Abstract
Alu retrotransposons, which form the largest family of mobile DNA elements in the human genome, have recently come to attention as a potential source of regulatory novelties, most notably by participating in enhancer function. Even though Alu transcription by RNA polymerase III is subjected to tight epigenetic silencing, their expression has long been known to increase in response to various types of stress, including viral infection. Here we show that, in primary human fibroblasts, adenovirus small e1a triggered derepression of hundreds of individual Alus by promoting TFIIIB recruitment by Alu-bound TFIIIC. Epigenome profiling revealed an e1a-induced decrease of H3K27 acetylation and increase of H3K4 monomethylation at derepressed Alus, making them resemble poised enhancers. The enhancer nature of e1a-targeted Alus was confirmed by the enrichment, in their upstream regions, of the EP300/CBP acetyltransferase, EP400 chromatin remodeler and YAP1 and FOS transcription factors. The physical interaction of e1a with EP400 was critical for Alu derepression, which was abrogated upon EP400 ablation. Our data suggest that e1a targets a subset of enhancer Alus whose transcriptional activation, which requires EP400 and is mediated by the e1a-EP400 interaction, may participate in the manipulation of enhancer activity by adenoviruses.
Graphical Abstract
Graphical Abstract.
Introduction
Alu elements are primate-specific members of the short-interspersed element (SINE) family of retrotransposons and constitute a large fraction (∼11%) of the human genome. Like other types of retrotransposons, Alus are subjected to tight epigenetic silencing, required to limit their inherently mutagenic mobilization (1,2). Alu transcription is thought to mostly depend on the RNA polymerase (Pol) III machinery, through the recognition of A- and B-box promoter DNA sequences within the transcribed region by the multi-subunit transcription factor TFIIIC. The TFIIIC-promoter DNA complex is bound, ∼30 bp upstream of the Alu transcription start site (TSS), by the Pol III initiation factor TFIIIB, which includes the TATA-binding protein (TBP) subunit required for initiation by all three of the eukaryotic nuclear RNA polymerases (3). Even though many Alu transcription units occur throughout the genome, the level of Alu transcription is normally kept very low by chromatin structure-based repression mechanisms (4). Recently, it has become possible to profile expression of specific Alus by determining the precise sequences of their transcripts, which allows mapping them to individual Alu genes, most of which have unique sequences due to sequence variation in a small percentage of positions (5–7). Similar advances have been reported for the expression profiling of other SINEs in mouse (8,9). Through these studies, some common features of Alu transcriptomes have emerged. First, in every human cell type analyzed the expressed Alu loci represent a very small fraction (∼0.3% at most) of the total Alu copies in the genome. Second, the majority of Alus with detectable transcripts are expressed in a cell type-specific manner, while only a limited number of Alu elements are expressed ubiquitously. Consequently, the total number of Alu elements expressed in at least one cell type/tissue amounts to ∼1.5% of the ∼1.2 million Alus annotated in the human genome. Third, histone post-translational modifications associated with expressed Alus tend to display epigenetic marks that are typically enriched at enhancers and promoters for RNA polymerase II. These include the histone H2A variant Z (H2A.Z), indicative of repeated disassembly and re-assembly of the TSS-region nucleosomes, and histone H3 lysine 4 mono-methylation (H3K4me1). This observation agrees with the hypothesis that a subset of Alu elements have evolved the properties of cell type-specific enhancers (7,10). Lastly, expressed Alu elements tend to be bound not only by the Pol III transcription machinery, but also by transcription factors typically involved in Pol II regulation, which potentially modulate Alu expression in a cell type-specific manner (5,7). This cell type-specific expression, as well as possible functions related to controlling Pol II enhancers, have also been reported for a small subset of mammalian-wide interspersed repeats (MIRs), the second most numerous family of SINEs in the human genome (11–13), and are increasingly recognized as a general property of transposable elements (14). A recent study further indicated that Alu elements contribute to human gene regulation by rewiring of the 3D chromatin architecture through an interaction network involving TFIIIC, ADNP, CTCF looping and TFIIIC-dependent histone H3 lysine 18 acetylation (H3K18ac) (15). Such properties may have contributed to the evolutionary impact of Alus and other SINEs (16).
Further pointing to the existence of highly organized chromatin at Alu elements, Alus and other SINEs typically are associated with translationally stable nucleosomes. In the case of full-length Alu elements, two nucleosomes are approximately centered on the left and right arms, where they might in principle mask access to the transcriptional machinery (17,18). Such a nucleosome arrangement is likely to be instrumental to Alu repression, together with DNA CpG methylation and H3K9 methylation (19–21), and represents a scaffold which every activation mechanism must confront.
Alu expression is known to increase in response to different types of cell stress, such as viral infection (22,23), heat shock and cycloheximide treatment (24), as well as under altered cellular conditions that occur with some pathologies including cancer (25). Induction of SINE expression by viral infection also occurs in mouse (8). The mechanisms leading to increased Alu RNA levels are still largely unexplored. According to previous studies, they may be both transcriptional, involving increased chromatin accessibility (26), and post-transcriptional, due to reduced DICER-dependent turnover of Alu RNA (27). Alu upregulation upon viral infection is of particular interest, because it occurs in response to different viruses, including the dsDNA adenovirus and herpes simplex virus (23,28,29), and it reflects a general property of SINEs in different mammals (8,30–33). In the case of human adenovirus type 5 (HAdV-C5), infection of cultured human cells was found to cause Alu activation in different cell lines (22), possibly as the result of overcoming chromatin-mediated repression (4). This observation came in the context of a more general phenomenon of Pol III transcription activation by the Ad5 immediate early protein E1A (34,35). Studies of the mechanisms of Pol III activation by E1A identified TFIIIC as a possible regulatory target of this viral oncoprotein (36,37), and identified an E1A-dependent increase in the number of active Pol III transcription complexes assembled in vitro with nuclear extracts from infected versus uninfected cells (38). To what extent these phenomena impact in vivo transcription of Pol III target genes coding for either canonical untranslated RNAs (e.g. tRNAs, 5S rRNA, U6 snRNA) or RNAs expressed from retrotransposons (Alu and other SINEs) has not been extensively investigated.
More recently, genome-wide studies of the effects of an E1A protein variant lacking the conserved region CR3 (referred to as small E1A, or ‘e1a’) revealed its surprising ability to alter global patterns of histone modifications as part of an epigenetic reprogramming process leading to cellular transformation (39,40). A pivotal role in this process is played by interactions of small e1a with key chromatin regulatory proteins such as the lysine acetylases EP300 and its paralog CREB Binding Protein (CREBBP, or CBP) and the tumor suppressor RB and related RB family members (41). Another chromatin regulator whose interaction with e1a is required for cell transformation is EP400, a SWI2/SNF2 DNA helicase-related chromatin remodeler (42). Other proteins found to be part of the same EP400 complex co-purified with E1A are the large multidomain protein TRRAP/PAF400, the TAP54 DNA helicase (RVB2), the oncogenic transcription factor MYC, actin-like proteins, the human homolog of the Polycomb protein (EPC1) (42), and the ∼20 subunit TIP60 lysine acetylase complexes (43). Importantly, cellular proteins bound by the N-terminus of e1a include EP400 and TRRAP associated with a variety of related cellular multiprotein complexes with histone acetyl-transferase activity supplied by the TIP60 subunits. The TIP60/EP400 complexes are often recruited by binding of their chromodomains to H3K4me1. This can increase chromatin accessibility at enhancers by promoting incorporation of the H2A.Z histone variant (44).
To explore the breadth of the epigenetic mechanisms through which Alus are likely to reconcile their predominant silencing and their frequent exaptation as regulatory elements, we investigated the mechanisms of their derepression in response to adenovirus e1a expression. We surmised that e1a, by virtue of its reported abilities to reprogram the epigenome of human IMR90 primary fibroblasts and to affect Pol III-dependent transcription, would prove to be sufficient for Alu upregulation, thus unveiling key regulatory interactions. Alu expression profiling at single locus resolution, ChIP-seq characterization of histone modification and regulatory protein enrichment at expressed and silenced Alus, and e1a mutants specifically affected in interactions with key chromatin regulators were used together to study the complex epigenetic context operating at Alu transcription units.
Materials and methods
Cell culture and viruses
IMR90 primary human fetal lung fibroblasts were purchased from the American Type Culture Collection (ATCC). Cells were grown in Dulbecco's modified medium supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin and 100 μg/ml streptomycin at 37°C in 5% CO2.
Ad5 dl1500 containing a 9-bp deletion removing the 13S E1A mRNA 5′-splice site was described (45). The ΔE1A deletion mutant dl312 was isolated as previously described (46). Small e1a binding mutants (e1a_RB-b−, e1a_p300-b−, e1a_p400-b−) were constructed as in (41) and afterwards incorporated into the dl1500 background. dl1500 and e1a binding mutant constructs were cloned into shuttle plasmid pAdlox. LoxP recombination between the HAdV-C5 backbone Ψ5 and the shuttle plasmid pAdlox, as well as propagation of viruses, were performed as described (47).
RNA extraction and total RNA-seq library preparation
IMR90 cells were grown to confluence in 60-mm Petri dishes and incubated two more days without changing the medium to arrest cells in G1/0. On the day of infection, the medium was collected (conditioned medium) and confluent cells were incubated with mock or with the indicated Ad5-based vectors for 1 h in PBS. At the end of the infection, cells were washed and transferred back to conditioned medium. The infections were performed at multiplicity of infection (MOI) 40 for dl1500 and dl312, 160 MOI for e1a_RB-b−, 60 MOI for e1a_p300-b− and 6 MOI for e1a_p400-b−, in order to achieve comparable amounts of wt and mutant e1a protein expression as assayed by Western blotting (48). 24 hours post-infection, cells were lysed with Trizol and total RNA was extracted using Direct-zol RNA MiniPrep Plus (Zymo research). After elution, RNA was subjected to DNaseI treatment (Invitrogen), followed by inactivation for 10 minutes at 65°C in the presence of 2 mM EDTA.
For the experiment in Figure 1, total RNA (1 μg) from dl1500, dl312 and mock infections (performed in duplicate) was processed using a Ribo-Zero rRNA Removal Kit (Epicentre) to remove rRNA. Total RNA-seq libraries were prepared using the Illumina TruSeq stranded RNA library Preparation kit. Libraries were sequenced using a HiSeq4000 Illumina Sequencer to obtain 100-base-long paired-end reads, using a sequencing depth of about 60 million reads per sample.
Figure 1.
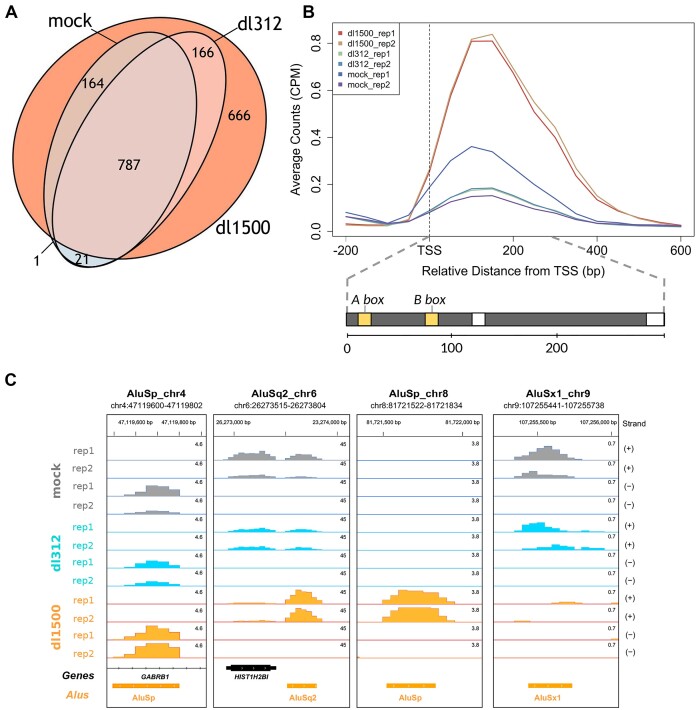
Upregulation of Alu expression by e1a in IMR90 cells. (A) Venn-diagram showing the number of expressed Alu elements in dl1500-, dl312- and mock-infected cells. The whole experiment was performed in duplicate. Expressed Alus included elements whose expression was detected by the pipeline in at least one replicate. (B) Average Alu expression profiles in the presence/absence of e1a, generated from normalized read counts (Counts Per Million, CPM) of the 1805 expression-positive Alus. Shown in the lower part of the panel is the structure of a typical full-length Alu element. The approximate positions of the A and B box internal control regions (yellow bars) are indicated above, those of internal and terminal poly(dA) motifs are indicated by white bars. The approximate position and extension of Alu internal sequence elements are indicated below (bp, base pairs). The upper graph reports the average read count for both replicates of each sample (dl1500, dl312 and mock), labelled by different colours as indicated. The vertical dashed line marks the position of the Alu transcription start site (TSS). (C) Base resolution expression profiles, shown as Integrated Genome Browser views (119), of 4 Alus representative of different types of response to e1a. For the Alu in the first view on the left (AluSp_chr4), no substantial expression changes were observed under the different conditions. The Alu in the second view from the left (AluSq2_chr6) is detected as expressed in dl312- and mock-infected cells and its expression is strongly increased by e1a. The expression of the Alu in the third view from the left (AluSp_chr8) is only detected in dl1500-infected cells. The rightmost view (AluSx1_chr9) illustrates one of the very few examples of e1a-dependent downregulation. Orange boxes represent the orientation of repetitive elements as evidenced by the RepeatMasker track. The chromosomal coordinates of each annotated Alu are shown above each view. Bigwig RNA-seq data are normalized as Counts Per Million. The profiles of both replicates on (+) and (–) strands are shown for each sample (mock-, dl312- and dl1500-infected cells).
For the experiments in Figure 5, total RNA (1 μg) from wt e1a (dl1500), e1a_RB-b−, e1a_p300-b−, e1a_p400-b− and mock infections (performed in duplicate) was processed using the RiboCop rRNA Depletion Kit (Lexogen), and total RNA-seq libraries were prepared as described above. Libraries were sequenced using a NovaSeq Illumina sequencer to obtain 150 base-long stranded paired-end reads, using a sequencing depth of about 100 million reads per sample.
Figure 5.
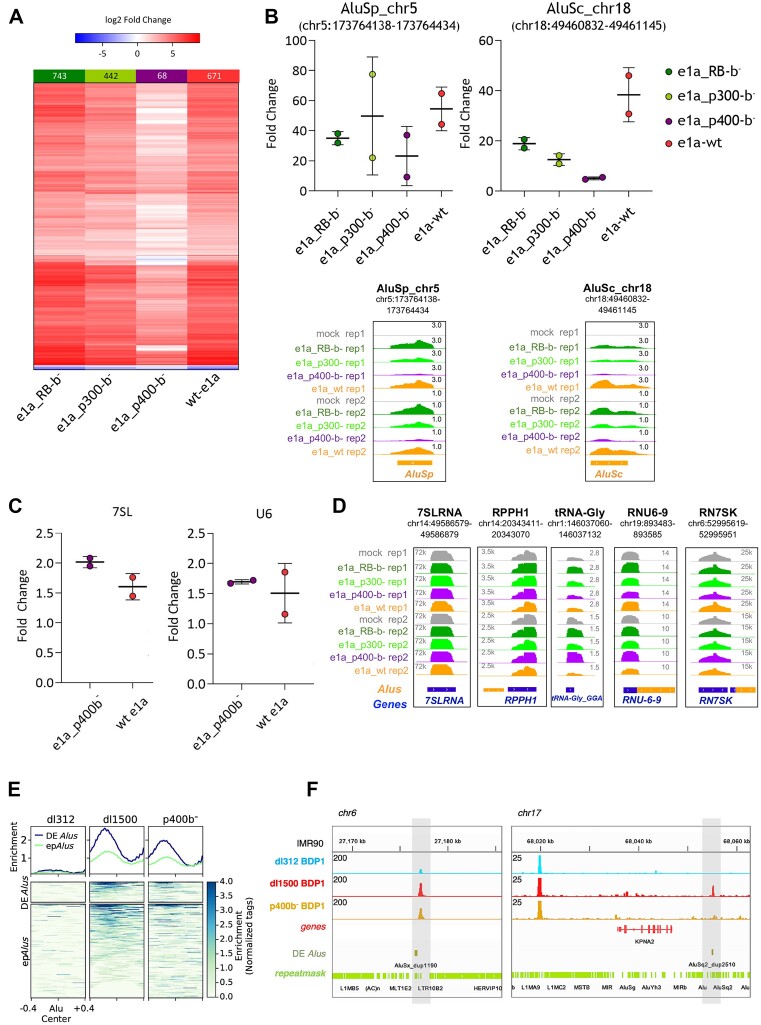
Dependence of Alu upregulation on e1a interaction with chromatin regulators. (A) Heatmap showing increased (red) or decreased (blue) expression of Alu elements triggered by wt e1a or e1a mutants defective in interaction with RB (e1a_RB-b−), p300 (e1a_p300-b−) or p400 (e1a_p400-b−), as compared to mock-infected cells. Boxed above each heatmap are the numbers of differentially expressed Alus (log2 fold-change ≥ 0.5 or ≤ –0.5 and an adjusted P-value < 0.05). The experiment was performed in two biological replicates. (B) Expression levels of two individual Alus as measured by RT-qPCR (upper graphs) and RNA-seq (lower views). Fold changes estimated by RT-qPCR are relative to the expression in mock-infected cells, after normalization to U1 snRNA gene expression. Primers were chosen to target the unique sequence of Alu elements within the 3′ trailer region. RT-qPCR data relative to each independent experiment are represented as dots. Indicated by horizontal bars are the means ± standard deviation between the replicates. RNA-seq data (lower subpanels) are presented as genome browser views of the same Alu elements analysed in the upper plots. Orange boxes represent the orientation of repetitive elements as evidenced by the RepeatMasker track. The chromosomal coordinates of each annotated Alu are shown in the upper part of each subpanel. Bigwig tracks are normalized per CPM. (C) Expression changes of 7SL RNA (left graph) and U6 snRNA (right graph) genes induced by either wt e1a or e1a_p400-b− mutant, as measured by RT-qPCR. Fold change is relative to mock-infected cells, after normalization to U1 snRNA gene expression. RT-qPCR data from each of two independent experiments are represented as dots. Indicated by horizontal bars are the means ± standard deviation between the replicates. (D) Genome browser views of the expression of RN7SL1, RPPH1, tRNA-His-GTG-1–1 (GtRNAdb), RNU6-9 and RN7SK genes, coding for 7SL RNA, Ribonuclease P RNA component H1, tRNAGly(GGA), U6 snRNA and 7SK RNA, respectively. Expression profiles are based on RNA-seq analysis of IMR90 cells infected as indicated on the left. (E) Heatmap and enrichment profiles (normalized read tags) of Bdp1 ChIP-seq occupancy at differentially expressed Alus (DE epAlus) and epAlus in IMR90 infected with dl312, dl1500 and p400-b− viruses. (F) Genome browser views Bdp1 ChIP-seq data of two highly dl1500-induced Alu elements as evidenced by the RepeatMasker track. The chromosomal coordinates of each annotated Alu are shown above each view. Bigwig tracks are normalized for the library size.
RNA-seq reads were aligned to the GRCh38 human reference genome using STAR (49). Only uniquely mapped reads were considered for downstream analyses and subjected to counting with the featureCounts tool of the SubRead Python package (50). The pipeline for Alu RNA profiling was essentially as previously described (6), with some improvements to reduce the rate of false positives. Specifically, we added parameters allowing for setting the cut-off value for the ratio of the expression coverage between the Alu body and its upstream and downstream regions. We also added a parameter controlling the fraction of the Alu body sequence that should be covered by reads to enable the identification of shorter, processed Alu transcripts. In the present study, Alus identified by the pipeline were subsequently filtered for an expression coverage of at least 1000 nt, corresponding to 5 paired-end reads of length 100 nt. The union of these Alus from all the samples in the experiment of Figure 1 constitutes the single, comprehensive list of expression-positive Alus (epAlus). Differentially expressed Alu sequences were identified using the DESeq2 package (51). Alu sequences with a log2 fold-change ≥0.5 or ≤−0.5 and an adjusted P-value <0.05 were deemed differentially expressed.
Alu proximity to protein-coding genes
GENCODE annotation v24 (human genome assembly GRCh38/hg38) was used to analyse Alu distance to protein-coding genes. RepeatMasker tracks were downloaded from the UCSC Genome Browser for human genome assembly hg38. Alu sequences that are positioned outside of protein-coding genes (intergenic Alu) and Alus that map to introns or exons of annotated genes in an antisense orientation (antisense Alu) were selected to obtain the intergenic/antisense Alu subset (802 571 Alu elements). The expression of intergenic/antisense Alus was considered only for the elements belonging to the single, comprehensive list of epAlus, whereas the remaining intergenic/antisense annotated Alu sequences were used as a control for unexpressed Alus. The presence of an Alu within protein-coding genes was analysed using the intersect tool of the BEDtools program package v2.29.1 (52) and custom R scripts. Enrichment analyses were performed using a two-tailed Fisher's exact test.
The transcription start site (TSS) position of protein-coding genes was extracted as the first or last nucleotide (forward or reverse strand, respectively) of the ‘gene’ feature in the gtf annotation file. The distance of Alu elements to the TSS was analysed using the closest tool from BEDtools (52) and custom R scripts. Gene expression was calculated as the average expression (TPM) across all samples. The difference in distribution of gene expression was determined to be statistically significant using a two-sided Wilcoxon rank-sum test. A two-tailed Fisher's exact test was used to analyse the difference between the fraction of the set of all of expression-positive Alus and the fraction of the set of all unexpressed Alus within each genomic range.
ChIP-seq analyses
ChIPs of TFIIIC (GTF3C2) and BDP1 were performed using procedures and antibodies described in (15) and references therein (53,54). All data was aligned to the hg38 human genome reference (GRCh38) and processed as in (15). The average ChIP signal and heatmap profiles were visualized using the tools plotHeatmap from the deepTools package v3.5.1 (55). Sources for publicly available ChIP-seq data are detailed in the Data Availability section.
Western blot
Cells infected with the adenovirus-based vectors were detached by scraping from a 60 mm plate and lysed in EBC buffer (50 mM Tris–Cl pH 8.0, 120 mM NaCl, 0.5% NP-40) with Roche cOmplete™ protease inhibitor cocktail. Samples were prepared in Laemmli buffer and heated for 5 minutes at 65°C. Protein extracts were resolved in a 9% SDS-polyacrylamide gel and electrotransferred to a polyvinylidene difluoride (PVDF) membrane. Blocking was performed in 5% skim milk in TBS-Tween 0.1% for 10 minutes. Extracts were probed with antibodies against E1A (anti-e1a MAb M73) (56), Ku86 H-300 (sc-9034; Santa Cruz) and p400 (Thermo Fisher A300-541A) at manufacturer recommended dilutions for 1 h at room temperature or O/N at 4°C. Membranes were washed 3 times with TBS–Tween 0.1% for 10 min at room temperature. Incubation with secondary antibodies was performed using anti-mouse or anti-rabbit IgG antibodies (Bio-Rad) in TBS–Tween 0.1% buffer with 5% skim milk for 1 h at room temperature. Membranes were washed three times with TBS–Tween 0.1% for 10 min at room temperature before visualization with the Pierce ECL Western Blotting substrate (Thermo Fisher).
siRNA EP400 knockdown
Dharmacon SMART ON-TARGETplus pool siRNA against human EP400 (L-021272-05-0005) and D-001810-10-05 ON-TARGETplus Non-Targeting Pool were used to carry out EP400 knockdown in IMR90 cells. Cells were seeded in the absence of antibiotics and culture for 16h prior to transfection with lipofectamine (Lipofectamine 3000, Invitrogen). siRNAs were used at 30 nM and cells were left in culture for 24 h in the presence of the siRNA, followed by adenoviral dl1500- and e1a_p400-b- and mock-infection for other 24 h. The knockdown efficiency was evaluated by RT-qPCR after total RNA extraction (Trizol) and reverse transcription to obtain cDNA. Knockdown efficiencies of EP400 were around 50–60% depletion in different experiments. Interferon response was monitored by RT-qPCR of STAT1 and IFIT2 expression.
Reverse transcription and real-time PCR
RNA extracted from infected cells (500 ng) was reverse-transcribed using SuperScript III Reverse Transcriptase (Thermo Fisher) with random hexamer primers. The Real Time PCR reaction was performed using 20 ng of cDNA and the PowerUp SYBR Green Master Mix (Applied Biosystems), in a 20 μl final volume with 300 nM primer concentration. Runs were performed using a 7500 Real-Time PCR System (Applied Biosystems). Expression levels were normalized using U1 snRNA as internal control and the ΔΔCt method was used to evaluate expression relative to mock-infections. Data are presented as an average of two replicates ± standard deviation. Expression levels of Alus for siRNA knockdown experiments were calculated using ΔΔCt (siEP400-siCTRL) method for each infection and compared to respective non-siRNA infections relative to mock. Primers used are listed in Supplementary Table S1.
TF binding motif analysis
DNA sequences upstream of expression-positive Alus (200 bp) were analysed using Analysis of Motif Enrichment (AME) from the MEME suite (57). The Jaspar 2020 collection was used as motif database (58) and the DNA sequences of 200 bp upstream of 20 000 random unexpressed Alus were used as a control.
Results
Small E1A upregulates Alu transcription in human fibroblasts
To investigate alterations in Alu expression caused by adenovirus e1a, we performed Alu expression profiling in contact-inhibited, G1-arrested IMR90 primary human fibroblasts infected with HAdV-C5 mutant dl1500, which expresses e1a with little or no expression of other viral genes, versus infection with HAdV-C5 mutant dl312, with deletion of the complete e1a coding region (46). Mock-infected cells subjected to the same changes in medium, but without the addition of virus, served as a further control. Alu expression before and after infection was evaluated by RNA-seq through an analysis enabling quantitation of the expression of individual Alu elements to profile Alu expression at single-locus resolution (5,6). To minimize detection of bystander Alu RNA embedded within longer, Pol II-dependent transcripts, the analysis was restricted to Alus that are not within any annotated protein-coding or ncRNA gene (intergenic) and Alus that map to introns or exons of annotated genes in an antisense orientation (antisense). Most of the members of this intergenic/antisense Alu subset, amounting to 802 431 Alu elements, are likely to represent independent Pol III transcription units, and are thus more suitable than the complete genomic Alu inventory for evaluating a possible Alu transcriptional response to e1a.
As shown in Figure 1A, the Alu transcriptome expanded considerably in response to e1a expression. 1783 different Alus were detected as expressed 24 h after infection with dl1500, versus 973 in mock-infected and 974 in dl312-infected cells. More precisely, 666 more Alus could be detected as expressed in dl1500-infected cells compared to dl312/mock-infected cells. From now on, we will refer to the entire set of Alus whose expression is detected in at least one of the three conditions as ‘expression-positive’ Alus (epAlus). We prefer this term, instead of simply ‘expressed’ Alus, because it indicates that an Alu element which is not expressed in just one or two conditions is nevertheless well distinguishable from the much larger number of Alus that are not expressed in any of the conditions. In general, we observed a >75% overlap between the two replicates of each infection (Supplementary Figure S1). The global effect of e1a on Alu expression can be observed in the profiles in Figure 1B, showing a ∼4-fold increase in average Alu expression in response to e1a expression. The viral e1a protein is thus able to derepress Alu elements genome-wide, likely as a part of its global epigenome reprogramming properties. As it is further evident from Figure 1A, dl312- and mock-infected cells are characterized by an incomplete overlap of expressed Alu sets, which may result from the interaction of cellular and viral receptors and the exposure of viral nucleic acids during the injection of viral DNA-protein complexes from the cell surface into the nucleus (59). However, most of these elements were also expressed in dl1500-infected cells, where many of them were upregulated. Of 1805 observed epAlus, almost 37% were exclusive to dl1500-infected cells, ∼62% were also expressed in dl312/mock cells, whereas only ∼1% were detected in dl312/mock-infected cells but not in dl1500-infected cells. Among the 953 Alus expressed in both dl312- and dl1500-infected cells, 385 were differentially expressed with FDR < 0.05. With just one exception, all differentially expressed Alus were upregulated in dl1500-infected cells. The coordinates of epAlus, including information on differentially expressed Alus and the corresponding expression values are reported in Supplementary Table S2.
In agreement with previous studies showing that only a small fraction of Alu elements is expression-positive in each cell type (5,7), e1a-dependent Alu upregulation was very limited on a genome-wide scale. Only 0.22% of the total intergenic/antisense Alu sequences could be detected as expressed after dl1500 infection, which nonetheless represented a remarkable increase with respect to the percentage of constitutively expressed Alu loci (0.12%) (see Table 1). Such low percentages might also reflect the detection limits of the technique. A few examples of individual Alu expression profiles, representative of different e1a effects on Alu expression, are shown in Figure 1C.
Table 1.
Subfamily distribution of expression-positive Alusa
| AluJ | AluS | AluY | No family | Tot | |
|---|---|---|---|---|---|
| Annotated | 213 441 (26.6%) | 484 941 (60.43%) | 100 903 (12.57%) | 3146 (0.39%) | 802 431 |
|
Expressed (dl1500) Subfamily enrichmentb P-valuec |
187 (10.49%) 0.39 <2.2 × 10−16 |
1499 (84.07%) 1.39 <2.2 × 10−16 |
96 (5.38%) 0.43 <2.2 × 10−16 |
1 (0.05%) | 1783 |
| Expressed (dl312) Subfamily enrichment P-value | 155 (15.91%) 0.60 2.3 × 10–15 |
748 (76.8%) 1.27 <2.2 × 10−16 | 70 (7.19%) 0.57 6.9 × 10−08 |
1 (0.1%) | 974 |
| Expressed (mock) Subfamily enrichment P-value | 153 (15.72%) 0.59 6.8 × 10−16 |
750 (77.08%) 1.28 <2.2 × 10−16 | 69 (7.09%) 0.56 3.9 × 10−08 |
1 (0.1%) | 973 |
aReported for each Alu subfamily (columns) is the absolute number of intergenic/antisense Alus and (in parentheses) their percentage. In the first row, the subfamily distribution of annotated Alus is reported. The other rows report the subfamily distribution of Alus expressed in dl1500, dl312 and mock-infected IMR90 cells.
bFor each sample, the subfamily enrichment was calculated as the ratio between the percentage of each subfamily out of all expressed Alus and the percentage of each subfamily out of all annotated Alus.
cThe P-value for subfamily enrichment (in italics) was calculated using a two-tailed Fisher's exact test.
Alu elements are classified into three main subfamilies, called AluJ, AluS and AluY, with AluY being the evolutionarily youngest, and thus the less degenerate in sequence. Accordingly, AluY is the only known subfamily which is at present actively retrotransposing in the human genome (60). Despite their retrotransposition activity, AluY elements were not found to be more expressed than the other two subfamilies in previous studies (5,7). We thus investigated the contribution (in terms of epAlu fraction) of AluJ, S and Y subfamilies to the total expression of Alu sequences in IMR90 cells subjected or not to e1a-dependent reprogramming. As detailed in Table 1, a highly significant enrichment in AluS subfamily elements was observed among expressed Alus in both control and dl1500-infected cells, while AluY- and AluJ-derived transcripts were significantly underrepresented in Alu transcriptomes. As recently suggested, the relatively low contribution of AluY to Alu transcriptomes is consistent with the possibility that specific repression systems tend to be more efficient on Alus with higher retrotransposition-dependent mutagenic potential (7). The underrepresentation of AluJ transcripts might be related to the higher number of mutations accumulated in these old elements (61), making them less able to support transcription, in particular in response to e1a. An evaluation of A- and B-box promoter elements of epAlus confirmed the results of previous analyses (5), showing a substantial match with canonical tRNA gene-derived consensus sequences for the two internal control regions (A-box: TRGYnnAnnnG; B-box: GWTCRAnnC), with the exception of the last position of the A-box, which in Alus is C instead of G (data not shown).
To check whether e1a-dependent derepression also occurs in other SINEs in addition to Alus, we analyzed the expression of mammalian-wide interspersed repeats (MIRs). With ∼500 000 copies in the human genome, MIRs are the second most numerous subgroup of human SINEs. As is the case for Alus, it has recently been shown that only a tiny percentage of MIRs are expressed in different cell lineages (11). Out of a total of 404 371 intergenic/antisense MIRs (defined as detailed above for Alus), only 137 were expression-positive, i.e. detected as expressed in at least one sample, with no evidence of dl1500-dependent upregulation (data not shown). The effect of e1a thus appears to be restricted to the Alu subgroup of human SINEs.
It was recently reported that expressed Alu elements tend to reside closer to the transcription start sites of Pol II-transcribed genes than unexpressed Alus, a property that may be associated with their ability to function as cell type-specific enhancers for nearby genes (7). To investigate the possible relevance of Alu expression for protein-coding gene expression in our experimental system, we first asked whether there is any bias in the distribution of epAlus between protein-coding genes and the remaining genomic regions. As shown in Supplementary Figure S2, the fraction of intragenic Alus (limited to those with antisense orientation) was significantly lower (and, correspondingly, the fraction of extra-genic elements was higher) for epAlus than for unexpressed Alus (panel A). Furthermore, there was a significant overrepresentation of epAlus within 32 kb from the TSSs of protein-coding genes (more marked upstream than downstream of the TSS), while at distances of more than 64 kb epAlus were significantly underrepresented (panel B). The expression levels (normalized for gene length) of protein-coding genes flanked by epAlus were generally higher than those of genes flanked by unexpressed Alus (panel C). Altogether, these data provide general support to the idea that epAlus are overrepresented in regions relatively close to the TSSs of protein-coding genes, where their expression tends to parallel the expression of nearby genes. However, we did not find evidence for differential expression of protein-coding genes paralleling the e1a-dependent induction of nearby Alus (data not shown).
e1a-dependent Alu derepression occurs through increased TFIIIB recruitment without changes in TFIIIC occupancy
Previous reports suggested that e1a affects TFIIIC activity (36,37) and that human TFIIIC can influence the epigenetic state of Alu elements genome-wide (15). We thus asked whether e1a induces changes in the genome-wide distribution of TFIIIC and TFIIIB in IMR90 cells, and whether such changes could explain at least in part the observed e1a-dependent Alu upregulation.
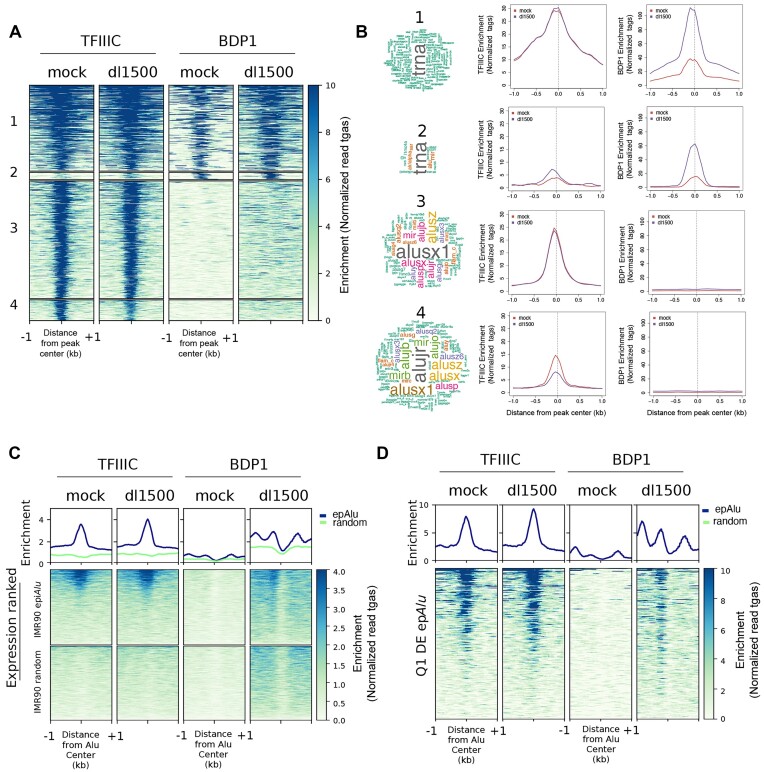
The genome-wide location of TFIIIC and TFIIIB in IMR90 cells in the presence or absence of e1a was assessed by ChIP-seq using antibodies targeting the 110 kDa subunit of TFIIIC and the Bdp1 component of TFIIIB. TFIIIC/TFIIIB-associated loci in IMR90 cells were classified into four combinatorial clusters according to their enrichment for TFIIIC and Bdp1 (Figure 2A). Loci of the first two clusters were characterized by the association of both TFIIIC and Bdp1, and their overlap with all repetitive elements was strongly enriched in tRNA genes (Figure 2B). Cluster 1 loci were characterized by an e1a-dependent increase in Bdp1 association, without any appreciable change in TFIIIC enrichment. An increase in both TFIIIC and TFIIIB association was instead observed at cluster 2 targets. Loci belonging to clusters 3 and 4 were generally found to be enriched in TFIIIC but not Bdp1. These two clusters turned out to be strongly enriched in SINEs, primarily Alu but also MIR elements (Figure 2B). The presence of human TFIIIC in the absence of other components of the Pol III machinery has previously been reported to occur at SINE loci (62,63). A distinctive feature of cluster 4 is an e1a-dependent decrease of TFIIIC association. Genomic Regions Enrichment of Annotations Tool (GREAT) analysis (64) found that the Alus of this cluster are significantly associated with genes involved in embryonic lethality (Supplementary Figure S3A), suggesting that e1a-dependent TFIIIC removal from these sites might participate in viral manipulation of cell identity and differentiation pathways (40,41,65).
Figure 2.
Genome-wide location analysis of TFIIIC and TFIIIB in the presence/absence of e1a. (A) Heatmap of TFIIIC (GTF3C2) and Bdp1 spanning ±1 kb across all TFIIIC-bound sites in mock- and dl1500-infected cells. Clusters 1 to 4 were created by combinatorial clustering of the two factors across all regions bound. Color bar scale with increasing shades of color stands for increasing enrichment (normalized read tags). (B) Shown on the left is the word cloud analysis of repetitive elements associated with regions occupied by TFIIIC and Bdp1 in the four clusters. Font size reflects enrichment for the indicated term. Reported on the right are the results of sitepro analysis (120) of TFIIIC and Bdp1 enrichment (normalized read tags) for each cluster reported in panel A. Enrichment is shown spanning 2 kb from the center of the peaks. (C) Shown in the upper part of the panel are the average ChIP-seq enrichment profiles (normalized read tags) of the TFIIIC 110 kDa subunit (left) or the Bdp1 component of TFIIIB (right) in either mock-infected or dl1500-infected IMR90 cells across the 1805 epAlus and across random Alus. Reported below the plots are heatmaps of TFIIIC and Bdp1 enrichment at the same Alus, sorted according to their expression level in dl1500-infected cells (top, high expression; bottom, low expression). (D) Enrichment profiles (normalized read tags) of TFIIIC and Bdp1, in either mock-infected or dl1500-infected IMR90 cells, at differentially expressed Alus whose expression levels in the presence of e1a falls in the first quartile (Q1 DE epAlu), sorted according to their expression level in dl1500-infected cells (top, high expression; bottom, low expression).
The analyses reported in Figure 2A and B related to TFIIIC and Bdp1 enrichment across the whole genome. To assess in a more accurate and sensitive way whether there was a correlation between the presence of TFIIIC/Bdp1 at individual Alu loci and their expression, we focused the ChIP-seq data analysis on the 1805 epAlus. We found that epAlus were globally characterized by a significant TFIIIC association independently from the presence of e1a (Figure 2C). Therefore, Alu upregulation in response to e1a does not appear to involve increased TFIIIC recruitment at epAlus. However, no comparable TFIIIC occupancy was observed in random Alu sets, considered to be representative of unexpressed Alus. As epAlus were ranked based on their levels of expression (from high to low) in Figure 2C, our results show marked TFIIIC occupancy for highly expressed epAlus for both dl1500- and mock-infected cells. This suggests that TFIIIC occupancy is higher at Alus with higher transcriptional potential and that the presence of e1a could eventually turn this potential into active transcription.
No specific signal was observed at epAlus in mock-infected cells through Bdp1 ChIP-seq analysis, while a specific Bdp1 association profile at epAlus was detected in dl1500-infected cells (Figure 2C), although it was weaker than the TFIIIC signal, possibly due to low efficiency of immunoprecipitation elicited by anti-Bdp1 antibodies and to the generally low level of Alu transcription activation. When the Bdp1 enrichment analysis was limited to differentially expressed Alus, and more specifically to those among them whose expression levels in the presence of e1a falls in the first quartile, a clear increase in Bdp1 enrichment was observed at an upstream position where TFIIIB is expected to be recruited by DNA-bound TFIIIC (Figure 2D). The Genome browser profiles of TFIIIC/Bdp1 occupancy along with expression profiles for three Alus included in the heatmap of Figure 2D are also reported (Supplementary Figure S3B). Altogether, the data suggest that Alu upregulation by e1a does not entail increased TFIIIC occupancy but does involve increased Bdp1 (and thus very likely TFIIIB) recruitment on an appreciable subset of epAlus.
Epigenetic signatures of expression-positive and e1a-upregulated Alu elements
Since Alu expression control is supposed to be mainly epigenetic, with the vast majority of Alus undergoing chromatin-mediated silencing (2), we asked whether some key histone modifications, as well as the association of chromatin regulatory proteins, could be associated with epAlus, and whether they are affected by e1a. We first made use of publicly available nucleosome mapping of IMR90 cells (66), which showed clear evidence for two strong nucleosomes positioned not only on epAlus, but also on random Alu sets (Supplementary Figure S4, panel A). As revealed by ATAC-seq analysis of the same cells (67), however, epAlus displayed more accessible chromatin upstream of TSS than random unexpressed Alus (Supplementary Figure S4, panel B). These preliminary observations prompted us to search for patterns of histone modifications at epAlus.
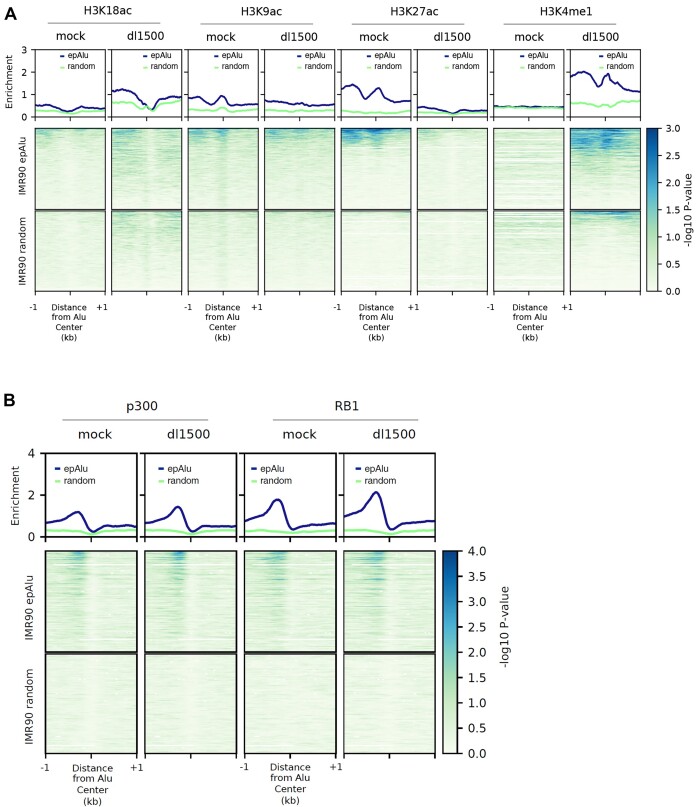
To gain insight into the epigenetic state of Alus and its possible e1a-induced changes, we employed ChIP-seq analysis of IMR90 cells, either mock- or dl1500-infected, for the following histone modifications: H3K9ac, H3K18ac, H3K27ac and H3K4me1 (the last three modifications being landmarks of enhancers). As shown in Figure 3A, the most noticeable e1a-dependent changes in the average profiles at epAlus were observed for H3K27ac, which was generally depleted in the presence of e1a, and for H3K4me1, whose presence upstream of epAlus was generally increased in response to e1a. More subtle and less specific changes in enrichment profiles were observed for H3K9ac and H3K18ac. The comparison of histone modification profiles of epAlus with random Alu sets revealed clear differences between expressed and unexpressed Alus in H3K4me1 and H3K27ac profiles. This supports the idea that epAlus bear enhancer-like chromatin marks and are epigenetically remodeled by e1a towards a state marked by H3K4me1 but lacking H3K27ac, reminiscent of poised enhancers (10).
Figure 3.
Histone modification and chromatin regulator enrichment profiles of epAlus in the presence/absence of e1a. (A) Shown in the upper graphs are the average ChIP-seq enrichment (–log10 of the Poisson P-value) profiles of (from left to right) H3K18ac, H3K9ac, H3K27ac and H3K4me1 across the 1805 epAlus in either mock-infected or dl1500-infected IMR90 cells. Reported below the plots are the heatmaps of the same histone modification enrichments, with epAlus and random Alus ranked according to enrichment expressed as –log10 of the Poisson P-value. (B) Shown in the upper part of the panel are the average ChIP-seq enrichment profiles (–log10 of the Poisson P-value) of EP300 (left) and RB1 (right) in either mock-infected or dl1500-infected IMR90 cells across the 1805 epAlus and across random Alus. Reported below the plots are heatmaps of EP300 and RB1 association to the same Alus, sorted according to their expression level in dl1500-infected cells (top, high expression; bottom, low expression).
Given the importance of e1a-EP300 and e1a-Rb interactions for e1a-dependent epigenome reprogramming (41), we also investigated by ChIP-seq the enrichment and possible redistribution of these two proteins at Alu loci. EpAlus were generally characterized by the presence of a p300 peak just upstream of the Alu body (Figure 3B). As in the case of TFIIIC, however, the average p300 association profile was not dramatically affected by e1a (Figure 3B). A similar behavior was observed for RB1, with a peak at the same position as the p300 peak (Figure 3B). Remarkably, the p300 and the RB1 association profiles were not present when random sets of Alus were compared with epAlus, thus pointing to a relevant role of these proteins in establishing the chromatin state of epAlus. Importantly, epAlus characterized by higher expression levels in dl1500-infected cells (top positions in the heatmaps) also generally displayed higher p300 and RB1 occupancy both in dl1500- and in mock-infected cells (Figure 3B). Therefore, as in the case of TFIIIC, the presence of p300 and Rb proteins appears to correlate with the Alu transcriptional potential.
Transcription factor signature of epAlus
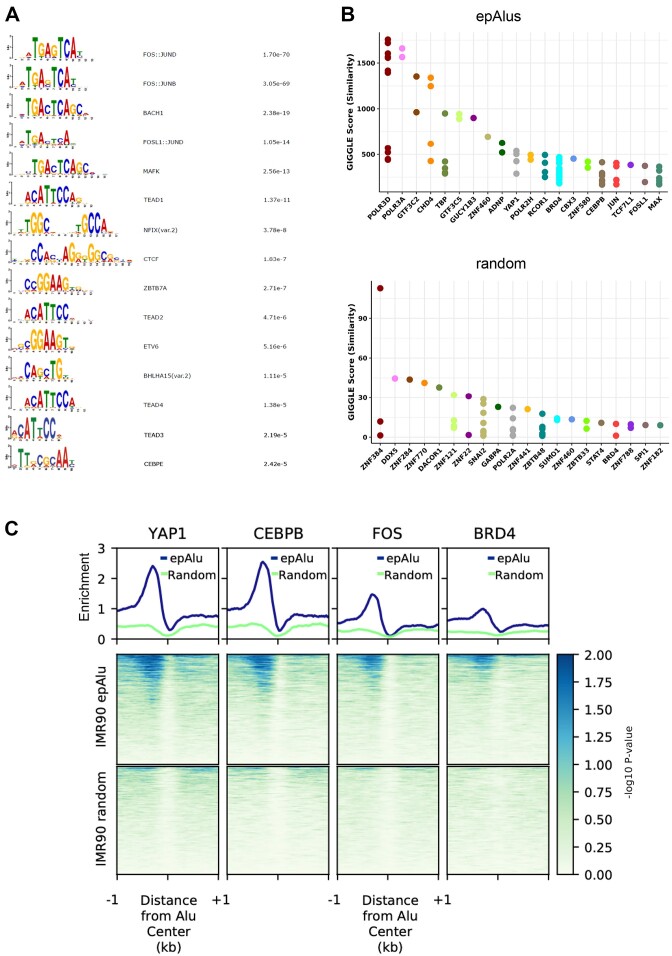
A number of transcription factors (TFs) have previously been shown to be enriched at expressed Alu elements (5,7), and the evolution of Alu elements towards enhancers has been suggested to be accompanied by the acquisition of TF binding sites over evolutionary timescales (10). To gain insight into the possible contribution of specific TFs to the epigenetic state of epAlus and their e1a-dependent upregulation, we searched for TF binding motifs enriched in the 200-bp region upstream of epAlus through the Analysis of Motif Enrichment (AME) tool (68). As shown in Figure 4A, the most significant enrichment was for motifs recognized by the Fos- and Jun-related factor heterodimers collectively referred to as AP-1 (69). In order of significance, the first AP-1-unrelated TF binding motif found to be enriched upstream of epAlus was the one recognized by TEAD1. Remarkably, TEAD2, TEAD3 and TEAD4 motifs were also significantly enriched. Other enriched motifs included those recognized by Nuclear Factor I (NFIX), CCCTC-binding factor (CTCF), Zinc finger and BTB domain-containing protein 7A (ZBTB7A), ETS Variant Transcription Factor 6 (ETV6), Basic Helix-Loop-Helix family member A15 (BHLHA15) and CCAAT Enhancer Binding Protein Epsilon (CEBPE). Some of these TFs or TFs of the same family (namely AP-1, CTCF, CEBPB), as well as the corresponding binding motifs, were previously found to occupy loci of expressed Alus in several cell lines (5,7). To further characterize the role of closely bound TFs in Alu expression, we took advantage of the Cistrome ToolKit (70) to perform an unbiased search for DNA binding proteins enriched, in any cell type or tissue, at the set of epAlus identified in this study (Figure 4B). We found that, in addition to components of the basal Pol III transcription machinery (POLR3D, POLR3A, GFT3C2, GTF3C5, TBP), the top 20 factors displaying the highest-ranking scores for epAlus included chromatin proteins and TFs whose presence suggests a complex regulatory scenario. Indeed, all three components of the ChAHP complex (CHD4, ADNP, CBX3), recently proposed to counteract CTCF binding by competing for the same sites in correspondence of mouse B2 SINEs (71), were found (Figure 4B). Importantly, we also identified TFs and chromatin regulators expected to be recruited to the enriched regulatory motifs of Figure 4A, such as: YAP1 (or YAP), a coactivator acting together with the paralogous TAZ protein to regulate target genes mainly through binding to DNA-bound TEAD TFs (72), and recently shown to be affected by e1a (73); bromodomain-containing protein 4 (BRD4), known to be physically engaged genome-wide by YAP/TAZ (74); the AP-1 subunits JUN and FOSL1; CEBPB, whose binding site specificity is the same as CEBPE (Figure 4B). As this analysis used ChIP-seq data from a wide range of cell types and tissues, we wanted to gain more insight into the relevance of these observations for the regulatory properties of our IMR90 epAlus. We thus further focused on publicly available CEBPB, FOS, YAP1 and BRD4 ChIP-seq data in IMR90 cells (75–79). We calculated the enrichment for all these ChIP-seq over the IMR90 epAlus. Our analysis unveiled a clear enrichment for YAP1, CEBPB and FOS peaking at the epAlu upstream region (Figure 4C), precisely where the corresponding binding sites were revealed by the DNA motif analysis. BRD4 was also found to be specifically enriched upstream of epAlus, while no corresponding signals were detected for random Alus (Figure 4C). Some representative profiles, showing the TF enrichment upstream of individual epAlus, are reported in Supplementary Figure S5.
Figure 4.
TF binding motifs and ChIP-seq enrichment at epAlus. (A) TF binding motifs enriched in the 200 bp upstream of the TSS of epAlus with (right column) their corresponding P-values adjusted for multiple testing with Bonferroni correction. (B) Cistrome Toolkit (http://dbtoolkit.cistrome.org/) analysis of epAlu and random Alu sets. Giggle score is calculated by using the genome coordinates of the two sets of Alus to retrieve which factors bind those intervals among all curated experiment in the Cistrome database (70). (C) Plotheatmap of ChIP-seq of YAP1, CEBPB, FOS and BRD4 at the 1805 epAlus and at random Alus based on data from (75–79). Ranking is according to enrichment of YAP1 reported as –log10 of the Poisson P-value.
Interaction of e1a with chromatin remodeler EP400 is required for Alu upregulation
The above analyses revealed a complex epigenetic infrastructure at epAlus but did not provide direct information on the molecular mechanism of e1a-dependent Alu upregulation. To address this issue, we analysed the functions of e1a interactions with host proteins by taking advantage of e1a mutants that are specifically impaired in their ability to interact with either RB-family proteins, EP300 and its paralog CREBBP, or EP400 (41,80,81). These mutants will henceforth be referred to as e1a_RB-b−, e1a_p300-b− and e1a_p400-b− (b− standing for ‘binding minus’).
Alu expression profiling was carried out through RNA sequencing of IMR90 cells infected with the three e1a mutant strains, compared to cells infected in parallel with mock- or dl1500. We first identified the proper MOI for the different mutants to obtain similar amounts of e1a protein with the different strains. RNA-seq and Western blot confirmed the proper expression of e1a binding mutants, with e1a_RB-b− expressed at significantly lower levels (Supplementary Figure S6). Samples from two rounds of infections performed as biological replicates were subjected to RNA-seq analysis. We then calculated Alu differential expression upon infection observed for each of the four e1a variants compared to mock-infected cells and reported the results as a heatmap (Figure 5A). Despite the lower expression levels of e1a_RB-b−, a slightly higher number of differentially expressed Alus were observed in its presence compared to wt e1a. A reduction in the number of differentially expressed Alus was instead observed with e1a_p300-b− and, much more markedly, with e1a_p400-b−. Therefore, RB-family proteins (RB, RBL1 and RBL2) appear to exert at best a negative modulatory effect on e1a-dependent Alu upregulation. By contrast, EP300/CREBBP appear to act positively in the same process, in accordance with their reported stimulatory activity on Pol III chromatin templates (82). But the major e1a function required for Alu derepression turned out to be the interaction with EP400, whose involvement in the regulation of Alu or any Pol III-transcribed gene has not been previously reported. Of the Alus activated by wt e1a, ∼90% were no longer activated by its mutant defective in EP400 binding.
The effect of the e1a mutations on Alu expression regulation was further confirmed by RT-qPCR on two individual epAlus whose unique 3′-trailer sequence was long enough to allow for unambiguous detection of Alu transcripts from the individual loci (Figure 5B, upper panel). Expression profiles of the same RT-qPCR tested loci reconstructed from RNA-seq data supported the same conclusion (Figure 5B, lower panel). For these two Alus, upregulation was strongly impaired with e1a_p400-b−, and it was also negatively affected, to a lower extent, by the loss of e1a interaction with RB and EP300/CREBBP (the behaviour of RB at these Alus thus deviates from the generally observed one). In contrast to these effects of e1a and its interaction mutants on Alu expression, no e1a-dependent expression modulation was observed for other non-Alu Pol III transcripts (7SL RNA, 7SK RNA, U6 snRNA, RNase P RNA, tRNAHis) whose genes are characterized by different types of promoter organization (83) (Figure 5C-D). As e1a-dependent Alu upregulation was found to entail increased Bdp1 recruitment (Figure 2), we wondered whether the weakening of Alu activation caused by the loss of e1a-EP400 interaction could be due to reduced Bdp1 recruitment. We thus employed Bdp1 ChIP-seq to systematically address whether Bdp1 enrichment at dl1500-differentially expressed Alus and epAlus was affected genome-wide by expression of e1a_p400-b−. By comparing Bdp1 enrichment profiles upon infection of IMR90 with dl312 (no e1a), dl1500 (wt e1a) and the e1a_p400-b− mutant, we found that infection with the latter led to a significant decrease in Bdp1 recruitment at both set of Alus (Figure 5E and F).
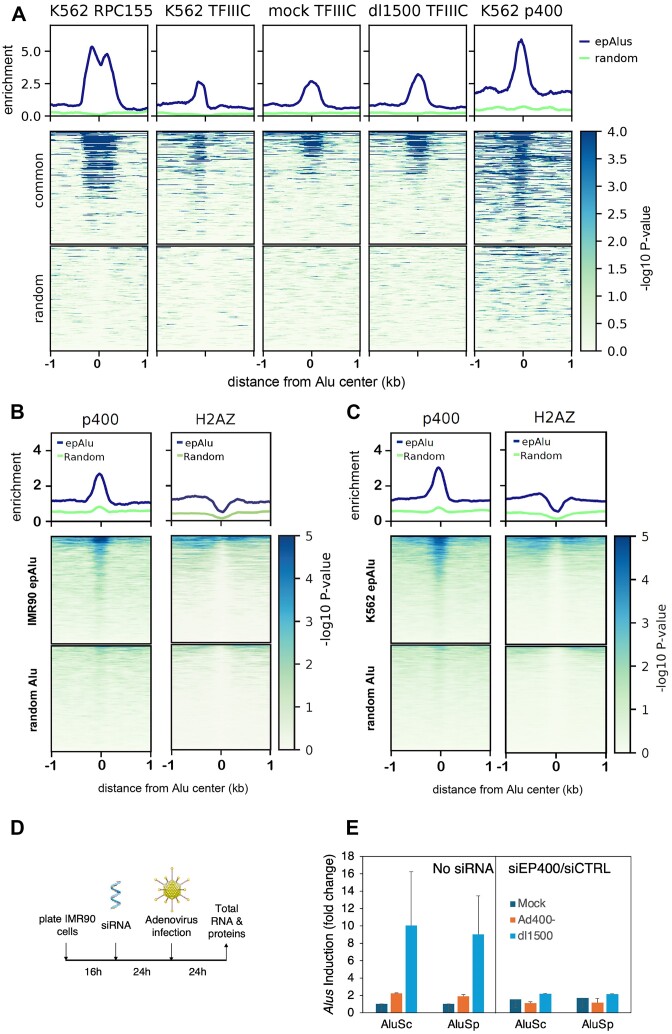
To find support for a general role of EP400 in Alu epigenetic control, we took advantage of the availability of ChIP-seq data for EP400 in K562 cells (84). Even though IMR90 and K562 represent different cell lineages and thus display largely different sets of expression-positive Alus, we observed nevertheless that 285 Alus are expression-positive in both cell lineages (Supplementary Figure S7A). Both the Pol III machinery and EP400 turned out to be strongly enriched at these Alus in K562 cells (Figure 6A). When the whole set of IMR90 epAlus was considered, EP400 enrichment was still significant (Mann–Whitney U test), while no enrichment signal was observed with random Alu subsets of similar size (Figure 6B, left subpanel). When the same analysis was carried out on the subset of Alus that are expression-positive in K562 cells, as expected EP400 was found to be even more enriched (Figure 6C, left subpanel). As EP400 has been implicated in the deposition of H2A.Z histone variant at nucleosomes (85), with the potential to influence both transcription and DNA repair, we compared the H2A.Z ChIP-seq profiles at expression-positive Alus of K562 cells and found an appreciable enrichment of H2A.Z compared to a random set (Figure 6B-C, right subpanels). To gain more mechanistic insight into the function of EP400 at Alus upregulated by e1a, we implemented siRNA knockdown of EP400 prior to infection with dl1500 and e1a_p400-b− adenovirus (Figure 6D-E, Supplementary Figure S7B and C). Our data show that siRNA depletion of EP400 did not cause any increase in epAlu expression in mock-infected cells, thus excluding that EP400 is a mere Alu repressor counteracted by e1a. In contrast, EP400 depletion from IMR90 cells abrogated e1a-mediated upregulation of AluSp and AluSc, thereby attesting that EP400 is actively involved in the induction of Alus caused by wt e1a expression (Figure 6E). Alu upregulation by e1a thus critically depends on previously unrecognized epigenetic features of epAlus involving the active role of p400 chromatin remodeler and possibly its histone exchange activity.
Figure 6.
Enrichment of EP400 and H2A.Z at epAlus and effects of EP400 depletion. (A) ChIP-seq enrichment profiles of Pol III (RPC155 subunit), TFIIIC and EP400 (p400) at the 285 Alu elements that are expression-positive in both IMR90 (this study) and K562 cells (76,78). Plotheatmap of ChIP-seq data. From left to right: Pol III enrichment in K562 cells, TFIIIC enrichment in K562 cells, TFIIIC enrichment in mock-infected and dl1500-infected IMR90 cells and EP400 enrichment in K562 cells. Ranking is according to enrichment of Pol III in K562 cells reported as -log10 of the Poisson P-value. (B) Plotheatmap of ChIP-seq enrichment of EP400 (left) and the H2A.Z histone variant (right) in K562 cells (76,78) across either the 1805 IMR90 epAlus or random Alus. Ranking is according to enrichment of EP400 in K562 cells reported as -log10 of the Poisson P-value. (C) Plotheatmap of ChIP-seq enrichment of EP400 (left) and the H2A.Z histone variant (right) in K562 cells across either the 3764 Alus detected as expressed in K562 cells or random Alus. Ranking is according to enrichment of p400 in K562 cells reported as -log10 of the Poisson P-value. (D) Schematic representation of the protocol of siRNA-mediated EP400 knock down (KD) followed adenoviral infection (time of each incubation is reported). (E) RT-qPCR for measuring expression of two epAlu loci (the same as in Figure 5B) comparing mock-, e1a_p400-b— and dl1500-infection in conditions of absence of silencing RNA (non-siRNA) or presence of siRNA against p400 (siEP400) compared to a scramble set of siRNA control (siCTRL). Standard error bars are indicated, as a result of two biological replicates.
Discussion
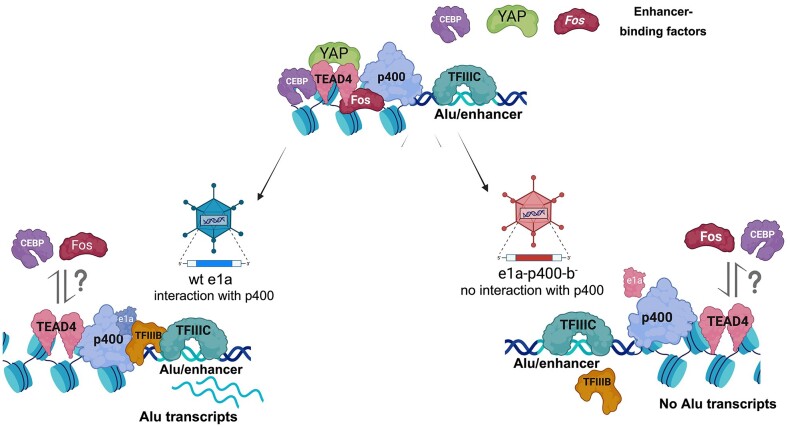
Alu expression has long been known to be upregulated in response to viral infection (86). Our study shows that the Adenovirus 5 e1a oncoprotein is responsible by itself for derepression of several hundred Alu elements across the human genome in IMR90 fibroblasts by virtue of its interaction with the host ATPase chromatin remodeler EP400. Our data show that many of the e1a-responsive Alus display features of YAP/TAZ- and AP-1-associated enhancers where e1a appears to reconfigure key histone modifications, most notably H3K27 acetylation. These effects can best be framed into the context of e1a epigenome reprogramming properties required for maximal viral DNA replication in permissive human cells and cell transformation of baby rat kidney cells (40). An impressive range of interactions with host proteins is exploited by e1a to induce cell cycling and dedifferentiation (87). Two of the best well-characterized e1a-host protein interactions take place with the tumour suppressor RB1 and the lysine acetyltransferases EP300/CREBBP. These interactions are key to the e1a-induced gene dysregulation underlying cell transformation. Their mechanistic contribution to this process has been widely studied and shown to involve, in addition to the displacement of RB1 from E2F transcription factors, a complex interplay with chromatin regulatory proteins (41,88–91). The less extensively investigated e1a-EP400 interaction has been known for twenty years to function in the e1a transforming process (42), and later was shown to stabilize MYC and to promote formation of MYC-EP400 complexes on chromatin leading to activation of MYC target genes (92). Our finding that derepression of a subset of epAlus strongly depends on the e1a-p400 interaction adds an important element to our knowledge of e1a-dependent epigenome reprogramming, by showing that it might also rely on the exploitation of a chromatin remodeler at a subset of retrotransposons. Whether such an epigenetic switch, operating at hundreds of loci throughout the genome, also involves the MYC family of transcription factors, or some other EP400-interacting proteins like TRRAP, remains to be established. In support of a possible involvement of Myc proteins, N-Myc was recently shown to interact with TFIIIC by proteomic analysis and to colocalize with TFIIIC at thousands of sites (93). c-Myc was also previously found to activate Pol III-dependent transcription of tRNA and 5S rRNA genes through a mechanism involving TRRAP and Gcn5 recruitment (94) and more generally to be present at Pol III-transcribed genes at the genome-wide scale (95). TRRAP recruitment was also previously reported to be required for cell transformation by E1A (96). Myc proteins could therefore participate in the complex interaction network expolited by e1a to epigenetically deregulate Alus.
Through its histone exchange activity, EP400 is likely to promote H2A.Z deposition at epAlus, whose enrichment was correlated with Alu expression in this and previous studies (7). Notably, we find that EP400 is actively involved in wt e1a induction of Alus in agreement with findings reporting EP400 to be necessary for H2A.Z and H3.3 deposition into enhancers and promoters in vivo (97). Another possible mechanistic facet of Alu activation through e1a-EP400 interaction is suggested by the ability of the TIP60/p400 complex to recognize H3K4me1 through its TIP60 component (44). As part of the Alu epigenetic switch, the e1a-EP400 interaction might favour H3K4me1-mediated TIP60/p400 recruitment, which would in turn promote Alu transcription. The presence at specific Alu loci of regulators of chromatin architecture might not be limited to p400, as another chromatin remodeler, CHD4, recently shown to associate to evolutionarily younger mouse SINEs as part of the ChAHP complex (71), was found to be enriched at a subset of Alu elements in human breast cancer cells (15).
The use of e1a mutants specifically defective in interaction with chromatin regulatory proteins also allowed us to exclude the possibility that e1a acts by relieving Rb-mediated repression at Alu elements. With an e1a mutant unable to interact with Rb, the number of upregulated Alus and the extent of upregulation was even higher than with wt e1a, thus suggesting that the e1a-Rb interaction impacts negatively on Alu expression. As to the e1a-p300 interaction, its loss only slightly weakened Alu expression upregulation. Collectively, these effects are in line with the complex interplay between e1a, Rb and p300 previously shown to lie behind activating and repressing chromatin conformations in Ad5-infected cells (41).
A possible key to understanding the epigenetic features of Alu elements, including their expression leading to Alu RNA, is their recently recognized nature of cis-regulatory elements serving as a repertoire for the de novo birth of enhancers (7,10). Such enhancer-like properties extend to different types of SINEs in different species, suggesting a relevant role of SINEs in the evolution of complex regulatory networks (98,99). Results in our study both confirm and expand the notion of Alus as enhancer-like elements. In mock-infected and e1a-expressing IMR90 cells, epAlus differed for two key histone modifications, H3K4me1 and H3K27ac, whose concomitant presence is an epigenetic hallmark of active enhancers. Specifically, we observed a general e1a-dependent increase of H3K4me1 and a general depletion of the H3K27ac marker. This epigenetic signature at epAlus in the presence of e1a roughly resembles the previously reported enhancer-like Alu epigenetic profile reminiscent of poised enhancers, marked by H3K4me1, but lacking H3K27ac (10). The association of p300 upstream of epAlus is also consistent with an enhancer-like state of these elements. At these sites, however, the acetyltransferase activity of p300 towards H3K27 appears to be inhibited by e1a, as it occurs at many enhancers and other genomic locations as part of the epigenome reprogramming effects of e1a (41). The ability of e1a to alter some Alus towards the epigenetic state of poised enhancers, corresponding to a cellular state preceding differentiation (100), might contribute to e1a's ability to promote cell dedifferentiation (73). On this regard, our finding that the YAP/TAZ coactivators associate to epAlus in unperturbed IMR90 cells (most likely through TEAD TFs recognizing their cognate DNA motif upstream of epAlus) appears as particularly relevant. As recently shown, e1a causes YAP/TAZ cytoplasmic sequestration, with consequent genome-wide loss of YAP/TAZ and H3K27ac at enhancers involved in cell differentiation (73). Even though we have not addressed the e1a-dependent YAP/TAZ dynamics at epAlus, it appears reasonable to hypothesize that e1a-dependent Alu derepression contributes, through unexplored mechanisms, to enhancer manipulation by e1a. Possibly related to these mechanisms are the previously reported interaction of YAP/TAZ with EP400 and the so far unreported enrichment at epAlus of BRD4, which is known to mediate YAP/TAZ-dependent transcriptional regulation in cancer cells (74).
Another related issue that deserves proper investigation is the role of two other families of enhancer-associated transcription factors, AP-1 and C/EBP. We found an overrepresentation of their cognate sites upstream of expression-positive Alus, as well as an enrichment of members of the two TF families at epAlus based on ChIP-seq data analysis, an observation also reported by previous studies (5,7). AP-1 and C/EBP TFs might play a role in Alu transcription and/or Alu enhancer-like function through recruitment of coactivators like p300/CBP (101,102). However, the interplay between the presence of these factors at epAlus and their e1a-dependent upregulation needs to be clarified, as e1a was previously shown to counteract Fos-dependent transcriptional activation by competing for p300/CBP binding (103). Of note, the concomitant presence of AP-1 and YAP/TAZ/TEAD at growth-controlling enhancers was previously associated to oncogenesis (104), thus reinforcing the idea that e1a-triggered Alu derepression participates in key cell-controlling enhancer functions. The identity of Alu-associated TF family members is another issue awaiting further clarification. The different isoforms of C/EBP (α, β, γ, δ, ϵ, ζ) have binding site specificities that are almost identical (with the exception of C/EBPζ) (105). Therefore, ChIP studies can tell more reliably than DNA sequence motif analysis which isoforms are more likely to be involved than others. Remarkably, C/EBPβ was previously reported to be significantly enriched at epAlus in HeLa, HepG2 and K562 cells based on analysis of ENCODE ChIP-seq data (5). The same association, again based on ENCODE ChIP-seq data, was observed in this study in the case of IMR90 cells. Moreover, C/EBPβ (alias CEBPB) has recently been shown to mediate the Pol III-dependent transcription of a microRNA transcription unit (miR-138) in human glioblastoma cell lines, and the same study provided evidence for a more general involvement of C/EBPβ in the recruitment of Pol III transcription complexes to their target genes (106). Notably, all the above mentioned epAlu-associated TFs were found to associate with target sites located upstream of the Alu body, suggesting a co-evolution of epAlus with these TF binding sites which may be relevant for Alu exaptation as regulatory elements, as well as for Alu transcription. In fact, the internal pol III elements (A and B boxes) are likely to synergize with the upstream flanking region for Alu transcription (5). The mostly random insertion of Alu elements into the genome creates a likelihood that the majority will land in regions that do not support transcription even under epigenetically open chromatin, due to the lack of appropriate upstream sequences. This could also explain why the relative number of epAlu loci is so small.
Viewing epAlus as enhancers or proto-enhancers suggests possible implications for their expression and their virus-dependent upregulation. A key to understanding these processes may in fact be provided by the tendency of enhancers to act as templates for noncoding enhancer RNA (eRNA) synthesis in a manner linked to their activity (107). EpAlu-encoded RNA may possibly be considered as a special case of eRNA, with the potential to influence transcription of genes placed under the control of the Alu enhancer, as shown for the regulation of FOS gene transcription by a SINE-encoded eRNA in mouse cortical neurons (98). Moreover, a recent work has revealed that ∼38% of the enhancer–promoter RNA interaction sites are overlapped with Alu sequences (108). It should be added, however, that Alu-associated TFIIIC has the potential to participate in organizing the landscape of chromatin loops, and thus in controlling gene expression at distance, just by virtue of its protein-protein interactions with transcription factors and/or architectural proteins such as ADNP and CTCF (15), even in the absence of RNA production. How frequently and to what extent Alu transcripts, or even the mere act of their transcription, participate in the enhancer-like activity of Alus still awaits further investigation (see below).
Another, more general issue that has still to be satisfactorily understood is the impact of epAlu derepression on the processes leading to cell transformation. In principle, Alu RNA accumulation might contribute to sustained cell proliferation via remodulation of the mRNA transcriptome which may occur both transcriptionally and post-transcriptionally (109,110). However, the actual amount of Alu transcripts elicited by e1a is probably low compared to the one obtained by artificial Alu RNA overexpression, and the duration and stability of e1a-dependent Alu RNA increase has not been evaluated. It is therefore somewhat difficult to attribute to increased Alu RNA a causative role in e1a-dependent cell transformation. Alternatively, the transcripts resulting from epAlu derepression might be considered to some extent as by-products of an epigenetic transition whose contribution to cell transformation is not due to the RNA products themselves but possibly to epAlu-triggered genome architectural rewiring (15).
Another property of e1a with the potential to contribute to cellular transformation is its previously reported ability to increase the activity of both TFIIIC and TFIIIB, at least in part by overcoming the repressive effect of Rb (35,37,111). Derepressed Pol III-dependent transcription of genes coding for RNAs required for protein synthesis and trafficking, such as tRNAs, 5S rRNA and 7SL RNA, would in turn contribute to cell transformation (112). Although our study did not address systematically the expression of canonical Pol III-transcribed genes, whose study at single-locus resolution is especially challenging for tRNA genes (113), expression analysis of a few of them showed the lack of an e1a-dependent upregulation comparable to the one of epAlus. At the same time, an e1a-dependent increase of Bdp1 (but not TFIIIC) association was generally observed for tRNA gene loci. According to a commonly accepted model, mainly based on in vitro studies, Rb sequesters a TFIIIB component without associating itself to DNA. In doing so, it impairs the TFIIIC-dependent recruitment of TFIIIB at promoters and subsequent transcription by Pol III. By interacting with Rb, e1a would release TFIIIB from repression (112). According to more recent analyses of ChIP-seq data, however, Rb stably associates to a relevant percentage of tRNA gene loci in IMR90 cells (114). This leaves open the possibility that Rb might participate in tRNA gene transcriptional regulation by influencing steps subsequent to TFIIIB recruitment, as it occurs in the case of Pol III-transcribed genes with a Type 3 promoter (e.g. the U6 snRNA gene) (115).
The most evident e1a-dependent change in TFIIIC enrichment occurred at a subset of TFIIIC genomic targets, strongly enriched in Alu elements, where marked depletion of TFIIIC occurred upon dl1500 infection. This Alu subset overlaps only marginally to the one of epAlus, thus implying that the modulation of TFIIIC association to these elements does not entail changes in Alu expression. Rather, TFIIIC depletion at these Alu loci might result in epigenetic changes possibly contributing to e1a interference with the cellular differentiation state, as suggested by the involvement of neighbouring genes in embryonic development.
In conclusion, molecular characterization of Alu upregulation in response to adenovirus infection led us to identify e1a as a key player in this phenomenon through its epigenome reprogramming properties. We uncovered an unexpectedly complex epigenetic landscape at epAlu elements. In some respects, our findings confirm the idea that epAlus represent a tiny subset of the whole Alu complement that have been -or are being- exapted to enhancer function (7,10). The observation that epAlus are enriched in older AluS element is also suggestive of evolutionary selection. We speculate that, among the >1 200 000 Alu elements in the human genome, such evolutionarily exapted Alus are likely to constitute a small set of non-silenced elements, sharing some basic epigenetic features that endow them with a certain transcriptional potential. Different subsets of such Alus would then be activated in different cell types due to further epigenetic changes accompanying cell differentiation. Whether and how Alu transcription and its modulation by different cues mechanistically contribute to enhancer function remains to be established. The fact that some derepressed Alus display features of YAP/TAZ enhancers, that are known to undergo e1a-dependent inactivation, supports an inverse relationship between Pol III transcription activation and enhancer activity. This would be in contrast with a previously proposed role of enhancer SINE Pol III transcription in activating Pol II-dependent transcription of the target gene (98). Other studies, however, provided evidence for a complex interplay between Pol III and Pol II transcription at regulatory SINEs (116,117), leaving room for the possibility that at enhancer Alus Pol III transcription counteracts Pol II-dependent eRNA synthesis and thus enhancer activity. In other respects, our study of the ability of e1a to turn on the Alu epigenetic switch revealed a possible unexpected role of chromatin remodelling in this process, with EP400 and possibly other chromatin remodelers performing a key activating function during induction of expression-prone Alus. As exemplified by the subtle control of promoter-bound nucleosome positioning by ATP-dependent remodeling complexes at rRNA genes (118), chromatin remodelers offer the possibility to rapidly switch between chromatin states of different transcriptional accessibility without radically changing the overall nucleosomal architecture. They are thus likely to be key players at exapted Alu loci where facilitation of accessibility changes may favour the evolutionary exploration of novel regulatory possibilities.
Supplementary Material
Acknowledgements
This work used computational and storage services associated with the Hoffman2 Shared Cluster provided by UCLA Institute for Digital Research and Education's Research Technology Group. RNA-seq was performed by the Broad Stem Cell Research Center High Throughput Sequencing Core at the University of California, Los Angeles, and by Fulgent Genetics, Temple City, California). BM, GD, MM and RF have benefited from the equipment and framework of the COMP-HUB and COMP-R Initiatives, funded by the ‘Departments of Excellence’ Program of the Italian Ministry for University and Research (MIUR, 2018–2022 and MUR, 2022–2027), and from the HPC (High-Performance Computing) facility of the University of Parma, Italy. The authors wish to thank the CIBIO NGS Facility of the University of Trento for sequencing samples. CIBIO Core Facilities are supported by the European Regional Development Fund (ERDF) 2014–2020. Graphical Abstract created with BioRender.com.
Notes
Present address: Simona Cantarella, Research Group RNA-protein complexes and Cell Proliferation, German Cancer Research Center (DKFZ), 69120 Heidelberg, Germany.
Present address: Nathan R. Zemke, Department of Cellular and Molecular Medicine, University of California, San Diego School of Medicine, La Jolla, CA, USA.
Present address: Coralie F. Daussy, University of Montpellier, CNRS UMR 9004, Institut de Recherche en Infectiologie de Montpellier (IRIM), Montpellier, France.
Contributor Information
Simona Cantarella, Department of Chemistry, Life Sciences and Environmental Sustainability, University of Parma, 43124 Parma, Italy.
Marco Vezzoli, Department of Chemistry, Life Sciences and Environmental Sustainability, University of Parma, 43124 Parma, Italy.
Davide Carnevali, Department of Chemistry, Life Sciences and Environmental Sustainability, University of Parma, 43124 Parma, Italy.
Marco Morselli, Department of Chemistry, Life Sciences and Environmental Sustainability, University of Parma, 43124 Parma, Italy.
Nathan R Zemke, Molecular Biology Institute, University of California at Los Angeles, Los Angeles, CA 90095, USA.
Barbara Montanini, Department of Chemistry, Life Sciences and Environmental Sustainability, University of Parma, 43124 Parma, Italy.
Coralie F Daussy, Bordeaux University, CNRS UMR 5234, Fundamental Microbiology and Pathogenicity, Bordeaux, France.
Harald Wodrich, Bordeaux University, CNRS UMR 5234, Fundamental Microbiology and Pathogenicity, Bordeaux, France.
Martin Teichmann, Bordeaux University, Inserm U 1312, Bordeaux Institute of Oncology, 33076 Bordeaux, France.
Matteo Pellegrini, Department of Molecular Cellular and Developmental Biology, University of California Los Angeles, Los Angeles, CA 90095, USA.
Arnold J Berk, Molecular Biology Institute, University of California at Los Angeles, Los Angeles, CA 90095, USA.
Giorgio Dieci, Department of Chemistry, Life Sciences and Environmental Sustainability, University of Parma, 43124 Parma, Italy.
Roberto Ferrari, Department of Chemistry, Life Sciences and Environmental Sustainability, University of Parma, 43124 Parma, Italy.
Data availability
Data of RNA-seq and ChIP-seq generated in this study are available at GEO under accession number GSE208717. Publicly available data were retrieved from the following sources: GSE32340 for H3K4me1 ChIP-seq (dl1500-infected IMR90 cells) and H3K9ac ChIP-seq (40); GSE43070 for H3K4me1 ChIP-seq (uninfected IMR90 cells) (40); GSE59693 for ChIP-seq of H3K27ac, EP300, Pol II and RB (mock- and dl1500-infected IMR90 cells) (41); GSM2828526 for EP400 ChIP-seq (K562 cells); GSM935372 for Pol III (RPC155 subunit) ChIP-seq (K562 cells); GSM935343 for TFIIIC (GTF3C2 subunit) ChIP-seq (K562 cells); GSM733786 for H2A.Z ChIP-seq (K562 cells); GSM3289968 for ATAC-seq (IMR90 cells); GSM935519 for CEBPB ChIP-seq (IMR90 cells); GSM818008 for H2A.Z ChIP-seq (IMR90 cells); GSM2825448 for FOS ChIP-seq (IMR90 cells); GSM1696207 for YAP1 ChIP-seq (IMR90 cells); GSM1915116 for BRD4 ChIP-seq (IMR90 cells).
Supplementary data
Supplementary Data are available at NAR Online.
Funding
Italian Association for Cancer Research [AIRC, IG2015-16877 to G.D., AIRC, IG2022-27712 to R.F.]; research in the A.J.B. laboratory was funded by the Professor June Lascelles Fund. Funding for open access charge: AIRC IG2022-27712; University of Parma.
Conflict of interest statement. None declared.
References
- 1. Goodier J.L. Restricting retrotransposons: a review. Mobile DNA. 2016; 7:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Varshney D., Vavrova-Anderson J., Oler A.J., Cairns B.R., White R.J.. Selective repression of SINE transcription by RNA polymerase III. Mobile Genet. Elem. 2015; 5:86–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dieci G., Conti A., Pagano A., Carnevali D.. Identification of RNA polymerase III-transcribed genes in eukaryotic genomes. Biochim. Biophys. Acta. 2013; 1829:296–305. [DOI] [PubMed] [Google Scholar]
- 4. Russanova V.R., Driscoll C.T., Howard B.H.. Adenovirus type 2 preferentially stimulates polymerase III transcription of Alu elements by relieving repression: a potential role for chromatin. Mol. Cell. Biol. 1995; 15:4282–4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Conti A., Carnevali D., Bollati V., Fustinoni S., Pellegrini M., Dieci G.. Identification of RNA polymerase III-transcribed alu loci by computational screening of RNA-seq data. Nucleic Acids Res. 2015; 43:817–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carnevali D., Dieci G.. Identification of RNA polymerase III-transcribed SINEs at single-locus resolution from RNA sequencing data. Noncoding RNA. 2017; 3:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang X.O., Gingeras T.R., Weng Z.. Genome-wide analysis of polymerase III-transcribed Alu elements suggests cell-type-specific enhancer function. Genome Res. 2019; 29:1402–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Karijolich J., Zhao Y., Alla R., Glaunsinger B.. Genome-wide mapping of infection-induced SINE RNAs reveals a role in selective mRNA export. Nucleic Acids Res. 2017; 45:6194–6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mori Y., Ichiyanagi K.. melRNA-seq for expression analysis of SINE RNAs and other medium-length non-coding RNAs. Mobile DNA. 2021; 12:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Su M., Han D., Boyd-Kirkup J., Yu X., Han J.D.. Evolution of Alu elements toward enhancers. Cell Rep. 2014; 7:376–385. [DOI] [PubMed] [Google Scholar]
- 11. Carnevali D., Conti A., Pellegrini M., Dieci G.. Whole-genome expression analysis of mammalian-wide interspersed repeat elements in human cell lines. DNA Res. 2017; 24:59–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jjingo D., Conley A.B., Wang J., Marino-Ramirez L., Lunyak V.V., Jordan I.K.. Mammalian-wide interspersed repeat (MIR)-derived enhancers and the regulation of human gene expression. Mobile DNA. 2014; 5:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ye M., Goudot C., Hoyler T., Lemoine B., Amigorena S., Zueva E.. Specific subfamilies of transposable elements contribute to different domains of T lymphocyte enhancers. Proc. Nat. Acad. Sci. U.S.A. 2020; 117:7905–7916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fueyo R., Judd J., Feschotte C., Wysocka J.. Roles of transposable elements in the regulation of mammalian transcription. Nat. Rev. Mol. Cell Biol. 2022; 23:481–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ferrari R., de Llobet Cucalon L.I., Di Vona C., Le Dilly F., Vidal E., Lioutas A., Oliete J.Q., Jochem L., Cutts E., Dieci G.et al.. TFIIIC binding to alu elements controls gene expression via chromatin looping and histone acetylation. Mol. Cell. 2020; 77:475–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ferrari R., Grandi N., Tramontano E., Dieci G.. Retrotransposons as drivers of mammalian brain evolution. Life (Basel). 2021; 11:376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li C., Luscombe N.M.. Nucleosome positioning stability is a modulator of germline mutation rate variation across the human genome. Nat. Commun. 2020; 11:1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tanaka Y., Yamashita R., Suzuki Y., Nakai K.. Effects of Alu elements on global nucleosome positioning in the human genome. Bmc Genomics [Electronic Resource]. 2010; 11:309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Englander E.W., Wolffe A.P., Howard B.H.. Nucleosome interactions with a human Alu element. Transcriptional repression and effects of template methylation. J. Biol. Chem. 1993; 268:19565–19573. [PubMed] [Google Scholar]
- 20. Kondo Y., Issa J.P.. Enrichment for histone H3 lysine 9 methylation at Alu repeats in human cells. J. Biol. Chem. 2003; 278:27658–27662. [DOI] [PubMed] [Google Scholar]
- 21. Varshney D., Vavrova-Anderson J., Oler A.J., Cowling V.H., Cairns B.R., White R.J.. SINE transcription by RNA polymerase III is suppressed by histone methylation but not by DNA methylation. Nat. Commun. 2015; 6:6569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Panning B., Smiley J.R.. Activation of RNA polymerase III transcription of human Alu repetitive elements by adenovirus type 5: requirement for the E1b 58-kilodalton protein and the products of E4 open reading frames 3 and 6. Mol. Cell. Biol. 1993; 13:3231–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Panning B., Smiley J.R.. Activation of RNA polymerase III transcription of human Alu elements by herpes simplex virus. Virology. 1994; 202:408–417. [DOI] [PubMed] [Google Scholar]
- 24. Liu W.M., Chu W.M., Choudary P.V., Schmid C.W.. Cell stress and translational inhibitors transiently increase the abundance of mammalian SINE transcripts. Nucleic Acids Res. 1995; 23:1758–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yeganeh M., Hernandez N.. RNA polymerase III transcription as a disease factor. Genes Dev. 2020; 34:865–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li T.H., Kim C., Rubin C.M., Schmid C.W.. K562 cells implicate increased chromatin accessibility in Alu transcriptional activation. Nucleic Acids Res. 2000; 28:3031–3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kaneko H., Dridi S., Tarallo V., Gelfand B.D., Fowler B.J., Cho W.G., Kleinman M.E., Ponicsan S.L., Hauswirth W.W., Chiodo V.A.et al.. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature. 2011; 471:325–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Panning B., Smiley J.R.. Activation of expression of multiple subfamilies of human Alu elements by adenovirus type 5 and herpes simplex virus type 1. J. Mol. Biol. 1995; 248:513–524. [DOI] [PubMed] [Google Scholar]
- 29. Jang K.L., Latchman D.S.. The herpes simplex virus immediate-early protein ICP27 stimulates the transcription of cellular Alu repeated sequences by increasing the activity of transcription factor TFIIIC. Biochem. J. 1992; 284:667–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dunker W., Zhao Y., Song Y., Karijolich J.. Recognizing the SINEs of infection: regulation of retrotransposon expression and modulation of host cell processes. Viruses. 2017; 9:386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Williams W.P., Tamburic L., Astell C.R.. Increased levels of B1 and B2 SINE transcripts in mouse fibroblast cells due to minute virus of mice infection. Virology. 2004; 327:233–241. [DOI] [PubMed] [Google Scholar]
- 32. Carey M.F., Singh K., Botchan M., Cozzarelli N.R.. Induction of specific transcription by RNA polymerase III in transformed cells. Mol. Cell. Biol. 1986; 6:3068–3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Singh K., Carey M., Saragosti S., Botchan M.. Expression of enhanced levels of small RNA polymerase III transcripts encoded by the B2 repeats in simian virus 40-transformed mouse cells. Nature. 1985; 314:553–556. [DOI] [PubMed] [Google Scholar]
- 34. Gaynor R.B., Feldman L.T., Berk A.J.. Transcription of class III genes activated by viral immediate early proteins. Science. 1985; 230:447–450. [DOI] [PubMed] [Google Scholar]
- 35. Hoeffler W.K., Roeder R.G.. Enhancement of RNA polymerase III transcription by the E1A gene product of adenovirus. Cell. 1985; 41:955–963. [DOI] [PubMed] [Google Scholar]
- 36. Hoeffler W.K., Kovelman R., Roeder R.G.. Activation of transcription factor IIIC by the adenovirus E1A protein. Cell. 1988; 53:907–920. [DOI] [PubMed] [Google Scholar]
- 37. Yoshinaga S., Dean N., Han M., Berk A.J.. Adenovirus stimulation of transcription by RNA polymerase III: evidence for an E1A-dependent increase in transcription factor IIIC concentration. EMBO J. 1986; 5:343–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kovelman R., Roeder R.G.. Sarkosyl defines three intermediate steps in transcription initiation by RNA polymerase III: application to stimulation of transcription by E1A. Genes Dev. 1990; 4:646–658. [DOI] [PubMed] [Google Scholar]
- 39. Horwitz G.A., Zhang K., McBrian M.A., Grunstein M., Kurdistani S.K., Berk A.J.. Adenovirus small e1a alters global patterns of histone modification. Science. 2008; 321:1084–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ferrari R., Pellegrini M., Horwitz G.A., Xie W., Berk A.J., Kurdistani S.K.. Epigenetic reprogramming by adenovirus e1a. Science. 2008; 321:1086–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ferrari R., Gou D., Jawdekar G., Johnson S.A., Nava M., Su T., Yousef A.F., Zemke N.R., Pellegrini M., Kurdistani S.K.et al.. Adenovirus small E1A employs the lysine acetylases p300/CBP and tumor suppressor Rb to repress select host genes and promote productive virus infection. Cell Host Microbe. 2014; 16:663–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fuchs M., Gerber J., Drapkin R., Sif S., Ikura T., Ogryzko V., Lane W.S., Nakatani Y., Livingston D.M.. The p400 complex is an essential E1A transformation target. Cell. 2001; 106:297–307. [DOI] [PubMed] [Google Scholar]
- 43. Sapountzi V., Logan I.R., Robson C.N.. Cellular functions of TIP60. Int. J. Biochem. Cell Biol. 2006; 38:1496–1509. [DOI] [PubMed] [Google Scholar]
- 44. Calo E., Wysocka J.. Modification of enhancer chromatin: what, how, and why?. Mol. Cell. 2013; 49:825–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Montell C., Courtois G., Eng C., Berk A.. Complete transformation by adenovirus 2 requires both E1A proteins. Cell. 1984; 36:951–961. [DOI] [PubMed] [Google Scholar]
- 46. Jones N., Shenk T.. An adenovirus type 5 early gene function regulates expression of other early viral genes. Proc. Nat. Acad. Sci. USA. 1979; 76:3665–3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hardy S., Kitamura M., Harris-Stansil T., Dai Y., Phipps M.L.. Construction of adenovirus vectors through cre-lox recombination. J. Virol. 1997; 71:1842–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hsu E., Zemke N.R., Berk A.J.. Promoter-specific changes in initiation, elongation, and homeostasis of histone H3 acetylation during CBP/p300 inhibition. eLife. 2021; 10:e63512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Liao Y., Smyth G.K., Shi W.. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014; 30:923–930. [DOI] [PubMed] [Google Scholar]
- 51. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang Z., Roeder R.G.. Three human RNA polymerase III-specific subunits form a subcomplex with a selective function in specific transcription initiation. Genes Dev. 1997; 11:1315–1326. [DOI] [PubMed] [Google Scholar]
- 54. Weser S., Gruber C., Hafner H.M., Teichmann M., Roeder R.G., Seifart K.H., Meissner W.. Transcription factor (TF)-like nuclear regulator, the 250-kDa form of homo sapiens TFIIIB", is an essential component of human TFIIIC1 activity. J. Biol. Chem. 2004; 279:27022–27029. [DOI] [PubMed] [Google Scholar]
- 55. Ramirez F., Dundar F., Diehl S., Gruning B.A., Manke T.. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014; 42:W187–W191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Harlow E., Franza B.R., Schley C. Monoclonal antibodies specific for adenovirus early region 1A proteins: extensive heterogeneity in early region 1A products. J. Virol. 1985; 55:533–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bailey T.L., Johnson J., Grant C.E., Noble W.S.. The MEME Suite. Nucleic Acids Res. 2015; 43:W39–W49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fornes O., Castro-Mondragon J.A., Khan A., van der Lee R., Zhang X., Richmond P.A., Modi B.P., Correard S., Gheorghe M., Baranasic D.et al.. JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2020; 48:D87–D92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Greber U.F., Flatt J.W.. Adenovirus entry: from infection to immunity. Annu. Rev. Virol. 2019; 6:177–197. [DOI] [PubMed] [Google Scholar]
- 60. Konkel M.K., Walker J.A., Hotard A.B., Ranck M.C., Fontenot C.C., Storer J., Stewart C., Marth G.T., Genomes C., Batzer M.A.. Sequence analysis and characterization of active Human alu subfamilies based on the 1000 genomes pilot project. Genome Biol. Evol. 2015; 7:2608–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bennett E.A., Keller H., Mills R.E., Schmidt S., Moran J.V., Weichenrieder O., Devine S.E.. Active Alu retrotransposons in the human genome. Genome Res. 2008; 18:1875–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Oler A.J., Alla R.K., Roberts D.N., Wong A., Hollenhorst P.C., Chandler K.J., Cassiday P.A., Nelson C.A., Hagedorn C.H., Graves B.J.et al.. Human RNA polymerase III transcriptomes and relationships to pol II promoter chromatin and enhancer-binding factors. Nat. Struct. Mol. Biol. 2010; 17:620–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Moqtaderi Z., Wang J., Raha D., White R.J., Snyder M., Weng Z., Struhl K.. Genomic binding profiles of functionally distinct RNA polymerase III transcription complexes in human cells. Nat. Struct. Mol. Biol. 2010; 17:635–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. McLean C.Y., Bristor D., Hiller M., Clarke S.L., Schaar B.T., Lowe C.B., Wenger A.M., Bejerano G.. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010; 28:495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ferrari R., Berk A.J., Kurdistani S.K.. Viral manipulation of the host epigenome for oncogenic transformation. Nat. Rev. Genet. 2009; 10:290–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhao Y., Wang J., Liang F., Liu Y., Wang Q., Zhang H., Jiang M., Zhang Z., Zhao W., Bao Y.et al.. NucMap: a database of genome-wide nucleosome positioning map across species. Nucleic Acids Res. 2019; 47:D163–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wang J., Zibetti C., Shang P., Sripathi S.R., Zhang P., Cano M., Hoang T., Xia S., Ji H., Merbs S.L.et al.. ATAC-Seq analysis reveals a widespread decrease of chromatin accessibility in age-related macular degeneration. Nat. Commun. 2018; 9:1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. McLeay R.C., Bailey T.L.. Motif Enrichment Analysis: a unified framework and an evaluation on ChIP data. BMC Bioinf. 2010; 11:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Shaulian E., Karin M.. AP-1 as a regulator of cell life and death. Nat. Cell Biol. 2002; 4:E131–E136. [DOI] [PubMed] [Google Scholar]
- 70. Zheng R., Wan C., Mei S., Qin Q., Wu Q., Sun H., Chen C.H., Brown M., Zhang X., Meyer C.A.et al.. Cistrome Data Browser: expanded datasets and new tools for gene regulatory analysis. Nucleic Acids Res. 2019; 47:D729–D735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kaaij L.J.T., Mohn F., van der Weide R.H., de Wit E., Buhler M.. The ChAHP complex counteracts chromatin looping at CTCF sites that emerged from SINE expansions in mouse. Cell. 2019; 178:1437–1451. [DOI] [PubMed] [Google Scholar]
- 72. Pocaterra A., Romani P., Dupont S.. YAP/TAZ functions and their regulation at a glance. J. Cell Sci. 2020; 133:jcs230425. [DOI] [PubMed] [Google Scholar]
- 73. Zemke N.R., Gou D., Berk A.J.. Dedifferentiation by adenovirus E1A due to inactivation of Hippo pathway effectors YAP and TAZ. Genes Dev. 2019; 33:828–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zanconato F., Battilana G., Forcato M., Filippi L., Azzolin L., Manfrin A., Quaranta E., Di Biagio D., Sigismondo G., Guzzardo V.et al.. Transcriptional addiction in cancer cells is mediated by YAP/TAZ through BRD4. Nat. Med. 2018; 24:1599–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Stein C., Bardet A.F., Roma G., Bergling S., Clay I., Ruchti A., Agarinis C., Schmelzle T., Bouwmeester T., Schubeler D.et al.. YAP1 Exerts its transcriptional control via TEAD-mediated activation of enhancers. PLoS Genet. 2015; 11:e1005465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Pope B.D., Ryba T., Dileep V., Yue F., Wu W., Denas O., Vera D.L., Wang Y., Hansen R.S., Canfield T.K.et al.. Topologically associating domains are stable units of replication-timing regulation. Nature. 2014; 515:402–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Lister R., Pelizzola M., Dowen R.H., Hawkins R.D., Hon G., Tonti-Filippini J., Nery J.R., Lee L., Ye Z., Ngo Q.M.et al.. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009; 462:315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. The ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Tasdemir N., Banito A., Roe J.S., Alonso-Curbelo D., Camiolo M., Tschaharganeh D.F., Huang C.H., Aksoy O., Bolden J.E., Chen C.C.et al.. BRD4 Connects enhancer remodeling to senescence immune surveillance. Cancer Discov. 2016; 6:612–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Pelka P., Ablack J.N., Fonseca G.J., Yousef A.F., Mymryk J.S.. Intrinsic structural disorder in adenovirus E1A: a viral molecular hub linking multiple diverse processes. J. Virol. 2008; 82:7252–7263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Avvakumov N., Kajon A.E., Hoeben R.C., Mymryk J.S.. Comprehensive sequence analysis of the E1A proteins of human and simian adenoviruses. Virology. 2004; 329:477–492. [DOI] [PubMed] [Google Scholar]
- 82. Mertens C., Roeder R.G.. Different functional modes of p300 in activation of RNA polymerase III transcription from chromatin templates. Mol. Cell. Biol. 2008; 28:5764–5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Orioli A., Pascali C., Pagano A., Teichmann M., Dieci G.. RNA polymerase III transcription control elements: themes and variations. Gene. 2012; 493:185–194. [DOI] [PubMed] [Google Scholar]
- 84. Davis C.A., Hitz B.C., Sloan C.A., Chan E.T., Davidson J.M., Gabdank I., Hilton J.A., Jain K., Baymuradov U.K., Narayanan A.K.et al.. The Encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res. 2018; 46:D794–D801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Gevry N., Hardy S., Jacques P.E., Laflamme L., Svotelis A., Robert F., Gaudreau L.. Histone H2A.Z is essential for estrogen receptor signaling. Genes Dev. 2009; 23:1522–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Tucker J.M., Glaunsinger B.A.. Host noncoding retrotransposons induced by DNA viruses: a SINE of infection?. J. Virol. 2017; 91:e00982–e00917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. King C.R., Zhang A., Tessier T.M., Gameiro S.F., Mymryk J.S.. Hacking the cell: network intrusion and exploitation by Adenovirus E1A. mBio. 2018; 9:e00390-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Ferrari R., Su T., Li B., Bonora G., Oberai A., Chan Y., Sasidharan R., Berk A.J., Pellegrini M., Kurdistani S.K.. Reorganization of the host epigenome by a viral oncogene. Genome Res. 2012; 22:1212–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Liu X., Marmorstein R.. Structure of the retinoblastoma protein bound to adenovirus E1A reveals the molecular basis for viral oncoprotein inactivation of a tumor suppressor. Genes Dev. 2007; 21:2711–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Lynch K.L., Gooding L.R., Garnett-Benson C., Ornelles D.A., Avgousti D.C.. Epigenetics and the dynamics of chromatin during adenovirus infections. FEBS Lett. 2019; 593:3551–3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Frisch S.M., Mymryk J.S.. Adenovirus-5 E1A: paradox and paradigm. Nat. Rev. Mol. Cell Biol. 2002; 3:441–452. [DOI] [PubMed] [Google Scholar]
- 92. Tworkowski K.A., Chakraborty A.A., Samuelson A.V., Seger Y.R., Narita M., Hannon G.J., Lowe S.W., Tansey W.P.. Adenovirus E1A targets p400 to induce the cellular oncoprotein Myc. Proc. Nat. Acad. Sci. U.S.A. 2008; 105:6103–6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Buchel G., Carstensen A., Mak K.Y., Roeschert I., Leen E., Sumara O., Hofstetter J., Herold S., Kalb J., Baluapuri A.et al.. Association with Aurora-A controls N-MYC-dependent promoter escape and pause release of RNA polymerase II during the cell cycle. Cell Rep. 2017; 21:3483–3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Kenneth N.S., Ramsbottom B.A., Gomez-Roman N., Marshall L., Cole P.A., White R.J.. TRRAP and GCN5 are used by c-Myc to activate RNA polymerase III transcription. Proc. Nat. Acad. Sci. U.S.A. 2007; 104:14917–14922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Walz S., Lorenzin F., Morton J., Wiese K.E., von Eyss B., Herold S., Rycak L., Dumay-Odelot H., Karim S., Bartkuhn M.et al.. Activation and repression by oncogenic MYC shape tumour-specific gene expression profiles. Nature. 2014; 511:483–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Deleu L., Shellard S., Alevizopoulos K., Amati B., Land H.. Recruitment of TRRAP required for oncogenic transformation by E1A. Oncogene. 2001; 20:8270–8275. [DOI] [PubMed] [Google Scholar]
- 97. Pradhan S.K., Su T., Yen L., Jacquet K., Huang C., Cote J., Kurdistani S.K., Carey M.F.. EP400 Deposits H3.3 into promoters and enhancers during gene activation. Mol. Cell. 2016; 61:27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Policarpi C., Crepaldi L., Brookes E., Nitarska J., French S.M., Coatti A., Riccio A.. Enhancer SINEs link pol III to pol II transcription in neurons. Cell Rep. 2017; 21:2879–2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Sasaki T., Nishihara H., Hirakawa M., Fujimura K., Tanaka M., Kokubo N., Kimura-Yoshida C., Matsuo I., Sumiyama K., Saitou N.et al.. Possible involvement of SINEs in mammalian-specific brain formation. Proc. Nat. Acad. Sci. U.S.A. 2008; 105:4220–4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Creyghton M.P., Cheng A.W., Welstead G.G., Kooistra T., Carey B.W., Steine E.J., Hanna J., Lodato M.A., Frampton G.M., Sharp P.A.et al.. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Nat. Acad. Sci. U.S.A. 2010; 107:21931–21936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Mink S., Haenig B., Klempnauer K.H.. Interaction and functional collaboration of p300 and C/EBPbeta. Mol. Cell. Biol. 1997; 17:6609–6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Bannister A.J., Oehler T., Wilhelm D., Angel P., Kouzarides T.. Stimulation of c-jun activity by CBP: c-jun residues Ser63/73 are required for CBP induced stimulation in vivo and CBP binding in vitro. Oncogene. 1995; 11:2509–2514. [PubMed] [Google Scholar]
- 103. Bannister A.J., Brown H.J., Sutherland J.A., Kouzarides T.. Phosphorylation of the c-fos and c-jun HOB1 motif stimulates its activation capacity. Nucleic Acids Res. 1994; 22:5173–5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Zanconato F., Forcato M., Battilana G., Azzolin L., Quaranta E., Bodega B., Rosato A., Bicciato S., Cordenonsi M., Piccolo S.. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat. Cell Biol. 2015; 17:1218–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Ramji D.P., Foka P.. CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem. J. 2002; 365:561–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Di Pascale F., Nama S., Muhuri M., Quah S., Ismail H.M., Chan X.H.D., Sundaram G.M., Ramalingam R., Burke B., Sampath P.. C/EBPbeta mediates RNA polymerase III-driven transcription of oncomiR-138 in malignant gliomas. Nucleic Acids Res. 2018; 46:336–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Hou T.Y., Kraus W.L.. Spirits in the material world: enhancer RNAs in transcriptional regulation. Trends Biochem. Sci. 2020; 46:138–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Liang L., Cao C., Ji L., Cai Z., Wang D., Ye R., Chen J., Yu X., Zhou J., Bai Z.et al.. Complementary Alu sequences mediate enhancer-promoter selectivity. Nature. 2023; 619:868–875. [DOI] [PubMed] [Google Scholar]
- 109. Cantarella S., Carnevali D., Morselli M., Conti A., Pellegrini M., Montanini B., Dieci G.. Alu RNA modulates the expression of cell cycle genes in Human fibroblasts. Int. J. Mol. Sci. 2019; 20:3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Di Ruocco F., Basso V., Rivoire M., Mehlen P., Ambati J., De Falco S., Tarallo V.. Alu RNA accumulation induces epithelial-to-mesenchymal transition by modulating miR-566 and is associated with cancer progression. Oncogene. 2018; 37:627–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Larminie C.G., Cairns C.A., Mital R., Martin K., Kouzarides T., Jackson S.P., White R.J.. Mechanistic analysis of RNA polymerase III regulation by the retinoblastoma protein. EMBO J. 1997; 16:2061–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. White R.J. RNA polymerase III transcription and cancer. Oncogene. 2004; 23:3208–3216. [DOI] [PubMed] [Google Scholar]
- 113. Orioli A. tRNA biology in the omics era: stress signalling dynamics and cancer progression. Bioessays. 2017; 39:1600158. [DOI] [PubMed] [Google Scholar]
- 114. Gjidoda A., Henry R.W.. RNA polymerase III repression by the retinoblastoma tumor suppressor protein. Biochim. Biophys. Acta. 2013; 1829:385–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Hirsch H.A., Jawdekar G.W., Lee K.A., Gu L., Henry R.W.. Distinct mechanisms for repression of RNA polymerase III transcription by the retinoblastoma tumor suppressor protein. Mol. Cell. Biol. 2004; 24:5989–5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Roman A.C., Gonzalez-Rico F.J., Molto E., Hernando H., Neto A., Vicente-Garcia C., Ballestar E., Gomez-Skarmeta J.L., Vavrova-Anderson J., White R.J.et al.. Dioxin receptor and SLUG transcription factors regulate the insulator activity of B1 SINE retrotransposons via an RNA polymerase switch. Genome Res. 2011; 21:422–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Petrie J.L., Swan C., Ingram R.M., Frame F.M., Collins A.T., Dumay-Odelot H., Teichmann M., Maitland N.J., White R.J.. Effects on prostate cancer cells of targeting RNA polymerase III. Nucleic Acids Res. 2019; 47:3937–3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Xie W., Ling T., Zhou Y., Feng W., Zhu Q., Stunnenberg H.G., Grummt I., Tao W.. The chromatin remodeling complex NuRD establishes the poised state of rRNA genes characterized by bivalent histone modifications and altered nucleosome positions. Proc. Nat. Acad. Sci. U.S.A. 2012; 109:8161–8166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Freese N.H., Norris D.C., Loraine A.E.. Integrated genome browser: visual analytics platform for genomics. Bioinformatics. 2016; 32:2089–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Shin H., Liu T., Manrai A.K., Liu X.S.. CEAS: cis-regulatory element annotation system. Bioinformatics. 2009; 25:2605–2606. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data of RNA-seq and ChIP-seq generated in this study are available at GEO under accession number GSE208717. Publicly available data were retrieved from the following sources: GSE32340 for H3K4me1 ChIP-seq (dl1500-infected IMR90 cells) and H3K9ac ChIP-seq (40); GSE43070 for H3K4me1 ChIP-seq (uninfected IMR90 cells) (40); GSE59693 for ChIP-seq of H3K27ac, EP300, Pol II and RB (mock- and dl1500-infected IMR90 cells) (41); GSM2828526 for EP400 ChIP-seq (K562 cells); GSM935372 for Pol III (RPC155 subunit) ChIP-seq (K562 cells); GSM935343 for TFIIIC (GTF3C2 subunit) ChIP-seq (K562 cells); GSM733786 for H2A.Z ChIP-seq (K562 cells); GSM3289968 for ATAC-seq (IMR90 cells); GSM935519 for CEBPB ChIP-seq (IMR90 cells); GSM818008 for H2A.Z ChIP-seq (IMR90 cells); GSM2825448 for FOS ChIP-seq (IMR90 cells); GSM1696207 for YAP1 ChIP-seq (IMR90 cells); GSM1915116 for BRD4 ChIP-seq (IMR90 cells).