mutat. 2009;30(4):688-694. doi: 10.1002/humu.20950 [DOI] [PubMed] [Google Scholar]
- 32.Chiò A, Borghero G, Pugliatti M, et al. ; Italian Amyotrophic Lateral Sclerosis Genetic ITALSGEN Consortium. Large proportion of amyotrophic lateral sclerosis cases in Sardinia due to a single founder mutation of the TARDBP gene. Arch Neurol. 2011;68(5):594-598. doi: 10.1001/archneurol.2010.352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Orru S, Manolakos E, Orrù N, et al. High frequency of the TARDBP p. Ala382Thr mutation in Sardinian patients with amyotrophic lateral sclerosis. Clin Genet. 2012;81(2):172-178. doi: 10.1111/j.1399-0004.2011.01668.x [DOI] [PubMed] [Google Scholar]
- 34.Hirokawa N, Takemura R. Kinesin superfamily proteins and their various functions and dynamics. Exp Cell Res. 2004;301(1):50-59. doi: 10.1016/j.yexcr.2004.08.010 [DOI] [PubMed] [Google Scholar]
- 35.Conforti L, Dell'Agnello C, Calvaresi N, et al. Kif1Bbeta isoform is enriched in motor neurons but does not change in a mouse model of amyotrophic lateral sclerosis. J Neurosci Res. 2003;71(5):732-739. doi: 10.1002/jnr.10517 [DOI] [PubMed] [Google Scholar]
- 36.Maximino JR, de Oliveira GP, Alves CJ, Chadi G. Deregulated expression of cytoskeleton related genes in the spinal cord and sciatic nerve of presymptomatic SOD1G93A amyotrophic lateral sclerosis mouse model. Front Cell Neurosci. 2014;8:148. doi: 10.3389/fncel.2014.00148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brenner D, Yilmaz R, Müller K, et al.; German ALS network MND-NET. Hot-spot KIF5A mutations cause familial ALS. Brain. 2018;141(3):688-697. doi: 10.1093/brain/awx370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Golia B, Moeller GK, Jankevicius G, et al. ATM induces MacroD2 nuclear export upon DNA damage. Nucleic Acids Res. 2017;45(1):244-254. doi: 10.1093/nar/gkw904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.The Autism Spectrum Disorders Working Group of The Psychiatric Genomics Consortium. Meta-analysis of GWAS of over 16,000 individuals with autism spectrum disorder highlights a novel locus at 10q24. 32 and a significant overlap with schizophrenia. Mol Autism. 2017;8:21. doi: 10.1186/s13229-017-0137-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ito H, Morishita R, Mizuno M, Kawamura N, Tabata H, Nagata K. Biochemical and morphological characterization of a neurodevelopmental disorder-related mono-ADP-ribosylhydrolase, MACRO domain containing 2. Dev Neurosci. 2018;40(3):278-287. doi: 10.1159/000492271 [DOI] [PubMed] [Google Scholar]
- 41.Xu B, Woodroffe A, Rodriguez-Murillo L, et al. Elucidating the genetic architecture of familial schizophrenia using rare copy number variant and linkage scans. Proc Natl Acad Sci U S A. 2009;106(39):16746-16751. doi: 10.1073/pnas.0908584106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McPherron AC, Lee S-J. GDF-3 and GDF-9: two new members of the transforming growth factor-beta superfamily containing a novel pattern of cysteines. J Biol Chem. 1993;268(5):3444-3449. doi: 10.1016/s0021-9258(18)53714-5 [DOI] [PubMed] [Google Scholar]
- 43.Knight PG, Glister C. TGF-beta superfamily members and ovarian follicle development. Reproduction. 2006;132(2):191-206. doi: 10.1530/rep.1.01074 [DOI] [PubMed] [Google Scholar]
- 44.Levine AJ, Brivanlou AH. GDF3, a BMP inhibitor, regulates cell fate in stem cells and early embryos. Development. 2006;133(2):209-216. doi: 10.1242/dev.02192 [DOI] [PubMed] [Google Scholar]
- 45.Wozney JM, Rosen V, Celeste AJ, et al. Novel regulators of bone formation: molecular clones and activities. Science. 1988;242(4885):1528-1534. doi: 10.1126/science.3201241 [DOI] [PubMed] [Google Scholar]
- 46.Sartori R, Gregorevic P, Sandri M. TGFβ and BMP signaling in skeletal muscle: potential significance for muscle-related disease. Trends Endocrinol Metab. 2014;25(9):464-471. doi: 10.1016/j.tem.2014.06.002 [DOI] [PubMed] [Google Scholar]
- 47.Titus MA. Myosin-driven intracellular transport. Cold Spring Harb Perspect Biol. 2018;10(3):a021972. doi: 10.1101/cshperspect.a021972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hammer JA 3rd, Sellers JR. Walking to work: roles for class V myosins as cargo transporters. Nat Rev Mol Cell Biol. 2011;13(1):13-26. doi: 10.1038/nrm3248 [DOI] [PubMed] [Google Scholar]
- 49.Miyashita A, Hatsuta H, Kikuchi M, et al. ; Japanese Alzheimer's Disease Neuroimaging Initiative. Genes associated with the progression of neurofibrillary tangles in Alzheimer's disease. Transl Psychiatry. 2014;4(6):e396. doi: 10.1038/tp.2014.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shannon M, Hamilton AT, Gordon L, Branscomb E, Stubbs L. Differential expansion of zinc-finger transcription factor loci in homologous human and mouse gene clusters. Genome Res. 2003;13(6A):1097-1110. doi: 10.1101/gr.963903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bu S, Lv Y, Liu Y, Qiao S, Wang H. Zinc finger proteins in neuro-related diseases progression. Front Neurosci. 2021;15:760567. doi: 10.3389/fnins.2021.760567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Popovici C, Zhang B, Grégoire MJ, et al. The t (6; 8)(q27; p11) translocation in a stem cell myeloproliferative disorder fuses a novel gene, FOP, to fibroblast growth factor receptor 1. Blood. 1999;93(4):1381-1389. doi: 10.1182/blood.v93.4.1381.404k30_1381_1389 [DOI] [PubMed] [Google Scholar]
- 53.Cassina P, Pehar M, Vargas MR, et al. Astrocyte activation by fibroblast growth factor-1 and motor neuron apoptosis: implications for amyotrophic lateral sclerosis. J Neurochem. 2005;93(1):38-46. doi: 10.1111/j.1471-4159.2004.02984.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
We are in the process of uploading the genotype data used in this study to dbGaP with the accession number phs000101. Once processed, the data will be available through application on dbGaP.