Abstract
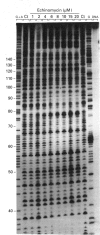
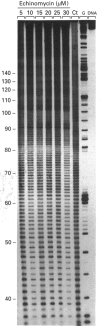
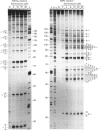
Four complementary footprinting and probing techniques utilizing DNAse I, methidiumpropyl EDTA (MPE).FeII, diethyl pyrocarbonate (DEPC) and KMnO4 as DNA-cleaving or DNA-modifying agents have been applied to investigate the sequence-specific binding to DNA of the antitumour antibiotic echinomycin. A 265 bp EcoRI-PvuII DNA restriction fragment excised from plasmid pBS was used as a substrate. Six regions of protection against DNAase I cleavage were located on the 265-mer: three sites encompass the sequences 5'-TCGA or 5'-GCGT and the three others contain 5'-GpG (CpC) dinucleotide sequences where the inhibition of DNAase I cutting by echinomycin is less pronounced. In contrast, MPE.FeII cleavage allows identification of only three echinomycin-binding sites on the 265-mer: two sites contain the sequence 5'-TCGA and one encompasses the sequence 5'-ACCA. Cleavage of DNA by MPE.FeII in the presence of echinomycin remains practically unaffected at the sequence 5'-GCGT, despite its identification by DNAase I as a strong site for binding the antibiotic, as well as at the two other sequences containing GpG steps. With both DNAase I and MPE.FeII, enhanced DNA cleavage is evident at AT-rich sequences in the presence of echinomycin. Enhanced reactivity towards KMnO4 and DEPC provides clear evidence for sequence-dependent conformational changes in DNA induced by the antibiotic. The experiments reveal that KMnO4 reacts most strongly with thymines located around, but not necessarily adjacent to, an echinomycin-binding site, whereas the carbethoxylation reactions caused by DEPC occur primarily at the adenine residues lying immediately 5' or 3' to the dinucleotide that denotes an echinomycin-binding site. The results reported here demonstrate that DEPC and KMnO4 serve as sensitive probes for different states of the DNA helix. It seems that the reaction with KMnO4 involves transient unstacking events, whereas the carbethoxylation reaction of DEPC requires larger-scale helix opening.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bailly C., OhUigin C., Rivalle C., Bisagni E., Hénichart J. P., Waring M. J. Sequence-selective binding of an ellipticine derivative to DNA. Nucleic Acids Res. 1990 Nov 11;18(21):6283–6291. doi: 10.1093/nar/18.21.6283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly C., Waring M. J. Preferential intercalation at AT sequences in DNA by lucanthone, hycanthone, and indazole analogs. A footprinting study. Biochemistry. 1993 Jun 15;32(23):5985–5993. doi: 10.1021/bi00074a009. [DOI] [PubMed] [Google Scholar]
- Dabrowiak J. C., Kissinger K., Goodisman J. Quantitative footprinting analysis of drug-DNA interactions: Fe(III) methidium-propyl-EDTA as a probe. Electrophoresis. 1989 May-Jun;10(5-6):404–412. doi: 10.1002/elps.1150100519. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Travers A. A. DNA structural variations in the E. coli tyrT promoter. Cell. 1984 Jun;37(2):491–502. doi: 10.1016/0092-8674(84)90379-9. [DOI] [PubMed] [Google Scholar]
- Fox K. R., Kentebe E. Echinomycin binding to the sequence CG(AT)nCG alters the structure of the central AT region. Nucleic Acids Res. 1990 Apr 25;18(8):1957–1963. doi: 10.1093/nar/18.8.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox K. R., Marks J. N., Waterloh K. Echinomycin binding to alternating AT. Nucleic Acids Res. 1991 Dec 25;19(24):6725–6730. doi: 10.1093/nar/19.24.6725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox K. R., Waring M. J. The use of micrococcal nuclease as a probe for drug-binding sites on DNA. Biochim Biophys Acta. 1987 Jul 14;909(2):145–155. doi: 10.1016/0167-4781(87)90036-4. [DOI] [PubMed] [Google Scholar]
- Furlong J. C., Sullivan K. M., Murchie A. I., Gough G. W., Lilley D. M. Localized chemical hyperreactivity in supercoiled DNA: evidence for base unpairing in sequences that induce low-salt cruciform extrusion. Biochemistry. 1989 Mar 7;28(5):2009–2017. doi: 10.1021/bi00431a008. [DOI] [PubMed] [Google Scholar]
- Gao X. L., Patel D. J. Antitumour drug-DNA interactions: NMR studies of echinomycin and chromomycin complexes. Q Rev Biophys. 1989 May;22(2):93–138. doi: 10.1017/s0033583500003814. [DOI] [PubMed] [Google Scholar]
- Gao X. L., Patel D. J. NMR studies of echinomycin bisintercalation complexes with d(A1-C2-G3-T4) and d(T1-C2-G3-A4) duplexes in aqueous solution: sequence-dependent formation of Hoogsteen A1.T4 and Watson--Crick T1.A4 base pairs flanking the bisintercalation site. Biochemistry. 1988 Mar 8;27(5):1744–1751. doi: 10.1021/bi00405a054. [DOI] [PubMed] [Google Scholar]
- Gilbert D. E., Feigon J. Proton NMR study of the [d(ACGTATACGT)]2-2echinomycin complex: conformational changes between echinomycin binding sites. Nucleic Acids Res. 1992 May 25;20(10):2411–2420. doi: 10.1093/nar/20.10.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert D. E., Feigon J. The DNA sequence at echinomycin binding sites determines the structural changes induced by drug binding: NMR studies of echinomycin binding to [d(ACGTACGT)]2 and [d(TCGATCGA)]2. Biochemistry. 1991 Mar 5;30(9):2483–2494. doi: 10.1021/bi00223a027. [DOI] [PubMed] [Google Scholar]
- Gilbert D. E., van der Marel G. A., van Boom J. H., Feigon J. Unstable Hoogsteen base pairs adjacent to echinomycin binding sites within a DNA duplex. Proc Natl Acad Sci U S A. 1989 May;86(9):3006–3010. doi: 10.1073/pnas.86.9.3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodisman J., Dabrowiak J. C. Structural changes and enhancements in DNase I footprinting experiments. Biochemistry. 1992 Feb 4;31(4):1058–1064. doi: 10.1021/bi00119a014. [DOI] [PubMed] [Google Scholar]
- Herr W. Diethyl pyrocarbonate: a chemical probe for secondary structure in negatively supercoiled DNA. Proc Natl Acad Sci U S A. 1985 Dec;82(23):8009–8013. doi: 10.1073/pnas.82.23.8009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppesen C., Nielsen P. E. Detection of intercalation-induced changes in DNA structure by reaction with diethyl pyrocarbonate or potassium permanganate. Evidence against the induction of Hoogsteen base pairing by echinomycin. FEBS Lett. 1988 Apr 11;231(1):172–176. doi: 10.1016/0014-5793(88)80725-7. [DOI] [PubMed] [Google Scholar]
- Jeppesen C., Nielsen P. E. Photofootprinting of drug-binding sites on DNA using diazo- and azido-9-aminoacridine derivatives. Eur J Biochem. 1989 Jun 15;182(2):437–444. doi: 10.1111/j.1432-1033.1989.tb14850.x. [DOI] [PubMed] [Google Scholar]
- Johnston B. H., Rich A. Chemical probes of DNA conformation: detection of Z-DNA at nucleotide resolution. Cell. 1985 Oct;42(3):713–724. doi: 10.1016/0092-8674(85)90268-5. [DOI] [PubMed] [Google Scholar]
- Leroy J. L., Gao X. L., Misra V., Guéron M., Patel D. J. Proton exchange in DNA-luzopeptin and DNA-echinomycin bisintercalation complexes: rates and processes of base-pair opening. Biochemistry. 1992 Feb 11;31(5):1407–1415. doi: 10.1021/bi00120a017. [DOI] [PubMed] [Google Scholar]
- Low C. M., Drew H. R., Waring M. J. Sequence-specific binding of echinomycin to DNA: evidence for conformational changes affecting flanking sequences. Nucleic Acids Res. 1984 Jun 25;12(12):4865–4879. doi: 10.1093/nar/12.12.4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand C., Bailly C., McLean M. J., Moroney S. E., Waring M. J. The 2-amino group of guanine is absolutely required for specific binding of the anti-cancer antibiotic echinomycin to DNA. Nucleic Acids Res. 1992 Nov 11;20(21):5601–5606. doi: 10.1093/nar/20.21.5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McCarthy J. G., Williams L. D., Rich A. Chemical reactivity of potassium permanganate and diethyl pyrocarbonate with B DNA: specific reactivity with short A-tracts. Biochemistry. 1990 Jun 26;29(25):6071–6081. doi: 10.1021/bi00477a027. [DOI] [PubMed] [Google Scholar]
- McLean M. J., Seela F., Waring M. J. Echinomycin-induced hypersensitivity to osmium tetroxide of DNA fragments incapable of forming Hoogsteen base pairs. Proc Natl Acad Sci U S A. 1989 Dec;86(24):9687–9691. doi: 10.1073/pnas.86.24.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean M. J., Waring M. J. Chemical probes reveal no evidence of Hoogsteen base pairing in complexes formed between echinomycin and DNA in solution. J Mol Recognit. 1988 Jun;1(3):138–151. doi: 10.1002/jmr.300010307. [DOI] [PubMed] [Google Scholar]
- Mendel D., Dervan P. B. Hoogsteen base pairs proximal and distal to echinomycin binding sites on DNA. Proc Natl Acad Sci U S A. 1987 Feb;84(4):910–914. doi: 10.1073/pnas.84.4.910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidle S., Abraham Z. Structural and sequence-dependent aspects of drug intercalation into nucleic acids. CRC Crit Rev Biochem. 1984;17(1):73–121. doi: 10.3109/10409238409110270. [DOI] [PubMed] [Google Scholar]
- Nielsen P. E. Chemical and photochemical probing of DNA complexes. J Mol Recognit. 1990 Feb;3(1):1–25. doi: 10.1002/jmr.300030102. [DOI] [PubMed] [Google Scholar]
- Palecek E. Local supercoil-stabilized DNA structures. Crit Rev Biochem Mol Biol. 1991;26(2):151–226. doi: 10.3109/10409239109081126. [DOI] [PubMed] [Google Scholar]
- Portugal J., Fox K. R., McLean M. J., Richenberg J. L., Waring M. J. Diethyl pyrocarbonate can detect a modified DNA structure induced by the binding of quinoxaline antibiotics. Nucleic Acids Res. 1988 May 11;16(9):3655–3670. doi: 10.1093/nar/16.9.3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers E. W., Waring M. J. Footprinting titration studies on the binding of echinomycin to DNA incapable of forming Hoogsteen base pairs. Biochemistry. 1993 Sep 7;32(35):9094–9107. doi: 10.1021/bi00086a014. [DOI] [PubMed] [Google Scholar]
- Smith J. M., Thomas D. J. Quantitative analysis of one-dimensional gel electrophoresis profiles. Comput Appl Biosci. 1990 Apr;6(2):93–99. doi: 10.1093/bioinformatics/6.2.93. [DOI] [PubMed] [Google Scholar]
- Ughetto G., Wang A. H., Quigley G. J., van der Marel G. A., van Boom J. H., Rich A. A comparison of the structure of echinomycin and triostin A complexed to a DNA fragment. Nucleic Acids Res. 1985 Apr 11;13(7):2305–2323. doi: 10.1093/nar/13.7.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyke M. M., Dervan P. B. Echinomycin binding sites on DNA. Science. 1984 Sep 14;225(4667):1122–1127. doi: 10.1126/science.6089341. [DOI] [PubMed] [Google Scholar]
- Van Dyke M. W., Dervan P. B. Methidiumpropyl-EDTA.Fe(II) and DNase I footprinting report different small molecule binding site sizes on DNA. Nucleic Acids Res. 1983 Aug 25;11(16):5555–5567. doi: 10.1093/nar/11.16.5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waring M. J., Wakelin L. P. Echinomycin: a bifunctional intercalating antibiotic. Nature. 1974 Dec 20;252(5485):653–657. doi: 10.1038/252653a0. [DOI] [PubMed] [Google Scholar]
- Waterloh K., Fox K. R. Interaction of echinomycin with An.Tn. and (AT)n regions flanking its CG binding site. Nucleic Acids Res. 1991 Dec 25;19(24):6719–6724. doi: 10.1093/nar/19.24.6719. [DOI] [PMC free article] [PubMed] [Google Scholar]