Abstract
OBJECTIVES
The goal of this study was to assess whether a deep learning estimate of age from a chest radiograph image (CXR-Age) can predict longevity beyond chronological age.
BACKGROUND
Chronological age is an imperfect measure of longevity. Biological age, a measure of overall health, may improve personalized care. This paper proposes a new way to estimate biological age using a convolutional neural network that takes as input a CXR image and outputs a chest x-ray age (in years) as a measure of long-term mortality risk.
METHODS
CXR-Age was developed using CXR from 116,035 individuals and validated in 2 held-out testing sets: 1) 75% of the CXR arm of PLCO (Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial) (N = 40,967); and 2) the CXR arm of NLST (National Lung Screening Trial) (N = 5,414). CXR-Age was compared to chronological age and a multivariable regression model of chronological age, risk factors, and radiograph findings to predict all-cause and cardiovascular mortality with a maximum 23 years and 13 years of follow-up, respectively. The primary outcome was observed mortality; results are provided for the testing datasets only.
RESULTS
In the PLCO testing dataset, a 5-year increase in CXR-Age carried a higher risk of all-cause mortality than a 5-year increase in chronological age (CXR-Age hazard ratio [HR]: 2.26 [95% confidence interval (CI): 2.24 to 2.29] vs. chronological age HR: 1.77 [95% CI: 1.75 to 1.78]; p < 0.001). A similar pattern was found for cardiovascular mortality (CXR-Age cause-specific HR: 2.45 per 5 years [95% CI: 2.34 to 2.56] vs. chronological age HR: 1.82 per 5 years [95% CI: 1.74 to 1.90]). Similar results were seen for both outcomes in the NLST external testing dataset. Adding CXR-Age to the multivariable model resulted in significant improvements for predicting both outcomes in both testing datasets (p < 0.001 for all comparisons).
CONCLUSIONS
Based on a CXR image, CXR-Age predicted long-term all-cause and cardiovascular mortality.
Keywords: biological age, cardiovascular risk prediction, chest radiographs, deep learning
Chronological age, defined as the number of years since birth, is a cornerstone of medical decision-making. We decide who is eligible for cancer screening based on chronological age; chronological age is among the most important inputs to current guidelines for primary prevention of atherosclerotic cardiovascular disease (ASCVD). Yet chronological age is an imperfect measure of health and longevity, as individuals age at different rates (1).
Biological age, defined as a cumulative measure of the effects of aging on an individual, has been proposed as a better measure of longevity and susceptibility to aging-related disease (2-4). Vascular age can be assessed by using imaging such as the coronary artery calcium score (5) and carotid intimal medial thickness (6). Other investigators have developed biological age measures using functional (e.g., gait speed [7], frailty [8]), physiological (e.g., vascular compliance) (9), and blood (e.g., DNA methylation [10], telomere length [11]) measures.
An advantage of framing risk in terms of biological age is that it can be easier for patients to grasp than risk score probabilities (5). In this study, biological age was defined as age-normalized mortality risk. For example, a chronologically 70-year-old individual with a biological age of 65 years has similar risk of mortality and expected longevity as the average 65-year-old. As proposed by Grundy (12), substituting a more accurate biological age for chronological age could improve the performance of existing risk scores. An accurate, noninvasive measure of biological age is desirable, to help inform decisions about prevention, screening, and treatment.
The current study proposes a new measure of biological age, based on a convolutional neural network (CNN) analysis of a chest radiograph (CXR or x-ray) image. Chest radiography is the most common diagnostic imaging test (13) and thus provides ample opportunity to assess aging from existing images. CNNs, a form of artificial intelligence, have made major advances in diagnosis (14,15) and, more recently, assessing prognosis (16) from CXRs. We hypothesized that the CNN’s estimate of chest x-ray age (CXR-Age) can predict long-term all-cause and cardiovascular mortality beyond chronological age (Central Illustration).
CENTRAL ILLUSTRATION. A Deep Learning-Based Chest X-Ray Age Predicts Long-Term All-Cause and Cardiovascular Mortality Using Only a Chest X-Ray Image.
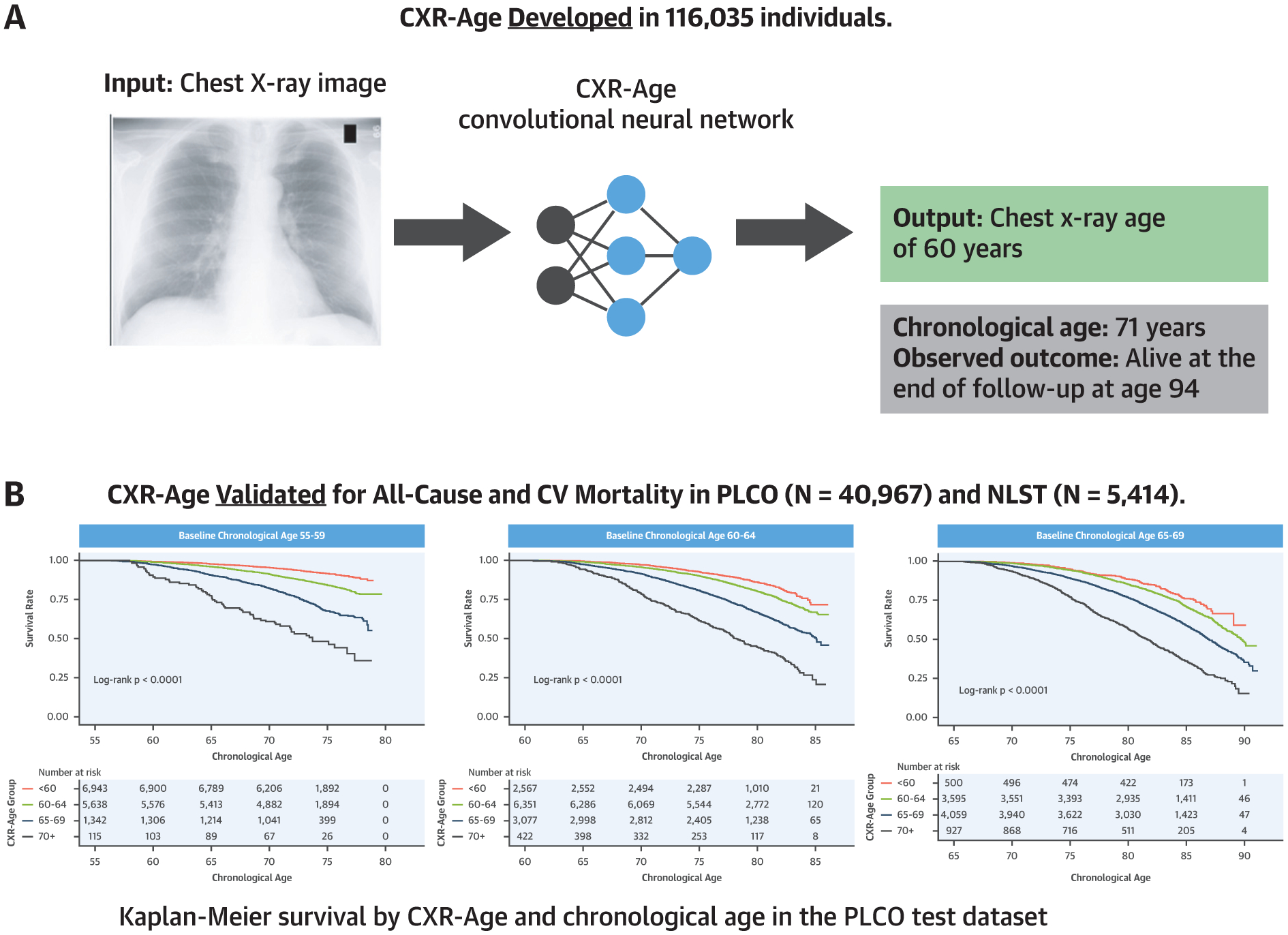
(A) We developed a convolutional neural network that takes a chest x-ray image as input and outputs a chest x-ray age (CXR-Age) in years. (B) Kaplan-Meier survival curves stratified by using baseline chronological age show that CXR-Age predicts all-cause mortality in an independent testing dataset from the PLCO (Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial; N = 40,967). Similar results were found for cardiovascular mortality and in an external testing dataset from the NLST (National Lung Screening Trial; N = 5,414). CV = cardiovascular.
METHODS
OVERVIEW OF CXR-AGE MODEL DEVELOPMENT AND TESTING.
The aim was to develop a CNN to estimate biological age from a chest x-ray image (CXR-Age). CXR-Age was developed in 2 stages (Figure 1). In Stage 1, we trained the model for one task (estimate chronological age from a CXR image) using large publicly available datasets. This model was used solely as a stepping stone for Stage 2 of model development; chronological age estimates are not reported or used at any point in the analysis. In Stage 2, the Stage 1 model was used as the starting point to train a second final model to estimate a biological age from a CXR image (CXR-Age) in a second smaller but more extensively phenotyped dataset with long-term mortality follow-up. This 2-stage technique of first pretraining a model using large publicly available datasets for one task, then using this foundation to fine-tune the model for a second related task, is called “transfer learning” and is frequently used to train CNNs when the second dataset is relatively small. Further details about image preprocessing, model architecture, and model development are provided in the Supplemental Methods. To encourage reproducible research, the CXR-Age model will be released as free open-source software (https://github.com/vineet1992/CXR-Age).
FIGURE 1. Overview of CXR-Age Model Development and Testing.
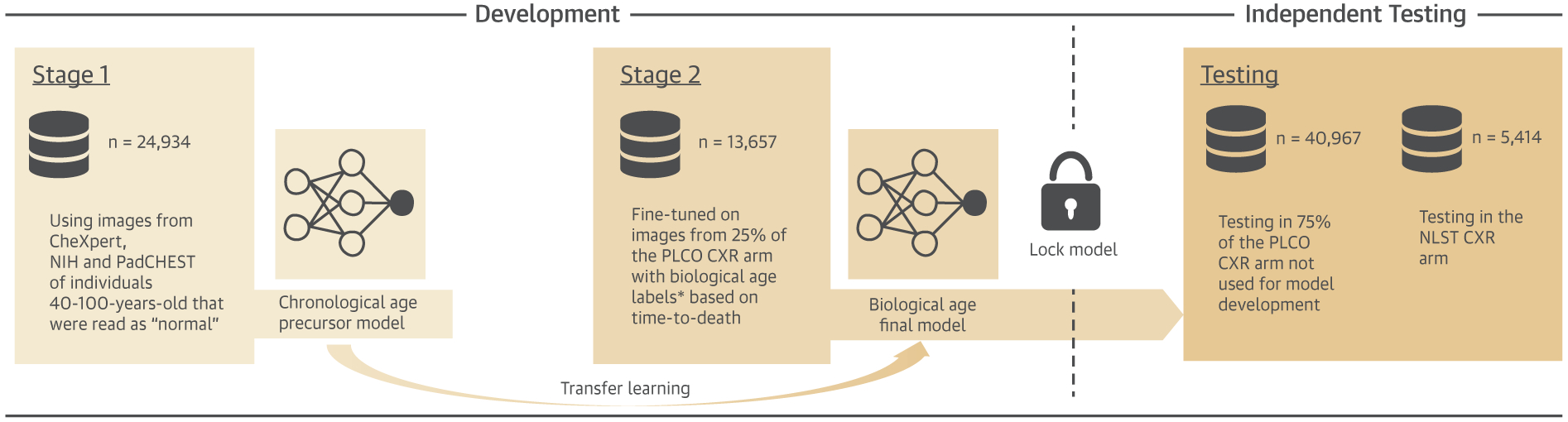
Development Stage 1: First, a deep learning model to estimate chronological age from a chest radiograph image (CXR-Age) was trained by using radiographs from 24,934 individuals from CheXpert, National Institutes of Health Chest X-ray 14, and PadCHEST. Development Stage 2: In the second step, this model was used as a foundation to train a second model to estimate biological age using 25% of the individuals (n = 13,657) from the PLCO (Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial) chest radiograph screening arm. Further details are provided in the Methods and Supplemental Methods. Testing: After Stage 2, the final CXR-Age model was locked and validated in the 2 independent testing datasets: 1) the remaining 75% of individuals from the PLCO chest radiograph arm not seen during development (n = 40,967); and 2) an external testing dataset in the NLST (National Lung Screening Trial) chest radiograph arm (n = 5,414). The outcome for model validation was actual, observed all-cause and cardiovascular mortality in PLCO and NLST.
* For the purposes of Development Stage 2 only, biological age labels were assigned as follows: A) if the persons died during follow-up, then biological age was defined by their current expected age relative to their age at death based on the Social Security Administration’s 2015 actuarial life table, and B) for those that did not die during follow-up, sex-specific Gaussian survival models developed in the control arm (n = 77,444; no radiography) of PLCO were used to predict age at death before applying A. Constructed biological age labels were used for model Development Stage 2 only. The independent testing results report actual observed mortality only.
The final CXR-Age model takes as input a CXR image and outputs an estimated chest x-ray age in years. CXR-Age was validated to predict actual observed all-cause and cardiovascular mortality in 2 independent held-out testing datasets of individuals who were not seen at any point during model training. Only these validation data are reported in the Results.
STUDY COHORTS.
Development Stage 1 chronological age datasets.
Data for the first stage of model development consisted of 34,012 frontal, posterior-anterior radiographs from 24,934 individuals from 3 publicly available cohorts: National Institutes of Health (NIH) Chest X-ray 14 (17), PadCHEST (18), and CheXpert (19) (age 57.2 ± 9.8 years). NIH Chest X-ray 14 consists of inpatient, frontal radiographs collected between 1992 and 2015 at the National Institutes of Health (NIH) Clinical Center (Bethesda, Maryland). PadCHEST consists of all available chest x-rays from the Hospital Universitario de San Juan (Alicante, Spain) between 2009 and 2017. CheXpert consists of both inpatient and outpatient radiographs from Stanford Hospital (Palo Alto, California) between 2002 and 2017. Only radiographs from individuals 40 to 100 years of age that were read as “normal” (i.e., no findings such as lung nodule or consolidation) were included. These data were used solely for the first stage of training of our model. No results are reported from these data.
Development Stage 2 biological age dataset.
The second stage of model development used radiographs from the Prostate, Lung, Colorectal, Ovarian (PLCO) Cancer Screening Trial (20,21). PLCO enrolled asymptomatic men and women 55 to 74 years of age at 10 U.S. sites from November 8, 1993, through July 1, 2001. Participants were randomized to cancer screening by CXR compared with controls. Intervention arm participants were given up to 4 annual radiographs (T0 to T3). The PLCO control arm (n = 77,444; age 62.6 ± 5.4 years; median follow-up 16.7 years) had no radiographs but was instead used to label biological age as described in the CXR-Age Model Development section. The PLCO intervention arm (191,506 radiographs from 54,258 individuals; age 63.2 ± 5.5 years; median follow-up 16.3 years) was divided into 25% of participants for Stage 2 of model development, with the remaining 75% of participants held out for independent testing of the final CXR-Age model (Figure 1).
TESTING DATASETS.
The final CXR-Age model was validated in 2 held-out independent testing datasets, and results are reported for these 2 testing datasets only. The final CXR-Age model was tested in the remaining 75% of individuals from the PLCO CXR arm not used for model development (n = 40,967; age 62.5 ± 5.4 years; median follow-up 16.9 years). For each person, only the first available radiograph was included.
NLST (National Lung Screening Trial) (22) was used as an external testing dataset. NLST was a randomized controlled trial of CXR versus low-dose chest computed tomography imaging for lung cancer screening. NLST enrolled a community cohort of asymptomatic heavy smokers aged 55 to 74 years at 21 U.S. sites between August 2002 and April 2004. The CXR arm of NLST was used for this analysis. Each individual had baseline (T0) and annual (T1 and T2) CXRs. Only baseline (T0) radiographs were included in this study (n = 5,414; age 61.7 ± 5.0 years; median follow-up 11.9 years).
Secondary use of PLCO and NLST data was approved by the National Cancer Institute (Bethesda, Maryland) and Partners Healthcare (Boston, Massachusetts) institutional review boards. Secondary use of CXRs from the NLST was also approved by the American College of Radiology Imaging Network. Participants consented to the parent trials; waiver of informed consent was approved for this retrospective study.
CXR-AGE MODEL DEVELOPMENT.
Development Stage 1: estimate chronological age using public CXR datasets.
The first stage of training leveraged large publicly available databases of chest x-rays where the only available outcome was chronological age. This “pre-training” approach is a common practice in machine learning. This pre-trained model was only used as an initial step before Stage 2 of model development. This first stage model took as input a CXR image and output an estimate of chronological age. We do not report any results based on this model or these public CXR datasets.
Development Stage 2: estimate biological age using PLCO training data.
The Stage 1 model was the starting point for a second model trained to estimate biological age from a CXR image. To train the model, it was necessary to assign a biological age label to each training dataset participant. For the purposes of training the model only, we defined the biological age labels based on a person’s expected risk of long-term mortality. For example, a person with a biological age of 65 years would have similar risk of long-term mortality as the average 65-year-old. We assigned these labels in the following manner. First, if the person died during follow-up, biological age was then defined as the difference between their expected age at death and their actual chronological age at death. Expected age at death was computed by using 2015 actuarial life tables published by the U.S. Social Security Administration (23). The formula for calculating test dataset age labels is as shown, where is the biological age label, is chronological age at the time of the x-ray, is expected age-at-death according to the U.S. Social Security Administration, and is actual, observed age at death.
An individual dying at the age the Social Security actuarial tables expect would have a biological age label equal to their chronological age. An individual who dies later than expected would have a lower biological age. Second, for those who did not die during follow-up, was estimated by using sex-specific Gaussian survival models (Supplemental Table 1) developed in the control arm (no radiograph) of PLCO (N = 77,444) to predict time-to-death.
These biological age labels were used only to train the CXR-Age model; they were not calculated in the testing datasets and are not reported in the Results. The Results validate the final CXR-Age model exclusively with actual, observed mortality in the testing datasets.
CXR-AGE MODEL VALIDATION IN INDEPENDENT TESTING DATASETS.
The final CXR-Age model takes as input a CXR image and outputs an estimated chest x-ray age in years. The CXR-Age model took no other inputs beyond the pixels on the CXR image; that is, it was blind to chronological age, prevalent risk factors, and the radiologist’s interpretation of the CXR. CXR-Age was validated in 2 held-out independent testing datasets from PLCO and NLST. These testing dataset participants were not seen during any part of the CXR-Age model development.
The chest x-ray age was validated to predict the primary outcome of observed, all-cause mortality over 23 years (PLCO) and 13 years (NLST) of follow-up. The secondary outcome was observed cardiovascular mortality over 13 years of follow-up, defined based on cause of death International Classification of Diseases-9 codes for ischemic heart disease, cerebrovascular accident, and other circulatory diseases (corresponding to International Classification of Diseases-9 codes 200XX, 300XX, and 400XX). Mortality and cause of death were determined via annual questionnaire, communication with next of kin, and the National Death Index in PLCO (20) and NLST (22).
PLCO and NLST participants self-reported prevalent risk factors such as smoking, diabetes, obesity, hypertension, and history of myocardial infarction or stroke. Radiographs were reviewed by centrally qualified radiologists who reported traditional radiographic findings such as a lung nodule and cardiomegaly. The added value of CXR-Age to these risk factors and radiographic findings was assessed.
Gradient-weighted class-activation maps for explainability.
Gradient-weighted class-activation maps (Grad-CAM) (24) were generated to localize anatomic regions contributing to CXR-Age. Feature maps from the last convolutional layer of the CXR-Age model were used to compute the Grad-CAM heatmap. Heatmaps were superimposed on the original images for visualization.
STATISTICAL ANALYSIS.
CXR-Age was evaluated in the PLCO testing dataset and externally in NLST. The concordance index (Harrell’s C-statistic) based on the Cox proportional hazards model for all-cause and cardiovascular mortality is reported for CXR-Age, chronological age, traditional radiograph findings, and prevalent risk factors. Kaplan-Meier curves were computed within CXR-Age strata (<60, 60 to 64, 65 to 69, and ≥70 years) and chronological age strata (<60, 60 to 64, 65 to 69, and ≥70 years). Cox proportional hazards regression was used to determine the association between CXR-Age and time to death. Univariate and multivariate hazard ratios (HRs) for all-cause mortality were computed by using Cox proportional hazards regression. Cause-specific HRs for cardiovascular mortality were computed by using competing risks regression via the cmprsk R package (25). Partial likelihood ratio tests between non-nested Cox regression models were used to compare CXR-Age against actual chronological age, defined as the number of years since birth. Mortality rate was compared by using a z-test between incident rate ratios. The continuous net reclassification improvement (NRI) (26) based on Kaplan-Meier risk estimates were calculated by using the nricens R package (27), and confidence intervals (CIs) were estimated by using 200 nonparametric bootstrap samples. The association between CXR-Age and clinical risk factors was assessed by using the chi-square test for binary variables and the Kruskal-Wallis test for continuous variables. For all comparisons; p < 0.05 was considered significant. All statistical analysis was performed in R (R Foundation for Statistical Computing, Vienna, Austria).
RESULTS
The final CXR-Age model was validated in 2 testing cohorts: 1) an internal testing cohort including the remaining 75% of PLCO individuals not seen during any part of training (n = 40,967; age 62.5 ± 5.4 years; median follow-up 16.9 years); and 2) an external testing cohort from the CXR arm of the NLST (22) (n = 5,414, age 61.7 ± 5.0 years; median follow-up 11.9 years). NLST had greater smoking burden and shorter length of follow-up than PLCO (Supplemental Table 2). Results are provided for the testing datasets only.
CXR-Age was correlated with chronological age in PLCO (r2 = 0.37; p < 0.001) and NLST (r2 = 0.25; p < 0.001) testing datasets. Most individuals had an estimated CXR-Age within 5 years of their chronological age (Supplemental Figure 1) in PLCO (73% [29,765 of 40,967]) and NLST (68% [3,688 of 5,414]). In PLCO, 17% (6,907 of 40,967) had a CXR-Age at least 5 years older than their chronological age, and 10% (4,295 of 40,967) had a CXR-Age at least 5 years younger. Similar results were found in NLST (5 years older: 23% [1,269 of 5,414]; 5 years younger: 8% [457 of 5,414]). NLST’s greater proportion of CXR-Age higher than chronological age may be attributed to the heavy (≥30 pack-year) smoking inclusion criterion in that trial.
To determine the relative predictive power of CXR-Age, chronological age, traditional radiograph findings such as lung nodules and cardiomegaly, and prevalent risk factors such as diabetes, we report the concordance index (Harrell’s c-statistic) based on the Cox proportional hazards model for all-cause and cardiovascular mortality (Supplemental Table 3). A univariable model with CXR-Age had a higher C-statistic than chronological age and traditional radiographic findings from the radiograph for all-cause and cardiovascular mortality in PLCO and NLST testing datasets (p < 0.05 for all comparisons) (Supplemental Table 3A). Adding CXR-Age to a multivariable model with risk factors, findings, and chronological age resulted in modest but statistically significant improvements for both all-cause and cardiovascular mortality in PLCO and NLST (p < 0.001 for all comparisons) (Supplemental Table 3B).
We further tested whether CXR-Age improved risk estimates for all-cause and cardiovascular (Supplemental Table 4) death using the continuous NRI between univariable Cox regression models of CXR-Age against chronological age. The Cox model using CXR-Age better estimated all-cause mortality risk than the model using chronological age in PLCO (NRI = 0.151; 95% CI: 0.10 to 0.20) and NLST (NRI = 0.173; 95% CI: 0.03 to 0.32) testing datasets. Similar results were found for cardiovascular mortality in PLCO.
To test how CXR-Age predicts time to death, CXR-Age predictions were stratified into 4 groups (CXR-Age <60, 60 to 64, 65 to 69, and ≥70 years). Kaplan-Meier survival estimates of CXR-Age groups partitioned according to baseline chronological age in the PLCO testing dataset are given in Figure 2, indicating a graded association of CXR-Age with mortality. This association was consistent in NLST (Supplemental Figure 2). CXR-Age predicted mortality better than chronological age in PLCO testing (CXR-Age HR: 2.26 per 5 years [95% CI: 2.24 to 2.29] vs. chronological age HR: 1.77 per 5 years [95% CI: 1.75 to 1.78]; p for comparison <0.001) and NLST testing datasets (CXR-Age HR: 1.82 per 5 years [95% CI: 1.76 to 1.89] vs. chronological age HR: 1.49 per 5 years [95% CI: 1.44 to 1.53]; p for comparison <0.001). These results were robust to adjustment for baseline clinical risk factors and radiograph findings (Supplemental Table 5).
FIGURE 2. Kaplan-Meier Survival Curves by CXR-Age Group.
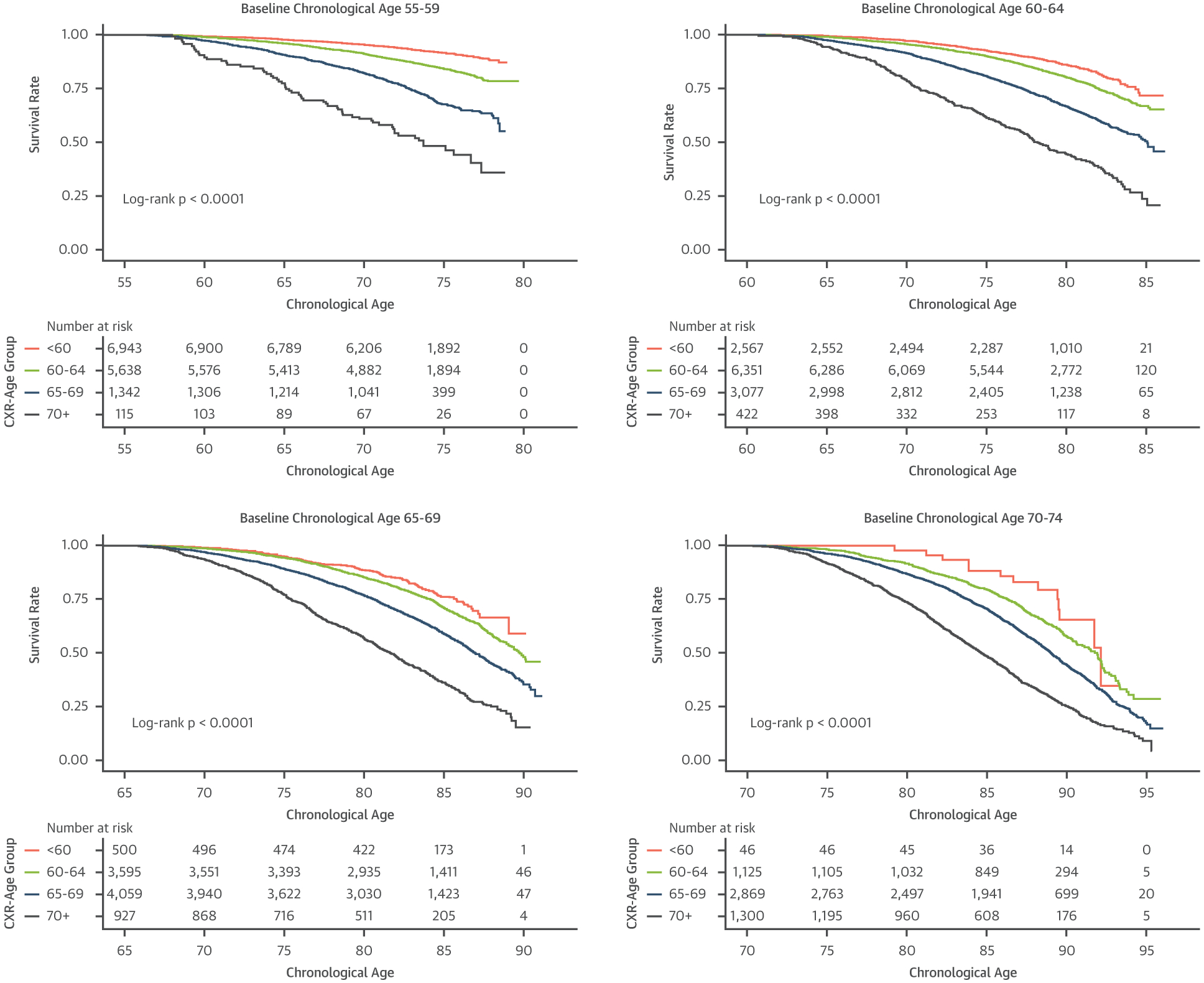
Kaplan-Meier survival curves in PLCO testing data for individuals with baseline chronological age as follows: 55 to 59 years, 60 to 64 years, 65 to 69 years, and 70 to 74 years. CXR-Age shows a graded association with all-cause mortality in individuals with similar baseline chronological age. Abbreviations as in Figure 1.
A similar pattern was found for 13-year cardiovascular mortality. Kaplan-Meier survival estimates for cardiovascular mortality according to CXR-Age groups in the PLCO testing dataset are given in Figure 3 and indicate a graded association. This association was consistent for all except those >70 years of chronological age in NLST (Supplemental Figure 3). Treating death from other causes as a competing risk, CXR-Age predicted cardiovascular mortality better than chronological age in PLCO testing (CXR-Age cause-specific HR: 2.45 per 5 years [95% CI: 2.34 to 2.56] vs. chronological age HR: 1.82 per 5 years [95% CI: 1.74 to 1.90]) and NLST testing (CXR-Age HR: 1.94 per 5 years [95% CI: 1.68 to 2.23] vs. chronological age HR: 1.59 per 5 years [95% CI: 1.40 to 1.82]) datasets. These results were robust to adjustment for baseline clinical risk factors (Supplemental Table 6).
FIGURE 3. Kaplan-Meier Survival Curves for Cardiovascular Mortality by CXR-Age Group.
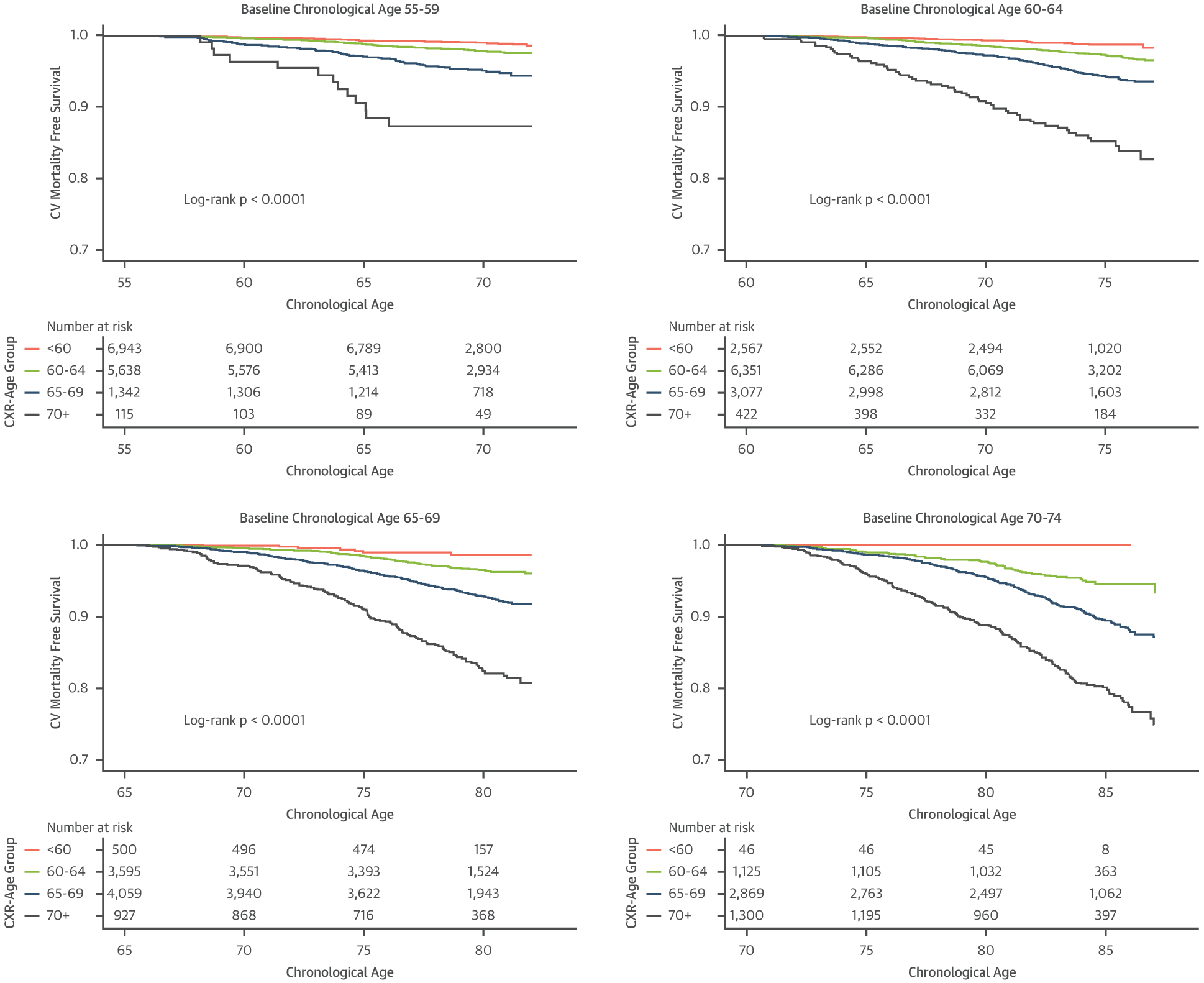
Kaplan-Meier survival curves in PLCO testing data for individuals with a baseline chronological age as follows: 55 to 59 years, 60 to 64 years, 65 to 69 years, and 70 to 74 years. CXR-Age shows a graded association with cardiovascular mortality in individuals with similar baseline chronological age. Abbreviations as in Figure 1.
To put these results in context, we give an example for individuals with a baseline chronological age of 65 years. In the PLCO testing set, there were 2,012 individuals aged 65 years, and 1,946 (96.7%) were followed up until they reached average U.S. life expectancy (78.7 years) or died prematurely. Among these 65-year-olds, those with a CXR-Age ≥70 years had a higher premature mortality rate (56 of 124 [45.2%]) than those with a CXR-Age of 60 to 69 years (314 of 1,643 [19.1%]; p < 0.001) and CXR-Age <60 years (19 of 179 [10.6%]; p < 0.001).
CXR-Age was associated with baseline cardiovascular risk factors (Supplemental Table 2) such as male sex, diabetes, hypertension, and obesity. In PLCO, which included both current, former, and never-smokers, CXR-Age was associated with smoking. Smoking was not significantly associated with CXR-Age in NLST, as only heavy smokers were enrolled in the trial. CXR-Age showed little association with traditional radiologist findings in PLCO and NLST, except for cardiovascular abnormalities (PLCO correlation 0.074; NLST correlation 0.057; both p < 0.001) (Supplemental Table 7). CXR-Age remained more predictive for longevity than chronological age in all subgroups of the PLCO testing dataset, including both men and women, Black and White race, smoking, obesity, past cancer, and past ASCVD (Figure 4).
FIGURE 4. Hazard Ratios for All-Cause Mortality per 5-Year Increase in Chronological Age and CXR-Age Within PLCO Testing Dataset Subgroups.
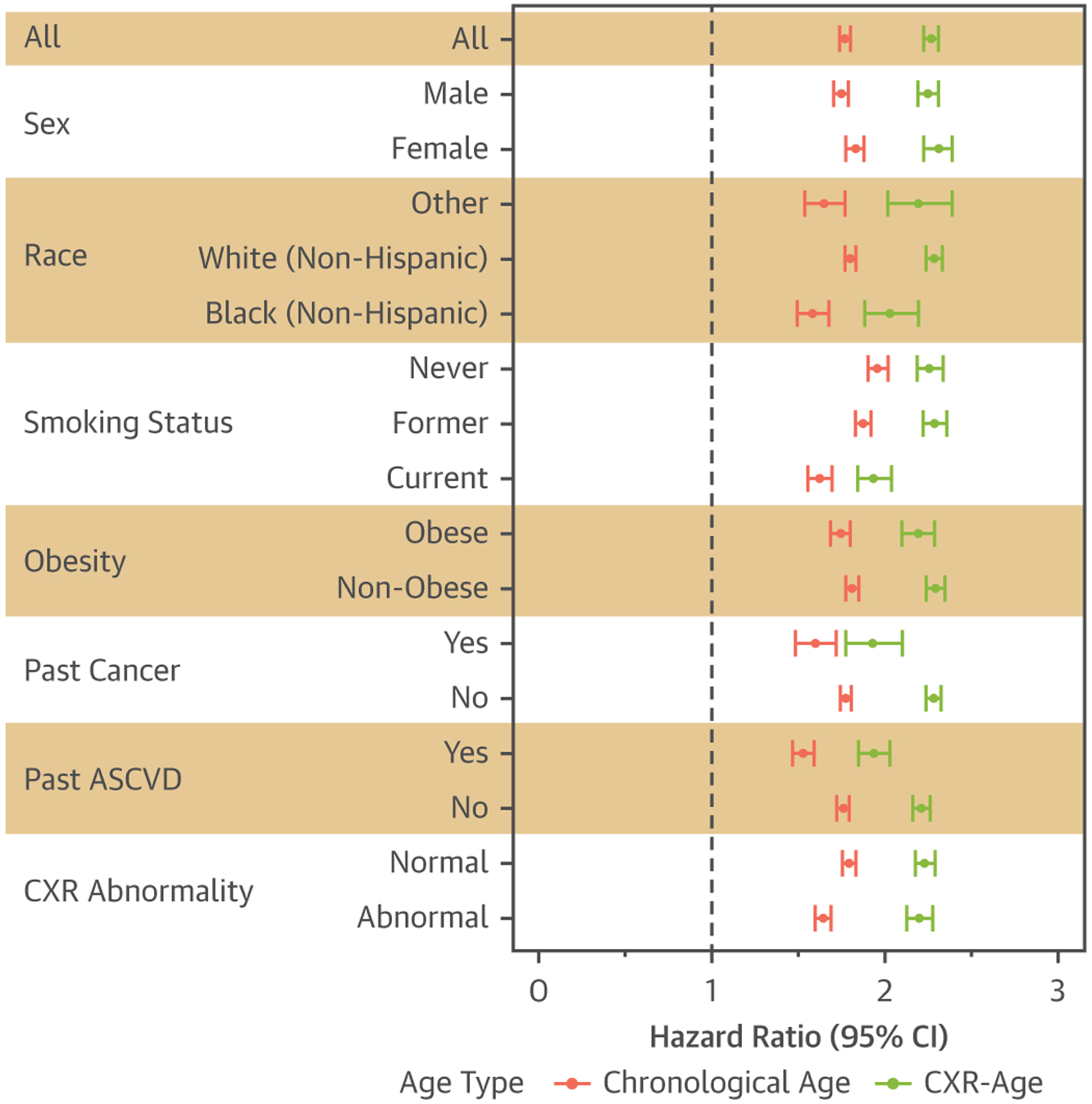
In all subgroups, a 5-year increase in CXR-Age (green) was associated with greater all-cause mortality than a 5-year increase in chronological age (red) (p < 0.01 for all groups except never-smokers, where p = 0.29). Obesity was defined as body mass index ≥30 kg/m2. Atherosclerotic cardiovascular disease (ASCVD) defined as atherosclerotic cardiovascular disease. CI = confidence interval; other abbreviations as in Figure 1.
To localize the anatomy contributing to CXR-Age, Grad-CAMs were generated (Figure 5) (24). The heatmaps indicate that CXR-Age commonly focuses on the mediastinum, the cardiac silhouette, and the aortic knob, all anatomy that dilates and becomes tortuous with aging (28,29). Activations were also seen over other anatomy, including the diaphragmatic silhouette, the upper mediastinum, and the low neck.
FIGURE 5. Grad-CAM Localize Anatomy Contributing to CXR-Age.
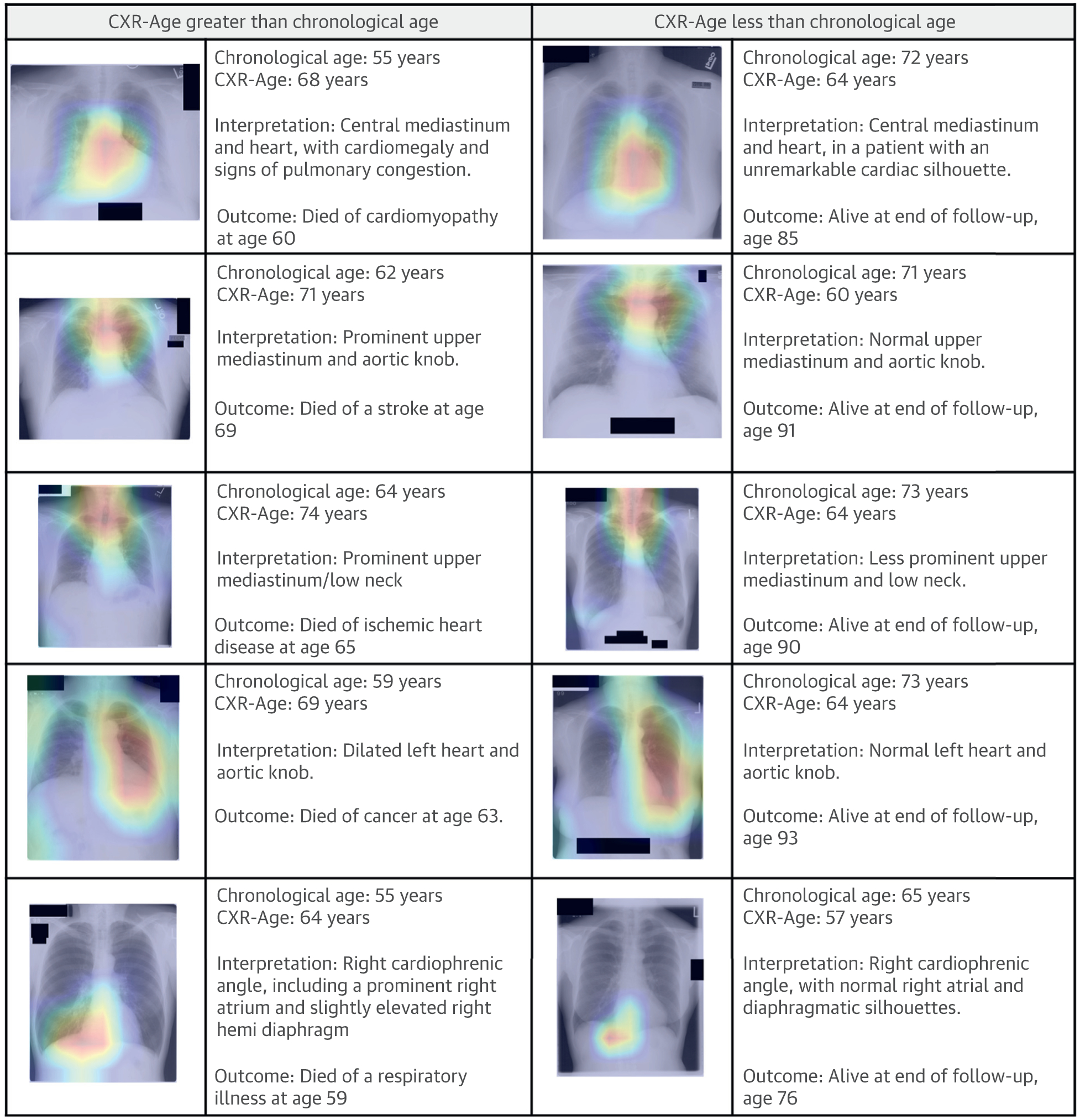
Common activation areas included the mediastinum, cardiac silhouette, and the aortic knob, anatomy that dilates and becomes tortuous with aging (29). CXR-Age = chest x-ray age; Grad-CAM = gradient-weighted class activation maps.
DISCUSSION
This study introduces a CNN (CXR-Age) developed in >115,000 individuals that takes a chest x-ray image as input and outputs an estimate of biological age based on this image. When validated in 2 independent held-out testing datasets from the PLCO (N = 40,967) and NLST (N = 5,414) trials, CXR-Age predicted all-cause and cardiovascular mortality beyond chronological age. CXR-Age was additive to traditional cardiovascular risk factors and radiographic findings.
To our knowledge, CXR-Age is the first CNN to predict biological age from CXRs. Karargyris et al. (30) estimated chronological age from CXRs, but they did not test the association with longevity or other health outcomes. Related to the current work, we previously developed a different modified Inception v4 CNN (CXR-Risk) to predict the probability (0 to 1) of all-cause mortality over 12 years in PLCO (16). In contrast to this previous work, CXR-Age provides a biological age in years, which is made possible by the recent public release of large CXR databases with chronological age labels and the release of extended follow-up in PLCO.
Chest x-rays are among the most common tests in medicine. A future implementation could compute CXR-Age from routine chest x-ray images, to give a better estimate of an individual’s biological age. The advantage of framing risk as a biological age (e.g., biological age of 55 years with a chronological age of 60 years) is that it is easier to grasp for patients than risk score probabilities (5). Furthermore, substituting biological age measures for chronological age could improve the performance of existing risk scores (12). In this way, CXR-Age could help inform decisions about prevention, screening, and treatment.
A common criticism of deep learning models is their “black-box” nature. In this work, we used Grad-CAMs to highlight regions of the CXR that had the greatest contribution to the CXR-Age estimate (Figure 5). Common activation areas were the mediastinum, aortic knob, and cardiac silhouette, anatomy known to become tortuous and dilate with age. Activations were also seen over the low neck and diaphragm; although this anatomy’s association with aging is less well defined, it may indicate diaphragmatic elevation related to obesity and degenerative disease of the low cervical spine. Older CXR-Age was associated with prevalent risk factors such as smoking, diabetes, hypertension, obesity, past myocardial infarction, and past stroke (Supplemental Table 2), which suggests that CXR-Age may identify the phenotypic effect of these risk factors on the chest anatomy. Of interest, CXR-Age estimates were not strongly associated with traditional radiographic findings such as lung nodules and fibrosis, although there was a consistent association with cardiac abnormalities such as cardiomegaly (Supplemental Table 7).
Further investigation of CXR-Age is planned. First, we intend to explore whether substituting the CXR-Age for chronological age can improve existing age-based risk scores such as the Framingham or ASCVD risk score and recommendations for cancer screening (31) (e.g., age ≥55 years for lung cancer screening). CXR-Age may also provide geroscience researchers a new aging measure; whether this proves complementary to existing blood and functional biological age markers will also require further investigation. To encourage reproducible research and future investigation, we will publish the CXR-Age model as free open source software.
STUDY LIMITATIONS.
Comparison with other, established biological age markers (e.g., frailty index, epigenetic clock) were not available. The model was validated in a population aged 55 to 74 years. CXR-Age was robust to adjustment for baseline cardiovascular risk factors (age, sex, diabetes, and hypertension); however, a comparison with ASCVD risk (32) and the Framingham Risk Score was not possible because cholesterol and blood pressure measurements were not available in PLCO and NLST. Only cardiovascular mortality was documented during follow-up, and thus we could not assess the association between CXR-Age and nonfatal cardiovascular events. PLCO and NLST CXRs were obtained in cancer screening trials; CXR-Age will need to be validated in radiographs obtained for other clinical indications. Cause of death was determined based on communication with next of kin and linkage to the National Death Index, and thus the potential for misclassification of cardiovascular death should be considered. The radiographs used in this study were down-sampled to low (224 × 224) resolution due to technical limitations on graphics processing unit random-access memory; future models may see higher performance with higher resolution images.
CONCLUSIONS
A CNN (CXR-Age) estimated a chest x-ray-based biological age that predicted long term all-cause and cardiovascular mortality. Further research is necessary to determine how CXR-Age may inform risk assessment and clinical decision-making.
Supplementary Material
PERSPECTIVES.
COMPETENCY IN PATIENT CARE AND PROCEDURAL SKILLS:
A deep learning model can estimate biological age from a chest x-ray image, and this CXR-Age predicts all-cause and cardiovascular mortality.
TRANSLATIONAL OUTLOOK:
Measures of biological age may help inform risk assessment and clinical decision-making.
FUNDING SUPPORT AND AUTHOR DISCLOSURES
This work was supported by the National Academy of Medicine Healthy Longevity Grand Challenge (2000011734). A graphics processing unit used for this research was donated to Dr Lu as an unrestricted gift through the Nvidia Corporation Academic Program. Original data collection for the ACRIN 6654 trial (NLST) was supported by National Cancer Institute Cancer Imaging Program grants. Dr Lu is supported by American Heart Association grants 18UNPG34030172 and 810966; has common stock in Nvidia and Advanced Micro Devices; reports research funding as a coinvestigator to Massachusetts General Hospital from Kowa Company Limited and Medimmune/AstraZeneca; and received personal fees from PQBypass unrelated to this work. Dr Hoffmann is supported by an NIH grant (5K24HL113128); has received research support on behalf of his institution from Duke University (Abbott), HeartFlow, Kowa Company Limited, and MedImmune/AstraZeneca; and has received consulting fees from Duke University (NIH) and Recor Medical unrelated to this research. Dr Raghu is supported by NIH grant T32HL076136; and has common stock in Nvidia, Alphabet, and Apple. Dr Aerts has received personal fees from Sphera, Genospace, and Onc. AI outside the submitted work. All other authors have reported that they have no relationships relevant to the contents of this paper to disclose.
ABBREVIATIONS AND ACRONYMS
- ASCVD
atherosclerotic cardiovascular disease
- CI
confidence interval
- CNN
convolutional neural network
- CXR
chest radiograph/x-ray
- Grad-CAM
gradient-weighted class activation map
- HR
hazard ratio
- NRI
net reclassification improvement
Footnotes
The authors attest they are in compliance with human studies committees and animal welfare regulations of the authors’ institutions and Food and Drug Administration guidelines, including patient consent where appropriate. For more information, visit the Author Center.
APPENDIX For supplemental Methods, tables, and figures, please see the online version of this paper.
REFERENCES
- 1.Christensen K, Thinggaard M, McGue M, et al. Perceived age as clinically useful biomarker of ageing: cohort study. BMJ. 2009;339:b5262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levine ME. Modeling the rate of senescence: can estimated biological age predict mortality more accurately than chronological age? J Gerontol A Biol Sci Med Sci. 2013;68:667–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jazwinski SM, Kim S. Examination of the dimensions of biological age. Front Genet. 2019;10:263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamczyk MR, Nevado RM, Barettino A, Fuster V, Andres V. Biological versus chronological aging: JACC Focus Seminar. J Am Coll Cardiol. 2020;75:919–930. [DOI] [PubMed] [Google Scholar]
- 5.McClelland RL, Nasir K, Budoff M, Blumenthal RS, Kronmal RA. Arterial age as a function of coronary artery calcium (from the Multi-Ethnic Study of Atherosclerosis [MESA]). Am J Cardiol. 2009;103:59–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stein JH, Fraizer MC, Aeschlimann SE, Nelson-Worel J, McBride PE, Douglas PS. Vascular age: integrating carotid intima-media thickness measurements with global coronary risk assessment. Clin Cardiol. 2004;27:388–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Studenski S, Perera S, Patel K, Rosano C, Faulkner K. Gait speed and survival in older adults. JAMA. 2011;305:50–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fried LP, Tangen CM, Walston J, et al. Frailty in older adults: evidence for a phenotype. J Gerontol A Biol Sci Med Sci. 2001;56:M146–M157. [DOI] [PubMed] [Google Scholar]
- 9.Bulpitt CJ, Rajkumar C, Cameron JD. Vascular compliance as a measure of biological age. J Am Geriatr Soc. 1999;47:657–663. [DOI] [PubMed] [Google Scholar]
- 10.Horvath S, Raj K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat Rev Genet. 2018;19:371–384. [DOI] [PubMed] [Google Scholar]
- 11.Sanders JL, Newman AB. Telomere length in epidemiology: a biomarker of aging, age-related disease, both, or neither? Epidemiol Rev. 2013;35:112–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grundy SM. Coronary plaque as a replacement for age as a risk factor in global risk assessment. Am J Cardiol. 2001;88:8E–11E. [DOI] [PubMed] [Google Scholar]
- 13.Ron E. Cancer risks from medical radiation. Health Phys. 2003;85:47–59. [DOI] [PubMed] [Google Scholar]
- 14.Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hannun AY, Rajpurkar P, Haghpanahi M, et al. Cardiologist-level arrhythmia detection and classification in ambulatory electrocardiograms using a deep neural network. Nat Med. 2019;25:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu MT, Ivanov A, Mayrhofer T, Hosny A, Aerts HJWL, Hoffmann U. Deep learning to assess long-term mortality from chest radiographs. JAMA Netw Open. 2019;2:e197416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Summers RM. ChestX-Ray8: Hospital-Scale Chest X-Ray Database and Benchmarks on Weakly-Supervised Classification and Localization of Common Thorax Diseases. In: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, July 21-26, 2017. Piscataway, NJ: IEEE, 2017;3462–3471. [Google Scholar]
- 18.Bustos A, Pertusa A, Salinas JM, de la Iglesia-Vayá M. Padchest: a large chest x-ray image dataset with multi-label annotated reports. Med Image Anal. 2020;66. [DOI] [PubMed] [Google Scholar]
- 19.Irvin J, Rajpurkar P, Ko M, et al. CheXpert: A large chest radiograph dataset with uncertainty labels and expert comparison. Proceedings of AAAI Conference on Artificial Intelligence. 2019;33:590–597. [Google Scholar]
- 20.Oken MM, Hocking WG, Kvale PA, et al. Screening by chest radiograph and lung cancer mortality: the Prostate, Lung, Colorectal, and Ovarian (PLCO) randomized trial. JAMA. 2011;306:1865–1873. [DOI] [PubMed] [Google Scholar]
- 21.Prorok PC, Andriole GL, Bresalier RS, et al. Design of the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial. Control Clin Trials. 2000;21:273S–309S. [DOI] [PubMed] [Google Scholar]
- 22.National Lung Screening Trial Research Team, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med. 2011;365:395–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.United States Social Security Administration. Actuarial Life Table. Vol. 2019; 2016. [Google Scholar]
- 24.Selvaraju RR, Cogswell M, Das A, Vendantam R, Parikh D, Batra D, et al. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. In Proceedings of 2017 IEEE International Conferenc on Computer Vision (ICCV), Venice, Oct 22–29, 2017. Piscataway, NJ: IEEE,2017:618–26. [Google Scholar]
- 25.Fine JP, Gray RJ. A proportional hazards model for the subdistribution of a competing risk. J Am Statistical Assoc. 1999;94:496–509. [Google Scholar]
- 26.Pencina MJ, D’Agostino RB Sr., Steyerberg EW. Extensions of net reclassification improvement calculations to measure usefulness of new biomarkers. Stat Med. 2011;30:11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Inoue E. R Package ‘nricens’; 2018. Accessed December 1, 2019. https://cran.r-project.org/web/packages/nricens/nricens.pdf [Google Scholar]
- 28.Redheuil A, Yu W, Mousseaux E, et al. Age-related changes in aortic arch geometry: relationship with proximal aortic function and left ventricular mass and remodeling. J Am Coll Cardiol. 2011;58:1262–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng S, Fernandes VRS, Bluemke DA, McClelland RL, Kronmal RA, Lima JAC. Age-related left ventricular remodeling and associated risk for cardiovascular outcomes: the Multi-Ethnic Study of Atherosclerosis. Circ Cardiovasc Imaging. 2009;2:191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karargyris A, Kashyap S, Wu JT, Sharma A, Moradi M, Syeda-Mahmood T. Age prediction using a large chest x-ray dataset. In: Medical Imaging 2019: Computer-Aided Diagnosis, Vol. 10950 109501U. Bellingham, WA: International Society for Optics and Photonics, 2019. [Google Scholar]
- 31.Moyer VA, US Preventive Services Task Force. Screening for lung cancer: U.S. Preventive Services Task Force recommendation statement. Ann Intern Med. 2014;160:330–338. [DOI] [PubMed] [Google Scholar]
- 32.Goff DC Jr., Lloyd-Jones DM, Bennett G, et al. 2013 ACC/AHA guideline on the assessment of cardiovascular risk: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. J Am Coll Cardiol. 2014;63:2935–2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


