Abstract
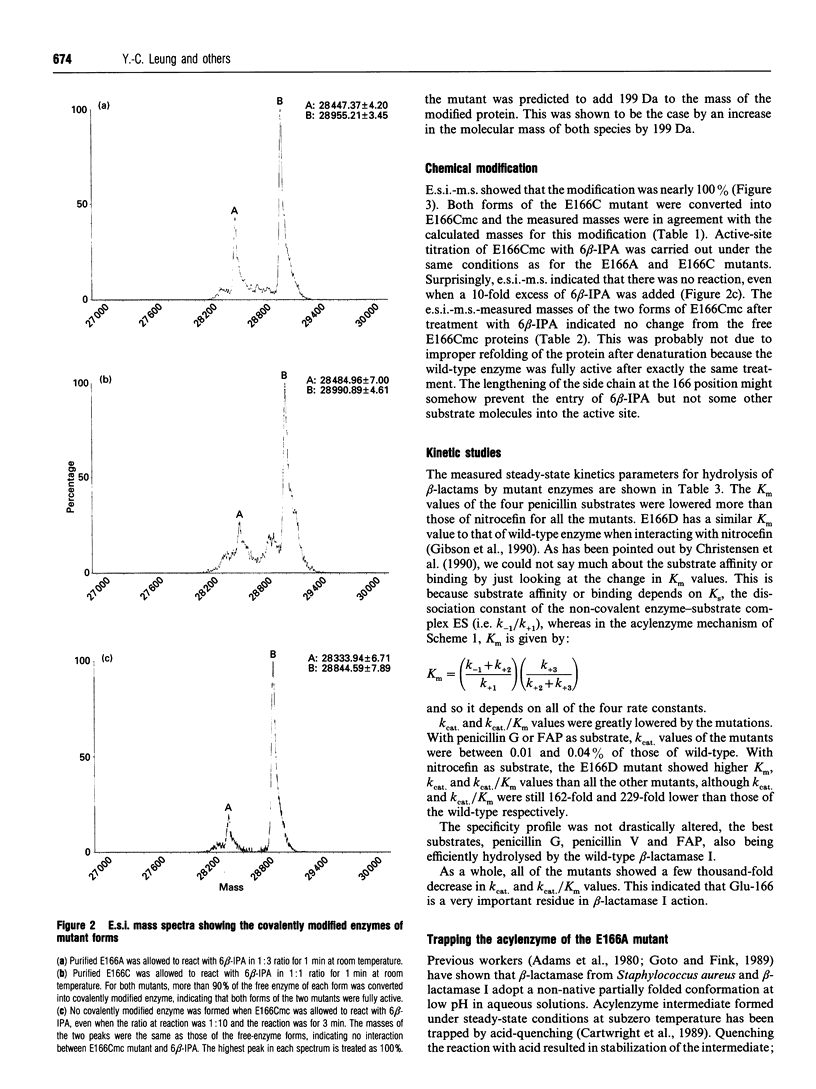
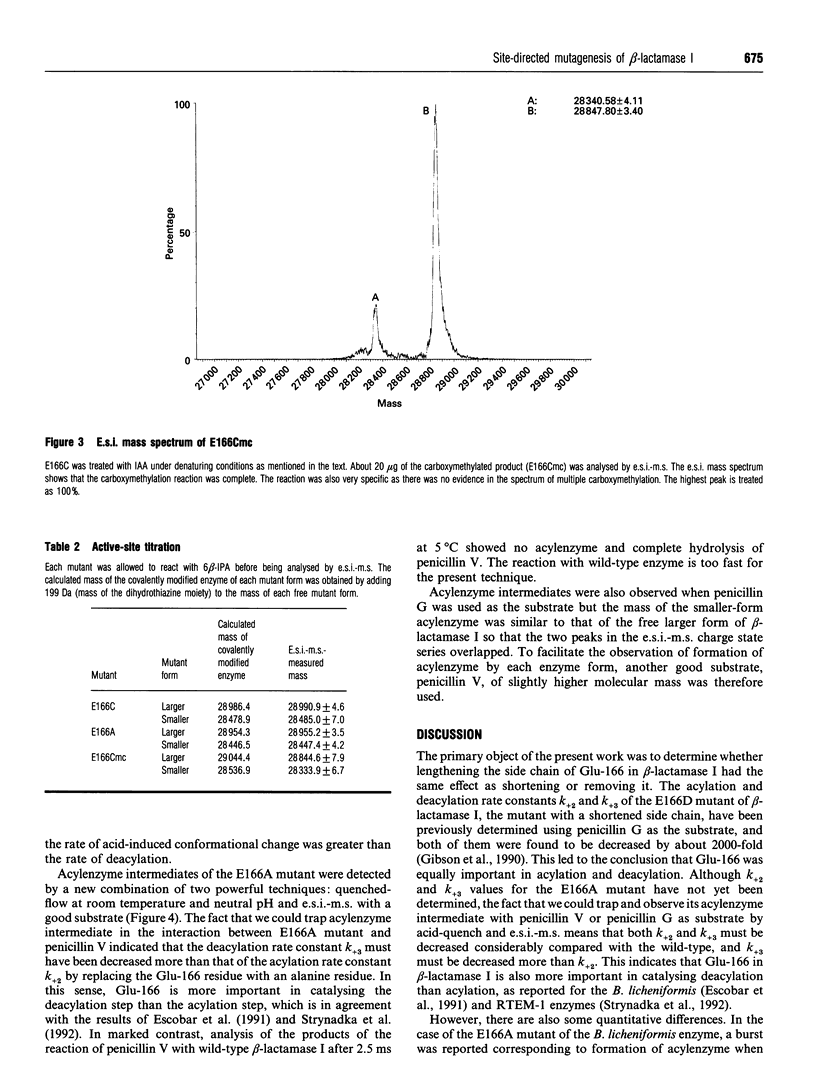
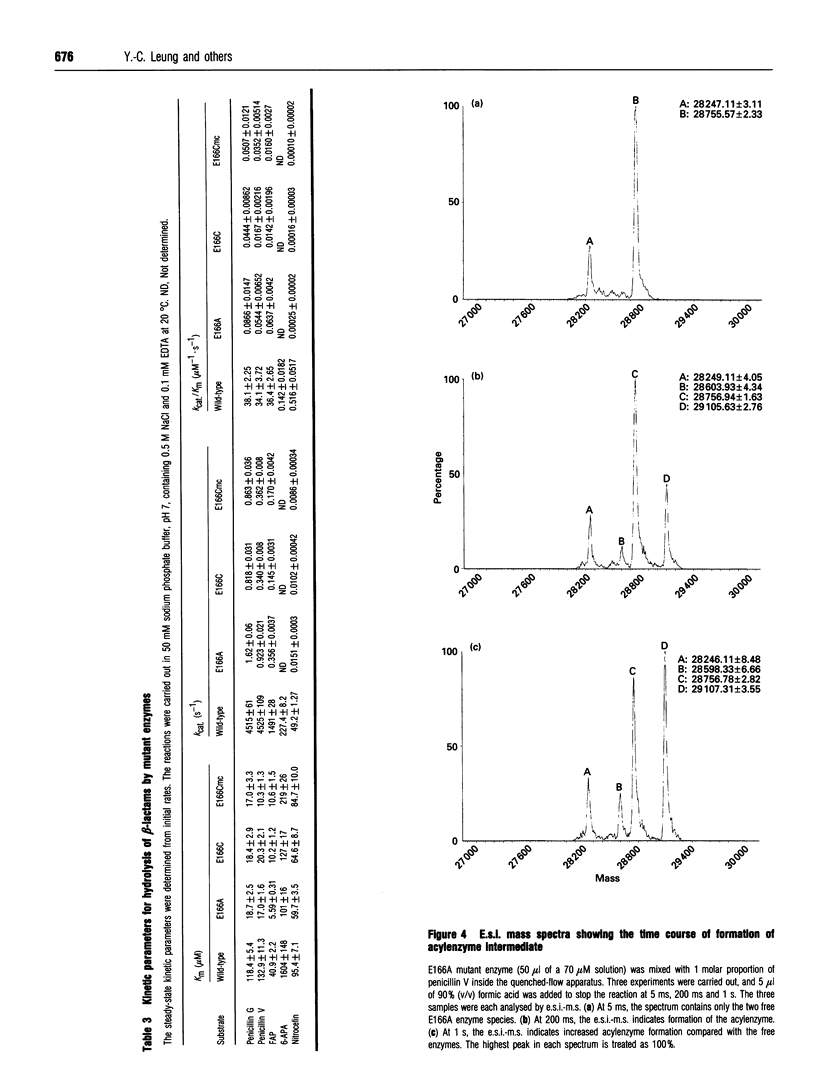
Two Glu-166 mutants of beta-lactamase I from Bacillus cereus 569/H were constructed: one with a lengthened side chain (E166Cmc, the S-carboxymethylcysteine mutant) and the other with the side chain shortened and made non-polar (E166A). Their kinetic properties were studied and compared with those of the wild-type and the E166D mutant (with a shortened side chain) previously made by Gibson, Christensen and Waley (1990) (Biochem. J. 272, 613-619). Surprisingly, with good penicillin substrates, Km, kcat. and kcat./Km of the two conservative mutants (E166Cmc and E166D) are similar to those of the non-conservative mutant E166A. Their kcat. values are 3000-fold lower than that of the wild-type enzyme, showing that Glu-166 is a very important residue. The acylenzyme intermediate of E166A and a good substrate, penicillin V, was trapped by acid-quench and observed by electrospray ionization mass spectrometry, suggesting that Glu-166 is more important in catalysing the deacylation step than the acylation step. The beta-lactamase I E166A mutant is about 200-fold more active than the Bacillus licheniformis E166A mutant with nitrocefin or 6 beta-furylacryloyl-amidopenicillanic acid as substrate. This suggested that other groups in the active site of the beta-lactamase I mutant may activate the catalytic water molecule for deacylation.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adachi H., Ohta T., Matsuzawa H. Site-directed mutants, at position 166, of RTEM-1 beta-lactamase that form a stable acyl-enzyme intermediate with penicillin. J Biol Chem. 1991 Feb 15;266(5):3186–3191. [PubMed] [Google Scholar]
- Ambler R. P., Coulson A. F., Frère J. M., Ghuysen J. M., Joris B., Forsman M., Levesque R. C., Tiraby G., Waley S. G. A standard numbering scheme for the class A beta-lactamases. Biochem J. 1991 May 15;276(Pt 1):269–270. doi: 10.1042/bj2760269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambler R. P. The structure of beta-lactamases. Philos Trans R Soc Lond B Biol Sci. 1980 May 16;289(1036):321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- Burnette W. N. "Western blotting": electrophoretic transfer of proteins from sodium dodecyl sulfate--polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem. 1981 Apr;112(2):195–203. doi: 10.1016/0003-2697(81)90281-5. [DOI] [PubMed] [Google Scholar]
- Carter P., Wells J. A. Dissecting the catalytic triad of a serine protease. Nature. 1988 Apr 7;332(6164):564–568. doi: 10.1038/332564a0. [DOI] [PubMed] [Google Scholar]
- Cartwright S. J., Tan A. K., Fink A. L. Trapping the acyl-enzyme intermediate in beta-lactamase I catalysis. Biochem J. 1989 Nov 1;263(3):905–912. doi: 10.1042/bj2630905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen H., Martin M. T., Waley S. G. Beta-lactamases as fully efficient enzymes. Determination of all the rate constants in the acyl-enzyme mechanism. Biochem J. 1990 Mar 15;266(3):853–861. [PMC free article] [PubMed] [Google Scholar]
- Cohen S. A., Pratt R. F. Inactivation of Bacillus cereus beta-lactamase I by 6 beta-bromopencillanic acid: mechanism. Biochemistry. 1980 Aug 19;19(17):3996–4003. doi: 10.1021/bi00558a017. [DOI] [PubMed] [Google Scholar]
- Davies R. B., Abraham E. P. Separation, purification and properties of beta-lactamase I and beta-lactamase II from Bacillus cereus 569/H/9. Biochem J. 1974 Oct;143(1):115–127. doi: 10.1042/bj1430115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugaiczyk A., Law S. W., Dennison O. E. Nucleotide sequence and the encoded amino acids of human serum albumin mRNA. Proc Natl Acad Sci U S A. 1982 Jan;79(1):71–75. doi: 10.1073/pnas.79.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar W. A., Tan A. K., Fink A. L. Site-directed mutagenesis of beta-lactamase leading to accumulation of a catalytic intermediate. Biochemistry. 1991 Nov 5;30(44):10783–10787. doi: 10.1021/bi00108a025. [DOI] [PubMed] [Google Scholar]
- Gibson R. M., Christensen H., Waley S. G. Site-directed mutagenesis of beta-lactamase I. Single and double mutants of Glu-166 and Lys-73. Biochem J. 1990 Dec 15;272(3):613–619. doi: 10.1042/bj2720613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto Y., Fink A. L. Conformational states of beta-lactamase: molten-globule states at acidic and alkaline pH with high salt. Biochemistry. 1989 Feb 7;28(3):945–952. doi: 10.1021/bi00429a004. [DOI] [PubMed] [Google Scholar]
- Herzberg O., Kapadia G., Blanco B., Smith T. S., Coulson A. Structural basis for the inactivation of the P54 mutant of beta-lactamase from Staphylococcus aureus PC1. Biochemistry. 1991 Oct 1;30(39):9503–9509. doi: 10.1021/bi00103a017. [DOI] [PubMed] [Google Scholar]
- Jacob F., Joris B., Frère J. M. Active-site serine mutants of the Streptomyces albus G beta-lactamase. Biochem J. 1991 Aug 1;277(Pt 3):647–652. doi: 10.1042/bj2770647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knap A. K., Pratt R. F. Inactivation of the RTEM-1 cysteine beta-lactamase by iodoacetate. The nature of active-site functional groups and comparisons with the native enzyme. Biochem J. 1991 Jan 1;273(Pt 1):85–91. doi: 10.1042/bj2730085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott-Hunziker V., Orlek B. S., Sammes P. G., Waley S. G. 6 beta-Bromopenicillanic acid inactivates beta-lactamase I. Biochem J. 1979 Jan 1;177(1):365–367. doi: 10.1042/bj1770365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott-Hunziker V., Orlek B. S., Sammes P. G., Waley S. G. Kinetics of inactivation of beta-lactamase I by 6 beta-bromopenicillanic acid. Biochem J. 1980 Jun 1;187(3):797–802. doi: 10.1042/bj1870797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox J. R., Moews P. C., Escobar W. A., Fink A. L. A catalytically-impaired class A beta-lactamase: 2 A crystal structure and kinetics of the Bacillus licheniformis E166A mutant. Protein Eng. 1993 Jan;6(1):11–18. doi: 10.1093/protein/6.1.11. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Madgwick P. J., Waley S. G. beta-lactamase I from Bacillus cereus. Structure and site-directed mutagenesis. Biochem J. 1987 Dec 15;248(3):657–662. doi: 10.1042/bj2480657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matagne A., Joris B., Van Beeumen J., Frère J. M. Ragged N-termini and other variants of class A beta-lactamases analysed by chromatofocusing. Biochem J. 1991 Feb 1;273(Pt 3):503–510. doi: 10.1042/bj2730503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mézes P. S., Blacher R. W., Lampen J. O. Processing of Bacillus cereus 569/H beta-lactamase I in Escherichia coli and Bacillus subtilis. J Biol Chem. 1985 Jan 25;260(2):1218–1223. [PubMed] [Google Scholar]
- Persaud K. C., Pain R. H., Virden R. Reversible deactivation of beta-lactamase by quinacillin. Extent of the conformational change in the isolated transitory complex. Biochem J. 1986 Aug 1;237(3):723–730. doi: 10.1042/bj2370723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strynadka N. C., Adachi H., Jensen S. E., Johns K., Sielecki A., Betzel C., Sutoh K., James M. N. Molecular structure of the acyl-enzyme intermediate in beta-lactam hydrolysis at 1.7 A resolution. Nature. 1992 Oct 22;359(6397):700–705. doi: 10.1038/359700a0. [DOI] [PubMed] [Google Scholar]
- Thatcher D. R. The partial amino acid sequence of the extracellular beta-lactamase I of Bacillus cereus 569/H. Biochem J. 1975 May;147(2):313–326. doi: 10.1042/bj1470313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornewell S. J., East A. K., Errington J. An efficient expression and secretion system based on Bacillus subtilis phage phi 105 and its use for the production of B. cereus beta-lactamase I. Gene. 1993 Oct 29;133(1):47–53. doi: 10.1016/0378-1119(93)90223-p. [DOI] [PubMed] [Google Scholar]
- Thornewell S. J., Waley S. G. Site-directed mutagenesis and substrate-induced inactivation of beta-lactamase I. Biochem J. 1992 Dec 15;288(Pt 3):1045–1051. doi: 10.1042/bj2881045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waley S. G. A spectrophotometric assay of beta-lactamase action on penicillins. Biochem J. 1974 Jun;139(3):789–790. doi: 10.1042/bj1390789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waley S. G. Beta-lactamases: a major cause of antibiotic resistance. Sci Prog. 1988;72(288 Pt 4):579–597. [PubMed] [Google Scholar]
- Waley S. G. Kinetic parameters from progress curves of competing substrates. Application to beta-lactamases. Biochem J. 1983 May 1;211(2):511–513. doi: 10.1042/bj2110511. [DOI] [PMC free article] [PubMed] [Google Scholar]


