Abstract
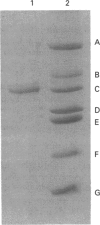
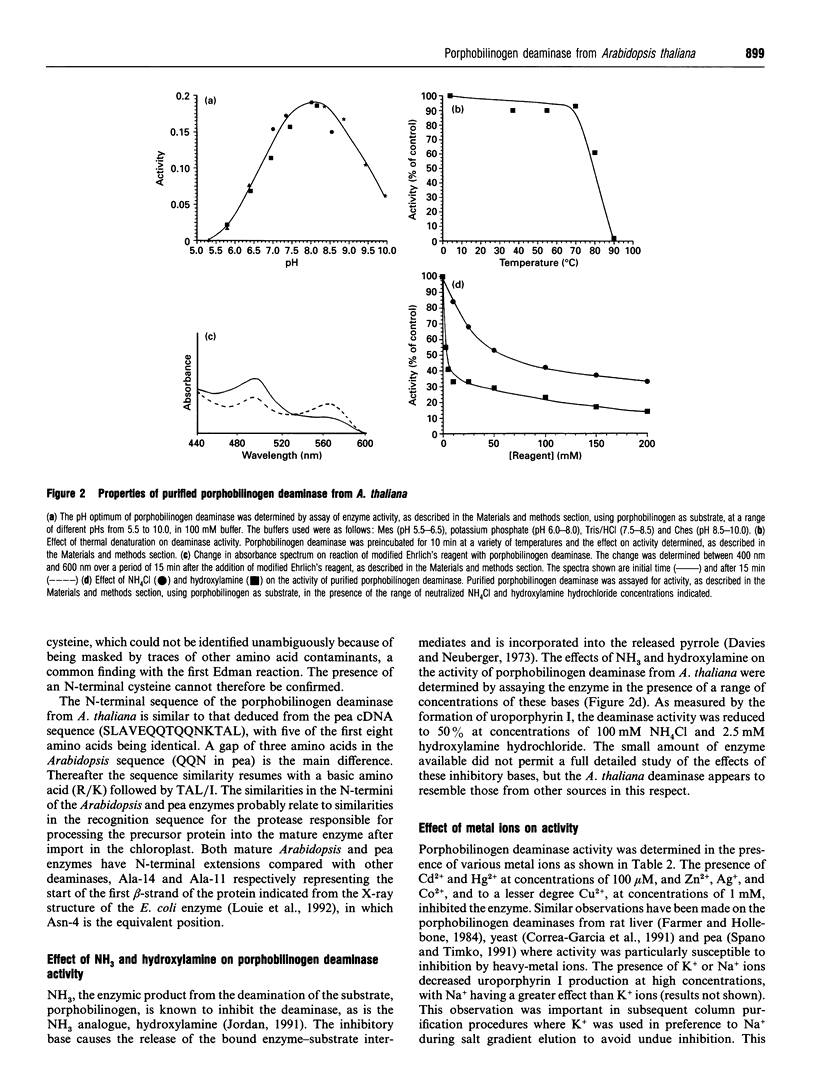
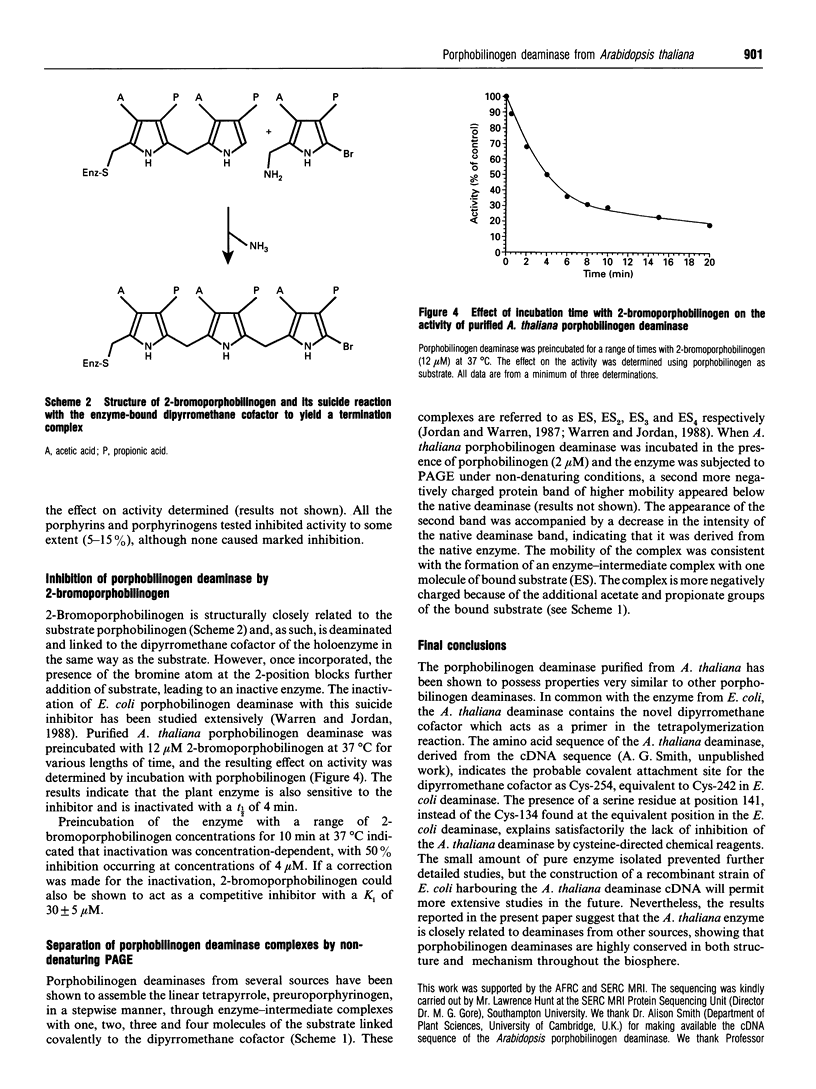
Porphobilinogen deaminase (EC 4.3.1.8) has been purified to homogeneity (16,000-fold) from the plant Arabidopsis thaliana in yields of 8%. The deaminase is a monomer of M(r) 35,000, as shown by SDS/PAGE, and 31,000, using gel-filtration chromatography. The pure enzyme has a Vmax. of 4.5 mumol/h per mg and a Km of 17 +/- 4 microM. Determination of the pI and pH optimum revealed values of 5.2 and 8.0 respectively. The sequence of the N-terminus was found to be NH2-XVAVEQKTRTAI. The deaminase is heat-stable up to 70 degrees C and is inhibited by NH3 and hydroxylamine. The enzyme is inactivated by arginine-, histidine- and lysine-specific reagents. Incubation with the substrate analogue and suicide inhibitor, 2-bromoporphobilinogen, results in chain termination and in inactivation.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson P. M., Desnick R. J. Purification and properties of uroporphyrinogen I synthase from human erythrocytes. Identification of stable enzyme-substrate intermediates. J Biol Chem. 1980 Mar 10;255(5):1993–1999. [PubMed] [Google Scholar]
- Beaumont C., Porcher C., Picat C., Nordmann Y., Grandchamp B. The mouse porphobilinogen deaminase gene. Structural organization, sequence, and transcriptional analysis. J Biol Chem. 1989 Sep 5;264(25):14829–14834. [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Correa García S. R., Rossetti M. V., Batlle A. M. Studies on porphobilinogen-deaminase from Saccharomyces cerevisiae. Z Naturforsch C. 1991 Nov-Dec;46(11-12):1017–1023. doi: 10.1515/znc-1991-11-1215. [DOI] [PubMed] [Google Scholar]
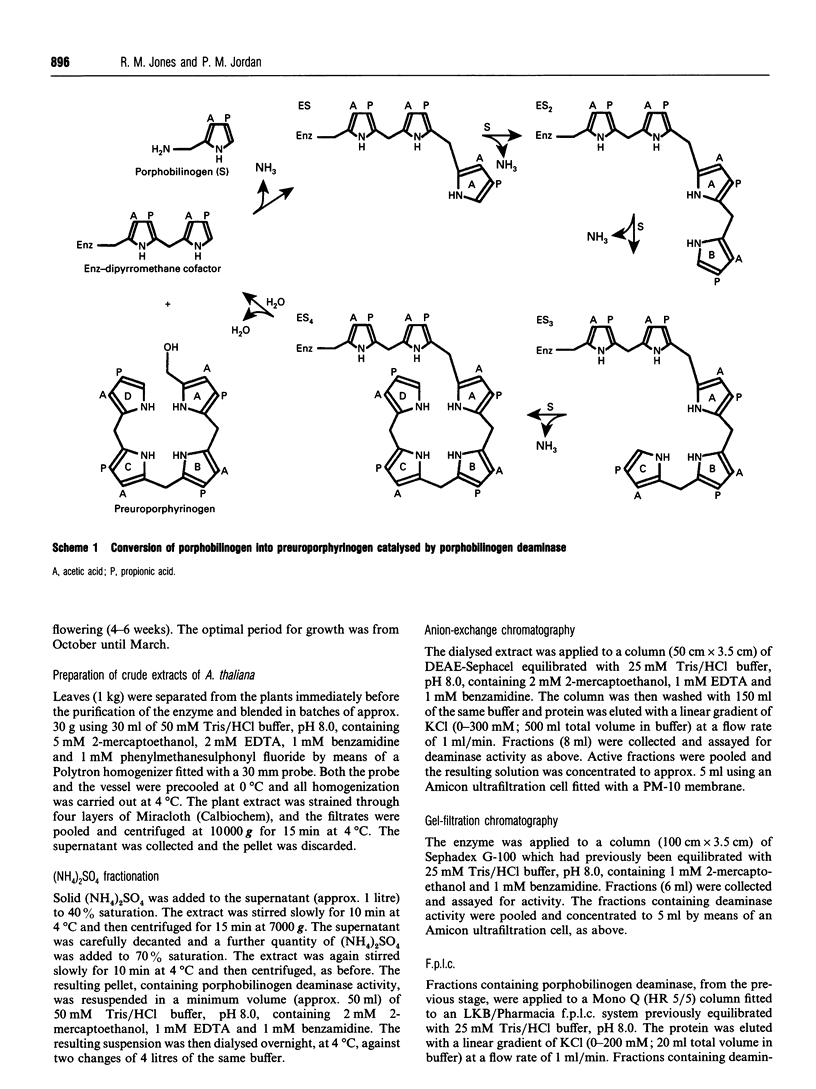
- Davies R. C., Neuberger A. Polypyrroles formed from porphobilinogen and amines by uroporphyrinogen synthetase of Rhodopseudomonas spheroides. Biochem J. 1973 Jul;133(3):471–492. doi: 10.1042/bj1330471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer D. J., Hollebone B. R. Comparative inhibition of hepatic hydroxymethylbilane synthase by both hard and soft metal cations. Can J Biochem Cell Biol. 1984 Jan;62(1):49–54. doi: 10.1139/o84-008. [DOI] [PubMed] [Google Scholar]
- Fumagalli S. A., Kotler M. L., Rossetti M. V., Batlle A. M. Human red cell porphobilinogen deaminase. A simpler method of purification and some unusual properties. Int J Biochem. 1985;17(4):485–494. doi: 10.1016/0020-711x(85)90144-2. [DOI] [PubMed] [Google Scholar]
- Hart G. J., Abell C., Battersby A. R. Purification, N-terminal amino acid sequence and properties of hydroxymethylbilane synthase (porphobilinogen deaminase) from Escherichia coli. Biochem J. 1986 Nov 15;240(1):273–276. doi: 10.1042/bj2400273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart G. J., Leeper F. J., Battersby A. R. Modification of hydroxymethylbilane synthase (porphobilinogen deaminase) by pyridoxal 5'-phosphate. Demonstration of an essential lysine residue. Biochem J. 1984 Aug 15;222(1):93–102. doi: 10.1042/bj2220093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart G. J., Miller A. D., Battersby A. R. Evidence that the pyrromethane cofactor of hydroxymethylbilane synthase (porphobilinogen deaminase) is bound through the sulphur atom of a cysteine residue. Biochem J. 1988 Jun 15;252(3):909–912. doi: 10.1042/bj2520909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi M., Bogorad L. The purification and properties of uroporphyrinogen I synthases and uroporphyrinogen III cosynthase. Interactions between the enzymes. Ann N Y Acad Sci. 1975 Apr 15;244:401–418. doi: 10.1111/j.1749-6632.1975.tb41545.x. [DOI] [PubMed] [Google Scholar]
- Jones R. M., Jordan P. M. Purification and properties of the uroporphyrinogen decarboxylase from Rhodobacter sphaeroides. Biochem J. 1993 Aug 1;293(Pt 3):703–712. doi: 10.1042/bj2930703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan P. M., Seehra J. S. Purification of porphobilinogen synthase from bovine liver. Methods Enzymol. 1986;123:427–434. doi: 10.1016/s0076-6879(86)23053-0. [DOI] [PubMed] [Google Scholar]
- Jordan P. M., Shemin D. Purification and properties of uroporphyrinogen I synthetase from Rhodopseudomonas spheroides. J Biol Chem. 1973 Feb 10;248(3):1019–1024. [PubMed] [Google Scholar]
- Jordan P. M., Thomas S. D., Warren M. J. Purification, crystallization and properties of porphobilinogen deaminase from a recombinant strain of Escherichia coli K12. Biochem J. 1988 Sep 1;254(2):427–435. doi: 10.1042/bj2540427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan P. M., Warren M. J. Evidence for a dipyrromethane cofactor at the catalytic site of E. coli porphobilinogen deaminase. FEBS Lett. 1987 Dec 10;225(1-2):87–92. doi: 10.1016/0014-5793(87)81136-5. [DOI] [PubMed] [Google Scholar]
- Jordan P. M., Warren M. J., Mgbeje B. I., Wood S. P., Cooper J. B., Louie G., Brownlie P., Lambert R., Blundell T. L. Crystallization and preliminary X-ray investigation of Escherichia coli porphobilinogen deaminase. J Mol Biol. 1992 Mar 5;224(1):269–271. doi: 10.1016/0022-2836(92)90590-g. [DOI] [PubMed] [Google Scholar]
- Jordan P. M., Warren M. J., Williams H. J., Stolowich N. J., Roessner C. A., Grant S. K., Scott A. I. Identification of a cysteine residue as the binding site for the dipyrromethane cofactor at the active site of Escherichia coli porphobilinogen deaminase. FEBS Lett. 1988 Aug 1;235(1-2):189–193. doi: 10.1016/0014-5793(88)81260-2. [DOI] [PubMed] [Google Scholar]
- Jordan P. M., Woodcock S. C. Mutagenesis of arginine residues in the catalytic cleft of Escherichia coli porphobilinogen deaminase that affects dipyrromethane cofactor assembly and tetrapyrrole chain initiation and elongation. Biochem J. 1991 Dec 1;280(Pt 2):445–449. doi: 10.1042/bj2800445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander M., Pitt A. R., Alefounder P. R., Bardy D., Abell C., Battersby A. R. Studies on the mechanism of hydroxymethylbilane synthase concerning the role of arginine residues in substrate binding. Biochem J. 1991 Apr 15;275(Pt 2):447–452. doi: 10.1042/bj2750447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louie G. V., Brownlie P. D., Lambert R., Cooper J. B., Blundell T. L., Wood S. P., Warren M. J., Woodcock S. C., Jordan P. M. Structure of porphobilinogen deaminase reveals a flexible multidomain polymerase with a single catalytic site. Nature. 1992 Sep 3;359(6390):33–39. doi: 10.1038/359033a0. [DOI] [PubMed] [Google Scholar]
- MAUZERALL D., GRANICK S. The occurrence and determination of delta-amino-levulinic acid and porphobilinogen in urine. J Biol Chem. 1956 Mar;219(1):435–446. [PubMed] [Google Scholar]
- Mazzetti M. B., Tomio J. M. Characterization of porphobilinogen deaminase from rat liver. Biochim Biophys Acta. 1988 Nov 2;957(1):97–104. doi: 10.1016/0167-4838(88)90161-6. [DOI] [PubMed] [Google Scholar]
- Miller A. D., Hart G. J., Packman L. C., Battersby A. R. Evidence that the pyrromethane cofactor of hydroxymethylbilane synthase (porphobilinogen deaminase) is bound to the protein through the sulphur atom of cysteine-242. Biochem J. 1988 Sep 15;254(3):915–918. doi: 10.1042/bj2540915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petricek M., Rutberg L., Schröder I., Hederstedt L. Cloning and characterization of the hemA region of the Bacillus subtilis chromosome. J Bacteriol. 1990 May;172(5):2250–2258. doi: 10.1128/jb.172.5.2250-2258.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluscec J., Bogorad L. A dipyrrylmethane intermediate in the enzymatic synthesis of uroporphyrinogen. Biochemistry. 1970 Nov 24;9(24):4736–4743. doi: 10.1021/bi00826a017. [DOI] [PubMed] [Google Scholar]
- RIMINGTON C., SVEINSSON S. L. The spectrophotometric determination of uroporphyrin. Scand J Clin Lab Invest. 1950;2(3):209–216. doi: 10.3109/00365515009049872. [DOI] [PubMed] [Google Scholar]
- Raich N., Romeo P. H., Dubart A., Beaupain D., Cohen-Solal M., Goossens M. Molecular cloning and complete primary sequence of human erythrocyte porphobilinogen deaminase. Nucleic Acids Res. 1986 Aug 11;14(15):5955–5968. doi: 10.1093/nar/14.15.5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott A. I., Clemens K. R., Stolowich N. J., Santander P. J., Gonzalez M. D., Roessner C. A. Reconstitution of apo-porphobilinogen deaminase: structural changes induced by cofactor binding. FEBS Lett. 1989 Jan 2;242(2):319–324. doi: 10.1016/0014-5793(89)80493-4. [DOI] [PubMed] [Google Scholar]
- Scott A. I., Roessner C. A., Stolowich N. J., Karuso P., Williams H. J., Grant S. K., Gonzalez M. D., Hoshino T. Site-directed mutagenesis and high-resolution NMR spectroscopy of the active site of porphobilinogen deaminase. Biochemistry. 1988 Oct 18;27(21):7984–7990. doi: 10.1021/bi00421a002. [DOI] [PubMed] [Google Scholar]
- Sharif A. L., Smith A. G., Abell C. Isolation and characterisation of a cDNA clone for a chlorophyll synthesis enzyme from Euglena gracilis. The chloroplast enzyme hydroxymethylbilane synthase (porphobilinogen deaminase) is synthesised with a very long transit peptide in Euglena. Eur J Biochem. 1989 Sep 15;184(2):353–359. doi: 10.1111/j.1432-1033.1989.tb15026.x. [DOI] [PubMed] [Google Scholar]
- Shioi Y., Nagamine M., Kuroki M., Sasa T. Purification by affinity chromatography and properties of uroporphyrinogen I synthetase from Chlorella regularis. Biochim Biophys Acta. 1980 Dec 4;616(2):300–309. doi: 10.1016/0005-2744(80)90147-3. [DOI] [PubMed] [Google Scholar]
- Spano A. J., Timko M. P. Isolation, characterization and partial amino acid sequence of a chloroplast-localized porphobilinogen deaminase from pea (Pisum sativum L.). Biochim Biophys Acta. 1991 Jan 8;1076(1):29–36. doi: 10.1016/0167-4838(91)90216-m. [DOI] [PubMed] [Google Scholar]
- Stubnicer A. C., Picat C., Grandchamp B. Rat porphobilinogen deaminase cDNA: nucleotide sequence of the erythropoietic form. Nucleic Acids Res. 1988 Apr 11;16(7):3102–3102. doi: 10.1093/nar/16.7.3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S. D., Jordan P. M. Nucleotide sequence of the hemC locus encoding porphobilinogen deaminase of Escherichia coli K12. Nucleic Acids Res. 1986 Aug 11;14(15):6215–6226. doi: 10.1093/nar/14.15.6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren M. J., Jordan P. M. Investigation into the nature of substrate binding to the dipyrromethane cofactor of Escherichia coli porphobilinogen deaminase. Biochemistry. 1988 Dec 13;27(25):9020–9030. doi: 10.1021/bi00425a021. [DOI] [PubMed] [Google Scholar]
- Warren M. J., Scott A. I. Tetrapyrrole assembly and modification into the ligands of biologically functional cofactors. Trends Biochem Sci. 1990 Dec;15(12):486–491. doi: 10.1016/0968-0004(90)90304-t. [DOI] [PubMed] [Google Scholar]
- Weber K., Osborn M. The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis. J Biol Chem. 1969 Aug 25;244(16):4406–4412. [PubMed] [Google Scholar]
- Williams D. C. Characterization of the multiple forms of hydroxymethylbilane synthase from rat spleen. Biochem J. 1984 Feb 1;217(3):675–683. doi: 10.1042/bj2170675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams D. C., Morgan G. S., McDonald E., Battersby A. R. Purification of porphobilinogen deaminase from Euglena gracilis and studies of its kinetics. Biochem J. 1981 Jan 1;193(1):301–310. doi: 10.1042/bj1930301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witty M., Wallace-Cook A. D., Albrecht H., Spano A. J., Michel H., Shabanowitz J., Hunt D. F., Timko M. P., Smith A. G. Structure and expression of chloroplast-localized porphobilinogen deaminase from pea (Pisum sativum L.) isolated by redundant polymerase chain reaction. Plant Physiol. 1993 Sep;103(1):139–147. doi: 10.1104/pp.103.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock S. C., Jordan P. M. Evidence for participation of aspartate-84 as a catalytic group at the active site of porphobilinogen deaminase obtained by site-directed mutagenesis of the hemC gene from Escherichia coli. Biochemistry. 1994 Mar 8;33(9):2688–2695. doi: 10.1021/bi00175a043. [DOI] [PubMed] [Google Scholar]