Abstract
Genomics allows the tracing of origin and evolution of cancer at molecular scale and underpin modern cancer diagnosis and treatment systems. Yet, molecular biomarker‐guided clinical decision‐making encounters major challenges in the realm of individualized medicine, consisting of the invasiveness of procedures and the sampling errors due to high tumor heterogeneity. By contrast, medical imaging enables noninvasive and global characterization of tumors at a low cost. In recent years, radiomics has overcomes the limitations of human visual evaluation by high‐throughput quantitative analysis, enabling the comprehensive utilization of the vast amount of information underlying radiological images. The cross‐scale integration of radiomics and genomics (hereafter radiogenomics) has the enormous potential to enhance cancer decoding and act as a catalyst for digital precision medicine. Herein, we provide a comprehensive overview of the current framework and potential clinical applications of radiogenomics in patient care. We also highlight recent research advances to illustrate how radiogenomics can address common clinical problems in solid tumors such as breast cancer, lung cancer, and glioma. Finally, we analyze existing literature to outline challenges and propose solutions, while also identifying future research pathways. We believe that the perspectives shared in this survey will provide a valuable guide for researchers in the realm of radiogenomics aiming to advance precision oncology.
Keywords: artificial intelligence, oncology, precision medicine, radiogenomics, radiomics
Radiogenomics is a new tool for precisive oncology by deeply integrating radiomics and genomics. This review deconstructs the current research status of radiogenomics through the multidimensional feature engineering of radiomics and the integration patterns of radiomics and genomics, and takes gliomas, lung cancers, breast cancers, and other common cancers as examples to show the advance of the application of radiogenomics in the diagnosis, treatment, and monitoring of tumors.

1. INTRODUCTION
In 2020, it is estimated that there were approximately 19.3 million new cancer cases and nearly 10.0 million cancer‐related deaths worldwide. 1 Cancers can arise from various cell types and organs in the human body. They are characterized by uncontrolled cell proliferation, confirmed to be caused by random somatic genomic abnormalities. Through the accumulation of heritable genetic mutations, and the interactions with the surrounding microenvironment, as well as natural selection from cancer therapies, advantageous mutations can accumulate over time, while deleterious ones are eliminated. 2 This evolutionary process enables cancer to develop phenotypes that promote survival and reproduction, leading to tumor progression, metastasis, and treatment resistance. 3 , 4
However, obtaining molecular information through invasive tissue sampling not only carries the risk of complications, posing dilemmas for pretreatment decision‐making and posttreatment patient monitoring, but is also quite time consuming. Meanwhile, the spatial heterogeneity within tumors causes a challenge for targeted therapies guided by regional sampling results. These therapies may only be effective against a subset of cancer cells, leaving other cancer subclones unaffected and potentially accelerating their growth, resulting in tumor evolution and recurrence. Besides, the high cost of advanced sequencing technology has limited its widespread use in clinical settings, making it particularly difficult for patients in medically disadvantaged areas to access easily.
Medical imaging represents a distinct and highly accessible method for acquiring tumor data compared with tissue sequencing. It allows for a macroscopic mapping of tumor cells, the microenvironment, and even the tissue surrounding the tumor at the voxel level, using noninvasive or minimally invasive multimodal imaging techniques. In recent years, computer technology has been integrated into medical imaging, enabling the high‐throughput extraction of quantitative features from medical images. Advanced machine learning (ML) and deep learning (DL) algorithms are then employed to analyze these features, facilitating a more effective assessment of large amounts of imaging data. In contrast to traditional imaging assessment methods, radiomic features offer a more objective and robust approach to capture tumor heterogeneity and reveal clinically significant higher‐order signatures that are not discernible to the human eye. 5 Radiomics have been utilized to construct radiomic models, which have
Shown superior performance in noninvasive tumor stratification and prognosis assessment. 6 Radiomics has emerged as a valuable addition to multiomics of cancer.
Radiogenomics, a new concept combines “radiomics” and “genomics,” has gained increasing attentions. 7 , 8 It involves the integration of advanced medical image analysis and multiomics data of tumors. Its goal is to uncover the relationship between radiomics and bio‐omics to pinpoint relevant biomarkers and build elaborate markers of disease and physiology and integrate multiple omics data for tumor diagnosis, classification, treatment decision, and prognosis.
In this review, we delineated the principal components of radiomics and genomics in oncology at the methodological level, elucidating their interconnections and integration mechanisms within the framework of radiogenomics with the aim of deconstructing the comprehensive landscape of radiogenomics. We also examine recent advancements in the application of radiogenomics to prevalent cancers such as glioma, lung cancer, and breast cancer, demonstrating its significant potential to address common clinical challenges. Finally, we highlight the current methodological challenges and limitations and discuss prospective directions for future research in the field.
2. RADIOMICS IN ONCOLOGY
Radiomics is the high‐throughput mining of quantitative image features from standard‐of‐care medical imaging that enables data to be extracted and applied within clinical‐decision support systems to improve diagnostic, prognostic, and predictive accuracy. 5 , 6 In the following, we present three perspectives to illustrate the types of tumoral radiomic features obtained through various methods that can be utilized in a radiogenomics framework (Figure 1).
FIGURE 1.
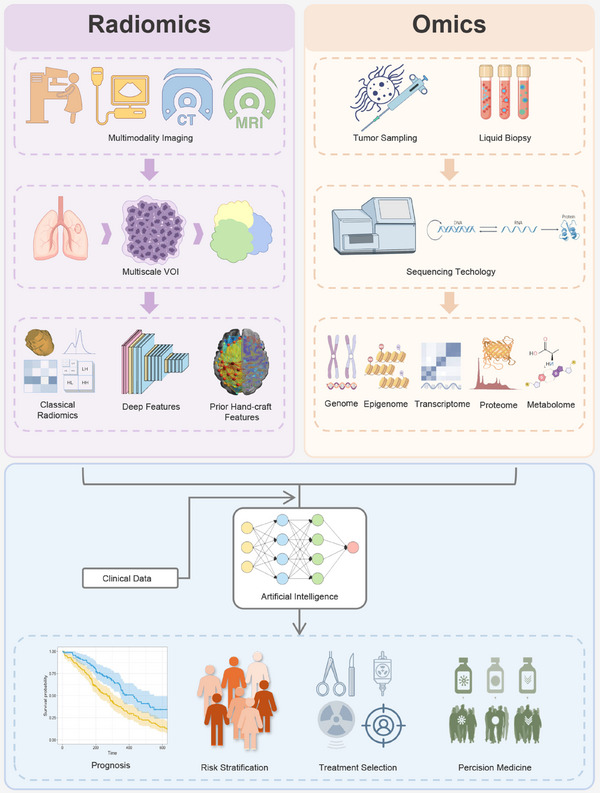
Schematic diagram illustrates the comprehensive integration of radiomics with omics data for precise cancer care. The first step involves collecting data resources, including imaging and biological samples. From these resources, various dimensions of radiomic features and molecular signatures of cancers are extracted and refined. Ultimately, radiomics and omics data are interconnected and integrated using advanced artificial intelligence algorithms to construct accurate clinical prediction models.
2.1. Features from multimodality images
Multimodality imaging technologies, such as digital radiography, computed tomography (CT), magnetic resonance imaging (MRI), nuclear medicine imaging such as positron emission tomography (PET), and others have been evolving for more than a century. These technologies employ different principles to capture various physical and chemical properties of tissues, offering a diverse range of imaging sequences. The radiomic features extracted from these multimodal images are often complementary, providing a multidimensional representation of tumors biology.
2.2. Features from multiscale of regions
Medical images acquired at different scales contain diverse biological information. Tumor‐level features, derived from both intratumor and peritumor regions, have been extensively utilized to characterize tumor heterogeneity. In addition, Subregion segmentation of cancers allows for multihabitat evaluation. However, it is important to note that cancer is not solely a localized disease; its occurrence, progression, and prognosis are often linked to the host organ or even the overall body condition. Radiogenomic studies have recently started incorporating the entire host organ, demonstrating predictive capabilities beyond tumor‐level profiling. 9 , 10 Anatomical multiscale radiomics enables a comprehensive assessment of cancer as a complex disease.
2.3. Approaches for feature extraction
2.3.1. Classical radiomics
Classical radiomic features are widely utilized hand‐crafted features that are extracted from preprocessed images using predefined programs and specifications (such as pyradiomics) to describe radiographic aspects of shape, intensity, and texture. 11 These features are derived from specific algorithms, which enhances their interpretability to some extent. Radiomic features are able to capture intratumor heterogeneity more effectively than the human eye, which is believed to explain the superiority of radiomics over traditional image analysis methods.
2.3.2. Deep learning
DL techniques were used to automatically learn feature representations from medical images, eliminating the need for manual feature detection. DL methods offer several advantages such as reducing the need for preprocessing steps, enabling collaborative analysis of large volumes of high‐dimensional data, and providing superior problem‐solving capabilities. DL also allows for multitasking, including tumor segmentation, classification, and prognosis. 12 , 13 , 14 Meanwhile, DL algorithms continue to evolve quickly and drive DL‐based radiomics forward.
2.3.3. Priori hand‐crafted radiomics
In recent studies, new hand‐crafted radiomic features have been extracted batchwise as biomarkers for cancers, such as brain structure connectomics, tumor location, and the tumor field effect. 15 , 16 , 17 , 18 These features differ from classical ones in that they incorporate clinical prior knowledge, enabling them to capture specific pathophysiological information. This makes them advantageous for specific clinical tasks, particularly when dealing with limited amounts of data during model training.
3. GENOMICS IN ONCOLOGY
Generally, genomics is the study of all genes and DNA sequences of an organism. However, in the context of radiogenomics, the term “genomics” is often broadened to include the analysis of RNA, proteins, and other critical biomarker data that can reflect the origin and progression of tumor cells at the molecular level. 7 Genomics should not be considered in isolation but rather in conjunction with transcriptomics, proteomics, metabolomics, and other “omics” disciplines. 19 , 20 Consequently, this review aligns with the prevailing academic perspective by integrating multiomics approaches within the framework of cancer radiogenomics. This integration is essential for radiogenomics to deliver a comprehensive biological and imaging‐based understanding of tumors.
3.1. Genomics from tumor tissue sampling
Obtaining samples directly from tumor tissues, including surgical resection, biopsy, and fine‐needle aspiration, is the most commonly used method of pathology sampling for genomics. 21 In recent years, molecular pathology has played a pivotal role in facilitating precise diagnosis and informed treatment decisions for cancer, leveraging techniques such as immunohistochemical staining, in situ hybridization, and gene sequencing. 22 , 23 Furthermore, the integrated analysis of multiomics data based on high‐throughput sequencing platforms, gene chips, and mass spectrometry, enables a more comprehensive elucidation of cancer mechanisms and aids in the discovery of novel biomarkers for early cancer detection, prognosis assessment, and the identification of therapeutic targets. 19 , 24 Notably, the recently proposed spatial genomics technology holds the potential to unravel the intricacies of tumor heterogeneity, promising to localize and define tumor boundaries, subclones, and microenvironments at the molecular level. 25
3.2. Genomics from liquid biopsy
Liquid biopsy has recently emerged as a promising sampling method for obtaining multiomics information from tumors. It primarily relies on blood samples to capture circulating tumor DNA, circulating tumor cells (CTCs), and exosomes present in the bloodstream, and detect the tumor‐derived multiomics biological information they carry. Liquid biopsy offers several advantages, including being noninvasive, highly reproducible, enabling early diagnosis, facilitating dynamic monitoring, and overcoming tumor heterogeneity. 26 , 27 Despite being in its infancy, liquid biopsy holds promise for promoting radiogenomics by furnishing highly time‐critical and continuously observable biomolecular data (Figure 1).
4. RADIOGENOMICS
4.1. Radiogenomics for precisive oncological molecular prediction
Investigating the association between radiomics and molecular biomarkers, with the aim of substituting invasive biomarkers with noninvasive and timely imaging markers for pre‐ and post‐treatment clinical decision‐making, plays a crucial role in radiogenomics (Figure 2).
FIGURE 2.
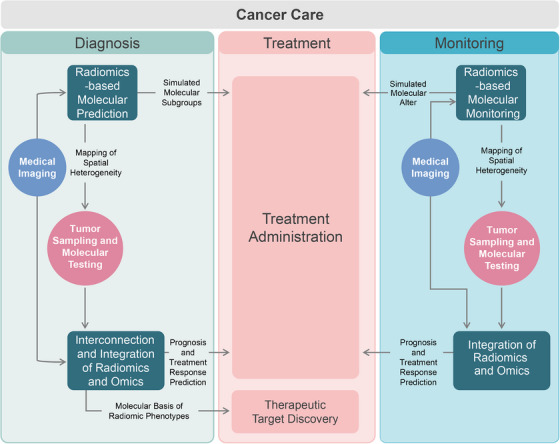
Potential enhanced clinical workflow with radiogenomics interventions. Radiogenomics offers the potential to noninvasively predict key molecular characteristics, including their temporal and spatial heterogeneity, at the initial diagnosis and posttreatment monitoring stages of cancer. This can help in discovering therapeutic targets, enhancing cancer prognosis, and predicting treatment response. Ultimately, radiogenomics can guide precision diagnosis and treatment of cancers, enhancing patient outcomes.
4.1.1. Prediction prior to treatment
Prior knowledge of tumor molecular subtypes before treatment is crucial for improved decision‐making in cancer care and is increasingly recognized as essential for neoadjuvant therapy (NAT). 28 , 29 Biopsy, the current gold standard, is inevitably invasive and prone to sampling errors due to tumor heterogeneity. Noninvasive Imaging, particularly with the emergence of radiomics, has demonstrated potential in predicting molecular subtypes throughout the entire tumor landscape, can provide a critical foundation for precise cancer treatment. In addition, by correlating with key biomolecules and pathways, biospecific radiomics features are likely to be more efficient and interpretable for determining tumor‐targeted treatment response and prognosis.
4.1.2. Spatial heterogeneity landscaping
The spatial heterogeneity of tumor molecules within tumor subregions and among metastases has been extensively documented. 30 , 31 The accuracy of biopsy results has been questioned due to the limited amount of tissue sampled. While multipoint biopsies and Sequencing may address this limitation, they also increase risks of complications. Predicting the spatial distribution of key molecules in tumors through radiogenomics has potential to improve the reliability and representativeness of tissue sampling, reduce associated risks, and even guide precision radiotherapy.
4.1.3. Molecular monitoring after treatment
Adaptive changes in the tumor genome from anticancer therapy are a key driver of treatment resistance. 32 Real‐time monitoring of these changes could underpin timely therapy adjustment and new targeted development. However, biopsy‐based molecular monitoring is often delayed and risky for repeated tests. Radiogenomic monitoring of tumor genomes noninvasively shows promise for precision medicine's timely implementation based on dynamic genomic shifts.
4.2. Radiogenomics for risk stratification and prognosis evaluation
Numbers of key biomarkers for tumor treatment and prognosis have been identified and partially implemented into clinical practice. However, even with the latest findings it is difficult to predict tumors perfectly. Radiomics’ universality, noninvasiveness, and ability to observe biological information at the tumor and even organ level make it a powerful complement to current molecular typing systems for cancer, which is expected to further facilitate precise and individualized treatment (Figure 2).
4.2.1. Prognosis stratification
Cancer outcomes, such as remission, progression, complications, death, and altered quality of life, are of paramount concern. Accurate prognosis stratification help to inform patients about the future course of disease and to guide doctors and patients in joint decisions on further treatment, as well as to facilitate of clinical research to develop new treatment options. The integration of multidimensional biological information, such as radiogenomics, promises a more comprehensive path to accurate prognosis.
4.2.2. Prediction of response to therapy
Advances in radiochemotherapy, targeted therapy, and immunotherapy has provided additional options for tumor treatment. These treatments elicit diverse responses within the patient population and are accompanied by varying degrees of side effects. Tumor genomics and other invasive biomarkers are utilized to stratify patients and select appropriate candidates for therapy. However, the limitations of regional biopsies hinder the accurate prediction of treatment benefit based on molecular information. Radiogenomics is expected to leverage the respective strengths of molecular markers and imaging to facilitate precision oncology decision making.
5. APPLICATION OF RADIOGENOMICS TO CANCER CARE
To provide a comprehensive overview of the advancements in radiogenomics across multiple facets of cancer care, this review focuses on the recent evidence from the past 5 years in three highly researched cancer types: glioma, lung cancer, and breast cancer (Tables 1, 2, 3). We also briefly outlined the recent evidence of radiogenomics in other cancers (Table 4). Additionally, we discuss the current challenges and future directions for further investigation.
TABLE 1.
Summary of recent key studies on radiogenomics decoding of gliomas.
| Subset | Application | Molecular data | Modality | Radiomic features | Training and validation cohorts | Public data sources | Results | References |
|---|---|---|---|---|---|---|---|---|
| Brainstem gliomas | Prediction of molecular | H3k27M | MRI (conventional, diffusion) | Classical radiomics, connectomics |
Training, n = 93 Temporal validation, n = 40 |
No | AUC of 95.31% from combined model, outperformed radiomics and connectomics alone | 16 |
| IDH‐mutant astrocytoma | Prediction of molecular | CDKN2A/B | MRI (conventional) | Deep learning |
Training, n = 234 Cross‐validation |
TCGA‐TCIA | Average AUC of 97.04% from deep learning model | 33 |
| Glioma | Prediction of molecular | IDH | MRI (conventional) | Classical radiomics, location features |
Training, n = 679 Intersite cross‐validation |
TCGA‐TCIA | Maximum AUC of 79.1% from the combined model, outperformed radiomic model | 18 |
| Intramedullary gliomas | Prediction of molecular | ATRX, P53 | MRI (conventional) | Classical radiomics |
Training, n = 229 External validation, n = 129 |
No | AUCs of 0.7622 and 0.7954 in predicting ATRX and P53, respectively | 34 |
| Glioma | Prediction of molecular | Transcriptome | MRI (conventional) | Classical radiomics |
Training, n = 130 External validation, n = 55 |
CGGA | AUC of 0.924 for identifying immune subtypes | 35 |
| IDH‐mutant low‐grade glioma | Prediction of molecular | ATRX | MRI (conventional, perfusion, diffusion), 18F‐FDG PET | Classical radiomics |
Training, n = 72 External validation, n = 30 |
No | AUC of 0.975 from PET+ADC+CE‐T1WI model | 36 |
| Glioma | Prediction of molecular | IDH, 1p19q | MRI (conventional) | Deep learning |
Training, n = 1508 External validation, n = 240 |
TCGA‐TCIA, BraTS | AUCs of 0.90 and 0.85 for the IDH and 1p/19q prediction | 12 |
| Glioma | Prediction of molecular | IDH | MRI (conventional, brain network) | Deep learning |
Training, n = 270 Internal validation, n = 117 |
TCGA‐TCIA | AUC of 96.2%, outperformed published baseline | 15 |
| Glioma | Prediction of molecular | IDH and MGMT | MRI (conventional) | Classical radiomics |
Training, n = 159 External validation, n = 189 |
TCGA‐TCIA | AUC of 0.866 | 37 |
| Glioma | Prediction of molecular | MGMT | MRI (conventional) | Deep learning | N up to 985 | BraTS | Most (80.2 and 60.0%) of the 420 developed models showed negative results in terms of test accuracy and test AUC | 38 |
| Glioma | Prediction of molecular | IDH, 1p19q | MRI (conventional, diffusion) | Deep learning |
Training, n = 384 External validation, n = 147 |
TCGA‐TCIA | Correctly classifying 95.2, 88.9, 60.0% of the three subtypes; better performance achieved using 3‐class structure and diffusion MRI | 39 |
| Midline pediatric high‐grade glioma | Prediction of molecular | H3k27M | MRI (conventional, diffusion) | Classical radiomics |
Training, n = 76 Internal validation, n = 31 |
No | AUC of 0.92 | 40 |
| Pediatric low‐grade glioma | Prediction of molecular | Braf | MRI (diffusion) | Classical radiomics |
Training, n = 299 External validation, n = 23 |
No | Average AUC of 0.74 | 41 |
| Glioma | Prediction of molecular | IDH, 1p19q | MRI (conventional) | Classical radiomics, deep learning |
Training, n = 780 Internal validation, n = 236 |
No | Maximum AUC of 4 tasks range from 0.68 to 0.89; deep learning outperformed radiomics in most tasks | 42 |
| Brainstem gliomas | Prediction of molecular | H3k27M | MRI (APTw) | Classical radiomics |
Training, n = 64 Temporal validation, n = 29 |
No | Accuracy of 0.86 | 43 |
| IDH‐wildtype glioma | Prediction of molecular | TERT | 18F‐FET PET | Classical radiomics |
Training, n = 112 Internal validation, n = 47 |
No | AUC of 0.61 | 44 |
| Glioblastoma | Radiogenomic correlation | Transcriptome | MRI (conventional) | Classical radiomics |
Training, n = 125 External validation, n = 22 |
TCIA | Difference found in radiomic phenotypes and signaling pathways between sexes | 45 |
| Glioma | Prediction of molecular | IDH, 1p19q, TERT | MRI (conventional, diffusion) | Classical radiomics |
Training, n = 238 Internal validation, n = 119 |
No | AUCs of 0.884, 0.815, and 0.669 for predicting IDH, 1p19q, and TERT status; similar prognosis shown with actual subtypes | 46 |
| Glioblastoma | Prediction of molecular | MGMT | 18F‐DOPA PET | Classical radiomics |
Training, n = 59 Internal validation, n = 10 |
No | AUC of 0.80 | 47 |
| Glioblastoma | Prediction of molecular | Genome | MRI (conventional, perfusion, diffusion) | Classical radiomics |
Training, n = 85 Internal validation, n = 35 |
No | AUCs of 0.88, 0.76, and 0.81 for the prediction of RTK, P53, and Rb pathways | 48 |
| Low‐grade glioma | Prediction of molecular | Genome | MRI (conventional) | Deep learning |
Training, n = 182 Cross‐validation |
TCGA‐TCIA | AUC of 0.698 for cluster coc1 vs. coc2 subtypes, 0.731 for cluster coc2 vs. coc3 | 49 |
| Glioma | Prediction of molecular | IDH | MRI (conventional, perfusion) | Deep learning |
Training, n = 395 Internal validation, n = 18 |
No | AUC of 0.95 | 50 |
| Glioblastoma | Prediction of molecular | POSTN | MRI (conventional) | Classical radiomics |
Training, n = 93 (patients)/40 (OXs) Cross‐validation |
No | AUC of 76.56% in patients and 92.26% in OXs; radiomic features in OXs were significantly associated with those in patients | 51 |
| Glioblastoma | Radiogenomic correlation | Genome and transcriptome | MRI (conventional) | Deep learning |
Training, n = 127 External validation, n = 389 |
TCGA‐TCIA, CGGA | Deep learning signature correlate with RTK, P53 and RB pathways, and CDKN2A deletion | 52 |
| Glioma | Prognosis: OS | IDH | MRI (conventional) of whole brain | Deep learning |
Training, n = 935 External validation, n = 465 |
TCIA | AUCs ranged between 0.77 and 0.94, outperformed model that require ROI | 9 |
| Glioma |
Prognosis: OS; Radiogenomic correlation |
Genome and transcriptome | MRI (diffusion) | Deep learning |
Training, n = 688/78 External validation, n = 1320 |
TCGA‐TCIA, CGGA | Deep learning signature improved prognosis of clinic‐molecular model, and correlated with five pathways | 53 |
| Low‐grade glioma |
Prognosis: OS; Benefit from chemotherapy |
IDH | MRI (conventional) | Classical radiomics |
Training, n = 149 External validation, n = 66 |
No | Radiomics joint with clinicopathologic data outperformed the clinicopathologic data alone (C‐index, 0.821 vs. 0.692) | 54 |
| Glioblastoma | Prognosis: OS | Genome | MRI (conventional, perfusion, diffusion) | Classical radiomics |
Training, n = 571 Cross‐validation |
No | Radiogenomics subtype for risk stratification at a hazard ratio of 1.64 | 55 |
| Glioma | Prognosis: OS | Multiomics | MRI (conventional) | Classical radiomics |
Training, n = 111 External validation, n = 53 |
TCGA‐TCIA, GEO, cbioportal, CCLE, GDSC, GO | Radiomics subtype for risk stratification at a hazard ratio of 2.70 | 56 |
Abbreviations: 18F‐DOPA, 18F‐dihydroxyphenylalanine; 18F‐FDG, 18F‐fluoro‐d‐glucose; 18F‐FET, 18F‐fluoro‐ethyl‐tyrosine; APTw, amide proton transfer‐weighted; AUPRC, area under the precision‐recall curve; BraTS, The Brain Tumor Segmentation challenge; CCLE, cancer cell line encyclopedia; CGGA, Chinese glioma genome atlas; GDSC, Genomics of drug sensitivity in cancer; GEO, the gene expression Omnibus; GO, the gene ontology; OS, Overall Survival; OXs, orthotopic xenografts; TCGA, the Cancer Genome Atlas Program; TCIA, the Cancer Image Archivexvv.
TABLE 2.
Summary of recent key studies on radiogenomics decoding of lung cancers.
| Subset | Application | Molecular data | Modality | Radiomic features | Training and validation cohorts | Public data sources | Results | References |
|---|---|---|---|---|---|---|---|---|
| NSCLC | Prediction of molecular | PD‐L1 | CT | Classical radiomics |
Training, n = 62 External validation, n = 109 |
TCGA‐TCIA | AUCs of 0.70, 0.72. and 0.66 at expression >1, >5, and >90%; potential prediction of response to PD‐1 or PD‐L1 treatment, pneumonia development, and patient survival | 57 |
| NSCLC |
Radiogenomic correlation; Prognosis: distant recurrence |
Genome associate with distant recurrence | 18F‐FDG PET | Classical radiomics |
Training, n = 34 Internal validation, n = 19 |
TCGA‐TCIA | AUC of 0.912 from combine model | 58 |
| NSCLC | Prediction of molecular | EGFR, ALK, ERBB2, BRAF, MET, ROS1, RET, KRAS, TP53, and PD‐L1 | CT | Classical radiomics, deep learning |
Training, n = 877 Internal validation, n = 110 |
No | AUCs of 0.856 to 0.877 for 4 tasks from radiomics and deep learning combined models | 59 |
| Lung cancer |
Prediction of molecular; Prognosis: after EGFR‐TKI treatment |
EGFR | CT (whole‐lung) | Deep learning |
Training, n = 10,427 External validation, n = 8375 |
TCIA | AUCs of 0.748–0.813; outperformed models based on ROI; helped identify TKI resistance; associated with genotype and pathways linked to drug resistance and progression | 10 |
| NSCLC | Prediction of molecular | EGFR, KRAS | CT, 18F‐FDG PET | Classical radiomics |
Training, n = 94 Internal validation, n = 42 |
TCIA | AUCs of 0.92 to 0.94 for EGFR and 0.91 to 0.94 for KRAS; combat harmonization improve performance | 60 |
| NSCLC | Prediction of molecular | PD‐L1 | CT | Classical radiomics, deep learning |
Training, n = 908 Internal validation, n = 227 |
No | AUCs of 0.950, 0.934, and 0.946 for predicting PD‐L1 expression signature <1, 1−49, and ≥50%; improved prognosis of clinical data | 61 |
| NSCLC | Prediction of molecular | EGFR status and subtypes, PD‐L1 | CT (tumor and whole lung) | Classical radiomics, deep learning |
Training, n = 3053 Internal validation, n = 763 |
No | AUCs of 0.841 to 0.905 for 5 predicting tasks from joint module | 62 |
| LUAD | Prediction of molecular | T790M mutation | CT | Classical radiomics |
Training, n = 186 Internal validation, n = 74 |
No | AUCs of 0.71 and 0.76 from radiomics, and nomogram models; | 63 |
| NSCLC | Radiogenomic correlation | Genome | CT | Deep learning |
Training, n = 142 External validation, n = 71 |
No | Deep learning score was associated with pathways and antitumor immune cell infiltration in the microenvironment | 64 |
| LUAD | Radiogenomic correlation | Genomic alterations | CT | Classical radiomics | n = 219 | No | Associations found between radiomic subset and clinical‐pathologic, genomic features, and outcomes | 65 |
| NSCLC | Prediction of molecular | ALK fusion | CT | Deep learning |
Training, n = 651 External validation, n = 286 |
No | AUCs of 0.775 and 0.848 from CT and CT‐clinicopathological combined model; stratified prognosis under ALK‐TKI treatment | 66 |
| NSCLC | Prediction of molecular | PD‐L1 | CT | Deep learning |
Training, n = 750 Internal validation, n = 96 |
No | AUC of 0.76, stratified prognosis under anti‐PD‐1 antibody treatment | 67 |
| NSCLC | Prediction of molecular | EGFR, PD‐L1 | 18F‐FDG PET/CT | Deep learning |
Training, n = 429 External validation, n = 65 |
No | AUCs of 0.81 and 0.84 from deep learning and combined model; stratified prognosis under TKIs and ICIs treatment | 68 |
| NSCLC | Prediction of molecular | TMB | CT | Deep learning |
Training, n = 236 Internal validation, n = 65 |
No | AUC = 0.81; stratified prognosis under ICIs treatment | 69 |
| LUAD | Prediction of molecular | Cytact | 18F‐FDG PET | Deep learning |
Training, n = 93 External validation, n = 59 |
TCGA‐TCIA | Predicted cytact positively correlated with ground truth; stratified prognosis under ICB treatment | 70 |
| NSCLC | Response to ICIs | Plasma extracellular vesicle PD‐L1 | CT | Classical radiomics |
Training, n = 27 Internal validation, n = 30 |
No | Radiogenomics model increase specificity, sensitivity, and accuracy of ICIs response prediction compared with genomics | 157 |
| NSCLC | Prediction of metastasis | Genome | 18F‐FDG PET | Deep learning |
Training, n = 93 Cross‐validation |
No | Highest AUC of 0.855, outperformed radiomic and genomic models | 71 |
Abbreviations: ALK, Anaplastic Lymphoma Kinase; cytact, cytolytic activity score; ICB, immune checkpoint blockade; ICIs, Immune Checkpoint Inhibitors; KRAS, Kirsten Rat Sarcoma Viral Oncogene Homologue; LUAD, lung adenocarcinoma; PD‐1, Programmed Death 1; PD‐L1, Programmed Cell Death Ligand 1; MET, Mesenchymal Epithelial Transition; ROS1, ROS proto‐oncogene 1; RET, Rearranged During Transfection; TKI, Tyrosine Kinase Inhibitors; TMB, Tumor Mutation Burden.
TABLE 3.
Summary of recent key studies on radiogenomics decoding of breast cancers.
| Subset | Application | Molecular data | Modality | Radiomic features | Training and validation cohorts | Public data sources | Results | References |
|---|---|---|---|---|---|---|---|---|
| Breast cancer | Prediction of molecular | HER2 | MRI (conventional) | Deep learning |
Training, n = 329 External validation, n = 61 |
No | AUCs of 0.76 and 0.75 for prediction of HER2‐overexpressing and HER2‐low‐positive; stratified prognosis | 72 |
| Breast cancer | Prediction of molecular change after NAT | ER‐/HER2‐ or ER‐low/HER2‐ | MRI (conventional, perfusion, diffusion) | Classical radiomics |
Training, n = 66 Internal validation, n = 19 |
No | AUC of 0.86 | 73 |
| Breast cancer | Prediction of molecular | TNBC and transcriptomic TNBC subtypes | MRI (conventional) | Classical radiomics |
Training, n = 420 External validation, n = 164 |
No | AUCs of 0.613 to 0.723 for identification of TNBC; AUCs of 0.598–0.796 for distinguishing TNBC subtypes; peritumoral radiomic features were associated with immune suppression and upregulated fatty acid synthesis | 74 |
| Breast cancer | Prediction of molecular | HR, HER2, Ki‐67 | MRI (conventional, perfusion) | Classical radiomics |
Training, n = 218 Internal validation, n = 73 |
No | Maximum AUC of 0.8 for prediction of molecular from texture features using random forest in SSF of 0 | 75 |
| Breast cancer | Prediction of molecular | ER, PR, PAM50 | MRI (conventional, perfusion) | Deep learning |
Training, n = 585 Cross‐validation |
No | AUCs of 0.942 and 0.920 for predicting ER and PR, 0.742 for PAM50; performance improved when peritumor region included | 76 |
| Breast cancer | Prediction of molecular | HR, HER2 | Contrast‐enhanced spectral mammography | Classical radiomics |
Training, n = 164 Internal validation, n = 18 |
No | ACCs of 0.89 and 0.85 for predicting HER2 and HR | 77 |
| Breast cancer | Prediction of molecular | CD8+ T cells‐based immunophenotype | MRI (conventional) | Classical radiomics |
Training, n = 137 Internal validation, n = 45 |
No | AUCs of 0.985 and 0.984; associated with complete response to NAC | 78 |
| Breast cancer | Prediction of molecular | Ki‐67, luminal A subtype | MRI (conventional, perfusion) | Deep learning |
Training, n = 122 Internal validation, n = 80 |
No | AUCs of 0.819 and 0.799 for predicting Ki‐67 and luminal A | 79 |
| TNBC | Prediction of molecular | Transcriptome | MRI (conventional) | Classical radiomics |
Training, n = 98 Internal validation, n = 41 |
No | AUC of 0.79, related to activated immune‐related pathways and hot immune microenvironment | 80 |
| Breast cancer | Prediction of molecular | Immunohistochemistry subtype | Ultrasound | Deep learning |
Training, n = 1275 External validation, n = 845 |
No | MCCs of 0.59–0.79 for predicting 4 subtypes; MCCs of 0.54 and 0.65 for discriminate luminal and nonluminal | 81 |
| Breast cancer | Prediction of molecular | Transcriptome | MRI (conventional) | Classical radiomics |
Training, n = 96 External validation, n = 155 |
TCGA‐TCIA | AUC of 0.815 for predicting immunoscore, radiomics signature associated with recurrence‐free and overall survival rates | 82 |
| Breast cancer | Prediction of molecular | PD‐L1 | MRI (conventional) | Classical radiomics |
Training, n = 62 Cross‐validation |
No | AUC of 0.904 | 83 |
| Breast cancer | Prediction of molecular | Tumor microenvironment subtype | MRI (perfusion) | Deep learning |
Training, n = 342 Cross‐validation |
TCGA‐TCIA | Radiomics features are correlated with markers of breast TME, such as ER, PR, HER2, PD‐1, PD‐L1, EGFR | 84 |
| Breast cancer | Prediction of molecular | Immunohistochemistry subtype | MRI (conventional, perfusion) | Classical radiomics |
Training, n = 211 Cross‐validation |
No | Maximum AUC of 0.832 from radiomic model based on tumor subregion related to fast‐flow kinetics, AUC increased to 0.897 in the tumor‐ and parenchyma‐based predictive modal | 85 |
| Breast cancer | Prediction of molecular | HER2 | Ultrasound (video) | Deep learning |
Training, n = 357 Internal validation, n = 88 |
No | AUCs of 0.72 and 0.81 from radiomic and combine model | 86 |
| Breast cancer | Prediction of molecular | Immunohistochemistry subtype | Mammography, ultrasound | Deep learning |
Training, n = 2688 Internal validation, n = 672 |
No | MCC of 0.837 for predicting 4‐category subtypes; AUC of 0.929 for discriminate luminal and nonluminal | 87 |
| Breast cancer | Prediction of molecular | HR | Mammography | Deep learning |
Training, n = 2083 Temporal validation, n = 190 |
No | Average AUC of 0.92 | 88 |
| Breast cancer | Response to NAC: pCR; prognosis: OS, RFS | IL‐17 and estrogen signaling pathways | MRI (perfusion) | Classical radiomics |
Training, n = 255 External validation, n = 174 |
TCGA‐TCIA | Radiomics predicted tumor shrinkage with an AUC of 0.886 and PCR with an AUC of 0.760, correlating with IL‐17 and estrogen signaling pathways | 89 |
| Breast cancer | Prognosis: PFS | Immunohistochemistry subtype, PD‐L1, Ki67 | CT | Classical radiomics |
Training, n = 171 Internal validation, n = 69 |
No | AUC of 0.961 for prognosis under ICIs‐based therapies from the integrated clinical‐radiomics model | 90 |
| TNBC | Response to NAC: pCR | Genome: the variant allele frequency features | MRI (pre‐ and posttreatment) | Classical radiomics |
Training, n = 75 Internal validation, n = 37 |
No | AUCs of 0.87 from radiogenomic model, outperformed radiomic model; two highly frequent mutations related to epirubicin resistance | 91 |
| TNBC | Response to NAC, Prognosis: DFS | 511 genes related to the development and targeted therapy | MRI (conventional, perfusion) | Classical radiomics |
Training, n = 413 Internal validation, n = 77 |
TCGA‐TCIA | AUC of 0.93 for predicting PCR from radiogenomic models; significantly stratify patients by disease‐free survival | 92 |
| Breast cancer | Predict lymph node metastasis and therapeutic response | Whole‐transcriptome | MRI (conventional) | Classical radiomics |
Training, n = 103 External validation, n = 924 |
TCGA‐TCIA | Radiogenomics nomogram identified axillary lymph node metastasis and drug therapeutic response at a statistically significant level (p < 0.05) | 93 |
Abbreviations: DFS, disease‐free survival; HR, Hormone Receptor; IL‐17, the Interleukin‐17; MCC, Matthews correlation coefficient; NAC, Neoadjuvant Chemotheropy; NAT, Neoadjuvant Theropy; pCR, pathologic Complete response; RFS, Relapse‐free Survival.
TABLE 4.
Summary of key studies on radiogenomics decoding of colorectal cancer, renal cell carcinoma and prostate cancer.
| Subset | Application | Molecular data | Modality | Radiomic features | Training and validation cohorts | Public data sources | Results | Reference |
|---|---|---|---|---|---|---|---|---|
| Colorectal cancer | Prediction of molecular | RAS | CT | Deep learning |
Training, n = 208 Internal validation, n = 23 |
No | AUC of 0.955 | 94 |
| Colorectal cancer | Prediction of molecular | DNA mismatch repair status | CT | Deep learning |
Training, n = 1124 External validation, n = 206 |
No | AUC of 0.915; similar satisfying prediction performance showed in subgroup analysis | 95 |
| Colorectal cancer (stage IV) | Prediction of molecular | TMB | 18F‐FDG PET | Classical radiomics |
Training, n = 91 Cross‐validation |
No | AUC of 0.719 | 96 |
| Colorectal cancers with liver metastasis | Prediction of molecular | CD73 expression | CT |
Classical radiomics; Deep learning (liver metastasis) |
Training, n = 125 Internal validation, n = 35 |
No | AUC of 0.79 from deep learning model; outperformed other models; prognostic value of radiogenomics was independent of the standard clinical risk score | 97 |
| Colorectal cancers with liver metastasis | Prediction of molecular | RAS and BRAF mutation | CT | Classical radiomics (liver metastasis) |
Training, n = 124 Internal validation, n = 35 |
No | AUC of 0.79 | 98 |
| Colorectal cancer | Prognosis: DFS | Four genomic subclones identified unsupervisedly | CT | Classical radiomics |
Training, n = 236 External validation, n = 69 |
Gene Expression Omnibus | Radiogenomic signatures were independent prognosis factor; associated with extracellular matrix and immune‐related pathways | 99 |
| ccRCC | Prediction of molecular | Immune‐related genomic signature | CT | Classical radiomics |
Training, n = 135 Internal validation, n = 58 |
TCGA‐TCIA | AUC of 0.72 for predicting immune‐related molecular subtypes | 100 |
| ccRCC | Prediction of molecular; Prognosis: OS | Transcriptome | CT | Classical radiomics |
Training, n = 127 External validation, n = 75 |
TCGA‐TCIA | Lipid metabolic pathway‐specific radiogenomics modeling is an independent risk factor for patient prognosis | 101 |
| ccRCC |
Prediction of molecular; Prognosis: OS |
Genome, transcriptome, proteome | CT | Classical radiomics |
Training, n = 104 Internal validation, n = 103 |
TCGA‐TCIA | AUCs of 0.949–0.973 for predicting 4 genetic mutations and 4 mRNA‐based subtypes; highest AUC achieved for prognosis using radiogenomic models | 102 |
| Prostate cancer | Radiogenomic correlation | CTCs count and plasma cfDNA level | CT | Classical radiomics (bone metastasis) | N = 8 | No | Radiomic features consistently and strongly positively correlated with CTCs count, plasma CFDNA CTCs clusters 6, 7, and 8 | 103 |
| Prostate cancer | Radiogenomic correlation | Genotypes for apoptosis, hypoxia, and androgen receptor expression | MRI (conventional, diffusion) | Classical radiomics | N = 8 | No | Significant correlation observed between radiomic features and CNV of genes associated with apoptosis, hypoxia, and androgen receptor (p ≤ 0.05) | 104 |
Abbreviations: ccRCC, Clear Cell Renal Cell Carcinoma; cfDNA, cell‐free DNA; CNV, copy number variation; CTCs, Circulating tumor Cells.
5.1. Glioma
Gliomas are heterogeneous entities, which characterized by specific gene alter. 105 Although some gliomas are benign and have a favorable prognosis, the majority, particularly glioblastoma, are highly fatal. 106 This is not only due to the direct impact of the tumor on the structure and function of the brain, but also because of the risk of serious complications during invasive procedures against the lesion. 107 , 108 , 109 Refining medical decisions with precision for optimal outcomes at minimal cost is vital. Medical imaging plays a pivotal role in assessing gliomas noninvasively. Radiogenomics holds significant potential in predicting the molecular subtypes of gliomas preoperatively and stratifying patients’ prognosis.
Isocitrate dehydrogenase (IDH) mutation and 1p19q chromosome codeletion serve as the key determinants for the classification of adult diffuse glioma. 105 These genetic alterations not only indicate distinct prognoses but also guide diverse treatment strategies. In several large‐scale studies, 12 , 13 , 18 , 42 radiomic models have shown the ability to predict IDH or 1p19q status either independently or within a comprehensive framework for adult diffuse glioma. DL models often exhibit superior performance than classical radiomics, even when applied to the same dataset. In particular, Van et al.’s DL model achieved high accuracy in externally validating molecular predictions, while also in performing multitask of tumor grading and segmentation. This highlights the significant advantages of high‐performance and multitasking capabilities in DL, especially when trained with ample data. Utilizing a single model to predict subtypes of IDH and 1p19q offers greater accessibility compared with predicting them individually. Cluceru et al. 39 concluded that a three‐class model for subtyping has superior generalization capability compared with a two‐tiered approach. This may be attributed to the significant reduction in training cases during the second step of the two‐tiered pattern, whereas the three‐class approach incorporates all data for model training.
Conventional MRI is superior in displaying anatomical structures, while other advanced imaging techniques, such as diffusion‐weighted imaging and perfusion‐weighted imaging, have been developed to display or amplify microenvironmental information, providing multidimensional data for molecular prediction of tumors. Radiogenomic studies have shown improved accuracy and stability of the model, although further external validation data are still required. 39 , 46 , 50 , 110 , 111 In addition to tumor signal, features from tumor location, which have been shown to correlate with IDH status and are less influenced by image acquisition and measurement variability, as well as brain network connectome, 112 which can identify disrupted white matter tracts and reveal hidden tumor invasion, have also been incorporated into radiogenomic models to enhance accuracy and generalization in predicting IDH status. 15 , 18
Substitution of lysine 27 to methionine in histone H3 (H3K27M) characterizes a subset of highly malignant pediatric gliomas that are unresectable and exhibit rapid progression with a dismal prognosis. 105 This has been proven to be a significant prognostic factor for overall survival, irrespective of age, tumor location, or histopathological grading in midline gliomas. 113 , 114 Radiomic models utilizing Conventional MRI have achieved area under the receiver operating characteristixc curves (AUCs) ranging from 0.78 to 0.85 in identifying H3K27M in midline glioma. 115 , 116 Moreover, the integration of brain structural connectomics or diffusion‐weighted imaging has shown the ability to further enhance the precision of the models. 16 , 40 Zhou et al. 43 have recently demonstrated the efficacy of radiomic models based on amide proton transfer weighted MRI, an emerging functional imaging technique, in predicting H3K27M in pontine gliomas, with an accuracy of 0.86 in an independent prospective cohort. 43 Although current radiogenomic predictions for H3K27M still lack multicenter external validation, these findings hold great promise, particularly with the application of multimodal imaging techniques.
O6‐methylguanine‐DNA‐methyltransferase (MGMT) promoter methylation serves as a significant molecular marker for assessing the therapeutic efficacy of alkylating agents like temozolomide, which is a first‐line chemotherapy drug for glioma. 117 Despite numerous attempts to construct radiogenomic models for the prediction of MGMT status, either the results were far from satisfactory or lacked adequate external validation. 118 The most recent systematic review indicates substantial heterogeneity in the results of MRI radiomics models for predicting the methylation status of MGMT in grade IV gliomas, with low performance observed in external validation. 119 Two large‐scale external validations of previous research findings published in 2022 and 2023 also demonstrate that these MRI‐based radiomics models are still insufficient in accurately predicting MGMT in gliomas prior to surgery. 38 , 120 A radiogenomic model based on PET indicated higher accuracy (AUC = 0.80 in cross‐validation) in predicting MGMT status, but further validation was required. 47 Nonetheless, it is worth exploring the use of multimodal images based radiomics to enhance prediction accuracy. Interestingly, the co‐occurrence of IDH mutation and MGMT methylation characterizes a subtype of gliomas with a favorable prognosis and potential benefits from temozolomide, and this can potentially be predicted using radiomic models. 37
Other molecular markers, such as alpha‐thalassemia mental retardation X‐linked (ATRX), telomerase reverse transcriptase (TERT), EGFR, tumor protein 53(TP53), cyclin‐dependent kinase inhibitor 2A/B (CDKN2A/B), proto‐oncogene B‐Raf and v‐Raf murine sarcoma viral oncogene homlog B (BRAF), cyclin D1 (CCND1), and cyclin‐dependent kinases 6 (CDK6), have also emerged as crucial factors in glioma classification, prognosis, and targeted therapy 121 , 122 and have become focal points for radiogenomic investigations. 33 , 34 , 36 , 41 , 44 , 46 , 123 , 124 , 125 , 126 , 127 However, currently, there is insufficient evidence to support the clinical application of radiogenomic models for these markers. Notably, Zinn et al. 51 developed a radiomics model to predict the expression level of periostin in glioblastoma, and importantly, they confirmed the causal relationship between radiomics subtypes and molecular expression through simultaneous radiomics analysis on orthotopic xenografts. As new molecular markers for gliomas are gradually integrated into clinical practice, further efforts are needed to establish substantial evidence regarding the application of radiogenomics to these relatively rare markers.
Thanks to microarray and next‐generation sequencing technologies, oncology research has made significant strides in comprehensively analyzing the molecular landscape of cancer cells and the tumor microenvironment, which goes beyond merely detecting specific genetic alterations. Several radiogenomic studies have revealed the intense associations between radiomic phenotypes and multiomics molecular subtypes and the tumor immune microenvironment (TIME). 71 , 72 , 73 Hu et al. 128 attempted to correlate radiomics with the genetic status of various subregions of the tumor. They collected 48 image‐guided biopsies from 13 glioblastomas and confirmed the spatial heterogeneity of genetic subtypes within the tumor, which correlated with radiomic features. Several studies have found correlations between MR radiomics prognostic phenotypes and specific molecular signaling pathways and intercellular communication in gliomas. 48 , 52 , 53 , 129 Recent studies have defined new phenotypic subtypes of gliomas based on radiomics or radiogenomics and found significant differences in survival, immune infiltration, and drug susceptibility among these subtypes, providing a better understanding of the molecular basis of phenotypic characterization of gliomas. 55 , 130 These findings reveal the underlying biological mechanisms behind radiomic models and may be used to identify potential therapeutic targets for gliomas. 53 In particular, Beig et al. 45 investigated the radiogenomic associations of MRI‐based phenotypes with transcriptomic data in male and female patients. Their aim was to identify the signaling pathways that drive sex‐specific tumor biology and treatment response in glioblastoma.
Radiomics is believed to capture tumor heterogeneity and provide additional biological information beyond the tumor, making it a valuable complement to molecular biomarkers used in clinical practice. 131 , 132 Whether in low‐grade gliomas, high‐grade gliomas, or overall diffuse gliomas, radiogenomic models showed superior performance in stratifying patient prognosis compared with classical radiomic or molecular‐clinical models. 9 , 52 , 133 By integrating genetic data such as IDH and MGMT status with radiomics, radiogenomic models can more accurately differentiate postoperative recurrence from pseudoprogression and assess the efficacy of chemotherapy. 54 , 134
5.2. Lung cancer
Lung cancer is one of the most frequently diagnosed cancers and the leading cause of cancer‐related deaths worldwide. 135 While surgical resection remains the preferred treatment modality, advancements in chemoradiotherapy, targeted therapy, and immunotherapy have significantly enhanced patient outcomes and quality of life, particularly in advanced non‐small cell lung cancer. 136 , 137 , 138 Furthermore, NAT has demonstrated its role in improving resectability, delaying recurrence and progression, and prolonging survival in select lung cancer patients. 139 However, the efficacy of these therapies varies across different populations. Molecular characteristics offer valuable insights into prognosis and therapeutic benefits, 140 yet the clinical application of these biomarkers obtained through tissue biopsy is limited. Given the widespread use of chest CT and PET/CT in the preoperative assessment of lung cancer, radiogenomics holds promise in addressing the limitations of molecular markers, enabling better patient stratification, and facilitating treatment decision‐making.
Epidermal growth factor receptor (EGFR) gene mutations are the most prevalent targeted driver mutations in lung cancer. 141 Constant updates are being made to EGFR‐tyrosine kinase inhibitor (TKI) targeted therapy regimens in order to combat drug resistance. 142 The choice of therapeutic agent has always relied on the accurate identification and subtyping of EGFR mutations. 143 Radiomic models have shown good to excellent performance in predicting EGFR mutations in lung cancer. 68 , 144 , 145 DL models appear to outperform classical radiomics, 146 , 147 and their combination may yield even better results. 62 Notably, an international multicenter study with a large cohort of cases developed a DL model based on the entire lung, which achieved an AUC of 0.812 in predicting EGFR status in lung cancer and successfully stratified progression‐free survival in patients treated with EGFR‐TKI. Furthermore, correlations were found between radiomic phenotypes and multiple genotypes, as well as gene pathways associated with drug resistance and cancer progression mechanisms, providing compelling evidence for the use of radiomics in predicting EGFR mutations. 148 Taking it a step further, Wang et al. 62 developed a radiomics‐DL joint model to determine EGFR mutation subtypes, including 19Del, L858R, and other mutations. Additionally, Yang et al. 63 constructed a radiomic model may aid in predicting the acquired drug‐resistant mutation T790M following targeted therapy for lung cancer, suggesting the potential application of radiogenomic models in optimizing EGFR‐targeted therapy decisions.
ALK fusion is another key therapeutic target in lung cancer, 149 and Song et al.’s CT‐based DL model yielded an AUC of 0.85 in external validation. The model also showed promising performance in predicting response to ALK‐TKI therapy, which was further validated. 66 Regarding Kirsten rat sarcoma viral oncogene homologue (KRAS) mutation, radiomic models based on low‐dose CT scan and PET‐CT have shown good predictive performance. However, additional validation using external data is necessary. 60 , 150
Over the past decade, immunotherapy has emerged as a pivotal breakthrough in the treatment of lung cancer, revolutionizing the therapeutic landscape. 151 Despite the significant advancements made in targeting immune checkpoints, particularly the programmed death receptor 1/programmed death ligand 1 (PD‐1/PD‐L1) axis, a substantial proportion of patients fail to derive benefits from PD‐1/PD‐L1 inhibitors. 151 Although CT‐based radiomic models have been developed to predict PD‐L1 expression in non‐small cell lung cancer, with AUCs ranging from 0.66 to 0.95, their performance in external validation has been suboptimal. Nonetheless, these models have demonstrated correlations with prognosis and immunotherapy response. 57 , 61 , 62 , 67 Mu et al. 68 constructed PET/CT‐based radiomic prediction models for PD‐L1 expression and EGFR mutation, and subsequently established treatment decision guidelines based on these models, along with Eastern Cooperative Oncology Group performance status scores. The clinical utility of these radiogenomic models was validated in external data, showcasing their effectiveness in guiding the selection of patients for TKIs and immune checkpoint inhibitors (ICIs) therapy. 68 , 152 In another study, a DL model was developed to predict tumor mutational burden (TMB), achieving an AUC of 0.81 69 and was further validated in stratifying survival outcomes following immunotherapy. Additionally, several studies have demonstrated associations between radiomic subtypes and immunophenotypes, such as CD8 expression and cytolytic activity score, as well as response to immunotherapy 70 , 153
For some rare molecular alterations of lung cancer, radiogenomic studies are restricted by data size. A few studies tried to simultaneously predict multiple molecular subtypes through radiomics based on CT or PET/CT to reflect more realistic clinical scenarios. However, external data are still required for validation. 59 , 154 Notably, for the first time, one of these studies discovered that the utilization of transformer algorithms, commonly used in large language models (LLMs), for constructing radiogenomic models outperformed those based on neural networks. Furthermore, robust correlations have been observed between radiomic features, genomic features, and tumor recurrence as well as response to neoadjuvant immunotherapy in lung cancer. 58 , 64 , 65 A comprehensive profiling of these radiogenomic associations contributes to improved decision‐making and the identification of novel therapeutic targets.
Recently, some studies have attempted to establish radiogenomics models to achieve better prognostic stratification of lung cancer, by combining radiomics with transcriptome, or CDK4 and TMB status. 155 , 156 Besides, Ju et al. 71 investigated the interaction between genomic and radiomic features and successfully achieved noninvasive prediction of lymph node metastasis in NSCLC. These studies show the clear potential of the integration of complementary multiscale information from imaging and genes in the stratification of prognosis and treatment response in lung cancer. Notably, by liquid biopsy, de Miguel‐Perez et al. 157 verified that dynamic expression of plasma extracellular vesicle PD‐L1 in the early stage of treatment correlated with sustained response to ICIs, and that radiogenomics modeling in conjunction with CT radiomics could further enhance the specificity, sensitivity, and accuracy of the model.
5.3. Breast cancer
Breast cancer is the most prevalent malignancy among women worldwide, and the characterization of its molecular markers has significantly contributed to the development of increasingly sophisticated diagnostic and treatment approaches. 158 In particular, the utilization of NAT, immunotherapies, and novel targeted therapeutic options has underscored the importance of accessing tumor biomarkers in a noninvasive manner. 159 Radiogenomics, which combines genetic and radiomic data, enhances genomics by providing voxel‐by‐voxel biological information for a heterogeneous tumor, enabling tailored therapy. Specifically, multiple imaging modalities, including mammography, ultrasound, MRI, CT, and PET/CT, are employed for diagnosis and treatment, thereby offering multidimensional data for accurate assessment of breast cancer. 160
Breast cancer is commonly classified into four subtypes based on the expression of estrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor receptor 2 (HER2), and Ki‐67, namely Luminal A, Luminal B, HER2‐enriched, and triple‐negative breast cancers (TNBC). 159 Ultrasound‐based DL models have shown good performance in identifying these subtypes and distinguishing between luminal and nonluminal diseases. 81 , 86 However, classical radiomic features have shown limited predictive efficacy for HER2. 161 On the other hand, mammography‐based radiomic models have shown promising results in predicting the hormone receptor and HER2 status of breast cancer. 77 , 88 , 162 Zhang et al. 87 developed a multimodal DL model using a large cohort that combined ultrasound and mammogram data. This model achieved an accuracy of 0.84 in the internal test set and an AUC of 0.92 for predicting luminal disease from nonluminal disease, significantly outperforming clinicians.
MRI is also used in radiogenomics to differentiate molecular subtypes; yet, there are few studies with large cohorts and external validation. MRI‐based radiomic models have been shown to successfully predict HER2 expression and pathologic complete response (PCR) of neoadjuvant chemotherapy (NAC) and disease‐free survival. 72 , 163 Some other studies have reported that radiomic models can identify ER, PR, Ki‐67 expression, or differentiate luminal A from other immunohistochemical subtypes, but all of them have only been internally or cross‐validated. 75 , 76 , 79 , 164 However, it has been observed that transfer learning can partially compensate for the lack of training data in DL, and ML algorithms may outperform DL algorithms when data are limited. Additionally, incorporating features from the peritumor region and perfusion images may improve the predictive ability of the model. Jiang et al. 74 validated in an external dataset that MRI‐based radiomic models could identify TNBC and distinguish internal subtypes of TNBC with favorable performance. Furthermore, an association was found between peritumoral radiomic features and immune suppression and upregulated fatty acid synthesis. Fan et al. 85 developed a radiomic model based on multiple hemodynamic subregions by referencing the unsupervised segmentation of intra‐ and extratumor regions on dynamic contrast‐enhanced images, which outperformed the simple whole‐tumor radiomic model in prediction of immunohistochemical subtypes, suggesting additional benefits obtained by delving deeper into intratumor heterogeneity from images. Interestingly, the new DL algorithm generates adversarial networks can synthesize realistic breast MRI images for training of radiomics models to predict breast cancer genotypes and mutational states. 165 Hormone receptor and/or HER2 status discordance after neoadjuvant treatment is a relatively common phenomenon and may require adjustments in post‐NAT strategies. 32 Liu et al. 73 constructed a radiomic model to predict post‐NAT discordance based on multimodality MRI. Although the sample size was small and external validation data were lacking, the results suggest that radiogenomics may provide guidance for retesting molecules and posttreatment alterations in the biology of cancer.
PAM50 subtyping of breast cancer, as opposed to immunohistochemical subtypes, offers superior stratification for disease progression, prognosis, and therapeutic resistance. 166 However, the clinical application of genomic assays is limited due to their high cost. To address this issue, a cost‐effective solution was proposed in the form of a deep transfer learning model that utilizes dynamic contrast‐enhanced images to predict PAM50 subtypes. 76 Additionally, Liang et al. 167 attempted to establish complex many‐to‐many associations between ultrasound radiomics and genomic features to screen for key radiomic and genomic features, providing clues for biological interpretation of radiomics and targeted therapeutic decisions. Gallivanone et al. 168 conducted a study correlating the MR radiomic phenotype of breast cancer with microRNAs, mRNAs, and regulatory networks to develop a radiomirnomic map. They found that the radiogenomic model provided better discrimination of breast cancer subtypes compared with miRNA or radiomics alone.
TIME is a crucial element in the progression and metastasis of breast cancer. 169 PD‐L1 and tumor‐infiltrating lymphocytes are strongly associated with immune evasion by tumors and serve as vital biomarkers for the effectiveness of ICIs. 170 MRI‐based radiomic models have been shown to predict PD‐L1 expression, tumor microenvironment phenotypes based on immune cell infiltration and omics. 78 , 80 , 82 , 83 , 171 Lv et al. 84 screened for genome‐related imaging features to construct interpretable imaging phenotypes that could predict different molecular features, including hormone receptor, epithelial growth factor receptor, and immune checkpoint protein expression. While still in the early stages, radiogenomics shows potential in enhancing noninvasive preoperative evaluation of the TIME and supporting the clinical implementation of immunotherapy.
Prediction of NAC response in breast cancer is a hot spot in radiogenomics. The addition of baseline MRI radiomics to molecular markers did not significantly improve the prediction of PCR after NAC. 172 However, radiomic models based on longitudinal MRI have shown improved predictive performance in comparison with molecular subtyping. 173 , 174 Similarly, a radiogenomic model combining five variant allele frequency features of nonsynonymous mutation sites and baseline MRI was able to predict PCR to NAC in TNBC patients, and a potential relationship was found between two high‐frequency mutations and epidoxorubicin resistance. 91 Huang et al. 175 developed a radiogenomic model incorporating MRI features, ER expression, and Ki‐67, which achieved an AUC of 0.94 in predicting tumor shrinkage patterns after NAC and maintained good predictive performance across different molecular subtypes. Recently, Radiogenomic models that united radiomics and transcriptomics were demonstrated to predict axillary lymph node metastasis as well as and response to drug therapy, while gene pathway enrichment analyses showed significant differences in signaling pathway activation across risk groups. 93 DCE‐based radiomics, reflecting intratumor and peritumor hemodynamic heterogeneity, in conjunction with genomics to constitutes a radiogenomics model also showed significant potential in predicting PCR and poor prognosis in TNBC patients. 92 These studies demonstrated the significant potential of radiogenomic models in predicting treatment response. Furthermore, radiogenomic models that incorporate CT, molecular subtyping, and clinical features from multicenter cohorts have shown promise in predicting immunotherapy response in breast cancer patients. 90
5.4. Other cancers
In recent years, significant advancements have been achieved in the field of radiogenomics for various types of cancers. For example, CT‐based radiomic models have been developed to predict specific gene mutations in clear cell renal cell carcinoma (ccRCC), such as VHL, Polybromo‐1 mutation, and Loss 9p21.3. 102 , 176 Radiomic models have also been used to differentiate omics‐based lipid metabolism or TIME subtypes and correlate them with patient survival. 100 , 101 Unsupervised clustering of radiomic and transcriptomic features of ccRCC allows for the identification of intrinsic subtypes which exhibit unique clinicopathological, prognostic, immunological, and molecular features, and this is expected to facilitate personalized diagnostic and treatment decisions. 177 Besides, It has been found that a radiogenomic model provided more accurate predictions of overall survival in kidney cancer compared with using radiomics alone. 102 Additionally, CT‐based DL models have been leveraged to predict the mutation status of the RAS gene and DNA mismatch repair in colorectal cancer. 94 , 95 Furthermore, a radiomic model based on PET images has shown promise in predicting tumor mutation burden and its correlation with prognosis. 96 Zhong et al. 99 showed that radiomic features were associated with tumor genome subcloning, and radiogenomic signatures could serve as independent predictors of prognosis in patients with colorectal cancer. In the case of liver metastases commonly found in colorectal cancer, radiomic scores have been used to determine RAS and BRAF mutation status, as well as CD73 expression. 97 , 98 For thyroid cancer, CT radiomic models could predict the status of cytokeratin 19, galectin 3, thyroperoxidase, and high‐molecular‐weight cytokeratin. 178 Recent studies showed radiomics features of prostate cancer and its bone metastases correlate with liquid biopsy monitoring of CTCs, free DNA, and genes related to apoptosis, hypoxia, and androgen receptor expression. 103 , 104 The potential of radiogenomics in cancer diagnosis and prognosis is immense; however, further works are warranted to explore its full scope, applicability, and clinical application.
6. CURRENT CHALLENGES AND FUTURE DIRECTIONS
Despite the reported successes of radiogenomics, several limitations and hurdles need to be addressed before widespread clinical adoption.
6.1. Stability and repeatability
The stability and repeatability of radiogenomic models is a key factor in their clinical translation. However, radiomic features are prone to instability caused by various factors, even though they sensitively characterize tumor heterogeneity. 179 Moreover, most of the results are based on retrospective studies with small sample sizes, which introduces bias in the data inclusion and analysis process. This makes it difficult to apply the findings to other centers and actual clinical scenarios. To mitigate these issues, standardization algorithms can be applied to reduce variations in medical images between machines and centers. 180 Besides, selecting stable features for constructing radiogenomic models is crucial. 181 In addition, improving the transparency and explainability of radiogenomics models helps to detect model hallucinations and biases and make adjustments before they are put into complex clinical scenarios. 182
More importantly, training the model using representative heterogeneous data with rigorous external validation rigorous external validation, especially in multicenter real‐world settings. We also need to assessing the general applicability of the model across different populations, because potential variation may exist in the efficacy of radiogenomic models due to differences in training data or biological factors between populations, such as race and gender. 183 This variation can potentially lead to social injustices. Therefore, it is important to effectively control bias in the training data and adequately assess and specify the scope of model applicability. 184 Finally, the open source of radiogenomics modeling code and the sharing of resources may facilitate the reproduction and further validation of modeling results by other researchers.
6.2. Explainability and interpretability
Although there are arguments that explainability of artificial intelligence (AI) is impractical, while rigorous validation of model efficacy and robustness is even more important. 185 However, we still believe that explainability of medical AI is critical and worth working towards. As radiogenomics models have become progressively complex to achieve greater predictive power, explainability decreased. DL is considered a “black box,” causing concern that it may make mistakes in complex clinical scenarios that exceed expectations, which can have a significant impact on medical decision‐making. As a tool, it is important for medical professionals to understand the scope of its use, the mistakes it can make, and the corresponding solutions. 186 In radiogenomics, there are different ways to achieve explainability and interpretability of models. One is in the feature extraction stage, where algorithms are used to extract features that reflect specific biological information about the cancer, and the association of this information with the prediction task is comprehensible, making the models constructed from features carrying biological information highly interpretable. For example, traditional radiomic features are used to quantify tumor morphology, signal intensity, and heterogeneity information, 187 and brain network features reflect damage to white matter fiber tracts. 16 DL model construction guided by biological information also has higher interpretability than direct models. 188 Second, some researchers have evaluated the model results by feature attribution (e.g., Shapley Additive Explanation), attention‐based (e.g., Class Activation Map), and example‐based methods to verify whether the model has an understandable inference mechanism. 120 , 189 These initiatives allow researchers to detect and control how radiogenomics models are performed, ensuring that they are always aligned with our clinical goals.
6.3. Data hunger
Radiogenomics increasingly craves for adequate matched images and multiomics data support. Molecular data on cancer are inherently limited due to its clinical importance, acquisition costs, and patient affordability. Fortunately, the availability of large‐volume omics and image data through increasing open datasets in recent years has facilitated the development of radiogenomics. 190 The Cancer Imaging Archive (TCIA) and The Cancer Genome Atlas (TCGA) are leading the way. TCIA is an extensive repository of medical images, including CT, MRI, and digital histopathology images, specifically curated for cancer research, while TCGA has cataloged genomic, epigenomic, transcriptomic, and proteomic data from thousands of cancer patients across more than 30 different cancer types. Importantly, their data are correlated. For many of the patients included in TCGA, corresponding imaging data are available in TCIA, allowing researchers to correlate omics and other molecular profiles with radiomics. TCIA and TCGA have supported numerous high‐impact radiogenomic studies that have led to new insights into cancer biology, improved diagnostic techniques, and the development of targeted therapies. Additionally, the privacy and data protection efforts of both TCIA and TCGA are exemplary for other data‐sharing projects. 191 , 192 , 193
Besides, cross‐institutional and even international collaborations are also significant in providing sufficiently heterogeneous data. Yet the distribution of costs and benefits of research and concerns about privacy are major obstacles. The establishment of equitable and mutually beneficial cooperative agreements to share research data, equipment and scientific findings facilitates reliable and stable partnerships. By adopting data anonymization techniques, such as differential privacy and federated learning, potential risks related to patient privacy during data sharing can be minimized. 194 , 195 , 196
Additionally, even with widely used imaging modalities, imaging data can be incomplete due to inconsistent imaging protocols or poor data management. Furthermore, the imbalance in data exacerbates the impact of overall insufficient data, making it challenging to train effective predictive models. Data augmentation algorithms and generative AI can be used to generate high‐quality synthetic data to compensate for unbalanced or incomplete data, enabling more effective model training. 197 , 198 Moreover, the utilization of DL algorithms such as transfer learning and self‐supervised learning holds the potential to fully leverage pretrained base models or large amounts of unsupervised data, resulting in high‐quality predictive models despite limited target samples. 15 , 199 , 200
6.4. Spatiotemporal registration in radiogenomics
The spatial and temporal genetic heterogeneity of cancers has been extensively studied and well documented. While imaging has shown potential in identifying these heterogeneities in some preliminary studies, there are still significant challenges in the field of radiogenomics, particularly in obtaining sufficient spatially and temporally based molecular data and aligning them with images. The ability to perform virtual biopsies of multiple regions or lesions and continuously monitor molecular changes within tumors is still a major challenge in terms of experimental design and execution. Fortunately, advances in spatiotemporal omics and liquid biopsy technology hold promise for the acquisition of spatiotemporal molecular data of cancers, and may present significant opportunities for decoding of spatiotemporal heterogeneity of cancers using radiogenomics.
6.5. Diversity of radiology
The key to radiogenomics lies in identifying valuable features that can accurately predict clinical outcomes. Although DL models automatically learn representations of image features and show superior results to classical radiomics in various tasks, hand‐crafted radiomics carry advantages including less data dependency, the ability to incorporate clinical prior knowledge, and higher interpretability. By integrating DL with radiomics, we can fully leverage the potential of imaging data. Furthermore, data obtained from multiple anatomical scales and multimodalities can provide multidimensional features that enable the characterization of cancer molecules and prognosis, with the potential to amplify the signals of specific molecules and microenvironment components.
Medical images commonly used are typically designed and generated for human visual interpretation. However, when it comes to radiomics, it is important to consider the differences between computer vision and human vision. These traditional forms of input may not always be optimal for achieving accurate and consistent outputs. A notable example is the need for quantization processing on medical images in classical radiomics, which is often performed prior to feature extraction. In recent years, the emergence of MR fingerprint technology has enabled the translation of current visual and qualitative MRI diagnostic criteria into a quantitative acquisition and analysis framework. 201 Additionally, raw data obtained from imaging equipment may contain biological information that can be utilized for computer processing, as opposed to medical images that have undergone graphical manipulations to enhance readability. 50 By altering the approach to image collection and processing, making them more suitable for ML, we can further advance the development of radiogenomics.
6.6. Multimodal AI
Finally, the integration and analysis of multiple types of medical data are considered crucial for advancing precision oncology. It is necessary to introduce multimodal ML algorithms capable of handling various data types to replace the linear regression methods commonly used for combining radiomic and molecular data. Additionally, the medical community has been inspired by LLMs and derived multimodal AI agents, which have gained significant attention. 202 , 203 In recent studies, multimodal LLMs can simultaneously interpret text and images to generate reports, closely mimicking current diagnostic pathways in radiology. 204 It is foreseeable that multimodal LLMs can be integrated in a variety of real‐world medical support scenarios including preoperative biomarker profiling of tumors with images, and postoperative clinical decision making based on radiogenomics, in a natural language‐based interaction. This process can be based on specific predictive capabilities obtained after targeted training, or the ability to call appropriate validated radiogenomics models autonomously, as possessed by the AI agent. In the future, AI agents based on medical foundation models may represent an ideal form of efficient automated precision medicine. These agents can assist in integrating patients' medical data and invoking appropriate validated specialized models for tasks such as patient counseling, disease classification, prognosis, and decision‐making. 205
7. CONCLUSIONS AND PROSPECTS
Recent high‐quality studies provide compelling evidence of its clinical utility by bridging fundamental research findings with precision medicine applications. Although there are still non‐negligible problems to overcome in radiogenomics. For example, the massive amounts of high‐quality imaging and biomolecular data for constructing strong enough radiogenomics models are still insufficient. Accurate matching of imaging genomics and multiomics data in time and space still faces great challenges at the technical level.
Nationally driven large‐scale public databases and collaborative projects, as well as the development of generative AI for virtual data synthesis, are expected to greatly facilitated development by consistently providing access to heterogeneous imaging and genomic data at scale. Incorporating structural, functional, and molecular radiomic at various anatomical scale levels and multiomics information from tissue and liquid samples enables more comprehensive and time‐critical tumor characterization, understanding, and prediction, providing determining basis for precision oncology. Advances in AI algorithms, such as attention mechanisms, transfer learning, and self‐supervised learning, are striving to utilize variable‐quality but highly accessible data to break through the limitation of insufficient high‐quality data. What is more, with the help of multimodal LLMs, it is expected to facilitate the efficient implementation of the latest results of radiogenomics into the existing clinical workflow.
Above all, standards for rigorous model evaluation are essential to assess validity, generalizability and clinical applicability. The promise of radiogenomics still warrants more real‐world validation and evaluation of its efficacy in diverse populations to ensure that these technologies are equitably delivering benefits to patients. Only through such evaluation can radiogenomic models be reliably applied to improve patient outcomes. Continued research efforts are moving the field closer to realizing radiogenomics' full potential for precision oncology.
AUTHOR CONTRIBUTIONS
We confirm contribution to the paper as follows. Study conception: Bin Zhang and Shuixing Zhang. Literature review: Wenle He, Bin Zhang, Shuixing Zhang, Xuewei Wu, Wenhui Huang, and Lu Zhang. Draft manuscript preparation: Wenhui Huang, Bin Zhang, and Shuixing Zhang. All authors reviewed the results and approved the final version of the manuscript.
CONFLICT OF INTEREST STATEMENT
The authors declare that they have no conflict of interest.
ETHICS STATEMENT
Not applicable.
ACKNOWLEDGMENTS
We thank draw.io for providing the tools used to create Figures 1 and 2. All other images and elements are original works created by the authors. This work was supported by grants from the National Key Research and Development Program of China (Grant number: 2023YFF1204600); the National Natural Science Foundation of China (Grant number: 82227802, 82302306, 82302336); the Science and Technology Projects in Guangzhou (Grant number: 202201020022, 2023A03J1036, 2023A03J1038); the Science and Technology Youth Talent Nurturing Program of Jinan University (Grant number: 21623209); and the Postdoctoral Science Foundation of China (Grant number: 2022M721349).
He W, Huang W, Zhang L, Wu X, Zhang S, Zhang B. Radiogenomics: Bridging the gap between imaging and genomics for precision oncology. MedComm. 2024;5:e722. 10.1002/mco2.722
Contributor Information
Shuixing Zhang, Email: shui7515@126.com.
Bin Zhang, Email: xld_Jane_Eyre@126.com.
DATA AVAILABILITY STATEMENT
Not applicable.
REFERENCESx
- 1. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209‐249. [DOI] [PubMed] [Google Scholar]
- 2. Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458(7239):719‐724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. De Visser KE, Joyce JA. The evolving tumor microenvironment: from cancer initiation to metastatic outgrowth. Cancer Cell. 2023;41(3):374‐403. [DOI] [PubMed] [Google Scholar]
- 4. Akhoundova D, Rubin MA. Clinical application of advanced multi‐omics tumor profiling: shaping precision oncology of the future. Cancer Cell. 2022;40(9):920‐938. [DOI] [PubMed] [Google Scholar]
- 5. Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278(2):563‐577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lambin P, Leijenaar RTH, Deist TM, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14(12):749‐762. [DOI] [PubMed] [Google Scholar]
- 7. Pinker K, Chin J, Melsaether AN, Morris EA, Moy L. Precision medicine and radiogenomics in breast cancer: new approaches toward diagnosis and treatment. Radiology. 2018;287(3):732‐747. [DOI] [PubMed] [Google Scholar]
- 8. Hartmann K, Sadée CY, Satwah I, Carrillo‐Perez F, Gevaert O. Imaging genomics: data fusion in uncovering disease heritability. Trends Mol Med. 2023;29(2):141‐151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li ZC, Yan J, Zhang S, et al. Glioma survival prediction from whole‐brain MRI without tumor segmentation using deep attention network: a multicenter study. Eur Radiol. 2022;32(8):5719‐5729. [DOI] [PubMed] [Google Scholar]
- 10. Wang S, Yu H, Gan Y, et al. Mining whole‐lung information by artificial intelligence for predicting EGFR genotype and targeted therapy response in lung cancer: a multicohort study. Lancet Digit Health. 2022;4(5):e309‐e319. [DOI] [PubMed] [Google Scholar]
- 11. Van Griethuysen JJM, Fedorov A, Parmar C, et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77(21):e104‐e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Van der Voort SR, Incekara F, Wijnenga MMJ, et al. Combined molecular subtyping, grading, and segmentation of glioma using multi‐task deep learning. Neuro‐Oncol. 2023;25(2):279‐289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Decuyper M, Bonte S, Deblaere K, Van Holen R. Automated MRI based pipeline for segmentation and prediction of grade, IDH mutation and 1p19q co‐deletion in glioma. Comput Med Imaging Graph. 2021;88:101831. October 2020. [DOI] [PubMed] [Google Scholar]
- 14. Zhang B, Wu X, Zhang S, et al. Biologically interpretable multi‐task deep learning pipeline predicts molecular alterations, grade, and prognosis in glioma patients. Published online February 20, 2024. [DOI] [PMC free article] [PubMed]
- 15. Wei Y, Chen X, Zhu L, et al. Multi‐modal learning for predicting the genotype of glioma. IEEE Trans Med Imaging. 2023. PP. [DOI] [PubMed] [Google Scholar]
- 16. Yang N, Xiao X, Gu G, et al. Diffusion MRI‐based connectomics features improve the noninvasive prediction of H3K27M mutation in brainstem gliomas. Radiother Oncol. 2023;186:109789. [DOI] [PubMed] [Google Scholar]
- 17. Ismail M, Prasanna P, Bera K, et al. Radiomic deformation and textural heterogeneity (R‐depth) descriptor to characterize tumor field effect: application to survival prediction in glioblastoma. IEEE Trans Med Imaging. 2022;41(7):1764‐1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu X, Zhang Q, Li J, et al. Coordinatized lesion location analysis empowering ROI‐based radiomics diagnosis on brain gliomas. Eur Radiol. 2023;33(12):8776‐8787. [DOI] [PubMed] [Google Scholar]
- 19. Baysoy A, Bai Z, Satija R, Fan R. The technological landscape and applications of single‐cell multi‐omics. Nat Rev Mol Cell Biol. 2023;24(10):695‐713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nam AS, Chaligne R, Landau DA. Integrating genetic and non‐genetic determinants of cancer evolution by single‐cell multi‐omics. Nat Rev Genet. 2021;22(1):3‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Siravegna G, Marsoni S, Siena S, Bardelli A. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol. 2017;14(9):531‐548. [DOI] [PubMed] [Google Scholar]
- 22. Passaro A, Al Bakir M, Hamilton EG, et al. Cancer biomarkers: emerging trends and clinical implications for personalized treatment. Cell. 2024;187(7):1617‐1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li MM, Cottrell CE, Pullambhatla M, et al. Assessments of somatic variant classification using the Association for Molecular Pathology/American Society of Clinical Oncology/College of American Pathologists Guidelines: a Report from the Association for Molecular Pathology. J Mol Diagn. 2023;25(2):69‐86. [DOI] [PubMed] [Google Scholar]
- 24. Jose A, Kulkarni P, Thilakan J, et al. Integration of pan‐omics technologies and three‐dimensional in vitro tumor models: an approach toward drug discovery and precision medicine. Mol Cancer. 2024;23(1):50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bressan D, Battistoni G, Hannon GJ. The dawn of spatial omics. Science. 2023;381(6657):eabq4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nikanjam M, Kato S, Kurzrock R. Liquid biopsy: current technology and clinical applications. J Hematol Oncol. 2022;15(1):131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang H, Zhang Y, Zhang H, et al. Liquid biopsy for human cancer: cancer screening, monitoring, and treatment. Medcomm. 2024;5(6):e564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Holder AM, Dedeilia A, Sierra‐Davidson K, et al. Defining clinically useful biomarkers of immune checkpoint inhibitors in solid tumours. Nat Rev Cancer. 2024;24(7):498‐512. [DOI] [PubMed] [Google Scholar]
- 29. Kagawa Y, Smith JJ, Fokas E, et al. Future direction of total neoadjuvant therapy for locally advanced rectal cancer. Nat Rev Gastroenterol Hepatol. 2024;21(6):444‐455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dagogo‐Jack I, Shaw AT. Tumour heterogeneity and resistance to cancer therapies. Nat Rev Clin Oncol. 2018;15(2):81‐94. [DOI] [PubMed] [Google Scholar]
- 31. Liang Y, Zhang H, Song X, Yang Q. Metastatic heterogeneity of breast cancer: molecular mechanism and potential therapeutic targets. Semin Cancer Biol. 2020;60:14‐27. [DOI] [PubMed] [Google Scholar]
- 32. Zhu S, Lu Y, Fei X, Shen K, Chen X. Pathological complete response, category change, and prognostic significance of HER2‐low breast cancer receiving neoadjuvant treatment: a multicenter analysis of 2489 cases. Br J Cancer. 2023;129(8):1274‐1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang L, Wang R, Gao J, et al. A novel MRI‐based deep learning networks combined with attention mechanism for predicting CDKN2A/B homozygous deletion status in IDH‐mutant astrocytoma. Eur Radiol. 2024;34(1):391‐399. [DOI] [PubMed] [Google Scholar]
- 34. Ma C, Wang L, Song D, et al. Multimodal‐based machine learning strategy for accurate and non‐invasive prediction of intramedullary glioma grade and mutation status of molecular markers: a retrospective study. BMC Med. 2023;21(1):198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Duan J, Zhang Z, Chen Y, et al. Imaging phenotypes from MRI for the prediction of glioma immune subtypes from RNA sequencing: a multicenter study. Mol Oncol. 2023;17(4):629‐646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang L, Pan H, Liu Z, et al. Multicenter clinical radiomics‐integrated model based on [(18)F]FDG PET and multi‐modal MRI predict ATRX mutation status in IDH‐mutant lower‐grade gliomas. Eur Radiol. 2023;33(2):872‐883. [DOI] [PubMed] [Google Scholar]
- 37. Sha Y, Yan Q, Tan Y, Wang X, Zhang H, Yang G. Prediction of the molecular subtype of IDH mutation combined with MGMT promoter methylation in gliomas via radiomics based on preoperative MRI. Cancers. 2023;15(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kim BH, Lee H, Choi KS, et al. Validation of MRI‐based models to predict MGMT promoter methylation in gliomas: brats 2021 radiogenomics challenge. Cancers. 2022;14(19). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cluceru J, Interian Y, Phillips JJ, et al. Improving the noninvasive classification of glioma genetic subtype with deep learning and diffusion‐weighted imaging. Neuro‐Oncol. 2022;24(4):639‐652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wu C, Zheng H, Li J, et al. MRI‐based radiomics signature and clinical factor for predicting H3K27M mutation in pediatric high‐grade gliomas located in the midline of the brain. Eur Radiol. 2022;32(3):1813‐1822. [DOI] [PubMed] [Google Scholar]
- 41. Soldatelli MD, Namdar K, Tabori U, et al. Identification of multiclass pediatric low‐grade neuroepithelial tumor molecular subtype with ADC MR imaging and machine learning. AJNR Am J Neuroradiol. 2024;45(6):753‐760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li Y, Wei D, Liu X, et al. Molecular subtyping of diffuse gliomas using magnetic resonance imaging: comparison and correlation between radiomics and deep learning. Eur Radiol. 2022;32(2):747‐758. [DOI] [PubMed] [Google Scholar]
- 43. Zhuo Z, Qu L, Zhang P, et al. Prediction of H3K27M‐mutant brainstem glioma by amide proton transfer‐weighted imaging and its derived radiomics. Eur J Nucl Med Mol Imaging. 2021;48(13):4426‐4436. [DOI] [PubMed] [Google Scholar]
- 44. Li Z, Kaiser L, Holzgreve A, et al. Prediction of tertp‐mutation status in IDH‐wildtype high‐grade gliomas using pre‐treatment dynamic [18F]FET PET radiomics. Eur J Nucl Med Mol Imaging. 2021;48(13):4415‐4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Beig N, Singh S, Bera K, et al. Sexually dimorphic radiogenomic models identify distinct imaging and biological pathways that are prognostic of overall survival in glioblastoma. Neuro‐Oncol. 2021;23(2):251‐263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yan J, Zhang B, Zhang S, et al. Quantitative MRI‐based radiomics for noninvasively predicting molecular subtypes and survival in glioma patients. NPJ Precis Oncol. 2021;5(1):72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Qian J, Herman MG, Brinkmann DH, et al. Prediction of MGMT status for glioblastoma patients using radiomics feature extraction from 18F‐DOPA‐PET imaging. Int J Radiat Oncol Biol Phys. 2020;108(5):1339‐1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Park JE, Kim HS, Park SY, et al. Prediction of core signaling pathway by using diffusionand perfusion‐based mri radiomics and next‐generation sequencing in isocitrate dehydrogenase wild‐type glioblastoma. Radiology. 2020;294(2):388‐397. [DOI] [PubMed] [Google Scholar]
- 49. Buda M, Albadawy EA, Saha A, Mazurowski MA. Deep radiogenomics of lower‐grade gliomas: convolutional neural networks predict tumor genomic subtypes using MR images. Radiol Artif Intell. 2020;2(1):e180050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Choi KS, Choi SH, Jeong B. Prediction of IDH genotype in gliomas with dynamic susceptibility contrast perfusion MR imaging using an explainable recurrent neural network. Neuro‐Oncol. 2019;21(9):1197‐1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zinn PO, Singh SK, Kotrotsou A, et al. A coclinical radiogenomic validation study: conserved magnetic resonance radiomic appearance of periostin‐expressing glioblastoma in patients and xenograft models. Clin Cancer Res. 2018;24(24):6288‐6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yan J, Sun Q, Tan X, et al. Image‐based deep learning identifies glioblastoma risk groups with genomic and transcriptomic heterogeneity: a multi‐center study. Eur Radiol. 2023;33(2):904‐914. [DOI] [PubMed] [Google Scholar]
- 53. Yan J, Zhao Y, Chen Y, et al. Deep learning features from diffusion tensor imaging improve glioma stratification and identify risk groups with distinct molecular pathway activities. Ebiomedicine. 2021;72:103583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang J, Zheng X, Zhang J, et al. An MRI‐based radiomics signature as a pretreatment noninvasive predictor of overall survival and chemotherapeutic benefits in lower‐grade gliomas. Eur Radiol. 2021;31(4):1785‐1794. [DOI] [PubMed] [Google Scholar]
- 55. Guo J, Fathi Kazerooni A, Toorens E, et al. Integrating imaging and genomic data for the discovery of distinct glioblastoma subtypes: a joint learning approach. Sci Rep. 2024;14(1):4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sun Y, Zhang Y, Gan J, et al. Comprehensive quantitative radiogenomic evaluation reveals novel radiomic subtypes with distinct immune pattern in glioma. Comput Biol Med. 2024;177:108636. [DOI] [PubMed] [Google Scholar]
- 57. Chen M, Lu H, Copley SJ, et al. A novel radiogenomics biomarker for predicting treatment response and pneumotoxicity from programmed cell death protein or ligand‐1 inhibition immunotherapy in NSCLC. J Thorac Oncol. 2023;18(6):718‐730. [DOI] [PubMed] [Google Scholar]
- 58. Ju HM, Kim BC, Lim I, Byun BH, Woo SK. Estimation of an image biomarker for distant recurrence prediction in NSCLC using proliferation‐related genes. Int J Mol Sci. 2023;24(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shao J, Ma J, Zhang S, et al. Radiogenomic system for non‐invasive identification of multiple actionable mutations and PD‐L1 expression in non‐small cell lung cancer based on CT images. Cancers. 2022;14(19):1‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Shiri I, Amini M, Nazari M, et al. Impact of feature harmonization on radiogenomics analysis: prediction of EGFR and KRAS mutations from non‐small cell lung cancer PET/CT images. Comput Biol Med. 2022;142:105230. [DOI] [PubMed] [Google Scholar]
- 61. Wang C, Ma J, Shao J, et al. Non‐invasive measurement using deep learning algorithm based on multi‐source features fusion to predict PD‐L1 expression and survival in NSCLC. Front Immunol. 2022;13:828560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wang C, Ma J, Shao J, et al. Predicting EGFR and PD‐L1 status in NSCLC patients using multitask AI system based on CT images. Front Immunol. 2022;13:813072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yang X, Fang C, Li C, et al. Can CT radiomics detect acquired T790M mutation and predict prognosis in advanced lung adenocarcinoma with progression after first‐ or second‐generation EGFR tkis? Front Oncol. 2022;12(July):1‐10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. She Y, He B, Wang F, et al. Deep learning for predicting major pathological response to neoadjuvant chemoimmunotherapy in non‐small cell lung cancer: a multicentre study. Ebiomedicine. 2022;86:104364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Perez‐Johnston R, Araujo‐Filho JA, Connolly JG, et al. CT‐based radiogenomic analysis of clinical stage I lung adenocarcinoma with histopathologic features and oncologic outcomes. Radiology. 2022;303(3):664‐672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Song Z, Liu T, Shi L, et al. The deep learning model combining CT image and clinicopathological information for predicting ALK fusion status and response to ALK‐TKI therapy in non‐small cell lung cancer patients. Eur J Nucl Med Mol Imaging. 2021;48(2):361‐371. [DOI] [PubMed] [Google Scholar]
- 67. Tian P, He B, Mu W, et al. Assessing PD‐L1 expression in non‐small cell lung cancer and predicting responses to immune checkpoint inhibitors using deep learning on computed tomography images. Theranostics. 2021;11(5):2098‐2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Mu W, Jiang L, Zhang JY, et al. Non‐invasive decision support for NSCLC treatment using PET/CT radiomics. Nat Commun. 2020;11(1):5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. He B, Dong D, She Y, et al. Predicting response to immunotherapy in advanced non‐small‐cell lung cancer using tumor mutational burden radiomic biomarker. J Immunother Cancer. 2020;8(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Park C, Na KJ, Choi H, et al. Tumor immune profiles noninvasively estimated by FDG PET with deep learning correlate with immunotherapy response in lung adenocarcinoma. Theranostics. 2020;10(23):10838‐10848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ju H, Kim K, Kim BI, Woo SK. Graph neural network model for prediction of non‐small cell lung cancer lymph node metastasis using protein‐protein interaction network and (18)F‐FDG PET/CT radiomics. Int J Mol Sci. 2024;25(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Guo Y, Xie X, Tang W, et al. Noninvasive identification of HER2‐low‐positive status by MRI‐based deep learning radiomics predicts the disease‐free survival of patients with breast cancer. Eur Radiol. 2024;. 34(2):899‐913. [DOI] [PubMed] [Google Scholar]
- 73. Liu HQ, Lin SY, Song YD, et al. Machine learning on MRI radiomic features: identification of molecular subtype alteration in breast cancer after neoadjuvant therapy. Eur Radiol. 2023;33(4):2965‐2974. [DOI] [PubMed] [Google Scholar]
- 74. Jiang L, You C, Xiao Y, et al. Radiogenomic analysis reveals tumor heterogeneity of triple‐negative breast cancer. Cell Rep Med. 2022;3(7):100694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lee JY, Lee KS, Seo BK, et al. Radiomic machine learning for predicting prognostic biomarkers and molecular subtypes of breast cancer using tumor heterogeneity and angiogenesis properties on MRI. Eur Radiol. 2022;32(1):650‐660. [DOI] [PubMed] [Google Scholar]
- 76. Ming W, Li F, Zhu Y, et al. Predicting hormone receptors and PAM50 subtypes of breast cancer from multi‐scale lesion images of DCE‐MRI with transfer learning technique. Comput Biol Med. 2022;150:106147. [DOI] [PubMed] [Google Scholar]
- 77. Petrillo A, Fusco R, Di Bernardo E, et al. Prediction of breast cancer histological outcome by radiomics and artificial intelligence analysis in contrast‐enhanced mammography. Cancers. 2022;14(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Jeon SH, Kim SW, Na K, Seo M, Sohn YM, Lim YJ. Radiomic models based on magnetic resonance imaging predict the spatial distribution of CD8+ tumor‐infiltrating lymphocytes in breast cancer. Front Immunol. 2022;13(December):1‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Fan M, Yuan C, Huang G, et al. A framework for deep multitask learning with multiparametric magnetic resonance imaging for the joint prediction of histological characteristics in breast cancer. IEEE J Biomed Health Inform. 2022;26(8):3884‐3895. [DOI] [PubMed] [Google Scholar]
- 80. Su GH, Xiao Y, Jiang L, et al. Radiomics features for assessing tumor‐infiltrating lymphocytes correlate with molecular traits of triple‐negative breast cancer. J Transl Med. 2022;20(1):471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Jiang M, Zhang D, Tang SCC, et al. Deep learning with convolutional neural network in the assessment of breast cancer molecular subtypes based on US images: a multicenter retrospective study. Eur Radiol. 2021;31(6):3673‐3682. [DOI] [PubMed] [Google Scholar]
- 82. Han X, Cao W, Wu L, Liang C. Radiomics assessment of the tumor immune microenvironment to predict outcomes in breast cancer. Front Immunol. 2022;12:1‐9. January. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Lo Gullo R, Wen H, Reiner JS, et al. Assessing pd‐l1 expression status using radiomic features from contrast‐enhanced breast mri in breast cancer patients: initial results. Cancers. 2021;13(24):1‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Lv T, Hong X, Liu Y, et al. AI‐powered interpretable imaging phenotypes noninvasively characterize tumor microenvironment associated with diverse molecular signatures and survival in breast cancer. Comput Methods Programs Biomed. 2024;243:107857. [DOI] [PubMed] [Google Scholar]
- 85. Fan M, Zhang P, Wang Y, et al. Radiomic analysis of imaging heterogeneity in tumours and the surrounding parenchyma based on unsupervised decomposition of DCE‐MRI for predicting molecular subtypes of breast cancer. Eur Radiol. 2019;29(8):4456‐4467. [DOI] [PubMed] [Google Scholar]
- 86. Quan MY, Huang YX, Wang CY, Zhang Q, Chang C, Zhou SC. Deep learning radiomics model based on breast ultrasound video to predict HER2 expression status. Front Endocrinol. 2023;14:1144812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Zhang T, Tan T, Han L, et al. Predicting breast cancer types on and beyond molecular level in a multi‐modal fashion. NPJ Breast Cancer. 2023;9(1):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Zhang M, Wang C, Cai L, et al. Developing a weakly supervised deep learning framework for breast cancer diagnosis with HR status based on mammography images. Comput Struct Biotechnol J. 2023;22:17‐26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Fan M, Wang K, Pan D, et al. Radiomic analysis reveals diverse prognostic and molecular insights into the response of breast cancer to neoadjuvant chemotherapy: a multicohort study. J Transl Med. 2024;22(1):637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Zhao J, Sun Z, Yu Y, et al. Radiomic and clinical data integration using machine learning predict the efficacy of anti‐PD‐1 antibodies‐based combinational treatment in advanced breast cancer: a multicentered study. J Immunother Cancer. 2023;11(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Zhang Y, You C, Pei Y, et al. Integration of radiogenomic features for early prediction of pathological complete response in patients with triple‐negative breast cancer and identification of potential therapeutic targets. J Transl Med. 2022;20(1):256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Zhou J, Bai Y, Zhang Y, et al. A preoperative radiogenomic model based on quantitative heterogeneity for predicting outcomes in triple‐negative breast cancer patients who underwent neoadjuvant chemotherapy. Cancer Imaging. 2024;24(1):98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Lai J, Chen Z, Liu J, et al. A radiogenomic multimodal and whole‐transcriptome sequencing for preoperative prediction of axillary lymph node metastasis and drug therapeutic response in breast cancer: a retrospective, machine learning and international multicohort study. Int J Surg. 2024;110(4):2162‐2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Lu N, Guan X, Zhu J, Li Y, Zhang J. A contrast‐enhanced CT‐based deep learning system for preoperative prediction of colorectal cancer staging and RAS mutation. Cancers. 2023;15(18):4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Cao W, Hu H, Guo J, et al. CT‐based deep learning model for the prediction of DNA mismatch repair deficient colorectal cancer: a diagnostic study. J Transl Med. 2023;21(1):214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Lee H, Moon SH, Hong JY, Lee J, Hyun SH. A machine learning approach using FDG PET‐based radiomics for prediction of tumor mutational burden and prognosis in stage IV colorectal cancer. Cancers. 2023;15(15):3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Saber R, Henault D, Messaoudi N, et al. Radiomics using computed tomography to predict CD73 expression and prognosis of colorectal cancer liver metastases. J Transl Med. 2023;21(1):507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Shi R, Chen W, Yang B, et al. Prediction of KRAS, NRAS and BRAF status in colorectal cancer patients with liver metastasis using a deep artificial neural network based on radiomics and semantic features. Am J Cancer Res. 2020;10(12):4513‐4526. [PMC free article] [PubMed] [Google Scholar]
- 99. Zhong ME, Duan X, li Ni‐jia‐tiMyidi, et al. CT‐based radiogenomic analysis dissects intratumor heterogeneity and predicts prognosis of colorectal cancer: a multi‐institutional retrospective study. J Transl Med. 2022;20(1):574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Gao J, Ye F, Han F, Jiang H, Zhang J. A radiogenomics biomarker based on immunological heterogeneity for non‐invasive prognosis of renal clear cell carcinoma. Front Immunol. 2022;13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. He H, Xie Y, Song F, Feng Z, Rong P. Radiogenomic analysis based on lipid metabolism‐related subset for non‐invasive prediction for prognosis of renal clear cell carcinoma. Eur J Radiol. 2024;175:111433. [DOI] [PubMed] [Google Scholar]
- 102. Zeng H, Chen L, Wang M, Luo Y, Huang Y, Ma X. Integrative radiogenomics analysis for predicting molecular features and survival in clear cell renal cell carcinoma. Aging. 2021;13(7):9960‐9975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Morrison G, Buckley J, Ostrow D, et al. Non‐invasive profiling of advanced prostate cancer via multi‐parametric liquid biopsy and radiomic analysis. Int J Mol Sci. 2022;23(5):2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Ogbonnaya CN, Alsaedi BSO, Alhussaini AJ, et al. Radiogenomics map‐based molecular and imaging phenotypical characterization in localised prostate cancer using pre‐biopsy biparametric MR imaging. Int J Mol Sci. 2024;25(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Louis DN, Perry A, Wesseling P, et al. The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro‐Oncol. 2021;23(8):1231‐1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Weller M, Wen PY, Chang SM, et al. Glioma. Nat Rev Dis Primer. 2024;10(1):33. [DOI] [PubMed] [Google Scholar]
- 107. Pellerino A, Caccese M, Padovan M, Cerretti G, Lombardi G. Epidemiology, risk factors, and prognostic factors of gliomas. Clin Transl Imaging. 2022;10(5):467‐475. [Google Scholar]
- 108. Sanai N, Berger MS. Surgical oncology for gliomas: the state of the art. Nat Rev Clin Oncol. 2018;15(2):112‐125. [DOI] [PubMed] [Google Scholar]
- 109. Gempt J, Förschler A, Buchmann N, et al. Postoperative ischemic changes following resection of newly diagnosed and recurrent gliomas and their clinical relevance. J Neurosurg. 2013;118(4):801‐808. [DOI] [PubMed] [Google Scholar]
- 110. Pei D, Guan F, Hong X, et al. Radiomic features from dynamic susceptibility contrast perfusion‐weighted imaging improve the three‐class prediction of molecular subtypes in patients with adult diffuse gliomas. Eur Radiol. 2023;33(5):3455‐3466. [DOI] [PubMed] [Google Scholar]
- 111. Yoo RE, Yun TJ, Hwang I, et al. Arterial spin labeling perfusion‐weighted imaging aids in prediction of molecular biomarkers and survival in glioblastomas. Eur Radiol. 2020;30(2):1202‐1211. [DOI] [PubMed] [Google Scholar]
- 112. Wei Y, Li C, Cui Z, et al. Structural connectome quantifies tumour invasion and predicts survival in glioblastoma patients. Brain. 2023;146(4):1714‐1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Karremann M, Gielen GH, Hoffmann M, et al. Diffuse high‐grade gliomas with H3 K27M mutations carry a dismal prognosis independent of tumor location. Neuro‐Oncol. 2018;20(1):123‐131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Ryall S, Krishnatry R, Arnoldo A, et al. Targeted detection of genetic alterations reveal the prognostic impact of H3K27M and MAPK pathway aberrations in paediatric thalamic glioma. Acta Neuropathol Commun. 2016;4(1):93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Pan CC CUN, Liu J, Tang J, et al. A machine learning‐based prediction model of H3K27M mutations in brainstem gliomas using conventional MRI and clinical features. Radiother Oncol. 2019;130:172‐179. [DOI] [PubMed] [Google Scholar]
- 116. Su X, Chen N, Sun H, et al. Automated machine learning based on radiomics features predicts H3 K27M mutation in midline gliomas of the brain. Neuro‐Oncol. 2020;22(3):393‐401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Esteller M, Garcia‐Foncillas J, Andion E, et al. Inactivation of the DNA‐repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343(19):1350‐4. [DOI] [PubMed] [Google Scholar]
- 118. Li L, Xiao F, Wang S, et al. Preoperative prediction of MGMT promoter methylation in glioblastoma based on multiregional and multi‐sequence MRI radiomics analysis. Sci Rep. 2024;14(1):16031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Doniselli FM, Pascuzzo R, Mazzi F, et al. Quality assessment of the MRI‐radiomics studies for MGMT promoter methylation prediction in glioma: a systematic review and meta‐analysis. Eur Radiol. 2024;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Saeed N, Ridzuan M, Alasmawi H, Sobirov I, Yaqub M. MGMT promoter methylation status prediction using MRI scans? An extensive experimental evaluation of deep learning models. Med Image Anal. 2023;90:102989. [DOI] [PubMed] [Google Scholar]
- 121. Śledzińska P, Bebyn MG, Furtak J, Kowalewski J, Lewandowska MA. Prognostic and predictive biomarkers in gliomas. Int J Mol Sci. 2021;22(19):10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Yang K, Wu Z, Zhang H, et al. Glioma targeted therapy: insight into future of molecular approaches. Mol Cancer. 2022;21(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Haubold J, Hosch R, Parmar V, et al. Fully automated MR based virtual biopsy of cerebral gliomas. Cancers. 2021;13(24). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Verduin M, Primakov S, Compter I, et al. Prognostic and predictive value of integrated qualitative and quantitative magnetic resonance imaging analysis in glioblastoma. Cancers. 2021;13(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Tak D, Ye Z, Zapaischykova A, et al. Noninvasive molecular subtyping of pediatric low‐grade glioma with self‐supervised transfer learning. Radiol Artif Intell. 2024;6(3):e230333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Sun C, Jiang C, Wang X, Ma S, Zhang D, Jia W. MR‐based radiomics predicts CDK6 expression and prognostic value in high‐grade glioma. Acad Radiol. 2024;S1076‐6332(24):00364‐00367. Published online July 3. [DOI] [PubMed] [Google Scholar]
- 127. Zhao K, Zhang H, Lin J, et al. Radiomic prediction of CCND1 expression levels and prognosis in low‐grade glioma based on magnetic resonance imaging. Acad Radiol. 2024;S1076‐6332(24):00196‐X. Published online. [DOI] [PubMed] [Google Scholar]
- 128. Hu LS, Ning S, Eschbacher JM, et al. Radiogenomics to characterize regional genetic heterogeneity in glioblastoma. Neuro‐Oncol. 2017;19(1):128‐137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Xu PF, Li C, Chen YS, et al. Radiomics‐based survival risk stratification of glioblastoma is associated with different genome alteration. Comput Biol Med. 2023;159:106878. [DOI] [PubMed] [Google Scholar]
- 130. Luan J, Zhang D, Liu B, et al. Immune‐related lncrnas signature and radiomics signature predict the prognosis and immune microenvironment of glioblastoma multiforme. J Transl Med. 2024;22(1):107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Hu LS, Hawkins‐Daarud A, Wang L, Li J, Swanson KR. Imaging of intratumoral heterogeneity in high‐grade glioma. Cancer Lett. 2020;477:97‐106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Yoon JH, Kim H. CT radiomics in oncology: insights into the tumor microenvironment of hepatocellular carcinoma. Radiology. 2023;307(1). [DOI] [PubMed] [Google Scholar]
- 133. Choi YS, Ahn SS, Chang JH, et al. Machine learning and radiomic phenotyping of lower grade gliomas: improving survival prediction. Eur Radiol. 2020;30(7):3834‐3842. [DOI] [PubMed] [Google Scholar]
- 134. Jang BSS, Park AJ, Jeon SH, et al. Machine learning model to predict pseudoprogression versus progression in glioblastoma using mri: a multi‐institutional study (krog 18‐07). Cancers. 2020;12(9):1‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Leiter A, Veluswamy RR, Wisnivesky JP. The global burden of lung cancer: current status and future trends. Nat Rev Clin Oncol. 2023;20(9):624‐639. [DOI] [PubMed] [Google Scholar]
- 136. Thai AA, Solomon BJ, Sequist LV, Gainor JF, Heist RS. Lung cancer. The Lancet. 2021;398(10299):535‐554. [DOI] [PubMed] [Google Scholar]
- 137. Lahiri A, Maji A, Potdar PD, et al. Lung cancer immunotherapy: progress, pitfalls, and promises. Mol Cancer. 2023;22(1):40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Tan AC, Tan DSW. Targeted therapies for lung cancer patients with oncogenic driver molecular alterations. J Clin Oncol. 2022;40(6):611‐625. [DOI] [PubMed] [Google Scholar]
- 139. Blumenthal GM, Bunn PA, Chaft JE, et al. Current status and future perspectives on neoadjuvant therapy in lung cancer. J Thorac Oncol. 2018;13(12):1818‐1831. [DOI] [PubMed] [Google Scholar]
- 140. Arbour KC, Riely GJ. Systemic therapy for locally advanced and metastatic non–small cell lung cancer. JAMA. 2019;322(8):764. [DOI] [PubMed] [Google Scholar]
- 141. Da Cunha Santos G, Shepherd FA, Tsao MS. EGFR mutations and lung cancer. Annu Rev Pathol Mech Dis. 2011;6(1):49‐69. [DOI] [PubMed] [Google Scholar]
- 142. Blaquier JB, Ortiz‐Cuaran S, Ricciuti B, Mezquita L, Cardona AF, Recondo G. Tackling osimertinib resistance in EGFR‐mutant non‐small cell lung cancer. Clin Cancer Res. 2023;29(18):3579‐3591. [DOI] [PubMed] [Google Scholar]
- 143. Passaro A, Jänne PA, Mok T, Peters S. Overcoming therapy resistance in EGFR‐mutant lung cancer. Nat Cancer. 2021;2(4):377‐391. [DOI] [PubMed] [Google Scholar]
- 144. Chang C, Zhou S, Yu H, et al. A clinically practical radiomics‐clinical combined model based on PET/CT data and nomogram predicts EGFR mutation in lung adenocarcinoma. Eur Radiol. 2021;31(8):6259‐6268. [DOI] [PubMed] [Google Scholar]
- 145. Yang X, Liu M, Ren Y, et al. Using contrast‐enhanced CT and non‐contrast‐enhanced CT to predict EGFR mutation status in NSCLC patients—a radiomics nomogram analysis. Eur Radiol. 2022;32(4):2693‐2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Zhao W, Yang J, Ni B, et al. Toward automatic prediction of EGFR mutation status in pulmonary adenocarcinoma with 3D deep learning. Cancer Med. 2019;8(7):3532‐3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Wang S, Shi J, Ye Z, et al. Predicting EGFR mutation status in lung adenocarcinoma on computed tomography image using deep learning. Eur Respir J. 2019;53(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Wang S, Yu H, Gan Y, et al. Mining whole‐lung information by artificial intelligence for predicting EGFR genotype and targeted therapy response in lung cancer: a multicohort study. Lancet Digit Health. 2022;4(5):e309‐e319. [DOI] [PubMed] [Google Scholar]
- 149. Schneider JL, Lin JJ, Shaw AT. ALK‐positive lung cancer: a moving target. Nat Cancer. 2023;4(3):330‐343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Le NQK, Kha QH, Nguyen VH, Chen YC, Cheng SJ, Chen CY. Machine learning‐based radiomics signatures for EGFR and KRAS mutations prediction in non‐small‐cell lung cancer. Int J Mol Sci. 2021;22(17):9254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Mountzios G, Remon J, Hendriks LEL, et al. Immune‐checkpoint inhibition for resectable non‐small‐cell lung cancer — opportunities and challenges. Nat Rev Clin Oncol. 2023;20(10):664‐677. [DOI] [PubMed] [Google Scholar]
- 152. Mu W, Tunali I, Gray JE, Qi J, Schabath MB, Gillies RJ. Abstract 868: prediction of clinical benefit to checkpoint blockade in advanced NSCLC patients using radiomics of PET/CT images. Cancer Res. 2020;80(16_Supplement):868‐868.31772036 [Google Scholar]
- 153. Sun R, Limkin EJ, Vakalopoulou M, et al. A radiomics approach to assess tumour‐infiltrating CD8 cells and response to anti‐PD‐1 or anti‐PD‐L1 immunotherapy: an imaging biomarker, retrospective multicohort study. Lancet Oncol. 2018;19(9):1180‐1191. [DOI] [PubMed] [Google Scholar]
- 154. Hinzpeter R, Kulanthaivelu R, Kohan A, et al. Predictive [18F]‐FDG PET/CT‐based radiogenomics modelling of driver gene mutations in non‐small cell lung cancer. Acad Radiol. 2024;S1076‐6332(24):00423‐00429. Published online July 11. [DOI] [PubMed] [Google Scholar]
- 155. Li L, Duan J, Gao Y, et al. Multi‐omics predictive model based on clinical, radiomic and genomic features for predicting the response of limited‐stage small cell lung cancer to definitive chemoradiotherapy. Clin Transl Med. 2024;14(1):e1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Verma S, Magazzù G, Eftekhari N, et al. Cross‐attention enables deep learning on limited omics‐imaging‐clinical data of 130 lung cancer patients. Cell Rep Methods. 2024;4(7):100817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. De Miguel‐Perez D, Ak M, Mamindla P, et al. Validation of a multiomic model of plasma extracellular vesicle PD‐L1 and radiomics for prediction of response to immunotherapy in NSCLC. J Exp Clin Cancer Res CR. 2024;43(1):81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Huppert LA, Gumusay O, Idossa D, Rugo HS. Systemic therapy for hormone receptor‐positive/human epidermal growth factor receptor 2‐negative early stage and metastatic breast cancer. CA Cancer J Clin. 2023;73(5):480‐515. [DOI] [PubMed] [Google Scholar]
- 159. Loibl S, Poortmans P, Morrow M, Denkert C, Curigliano G. Breast cancer. The Lancet. 2021;397(10286):1750‐1769. [DOI] [PubMed] [Google Scholar]
- 160. Acciavatti RJ, Lee SH, Reig B, et al. Beyond breast density: risk measures for breast cancer in multiple imaging modalities. Radiology. 2023;306(3):e222575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Cui H, Sun Y, Zhao D, et al. Radiogenomic analysis of prediction HER2 status in breast cancer by linking ultrasound radiomic feature module with biological functions. J Transl Med. 2023;21(1):44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Nicosia L, Bozzini AC, Ballerini D, et al. Radiomic features applied to contrast enhancement spectral mammography: possibility to predict breast cancer molecular subtypes in a non‐invasive manner. Int J Mol Sci. 2022;23(23). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Bitencourt AGV, Gibbs P, Rossi Saccarelli C, et al. MRI‐based machine learning radiomics can predict HER2 expression level and pathologic response after neoadjuvant therapy in HER2 overexpressing breast cancer. Ebiomedicine. 2020;61:103042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Zhu Z, Albadawy E, Saha A, Zhang J, Harowicz MR, Mazurowski MA. Deep learning for identifying radiogenomic associations in breast cancer. Comput Biol Med. 2019;109:85‐90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Huang ZH, Chen L, Sun Y, Liu Q, Hu P. Conditional generative adversarial network driven radiomic prediction of mutation status based on magnetic resonance imaging of breast cancer. J Transl Med. 2024;22(1):226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Mathews JC, Nadeem S, Levine AJ, Pouryahya M, Deasy JO, Tannenbaum A. Robust and interpretable PAM50 reclassification exhibits survival advantage for myoepithelial and immune phenotypes. Npj Breast Cancer. 2019;5(1):30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Liang S, Xu S, Zhou S, et al. IMAGGS: a radiogenomic framework for identifying multi‐way associations in breast cancer subtypes. J Genet Genomics Yi Chuan Xue Bao. 2024;51(4):443‐453. [DOI] [PubMed] [Google Scholar]
- 168. Gallivanone F, Cava C, Corsi F, Bertoli G, Castiglioni I. In silico approach for the definition of radiomirnomic signatures for breast cancer differential diagnosis. Int J Mol Sci. 2019;20(23). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Harris MA, Savas P, Virassamy B, et al. Towards targeting the breast cancer immune microenvironment. Nat Rev Cancer. 2024;24(8):554‐577. [DOI] [PubMed] [Google Scholar]
- 170. Wang H, Ding XH, Liu CL, et al. Genomic alterations affecting tumor‐infiltrating lymphocytes and PD‐L1 expression patterns in triple‐negative breast cancer. J Natl Cancer Inst. 2023;115(12):1586‐1596. [DOI] [PubMed] [Google Scholar]
- 171. Han X, Gong Z, Guo Y, Tang W, Wei X. Exploration of a noninvasive radiomics classifier for breast cancer tumor microenvironment categorization and prognostic outcome prediction. Eur J Radiol. 2024;175:111441. [DOI] [PubMed] [Google Scholar]
- 172. Pesapane F, Rotili A, Botta F, et al. Radiomics of MRI for the prediction of the pathological response to neoadjuvant chemotherapy in breast cancer patients: a single referral centre analysis. Cancers. 2021;13(17). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Sutton EJ, Onishi N, Fehr DA, et al. A machine learning model that classifies breast cancer pathologic complete response on MRI post‐neoadjuvant chemotherapy. Breast Cancer Res. 2020;22(1):57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Fan M, Chen H, You C, et al. Radiomics of tumor heterogeneity in longitudinal dynamic contrast‐enhanced magnetic resonance imaging for predicting response to neoadjuvant chemotherapy in breast cancer. Front Mol Biosci. 2021;8:622219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Huang Y, Chen W, Zhang X, et al. Prediction of tumor shrinkage pattern to neoadjuvant chemotherapy using a multiparametric MRI‐based machine learning model in patients with breast cancer. Front Bioeng Biotechnol. 2021;9:662749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Kocak B, Durmaz ES, Ates E, Ulusan MB. Radiogenomics in clear cell renal cell carcinoma: machine learning–based high‐dimensional quantitative CT texture analysis in predicting PBRM1 mutation status. Am J Roentgenol. 2019;212(3):W55‐W63. [DOI] [PubMed] [Google Scholar]
- 177. Gao R, Pang J, Lin P, et al. Identification of clear cell renal cell carcinoma subtypes by integrating radiomics and transcriptomics. Heliyon. 2024;10(11):e31816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Gu J, Zhu J, Qiu Q, Wang Y, Bai T, Yin Y. Prediction of immunohistochemistry of suspected thyroid nodules by use of machine learning‐based radiomics. Am J Roentgenol. 2019;213(6):1348‐1357. [DOI] [PubMed] [Google Scholar]
- 179. Roy S, Whitehead TD, Quirk JD, et al. Optimal co‐clinical radiomics: sensitivity of radiomic features to tumour volume, image noise and resolution in co‐clinical T1‐weighted and T2‐weighted magnetic resonance imaging. Ebiomedicine. 2020;59:102963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Moher Alsady T, Voskrebenzev A, Behrendt L, et al. Multicenter standardization of phase‐resolved functional lung MRI in patients with suspected chronic thromboembolic pulmonary hypertension. J Magn Reson Imaging. 2024;59(6):1953‐1964. [DOI] [PubMed] [Google Scholar]
- 181. Whybra P, Zwanenburg A, Andrearczyk V, et al. The image biomarker standardization initiative: standardized convolutional filters for reproducible radiomics and enhanced clinical insights. Radiology. 2024;310(2):e231319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Varriano G, Guerriero P, Santone A, Mercaldo F, Brunese L. Explainability of radiomics through formal methods. Comput Methods Programs Biomed. 2022;220:106824. [DOI] [PubMed] [Google Scholar]
- 183. Liu M, Ning Y, Teixayavong S, et al. A translational perspective towards clinical AI fairness. Npj Digit Med. 2023;6(1):172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Andaur Navarro CL, Damen JAA, Takada T, et al. Risk of bias in studies on prediction models developed using supervised machine learning techniques: systematic review. BMJ. 2021:n2281. Published online October 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Ghassemi M, Oakden‐Rayner L, Beam AL. The false hope of current approaches to explainable artificial intelligence in health care. Lancet Digit Health. 2021;3(11):e745‐e750. [DOI] [PubMed] [Google Scholar]
- 186. Azodi CB, Tang J, Shiu SH. Opening the black box: interpretable machine learning for GenetICIsts. Trends Genet. 2020;36(6):442‐455. [DOI] [PubMed] [Google Scholar]
- 187. Tomaszewski MR, Gillies RJ. The biological meaning of radiomic features. Radiology. 2021;298(3):505‐516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Jiang Y, Zhang Z, Wang W, et al. Biology‐guided deep learning predicts prognosis and cancer immunotherapy response. Nat Commun. 2023;14(1):5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Liu Q, Hu P. Extendable and explainable deep learning for pan‐cancer radiogenomics research. Curr Opin Chem Biol. 2022;66:102111. [DOI] [PubMed] [Google Scholar]
- 190. Zanfardino M, Pane K, Mirabelli P, Salvatore M, Franzese M. TCGA‐TCIA impact on radiogenomics cancer research: a systematic review. Int J Mol Sci. 2019;20(23):6033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Prior F, Smith K, Sharma A, et al. The public cancer radiology imaging collections of The Cancer Imaging Archive. Sci Data. 2017;4(1):170124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Ganini C, Amelio I, Bertolo R, et al. Global mapping of cancers: the Cancer Genome Atlas and beyond. Mol Oncol. 2021;15(11):2823‐2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Kirby J, Tarbox L, Freymann J, Jaffe C, Prior F. TU‐AB‐BRA‐03: the cancer imaging archive: supporting radiomic and imaging genomic research with open‐access data sets. Med Phys. 2015;42(6Part31):3587‐3587. [Google Scholar]
- 194. Khalid N, Qayyum A, Bilal M, Al‐Fuqaha A, Qadir J. Privacy‐preserving artificial intelligence in healthcare: techniques and applications. Comput Biol Med. 2023;158:106848. [DOI] [PubMed] [Google Scholar]
- 195. Tang R, Liang H, Guo Y, et al. Pan‐mediastinal neoplasm diagnosis via nationwide federated learning: a multicentre cohort study. Lancet Digit Health. 2023;5(9):e560‐e570. [DOI] [PubMed] [Google Scholar]
- 196. Zhou J, Chen S, Wu Y, et al. PPML‐Omics: a privacy‐preserving federated machine learning method protects patients’ privacy in omic data. Sci Adv. 2024;10(5):eadh8601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Huang P, Li D, Jiao Z, et al. Common feature learning for brain tumor MRI synthesis by context‐aware generative adversarial network. Med Image Anal. 2022;79:102472. [DOI] [PubMed] [Google Scholar]
- 198. Ahmadian M, Bodalal Z, van der Hulst HJ, et al. Overcoming data scarcity in radiomics/radiogenomics using synthetic radiomic features. Comput Biol Med. 2024;174:108389. [DOI] [PubMed] [Google Scholar]
- 199. Hosny A, Parmar C, Quackenbush J, Schwartz LH, Aerts HJWL. Artificial intelligence in radiology. Nat Rev Cancer. 2018;18(8):500‐510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Mitra S. Deep learning with radiogenomics towards personalized management of gliomas. IEEE Rev Biomed Eng. 2023;16:579‐593. [DOI] [PubMed] [Google Scholar]
- 201. Tippareddy C, Onyewadume L, Sloan AE, et al. Novel 3D magnetic resonance fingerprinting radiomics in adult brain tumors: a feasibility study. Eur Radiol. 2023;33(2):836‐844. [DOI] [PubMed] [Google Scholar]
- 202. Wu X, Zhang B. Chatgpt promotes healthcare: current applications and potential challenges. Int J Surg. 2024;110(1):606‐608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Mehandru N, Miao BY, Almaraz ER, Sushil M, Butte AJ, Alaa A. Evaluating large language models as agents in the clinic. Npj Digit Med. 2024;7(1):84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Bhayana R. Chatbots and large language models in radiology: a practical primer for clinical and research applications. Radiology. 2024;310(1):e232756. [DOI] [PubMed] [Google Scholar]
- 205. Li J, Wang S, Zhang M, et al. Agent Hospital: A Simulacrum of Hospital with Evolvable Medical Agents. Published online May 5, 2024.
- 206. Hsu JBK, Lee GA, Chang TH, et al. Radiomic immunophenotyping of GSEA‐assessed immunophenotypes of glioblastoma and its implications for prognosis: a feasibility study. Cancers. 2020;12(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.


