Abstract
Colibacillosis, a bacterial disease caused by avian pathogenic E. coli (APEC), is a prevalent condition in the poultry industry, resulting in substantial economic losses annually. Previously, we identified PTEN as a crucial candidate gene that may play a significant role in chicken's immune response to APEC infection. Bioinformatics analysis indicated that the PTEN protein was unstable, hydrophilic and nuclear localization, with multiple putative phosphorylation sites and a high degree of similarity to duck and goose PTEN. Moreover, PTEN exhibited high expression levels in various tissues such as the stomach, cecum, small intestine, spleen, thymus, harderian gland, muscle, cerebrum, cerebellum, lung, and liver in comparison to heart tissue. Overexpression of PTEN resulted in a significant promotion of the expression level of pro-apoptosis genes and inflammatory mediators, as well as the production of NO, with or without APEC infection, which led to cellular injury. Furthermore, overexpression of PTEN was found to regulate the expression levels of autophagy related genes, regardless of APEC infection. Additionally, PTEN was a target gene of gga-miR-20a-5p and regulated by gga-miR-20a-5p upon APEC infection. Taken together, these findings establish a foundation for investigating the biological function of chicken PTEN, providing a potential target for future treatments against APEC infection as well as the breeding of genetically resistant poultry.
Key words: Avian pathogenic E. coli, PTEN, inflammatory response, autophagy, gga-miR-20a-5p
INTRODUCTION
Avian pathogenic E. coli (APEC), is a systemic disease caused by gram-negative bacteria that primarily affects chickens. APEC enters the birds' respiratory tract and spreads throughout the body, leading to various symptoms such as airsacculitis, perihepatitis, colisepticaemia, and mortality (Stokholm et al., 2010; Nolan et al., 2013). This condition poses a significant economic burden on poultry production worldwide, resulting in substantial losses each year. Currently, the primary methods used to combat APEC infections are antibiotic treatment and vaccination (Azam et al., 2020; Nguyen et al., 2021). However, due to the wide diversity of APEC strains, vaccines are only effective against specific strains, limiting their overall effectiveness. Prolonged antibiotic use also carries the risk of drug resistance development, making it challenging to control APEC infections. Therefore, exploring host genetic factors that confer resistance to APEC is an exciting research direction. Host genetic control of resistance is not restricted by bacterial serotypes and does not rely on external agents, offering a promising approach to enhance resistance against APEC infections.
Phosphatase and tensin homolog (PTEN) is a significant oncogenic factor involved in regulating crucial cellular processes such as cell growth, proliferation, and metabolism. For example, PTEN was able to regulate the development and functionality of T cells and B cells (Suzuki et al., 2003; Buckler et al., 2006; Shojaee et al., 2016). Moreover, Li et al. (2016) found that PTEN can participate in antiviral immunity by regulating the production of type I interferon (IFN) through IRF3. Additionally, PTEN acts as a vital antagonist of PI3K, modulating immune response via the PI3K/Akt/mTOR signaling pathway (Haddadi et al., 2018). These findings highlight the crucial role of PTEN in immune response. In previous study, we used chicken macrophages as experimental material and performed RNA sequencing before and after APEC infection (Sun et al., 2022). Results showed that thousands of differentially expressed genes were identified during APEC infection, of which PTEN emerged as key candidate gene. Further investigations are necessary to elucidate the specific functions of PTEN during APEC infection.
Recently, many researches have been demonstrated the significant role of miR-20a-5p in the chicken immune response. For instance, Su et al. (2021a; 2021b) observed a significant down-regulation of chicken miR-20a-5p in the bursa of Fabricius and spleen during avian influenza virus infection. Moreover, miR-20a-5p has been shown to regulate the immune response of chicken macrophages by targeting IFNGR2, MAPK1, MAP3K5, and MAP3K14 (Hong et al., 2023). Additionally, previous miRNA sequencing data showed a significant decrease expression level of chicken miR-20a-5p upon APEC infection (Yang et al., 2023). Bioinformatics analysis further suggests a target relationship between PTEN and miR-20a-5p. However, additional experimental verification is needed to determine whether PTEN is regulated by miR-20a-5p.
This study aimed to investigate the functions of PTEN on chicken macrophages immune response toward APEC infection and to elucidate the relationship between PTEN and miR-20a-5p. The findings of this research will enhance our understanding of the role of PTEN in chicken immune response to APEC infection and establish a theoretical foundation for the control of avian colibacillosis.
MATERIALS AND METHODS
Ethical Statement
The animal care procedures were conducted in strict compliance with the guidelines of the U.S. National Institute of Health (NIH Pub. No. 85-23, revised 1996). The experiments were approved by Ethics Committee of Yangzhou University for Laboratory and Experimental Animals (Permit Number: YZUDWSY, Government of Jiangsu Province, China).
Bioinformatics Analysis of PTEN
ProtParam (http://web.expasy.org/protparam/) (Gasteiger et al., 2005) was used to predict the molecular formula, molecular weight, isoelectric point (pI), and instability coefficient of the PTEN protein. SignalP 4.1 (http://www.cbs.dtu.dk/services/SignalP-4.1/) (Petersen et al., 2011) was performed to analyze the signal peptide of PTEN. The online software TMHMM 2.0 (http://www.cbs.dtu.dk/services/TMHMM/) (Möller et al., 2001) was utilized to evaluate the localization signal, secretory protein and protein transmembrane region. NetPhos 3.1 Server (http://www.cbs.dtu.dk/services/NetPhos/) (Blom et al., 1999) was used to predict the potential threonine, serine, or tyrosine phosphorylation sites of the protein. Potential O-glycosylation sites and potential N-glycosylation sites were separately predicted by the O-glycosylation sites (https://services.healthtech.dtu.dk/services/NetOGlyc-4.0/) (Steentoft et al., 2013) and NetNGlyc 3.1 Server (http://www.cbs.dtu.dk/services/NetNGlyc/). DNAMAN software was utilized to construct the phylogenetic tree by using the neighbor-joining method. The protein secondary structure, the conserved domain and 3-dimensional homology of PTEN were predicted by SOPMA and SWISS-MODEL (Waterhouse et al., 2018). Finally, the protein-protein interaction (PPI) network of the PTEN protein was analyzed by the STRING database.
Cell Culture
The chicken macrophage-like cell line HD11 was purchased from the American Type Culture Collection (ATCC, Manassas, Virginia, VA, U.S.A.). Chicken macrophages were cultured in DMEM medium supplemented with 10% fetal bovine serum (FBS). The macrophages were maintained in a cell culture incubator at a temperature of 37°C, with a 5% CO2 atmosphere and a humidity level of 60% to 70%.
Experimental Animals
One-day-old Rugao yellow chicks with uniform body weight were obtained from the Poultry Research Institute of the Chinese Academy of Agricultural Sciences (Yangzhou, China). These chicks were raised under standard housing conditions without receiving any vaccinations. A total of 8 chicks were euthanized by CO2 inhalation (Baker et al., 2020). Thirteen tissues, including the heart, liver, spleen, lung, stomach, cecum, small intestine, muscle, cerebrum, cerebellum, harderian gland, hypothalamus, and thymus, were collected. All harvested tissues were immediately immersed in RNA preservation solution and frozen at -80°C for subsequent total RNA extraction.
Total RNA Extraction and cDNA Synthesis
Total RNA from tissues or cells was extracted by TRIzol reagent (Life Technologies, Carlsbad, CA). The purity and concentration of the isolated total RNA were assessed by a NanoDrop-1000 micro nucleic acid analyzer. For reverse transcription, the total RNA from various experiments was converted into cDNA using the FastKing 1-Step Reverse Transcription-Fluorescence Quantitative Kit (Tiangen, Beijing, China). The reaction mixture volume was 20 μL, and the reverse transcription reaction was performed at 42°C for 15 min.
Real-Time Quantitative PCR
To evaluate the expression level of the candidate genes, Real-time quantitative (RT-qPCR) was performed using a SYBR Premix Ex Taq II kit (Takara, Dalian, China) following the manufacturer's instructions. The primer sequences for the candidate genes can be found in Table 1. The RT-qPCR reaction program consisted of an initial denaturation step at 95°C for 3 min, followed by 40 cycles of denaturation at 95°C for 10 s, annealing at 58°C for 30 s, and then 72°C for 30 s. RT-qPCR was performed by the Applied Biosystems 7500 Fast DX real-time PCR (ABI, Los Angeles, California, USA). The chicken β-actin gene was selected as the internal reference. The RT-qPCR data were analyzed using the 2–ΔΔCT method, with 3 technical replicates for each sample. The ΔΔCt value was calculated as (Ct of gene in test group - Ct of β-actin in test group) - (Ct of gene in control group - Ct of β-actin in control group).
Table 1.
Primers for candidate genes/miRNAs.
| Name | Forward (5′–3′) | Reverse (5′–3′) |
|---|---|---|
| PTEN (CDS) | ctggctagcgtttaaacttaATGGGTTTTCCTGCGGAG | tccaccacactggactagtgTCAGACTTTTGTAATCTGTGTATGC |
| PTEN (wild type 3′UTR) | GACGCGTAAACCCACGAAGAACGAC | CGAGCTCGACCAGGGATGCTTAGGG |
| PTEN (mutant 3′UTR) | GGAACTATAAATATGCCCATTTTATTCCAGTTTTATAAAAGTG | GGGCATATTTATAGTTCCTAATTGAAGTTTTAATGT |
| PTEN (RT-qPCR) | GCACTGAATGGGATGAGG | TTACGGAAACTGCTGGGT |
| β-actin | CAGCCAGCCATGGATGATGA | ACCAACCATCACACCCTGAT |
| IL1β | GCCGAGGAGCAGGGACTTT | ACTGTGAGCGGGTGTAGCG |
| IL8 | GAGTTCACTGACCACCCT | TGCCTGAGCCATACCTTT |
| IL6 | TTATGGAGAAGACCGTGAG | GTGGCAGATTGGTAACAGA |
| TNFα | CGTTCGGGAGTGGGCTTTA | TTGTGGGACAGGGTAGGG |
| IL18 | ATCTGGCAGTGGAATGTAC | CAGGAATGTCTTTGGGAAC |
| NFκB | CAGAGGAAGAGGCAGAAGC | AGGAAGGATAACTGAACCC |
| BCL2 | CGTGCGTCTGGAAGTAAAG | TAAAGCAGCCAACGGTAAT |
| BCL2L1 | CTTTCAGCGACCTCACCTC | ACAATGCGTCCCACCAGTA |
| CASP1 | GAGGAAGCAGGAAGCAAAC | CCGTCCCGCTGTCTCAAGT |
| CASP3 | CTGAAGGCTCCTGGTTTAT | CTGCCACTCTGCGATTTAC |
| CASP9 | AAAACCCAAACTCTTCTTCA | CGTCTGGCTCGTCCTCATT |
| BAG2 | ATTTCAGTCCGTTGTCATTG | TGGTTTGCCAGAAGTAGCC |
| BECN1 | GTATGGCAACCACTCGTAT | CAATCTTTCCCTTCTCCAC |
| ULK1 | GAACAACGAGAAGCCAATG | TGCCTGCCAGTGAATGAGC |
| ATG5 | GGTTTGCTGTTTGATTTGC | TGACTTGACTTTTGTGCTT |
| mTOR | GAAAGGAATGAACCGTGAT | TAGTGAAGGGAGTGATGTG |
| gga-miR-20a-5p | TAAAGTGCTTATAGTGCAGGTAG | CAGTGCGTGTCGTGGAGT |
| GAPDH (Convergent) | GACCTGCCGTCTGGAGAAA | ATCAAAGGTGGAGGAATGG |
| U6 | CAAGGACCCATCGTTCCACA | CCATTGGACACGCAGAATGC |
Construction of PTEN Overexpression Vector
To generate the overexpression vector of PTEN, the restriction sites Bam HI and Hind III were employed based on the full-length coding domain sequence (CDS) of PTEN available in the NCBI database. The specific primer for PTEN CDS region can be found in Table 1. PTEN was amplified from chicken macrophage cDNA. The purified PCR product and pcDNA3.1 plasmid were separately digested with Bam HI and Hind III for 30 min at 37°C. Following digestion, the purified pcDNA3.1 plasmid and PTEN fragments were mixed at a ratio of 1:3, and ligated with SoSoo ligase at 50°C for 15 min. The ligation product was then transferred into DH5α competent cells. The cells were cultured in an incubator at 37°C for 12 h after plating. Monoclonal colonies were selected and identified by PCR amplification using PTEN full-length primers.
Cell Transfection and APEC Infection
Cell transfection was performed using the Lipofectamine 8000 reagent (Invitrogen, Carlsbad, CA) following the manufacturer's instructions. Prior to transfection, 6×105 cells/well were seeded into 24-well plates. Transfection was carried out when cell confluence reached 70∼80%. A transfection mixture was prepared, consisting of 500 ng pcDNA3.1 or pcDNA-PTEN, 25 μL DMEM, and 0.8 μL Lipofectamine 8000. Subsequently, 25 μL of the mix was added to each well. After transfection with pcDNA-PTEN for 48 h, cells were either infected with or without 0.1 mL 1×108 cfu/mL of APEC O78 for 24 h. Total RNA was then collected for subsequent experiments. The cultivation of APEC O78 bacteria and the optimal infection concentration were previously described in studies by Sun et al. (2022) and Yang et al. (2023).
Dual-Luciferase Reporter Assay
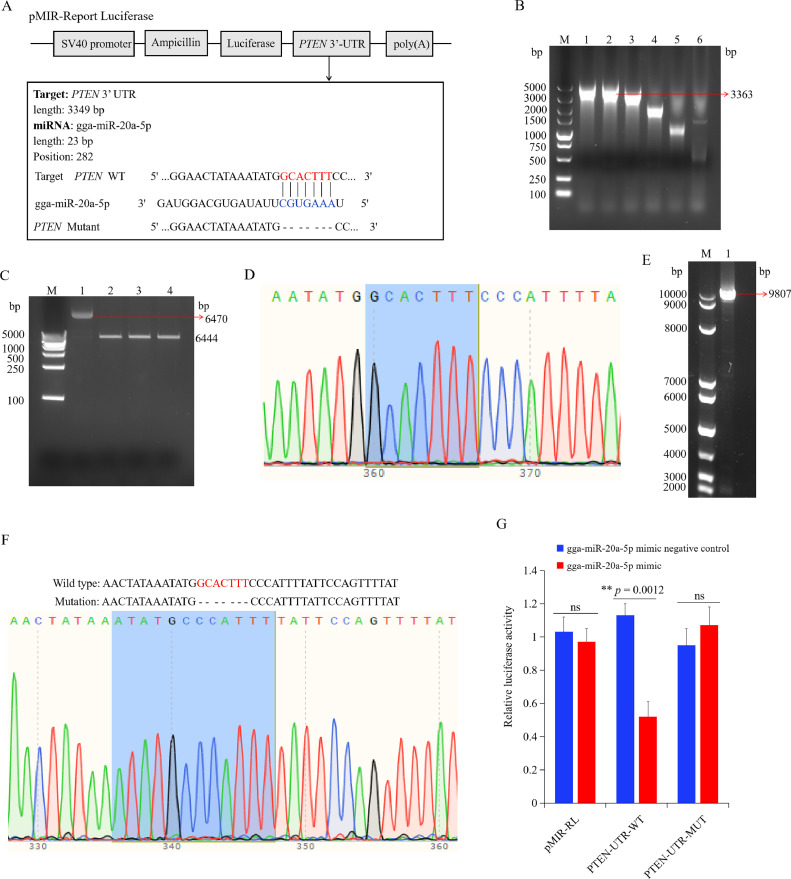
To construct the dual luciferase reporter vector, PCR was performed to amplify the PTEN 3′ UTR regions containing the predicted binding sites of miR-20a-5p and PTEN. For the construction of the PTEN 3′ UTR mutant (pMIR-PTEN-3′ UTR-MT) vector, the predicted binding sites (GCACTTT) were eliminated using the Mut Express II Fast Mutagenesis Kit V2 (Vazyme, Nanjing, China) according to the manufacturer's instructions. The specific primers for homologous recombination of the PTEN gene can be found in Table 1. The homologous recombinant primers for the PTEN gene can be found in Table 1. Subsequently, The amplified fragments were ligated into pMIR-REPORTER using the homologous recombination method. The recombinant plasmid was cleaved with Sac I and Mlu I restriction enzymes. To confirm successful insertion of the target fragment, the constructed recombinant plasmids were sent to company for sanger sequencing.
Cell Viability Assay
The viability of cells from different groups (Control, pcDNA-PTEN, APEC infection, and pcDNA-PTEN + APEC infection) was evaluated using the Cell Counting Kit-8 (CCK-8). Cells were seeded in triplicate at a density of 1×106 cells per well in a 96-well plate. After PTEN overexpression vector transfection and APEC infection, cells were treated with 100 µL of medium and incubated for 48 h. Subsequently, the cells were treated with 10 µL of CCK-8 solution and incubated for an additional 2 h. The absorbance (optical density, OD) was measured at 450 nm using a spectrophotometer.
Nitric oxide Production Assay
The measurement of Nitric oxide (NO) production in the cell supernatant from different groups (Control, pcDNA-PTEN, APEC infection, and pcDNA-PTEN + APEC infection) was conducted by using the Griess reagent kit (Molecular Probes, Carlsbad, CA). The cell supernatant was mixed with Griess reagents and incubated for 30 min in the absence of light. Following incubation, the absorbance at 540 nm was measured using a spectrophotometer. The absorbance values were then compared to the sodium nitrite standard curve to determine the concentration of nitrite (μM) in each respective group.
Statistical Analysis
All data are presented as mean ± SD, and all experiments were conducted in triplicate. Multiple comparative analyses were performed using 1 way ANOVA in JMP statistical software (Version 15.2.1, SAS Institute). A P-value less than 0.05 was considered statistically significant.
RESULTS
Bioinformatics Analysis of PTEN
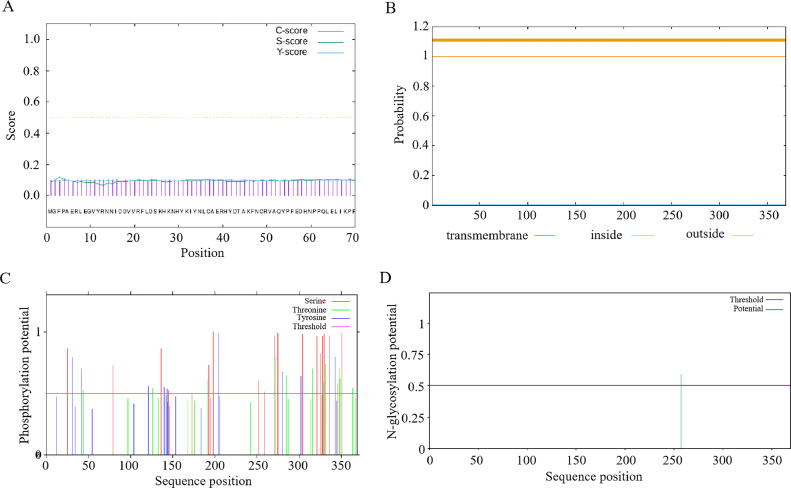
The coding sequence (CDS) of PTEN was 1110 bp, corresponding to the synthesis of 369 amino acids. The ProtParam software analysis showed that the PTEN protein has a molecular weight of 43 kDa and a molecular formula of C1940H2931N513O577S15. The protein consists of a total of 5976 atoms. The theoretical isoelectric point (pI) of PTEN was calculated to be 5.93. The protein contains 57 negatively charged residues (Asp+Glu) and 49 positively-charged residues (Arg+Lys). The instability index (II) of PTEN was determined to be 41.39, indicating that it is an unstable protein. Furthermore, the protein possesses an aliphatic index of 64.42 and a grand average of hydropathicity (GRAVY) value of −0.719. No putative signal sequence was identified using the peptide SignalP 4.1 server (Figure 1A). Analysis by the TMHMM server indicated the absence of transmembrane domains in PTEN, and the total probability of the N-terminal being located in the cytoplasm was 0.0022 (Figure 1B). The NetPhos server predicted the presence of putative phosphorylation sites at 15 threonine, 34 serine, and 12 tyrosine amino acids in the PTEN protein (Figure 1C). Additionally, the NetOGlyc server predicted the occurrence of 12 O-glycosylation sites in PTEN protein, while the NetNGlyc server identified a potential N-glycosylation site at the 257th amino acid residue, with a predicted rate of 0.5856 (Figure 1D). In conclusion, the PTEN protein is characterized as a hydrophilic and unstable protein, with multiple putative phosphorylation sites.
Figure 1.
Bioinformatics analysis of PTEN. (A) Signal peptide prediction of the PTEN protein. (B) Transmembrane domain prediction of the PTEN protein. (C) Prediction of phosphorylation sites of chicken PTEN protein. (D) Prediction of glycosylation sites in chicken PTEN protein.
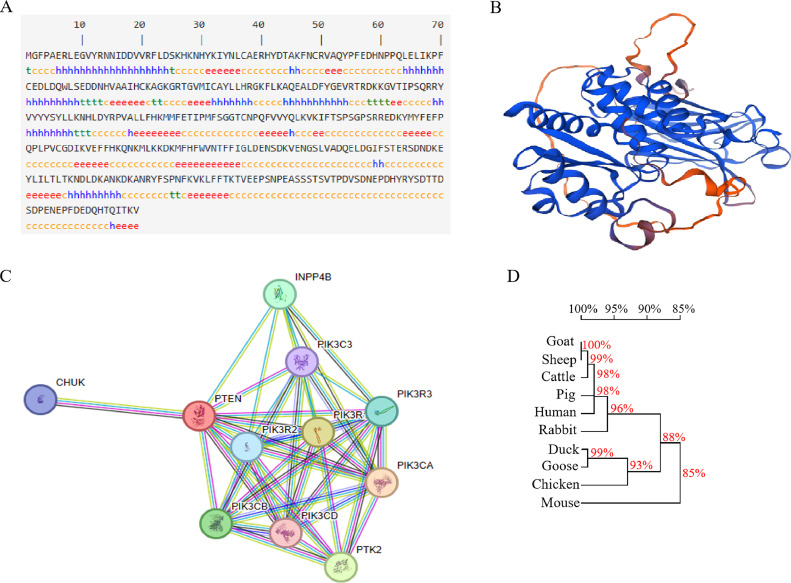
The secondary structure of the PTEN protein was predicted, and the analysis revealed that 198 amino acids (53.66%) formed a random coil, while 79 amino acids (21.41%) formed an α-helix, and 75 amino acids (20.33%) formed an extended strand configuration (Figure 2A). Utilizing the SWISS-MODEL software, the tertiary structure of PTEN was predicted, providing insights into its 3-dimensional arrangement (Figure 2B). To explore protein interactions, a protein-protein interaction network for PTEN was constructed using the STRING database, employing various criteria such as co-expression, co-occurrence, text mining, experiment databases, neighborhood and gene fusion. Through this analysis, ten genes were identified as associated with PTEN (Figure 2C). The evolutionary relationship of the PTEN gene among different animal species was analyzed using DNAMAN software. The resulting phylogenetic tree map (Figure 2D) demonstrated that chicken and duck/goose clustered together, suggesting a high degree of homology among the 3 species.
Figure 2.
PTEN protein structure, interaction network, and homologous among different species. (A) Secondary structure prediction of the PTEN protein. Note: h represents α-helix, e represents extended strand and c represents random coil. (B) Tertiary structure prediction of the PTEN protein. (C) Protein interaction network of PTEN. (D) Phylogenetic tree of PTEN and other homologous sequences.
Expression Pattern and Subcellular Localization of Chicken PTEN
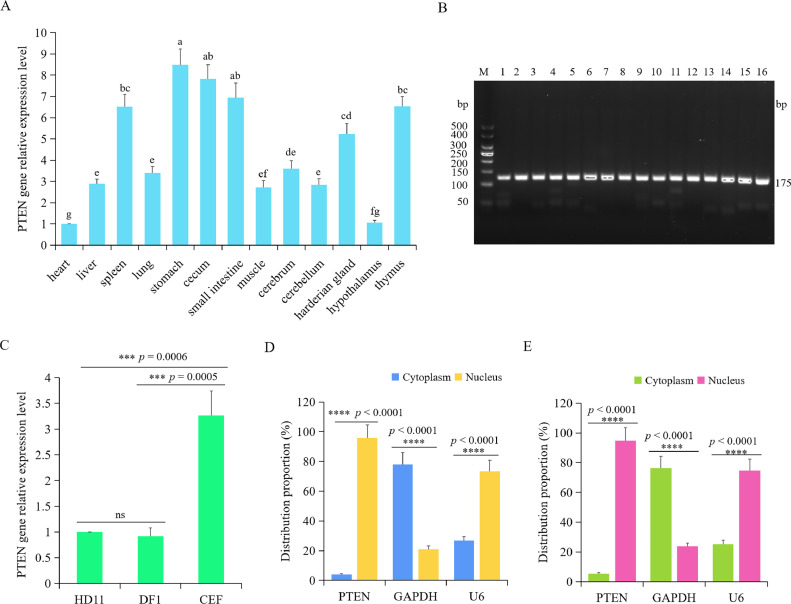
To investigate the tissue-specific expression patterns of chicken PTEN, total RNA was extracted from various tissues, including the heart, liver, spleen, lung, stomach, cecum, small intestine, muscle, cerebrum, cerebellum, harderian gland, hypothalamus, and thymus. RT-qPCR was performed to assess the relative expression levels of PTEN in these tissues. It was found that the PTEN expression level was significantly higher in the stomach (P < 0.0001), cecum (P < 0.0001), small intestine (P < 0.0001), spleen (P < 0.0001), thymus (P < 0.0001), harderian gland (P < 0.0001), muscle (P = 0.0404), cerebrum (P = 0.0004), cerebellum (P = 0.0207), lung (P = 0.0011), and liver (P = 0.0171) compared to heart tissue (Figure 3A). However, no significant difference in PTEN expression was observed between the hypothalamus and heart (P > 0.05) (Figure 3A). Furthermore, the expression pattern of PTEN was examined in HD11 macrophages, DF1 cells, and CEF cells. The results showed that CEF cells exhibited a significantly higher expression level of PTEN compared to HD11 macrophages (P = 0.0006) and DF1 cells (P = 0.0005) (Figure 3C). However, no significant difference in PTEN expression was observed between HD11 macrophages and DF1 cells (P > 0.05) (Figure 3C). Additionally, agarose gel electrophoresis was employed to verify the RT-qPCR products of PTEN, confirming the amplification of the correct PTEN product (Figure 3B).
Figure 3.
Relative expression pattern of PTEN in chicken different tissues and subcellular localization of PTEN. (A) The relative expression level of PTEN gene in heart, liver, spleen, lung, stomach, cecum, small intestine, muscle, cerebrum, cerebellum, harderian gland, hypothalamus, and thymus were measured by using RT-qPCR. The result was normalized with β-actin gene and relative to gene expression in the heart group. Data are shown as mean ± SD; n = 8; ANOVA test; Different letters represent significant differences (P < 0.05); The same letters represent insignificant differences (P > 0.05). (B) The agarose gel electrophoresis results of RT-qPCR amplification products of PTEN. Abbreviations: 1, heart; 2, liver; 3, spleen; 4, lung; 5, stomach; 6, cecum; 7, small intestine; 8, muscle; 9, cerebrum; 10, cerebellum; 11, harderian gland; 12, hypothalamus; 13, thymus; 14, HD11 macrophages; 15, DF1 cells; 16, CEF cells. (C) The relative expression level of PTEN gene in HD11 macrophages, DF1 cells, and CEF cells. Data are shown as mean ± SD; n = 4 independent experiments; ANOVA test; ns, not significant; ***P < 0.001. D-E. Distribution of PTEN gene in the nucleus and cytoplasm of wild type macrophages (D) and APEC infected macrophages. (E) GAPDH and U6 are the marker genes of cytoplasm and nucleus, respectively; n = 4 independent experiments; paired t-test; ****P < 0.0001.
Following the utilization of the NE-PER kit to isolate the nucleus and cytoplasm fractions of chicken macrophages, the corresponding total RNA from each fraction was extracted to perform subsequent RT-qPCR assays aimed at assessing the expression level of PTEN. Results showed that chicken PTEN was predominantly localized within the nucleus of both wild-type macrophages and macrophages infected with APEC (Figures 3D–E). These results suggest that PTEN may be primarily located in the nucleus of chicken macrophages.
Construction of PTEN Overexpression Vector
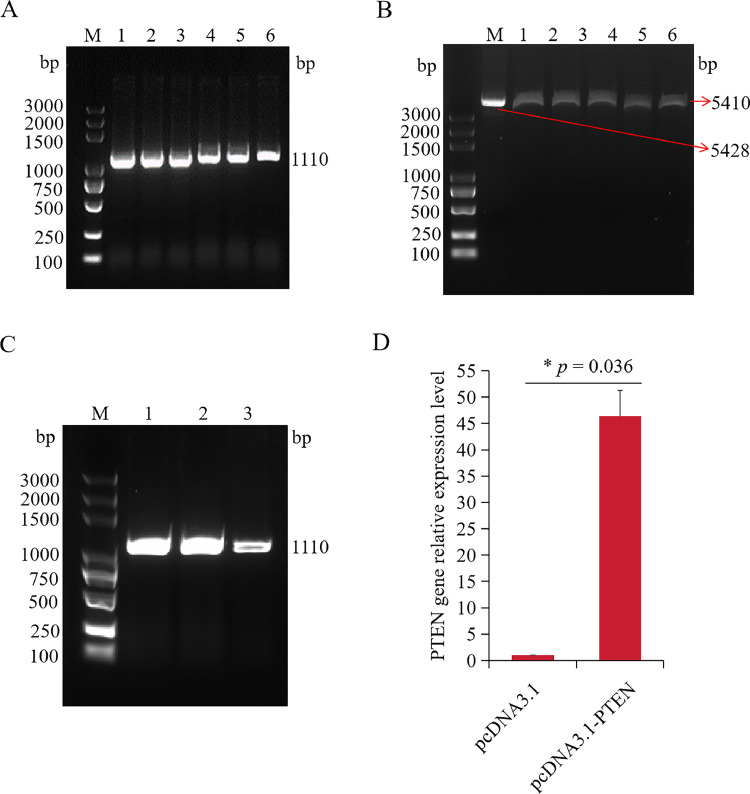
The target fragment was amplified using the designed pcDNA3.1-PTEN primers (Figure 4A). Successful digestion of the circular pcDNA3.1 plasmid was achieved using Bam HI and Hind III restriction enzymes (Figure 4B). The bacterial solution contained massive amounts of the recombinant vector pcDNA3.1-PTEN (Figure 4C), and the extracted plasmids were subsequently subjected to sequencing. The sequencing result exhibited a complete match with the PTEN sequence, with a 100% match rate (Supplementary file 1). Therefore, the pcDNA3.1-PTEN overexpression plasmid was successfully constructed.
Figure 4.
Construction and activity verification of PTEN overexpression vector. (A) The PCR product of PTEN with different values of Tm. M, Marker; 1 to 6, PTEN production with Tm of 55°C, 57°C, 59°C, 61°C, 63°C, 65°C. (B) Double enzyme digestion of the pcDNA3.1 plasmid. M, Marker; 1, circular pcDNA3.1 plasmid; 2-6, the linear pcDNA3.1 plasmid after double enzyme digestion. (C) the PCR product of pcDNA3.1-PTEN DH5α bacterial liquid. M, Marker; 1 to 3, PTEN. (D) Relative expression of PTEN gene in macrophages transfected with overexpression vector for 48 h as measured by RT-qPCR. Data are shown as mean ± SD; n = 4 independent experiments; Wilcoxon test; ns, not significant; **P < 0.01.
Chicken macrophages were transfected with the successfully constructed PTEN overexpression vector, pcDNA3.1-PTEN, as well as the control vector, pcDNA3.1, for 48 h. Subsequent RT-qPCR analysis revealed a significant elevation in PTEN mRNA expression in the pcDNA3.1-PTEN group compared to the control group (P < 0.0001) (Figure 4D). The successfully constructed pcDNA3.1-PTEN overexpression plasmid can be used for subsequent cell experiments to explore the regulatory mechanism of PTEN.
Overexpression of PTEN Caused Cell Damages During APEC Infection
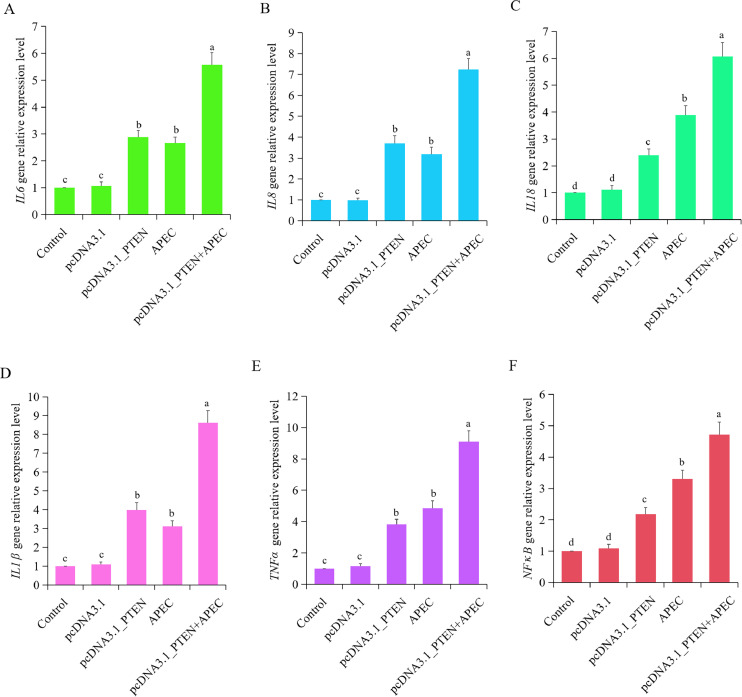
The expression level of IL1β, IL8, IL18, IL6, TNFα, and NFκB were assessed by RT-qPCR after transfection with the PTEN overexpression plasmid for 48 h, both in the presence and absence of APEC infection. It was found that a significant increase in the expression of IL1β, IL8, IL18, IL6, TNFα, and NFκB was observed in macrophages transfected with PTEN overexpression plasmid in comparison to the control and empty vector group (Figures 5A–F). Furthermore, in the PTEN overexpression combined with APEC infection group, these 6 inflammatory cytokines exhibited a substantial increase in expression levels compared to both the control group and the APEC infection group (Figures 5A–F). These findings suggested that the overexpression of PTEN can increase the expression of inflammatory mediators, regardless of the presence or absence of APEC infection.
Figure 5.
Overexpression of PTEN caused cellular inflammatory response during APEC infection. A-F. The expression levels of 6 pro-inflammatory mediators, including IL6 (A), IL8 (B), IL18 (C), IL1β (D), TNFα (E), and NFκB (F) were analyzed using RT-qPCR after chicken macrophages were transfected with the overexpression of PTEN vector associated with or without APEC infection. Data are shown as mean ± SD; n = 4 independent experiments; ANOVA test; Different letters represent significant differences (P < 0.05); The same letters represent insignificant differences (P > 0.05).
Overexpression of PTEN Influenced Apoptosis Related Genes in Chicken Macrophages With or Without APEC Infection
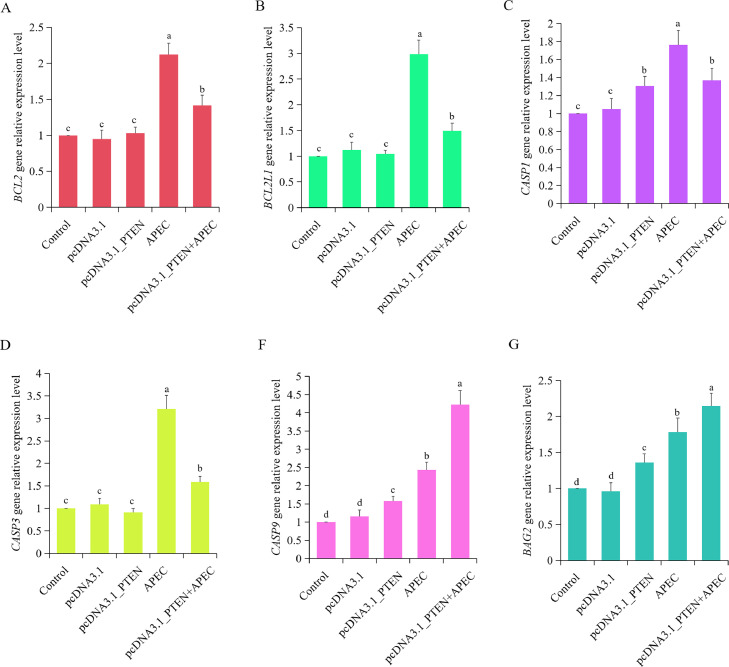
The expression level of apoptosis related genes, including BCL2, BCL2L1, CASP1, CASP3, CASP9, and BAG2, were assessed in chicken macrophages transfacted with the PTEN overexpression vector, with or without APEC infection. Results showed that compared to APEC group, overexpression of PTEN + APEC group can significantly decrease the expression of the antiapoptotic genes BCL2 and BCL2L1 (Figures 6A and 6B). Moreover, overexpression of PTEN can significantly up-regulate the expression levels of pro-apoptosis genes CASP1, CASP9, and BAG2 in comparison to the control and empty vector group, regardless of APEC infection (Figures 6C, 6F, and 6G). These results suggested that PTEN can influence the expression of pro-apoptosis and antiapoptosis related genes, upon APEC infection.
Figure 6.
Effects of PTEN on the expression of apoptosis related genes. A-F. The mRNA expression level of BCL2 (A), BCL2L1 (B), CASP1 (C), CASP3 (D), CASP9 (E) and BAG2 (F) in chicken macrophages transfected with overexpressed PTEN plasmid with or without APEC infection. Data are shown as mean ± SD; n=4 independent experiments; ANOVA test; Different letters represent significant differences (P < 0.05); The same letters represent insignificant differences (P > 0.05).
Overexpression of PTEN Decreased Cell Viability and Increased NO Production During APEC Infection
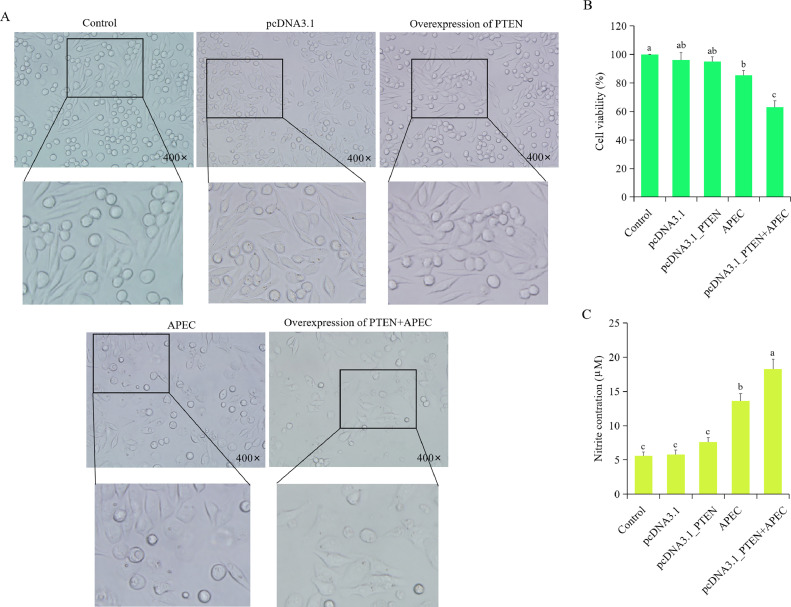
Morphological changes were examined in the groups of control, overexpression of PTEN, APEC infection, and overexpression of PTEN + APEC infection. Results showed no significant differences among the control, empty vector, and overexpression of PTEN groups in terms of morphological changes. However, both the APEC infection and overexpression of PTEN + APEC infection groups exhibited evident cytopathic effects (Figure 7A). Furthermore, no significant differences in cell viability were observed between the control/empty vector and PTEN overexpressed macrophages (Figure 7B). Notably, the cell viability in the PTEN overexpression combined with APEC infection group was significantly lower than that in the control, empty vector, and APEC infection groups (Figure 7B).
Figure 7.
Overexpression of PTEN decreased cell viability and increased NO production during APEC infection. (A) The morphology of chicken macrophages in the groups of Control, empty vector, ovexpression of PTEN, APEC, and overexpression of PTEN + APEC. (B) The cell viability of chicken macrophages in the groups of Control, empty vector, overexpression of PTEN, APEC, and overexpression of PTEN + APEC. Data are shown as mean ± SD; n = 4 independent experiments; ANOVA test; Different letters represent significant differences (P < 0.05); The same letters represent insignificant differences (P > 0.05). (C) The nitric oxide (NO) production of chicken macrophages in the group of Control, empty vector, overexpression of PTEN, APEC, and overexpression of PTEN + APEC. Data are shown as mean ± SD; n = 4 independent experiments; ANOVA test; Different letters represent significant differences (P < 0.05); The same letters represent insignificant differences (P > 0.05).
There was no significant difference in NO production among the control, empty vector, and the PTEN overexpression group (Figure 7C). However, APEC infection and PTEN overexpression combined with APEC infection resulted in a significant increase in NO production in chicken macrophages in comparison to the control and empty vector groups, with the PTEN overexpression combined with APEC infection group giving higher levels of NO compared to the APEC infection group (Figure 7C). These results suggested that the overexpression of PTEN can significantly increase the NO production upon APEC infection.
Overexpression of PTEN Promoted the Expression of Autophagy Related Genes With or Without APEC Infection
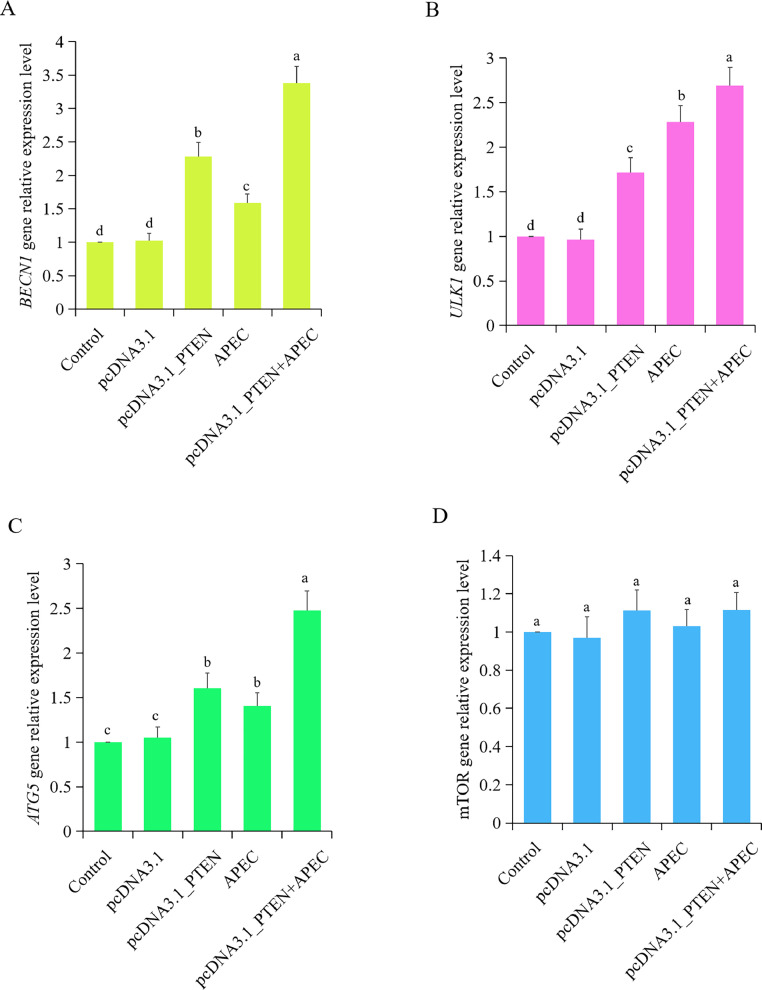
To investigate the role of PTEN in cellular autophagy during APEC infection, RT qPCR was employed to measure the mRNA expression levels of key autophagy-related genes, namely mTOR, BECN1, ATG5, and ULK1. As shown in Figure 8, results showed a significant elevation in the mRNA expression levels of BECN1, ATG5, and ULK1 following APEC infection, when compared to the control and empty vector groups. However, there was no notable difference observed in the mRNA expression of mTOR between the control/empty vector and the APEC group (Figures 8A–D). Interestingly, when comparing with the control and empty vector groups, we observed that overexpression of PTEN substantially enhanced the mRNA expression levels of BECN1, ATG5, and ULK1, both in the presence and absence of APEC infection (Figures 8A–C). These results collectively indicated that overexpression of PTEN promoted the expression of autophagy related genes, irrespective of APEC infection.
Figure 8.
Overexpression of PTEN promoted autophagy with or without APEC infection. (A–D) The mRNA expression of BECN1 (A), ATG5 (B), ULK1 (C) and mTOR (D) in the group of Control, empty vector, overexpression of PTEN, APEC, and overexpression of PTEN + APEC. Data are shown as mean ± SD; n = 4 independent experiments; ANOVA test; Different letters represent significant differences (P < 0.05); The same letters represent insignificant differences (P > 0.05).
PTEN was the Directly Target Gene of gga-miR-20a-5p
Based on the results of miRanda and targetscan, PTEN was predicted to be a potential target of gga-miR-20a-5p (Figure 9A). The wild type 3′ UTR of PTEN was first amplified (Figure 9B). The pMIR-Report luciferase plasmid was successfully digested by Mlu I and Sac I (Figure 9C). The wild type 3′ UTR of the PTEN fragment was ligated with the digested pMIR-Report luciferase plasmid. After sequencing the production, the recombination plasmid (pMIR-PTEN-WT) was successfully obtained. The recombinant plasmid was digested using Dpn I. Then, the digested pMIR-PTEN-WT plasmid was used as a DNA template, amplified by Vazyme point mutation kit (Figure 9E), and sequenced (Figure 9F). The sequencing result showed that the binding site of PTEN 3′UTR to miR-20a-5p were successfully deleted (Figure 9F), and the PTEN 3′UTR mutant plasmid (pMIR-PTEN-MUT) was successfully constructed.
Figure 9.
PTEN was a directly target of gga-miR-20a-5p. (A) The sequence alignment of gga-miR-20a-5p and 1 target site in the 3′ UTR of PTEN is shown. (B) The PCR product of wild type 3′ UTR of PTEN with different values of Tm. M, Marker; 1 to 6, wild type 3′ UTR of PTEN production with Tm of 55°C, 57°C, 59°C, 61°C, 63°C, 65°C. (C) Double enzyme digestion of the pMIR-report luciferase plasmid. M, Marker; 1, circular pMIR-Report Luciferase plasmid; 2 to 4, the linear pMIR-report luciferase plasmid after double enzyme digestion. (D) The sequencing result of wild type 3′ UTR of PTEN. (E) Construction of the mutant of PTEN 3′ UTR. M, Marker; 1, The PCR product amplified by the mutant 3′UTR of PTEN primers using the wild type 3′UTR of PTEN plasmid as a template. (F) The sequencing result of mutant 3′UTR of PTEN plasmid. (G) Chicken macrophages were co-transfected with pMIR-report luciferase or 3′UTR of PTEN luciferase reporter plasmid (wild-type or mutation), along with mimics negative control or gga-miR-20a-5p mimics as indicated. Firefly luciferase activity was measured and normalized using renilla luciferase activity to finally calculate the relative luciferase activity 48 h after transfection. Data are shown as mean ± SD; n = 4 independent experiments; paired t test; ns, not significant; **P < 0.01.
To validate the relationship between PTEN and gga-miR-20a-5p, the dual-luciferase reporter gene system was performed by transfecting chicken macrophages with the wild type or mutant 3′ UTR of PTEN, along with mimic of gga-miR-20a-5p or mimic negative control. As shown in Figure 9G, the luciferase activity of the cells transfected with wild type PTEN was significantly attenuated by mimic of gga-miR-20a-5p in comparison to mimic negative control, whereas the luciferase activity of the cells transfected with mutant PTEN was not affected by mimic of miR-20a-5p. In summary, these results suggested that PTEN was a target gene of gga-miR-20a-5p.
PTEN Gene was Regulated by gga-miR-20a-5p With APEC Infection
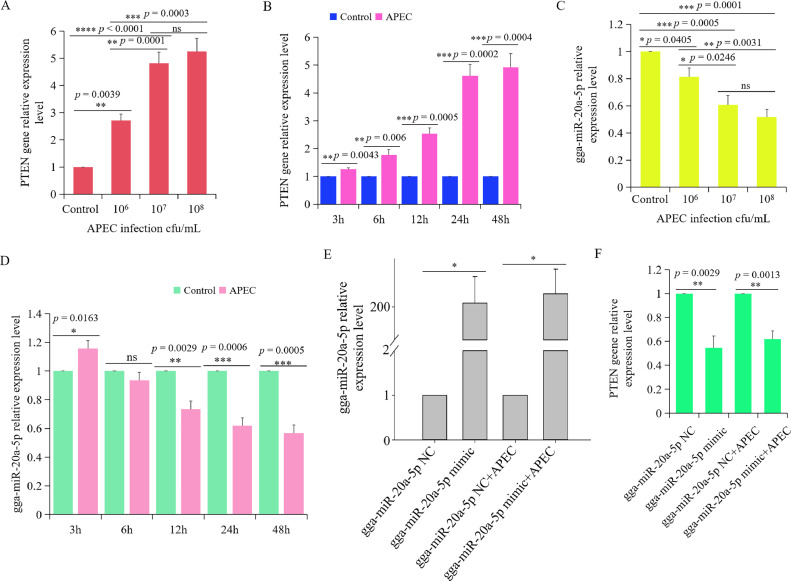
The relationship between gga-miR-20a-5p and PTEN was investigated by examining their transcript levels in chicken macrophages infected with APEC at different time points and dosages. The expression of PTEN showed a significant increase in a dose- and time-dependent manner upon APEC infection. After 24 h of infection, the expression level of PTEN was significantly elevated when infected with APEC at concentration of 1×106 cfu/mL, 1×107 cfu/mL, and 1×108 cfu/mL (Figure 10A). Furthermore, the expression level of PTEN reached its peak with an APEC infection concentration of 1×107 cfu/mL. There was no significant difference in PTEN expression between 1×107 cfu/mL and 1×108 cfu/mL of APEC infection. At a concentration of 1×107 cfu/mL of APEC, the expression level of PTEN was significantly increased at 6 h, 12 h, and reached its peak at 24-48 h (Figure 10B). However, gga-miR-20a-5p exhibited an opposite pattern to PTEN in a dose-dependent manner (Figure 10C) and a time-dependent manner upon APEC infection (Figure 10D). These results indicated a negative correlation between PTEN and gga-miR-20a-5p upon APEC infection.
Figure 10.
PTEN gene was regulated by gga-miR-20a-5p with APEC infection. (A and C) The expression level of PTEN (B) and gga-miR-20a-5p (D) after chicken macrophages infected with APEC at different concentrations (0, 106 cfu/mL, 107 cfu/mL, and 108 cfu/mL) for 24 h via RT-qPCR. Data represent the mean±SD; n = 4 independent experiments; ANOVA test; ns, not significant; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. (B and D) The expression level of PTEN (B) and gga-miR-20a-5p (D) after chicken macrophages infected with APEC (1× 107 cfu/mL) for 3 h, 6 h, 12 h, 24 h, and 48 h by using RT-qPCR. Data represent the mean ± SD; n = 4 independent experiments; ANOVA test; ns, not significant; *P < 0.05; **P < 0.01; ***P < 0.001. (E) The relative expression of gga-miR-20a-5p in chicken macrophages transfected with gga-miR-20a-5p mimic for 48 h. Data are shown as mean ± SD; n = 4 independent experiments; Wilcoxon test; *P < 0.05. (F) The relative expression of PTEN in chicken macrophages transfected with gga-miR-20a-5p mimic for 48 h. Data are shown as mean ± SD; n = 4 independent experiments; ANOVA test; ns, not significant; **P < 0.01.
The overexpression efficiency of gga-miR-20a-5p was assessed by transfecting the gga-miR-20a-5p mimic into chicken macrophages, both with and without APEC infection. The expression level of gga-miR-20a-5p was significantly elevated in both aforementioned conditions (Figure 10E). Subsequently, the expression of PTEN was also evaluated by RT-qPCR after transfecting with the gga-miR-20a-5p mimic into chicken macrophages with APEC infection.
Results showed a significant decrease of PTEN expression in both the gga-miR-20a-5p mimic-transfected macrophages and the APEC-infected macrophages transfected with the gga-miR-20a-5p mimic (Figure 10F). These findings collectively demonstrate that gga-miR-20a-5p was the regulator of PTEN gene during APEC infection.
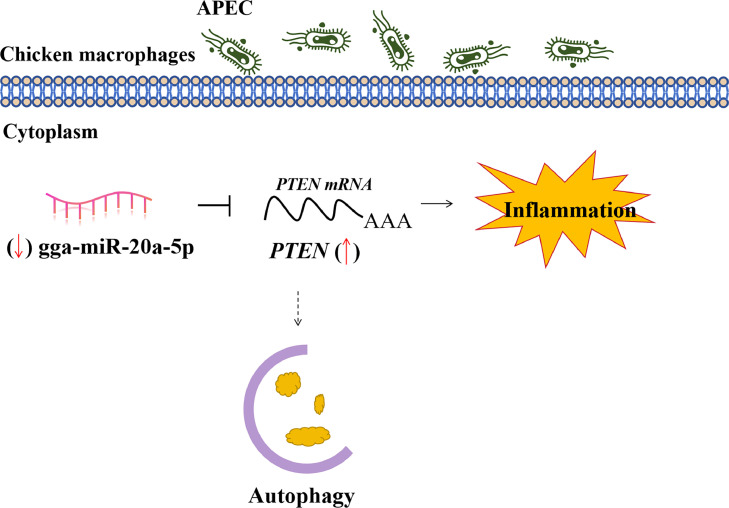
A Model to Explain the Role of PTEN in APEC Infection
In summary, PTEN was identified as involved in the cellular immune and inflammatory response against APEC infection (Figure 11). Firstly, overexpression of PTEN was found to decrease cell viability and increase the production of NO in response to APEC infection. Furthermore, mechanistic investigations demonstrated that overexpression of PTEN regulated the expression levels of autophagy-related genes, inflammatory-related genes, and pro-apoptosis-related genes upon APEC infection. Additionally, our findings confirmed that PTEN was a direct target gene of gga-miR-20a-5p, regardless of APEC infection. Taken together, our results provided evidence that the PTEN regulated by gga-miR-20a-5p played a crucial role in response to APEC infection via modulating autophagy.
Figure 11.
A model to explain the role of PTEN in assisting cellular immune and inflammatory response against APEC infection.
DISCUSSION
The chicken's antibacterial response is a highly intricate and interactive signaling and regulatory network. Colibacillosis, caused by APEC, is a complex disease under the control of multiple genes. Recent studies on APEC have highlighted the significant role of PTEN in the process of APEC infection (Sun et al., 2022). PTEN, a candidate tumor suppressor gene, was the first gene discovered to possess both oncogenic and lipid/protein phosphatase activities. While much attention has been given to PTEN's role in cancer, it is also involved in various cellular processes related to cell polarity, migration, growth, and metabolism (Song et al., 2012; Lee et al., 2018). However, the function of PTEN in poultry, particularly in response to APEC infection, remains unexplored.
Recently, several studies have highlighted the role of PTEN in determining the cell fate of macrophages (Sahin et al., 2014; Shang et al., 2023). It has been demonstrated that macrophages from mice with myeloid-specific PTEN knockout exhibit a hyporesponsive phenotype to LPS, as evidenced by decreased mRNA expression level of TNFα and reduced NO release (Cao et al., 2004; Kuroda et al., 2008). Consistent with these findings, our study also revealed a significant elevation in the mRNA expression levels of TNFα and increase NO production in chicken macrophages transfected with overexpression of PTEN plasmid, both with or without APEC infection. Furthermore, knockout of PTEN has been shown to result in the reduction production of IL1β and IL6 in mice (Sahin et al., 2014; Cui et al., 2020), which is also consistent with our observations of significantly elevated mRNA expression levels of IL1β and IL6 in chicken macrophages overexpressing of PTEN, regardless of APEC infection. In summary, our findings demonstrated that overexpression of PTEN promoted cellular inflammation and NO production in chicken macrophages, irrespective of APEC infection.
Apoptosis plays a crucial role in the immune defense system and is essential for maintaining immune system homeostasis. The relationship between bacterial infections and apoptosis has been well-established. Notably, the involvement of PTEN in apoptosis is well-documented. Zhu et al. (2006) identified that PTEN was able to regulate the expression of pro-apoptosis genes, including Bax, cytochrome c (Cytc), and caspase 3 (CASP3), in mice, indicating that PTEN promoted cellular apoptosis. Similarly, 3 other studies in mammals reported that overexpression of PTEN enhanced the expression of pro-apoptosis related genes, such as CASP3, caspase 1 (CASP1), and caspase9 (CASP9), while decreasing the expression of the antiapoptotic gene BCL2 (Dupont et al., 2002; Tong et al., 2016; Cui et al., 2020; He et al., 2020). These findings are partially consistent with our observations. In our study, we found that compared to APEC group, overexpression of PTEN + APEC group can significantly increase the mRNA expression of pro-apoptosis related genes CASP9 and BAG2 and decrease the expression of the antiapoptotic genes BCL2 and BCL2L1. However, the expression of CASP1 and CASP3 was lower in the overexpression of PTEN + APEC group compared to the APEC group. Further studies are needed to verify the specific mechanism of PTEN on the apoptosis during APEC infection.
Autophagy, a cellular process, is induced by various stressors, including bacterial infections, and plays diverse physiological and pathophysiological roles. To assess cellular autophagic levels, specific markers such as LC3-II, p62, and ATG5 are commonly employed (Zhu et al., 2021). ATG5 is located in the precursor membrane of autophagosomes and is a crucial for their formation and development. ATG5 recruits LC3 in autophagosomes, which is later detached, leaving behind LC3b-II (Ribas et al., 2015; Liu et al., 2017). BECN1 is a key factor in autophagic process and collaborates with ATG5, LC3b, and p62 to form autophagosomes (Vucicevic et al., 2014; Kruse et al., 2015). mTOR negatively regulates autophagy, and inhibition of mTOR with rapamycin, a mTOR inhibitor, can increase BECN1 expression and induce autophagy (Yang et al., 2015). ULK1 and its complexes serve as initiators of autophagy and are involved in the regulation of numerous downstream autophagy-related signaling pathways.
In this study, we observed that overexpression of PTEN can significantly promote autophagy, as indicated by increased mRNA expression levels of BECN1, ULK1, and ATG5 with or without APEC infection. These findings were consistent with the majority of previous studies on PTEN and autophagy. For instance, Ueno et al. (2008) demonstrated that PTEN deficiency suppressed autophagy at the formation and maturation steps of autophagosomes, particularly in response to increased insulin signaling. Furthermore, Wang et al. (2021) reported that overexpression of PTEN can activate autophagy via PI3K/Akt/mTOR pathway in hypoxia/reoxygenation induced HK-2 cells in mice. However, it is worth noting that Huang et al. (2023) found that overexprssion of PTEN can significantly suppress autophagy induced by sodium arsenite. These divergent findings suggest that the effect of PTEN on autophagy is highly dependent on the specific cellular environment rather than technical discrepancies.
CONCLUSIONS
In summary, the coding sequence of PTEN was obtained and the physicochemical properties and molecular functions of PTEN were analyzed. It was found that PTEN, regulated by gga-miR-20a-5p, played a crucial role in modulating host immune and inflammatory response to APEC infection via autophagy. These findings will contribute to a deeper understanding of the mechanisms underlying the host immune and inflammatory response during APEC infection in chickens macrophages, with PTEN serving as a key player. It is worth mentioning that PTEN, as a direct target gene of gga-miR-20a-5p, might provide a potential target for the treatment of APEC infection. Furthermore, the insights gained from this study may aid in the development of strategies for breeding poultry with genetic resistance to APEC in the future.
DISCLOSURES
The authors declare that they have no conflict of interest.
ACKNOWLEDGMENTS
This research was funded by Yangzhou International Science and Technology Cooperation Project (YZ2023260), Natural Science Foundation of the Jiangsu Higher Education Institutions of China (24KJA230003), and “JBGS” Project of Seed Industry Revitalization in Jiangsu Province (JBGS[2021] 029).
Author contributions: Conceptualization, H.S.; validation, Y.M. and X.C.; formal analysis, Y.M.; original draft preparation, H.S.; review and editing, H.L., W.H., L.Q., and S.L. All authors have read and agreed to the published version of the manuscript.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.psj.2024.104170.
Appendix. Supplementary materials
REFERENCES
- Azam M., Mohsin M., Johnson T.J., Smith E.A., Johnson A., Umair M., Saleemi M.K., Sajjad-Ur-Rahman Genomic landscape of multi-drug resistant avian pathogenic Escherichia coli recovered from broilers. Vet. Microbiol. 2020;247 doi: 10.1016/j.vetmic.2020.108766. [DOI] [PubMed] [Google Scholar]
- Baker B.I., Torrey S., Widowski T.M., Turner P.V., Knezacek T.D., Nicholds J., Crowe T.G., Schwean-Lardner K. Defining characteristics of immersion carbon dioxide gas for successful euthanasia of neonatal and young broilers. Poult. Sci. 2020;99:4408–4416. doi: 10.1016/j.psj.2020.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom N., Gammeltoft S., Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 1999;294:1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- Buckler J.L., Walsh P.T., Porrett P.M., Choi Y., Turka L.A. Cutting edge: T cell requirement for CD28 costimulation is due to negative regulation of TCR signals by PTEN. J. Immunol. 2006;177:4262–4266. doi: 10.4049/jimmunol.177.7.4262. [DOI] [PubMed] [Google Scholar]
- Cao X., Wei G., Fang H., Guo J., Weinstein M., Marsh C.B., Ostrowski M.C., Tridandapani S. The inositol 3-phosphatase PTEN negatively regulates Fcγ receptor signaling, but supports Toll-like receptor 4 signaling in murine peritoneal macrophages. J. Immunol. 2004;172:4851–4857. doi: 10.4049/jimmunol.172.8.4851. [DOI] [PubMed] [Google Scholar]
- Cui Q., Wang J., Liu X., Wang X., Su G. Knockout of PTEN improves cardiac function and inhibits NLRP3-mediated cardiomyocyte pyroptosis in rats with myocardial ischemia-reperfusion. Xi bao yu fen zi Mian yi xue za zhi= Chinese. J. Cell. Mol. Immunol. 2020;36:205–211. [PubMed] [Google Scholar]
- Dupont J., Renou J.P., Shani M., Hennighausen L., LeRoith D. PTEN overexpression suppresses proliferation and differentiation and enhances apoptosis of the mouse mammary epithelium. J. Clin. Invest. 2002;110:815–825. doi: 10.1172/JCI13829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M.R., Appel R.D., Bairoch A. Springer; New York: 2005. Protein identification and analysis tools on the ExPASy server. [DOI] [PubMed] [Google Scholar]
- Haddadi N., Lin Y., Travis G., Simpson A.M., Nassif N.T., McGowan E.M. PTEN/PTENP1:‘Regulating the regulator of RTK-dependent PI3K/Akt signalling’, new targets for cancer therapy. Mol. Cancer. 2018;17:1–14. doi: 10.1186/s12943-018-0803-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He P., Ma J., Liu Y., Deng H., Dong W. Hesperetin promotes cisplatin- induced apoptosis of gastric cancer in vitro and in vivo by upregulating PTEN expression. Front. Pharmacol. 2020;11:1326. doi: 10.3389/fphar.2020.01326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y., Heo J., Kang S., Vu T.H., Lillehoj H.S., Hong Y.H. Exosome-mediated delivery of gga-miR-20a-5p regulates immune response of chicken macrophages by targeting IFNGR2, MAPK1, MAP3K5, and MAP3K14. Anim. Biosci. 2023;36:851. doi: 10.5713/ab.22.0373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F., Ding G., Yuan Y., Zhao L., Ding W., Wu S. PTEN Overexpression alters autophagy levels and slows sodium arsenite-induced hepatic stellate cell fibrosis. Toxics. 2023;11:578. doi: 10.3390/toxics11070578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse R., Vind B.F., Petersson S.J., Kristensen J.M., Højlund K. Markers of autophagy are adapted to hyperglycaemia in skeletal muscle in type 2 diabetes. Diabetologia. 2015;58:2087–2095. doi: 10.1007/s00125-015-3654-0. [DOI] [PubMed] [Google Scholar]
- Kuroda S., Nishio M., Sasaki T., Horie Y., Kawahara K., Sasaki M., Natsui M., Matozaki T., Tezuka H., Ohteki T. Effective clearance of intracellular Leishmania major in vivo requires Pten in macrophages. Eur. J. Immunol. 2008;38:1331–1340. doi: 10.1002/eji.200737302. [DOI] [PubMed] [Google Scholar]
- Lee Y.-R., Chen M., Pandolfi P.P. The functions and regulation of the PTEN tumour suppressor: new modes and prospects. Nat. Rev. Mol. cell Biol. 2018;19:547–562. doi: 10.1038/s41580-018-0015-0. [DOI] [PubMed] [Google Scholar]
- Li S., Zhu M., Pan R., Fang T., Cao Y.-Y., Chen S., Zhao X., Lei C.-Q., Guo L., Chen Y.U. The tumor suppressor PTEN has a critical role in antiviral innate immunity. Nat. Immunol. 2016;17:241–249. doi: 10.1038/ni.3311. [DOI] [PubMed] [Google Scholar]
- Liu P., Zhang Z., Wang Q., Guo R., Mei W. Lithium chloride facilitates autophagy following spinal cord injury via ERK-dependent pathway. Neurotox. Res. 2017;32:535–543. doi: 10.1007/s12640-017-9758-1. [DOI] [PubMed] [Google Scholar]
- Möller S., Croning M.D.R., Apweiler R. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics. 2001;17:646–653. doi: 10.1093/bioinformatics/17.7.646. [DOI] [PubMed] [Google Scholar]
- Nguyen L.T., Thuan N.K., Tam N.T., Huyen Trang C.T., Khanh N.P., Bich T.N., Taniguchi T., Hayashidani H., Lien Khai L.T. Prevalence and genetic relationship of predominant Escherichia coli serotypes isolated from poultry, wild animals, and environment in the Mekong Delta, Vietnam. Vet. Med. Int. 2021;2021 doi: 10.1155/2021/6504648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan L.K., Barnes H.J., Vaillancourt J.-P., Abdul-Aziz T., Logue C.M. John Wiley & Sons, Inc.; Hoboken, NJ, USA: 2013. Colibacillosis, Diseases of Poultry; pp. 751–805. [Google Scholar]
- Petersen T.N., Brunak S., Von Heijne G., Nielsen H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- Ribas V.T., Schnepf B., Challagundla M., Koch J.C., Bähr M., Lingor P. Early and sustained activation of autophagy in degenerating axons after spinal cord injury. Brain Pathol. 2015;25:157–170. doi: 10.1111/bpa.12170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin E., Haubenwallner S., Kuttke M., Kollmann I., Halfmann A., Dohnal A.B., Chen L., Cheng P., Hoesel B., Einwallner E. Macrophage PTEN regulates expression and secretion of arginase I modulating innate and adaptive immune responses. J. Immunol. 2014;193:1717–1727. doi: 10.4049/jimmunol.1302167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang M., Ni L., Shan X., Cui Y., Hu P., Ji Z., Shen L., Zhang Y., Zhou J., Chen B. MTHFD2 reprograms macrophage polarization by inhibiting PTEN. Cell Rep. 2023;42 doi: 10.1016/j.celrep.2023.112481. [DOI] [PubMed] [Google Scholar]
- Shojaee S., Chan L.N., Buchner M., Cazzaniga V., Cosgun K.N., Geng H., Qiu Y.H., Von Minden M.D., Ernst T., Hochhaus A. PTEN opposes negative selection and enables oncogenic transformation of pre-B cells. Nat. Med. 2016;22:379–387. doi: 10.1038/nm.4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M.S., Salmena L., Pandolfi P.P. The functions and regulation of the PTEN tumour suppressor. Nat. Rev. Mol. Cell Biol. 2012;13:283–296. doi: 10.1038/nrm3330. [DOI] [PubMed] [Google Scholar]
- Steentoft C., Vakhrushev S.Y., Joshi H.J., Kong Y., Vester-Christensen M.B., Schjoldager K.T., Lavrsen K., Dabelsteen S., Pedersen N.B., Marcos-Silva L., Gupta R., Bennett E.P., Mandel U., Brunak S., Wandall H.H., Levery S.B., Clausen H. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 2013;32:1478–1488. doi: 10.1038/emboj.2013.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokholm N.M., Permin A., Bisgaard M., Christensen J.P. Causes of mortality in commercial organic layers in Denmark. Avian Dis. 2010;54:1241–1250. doi: 10.1637/9375-041910-Reg.1. [DOI] [PubMed] [Google Scholar]
- Su A., Guo Y., Tian H., Zhou Y., Li W., Tian Y., Li K., Sun G., Jiang R., Yan F. Analysis of miRNA and mRNA reveals core interaction networks and pathways of dexamethasone-induced immunosuppression in chicken bursa of Fabricius. Mol. Immunol. 2021;134:34–47. doi: 10.1016/j.molimm.2021.02.022. [DOI] [PubMed] [Google Scholar]
- Su A., Zhou Y., Guo Y., Yang X., Zhang Y., Li W., Tian Y., Li K., Sun G., Jiang R. Identification and expression analysis of MicroRNAs in chicken spleen in a corticosterone-induced stress model. Res. Vet. Sci. 2021;136:287–296. doi: 10.1016/j.rvsc.2021.02.023. [DOI] [PubMed] [Google Scholar]
- Sun H., Yang Y., Cao Y., Li H., Qu L., Lamont S.J. Gene expression profiling of RIP2-knockdown in HD11 macrophages—elucidation of potential pathways (gene network) when challenged with avian pathogenic E. coli (APEC) BMC Genom. 2022;23:341. doi: 10.1186/s12864-022-08595-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A., Kaisho T., Ohishi M., Tsukio-Yamaguchi M., Tsubata T., Koni P.A., Sasaki T., Mak T.W., Nakano T. Critical roles of Pten in B cell homeostasis and immunoglobulin class switch recombination. J. Exp. Med. 2003;197:657–667. doi: 10.1084/jem.20021101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong J., Zhang H., Sun D., Wang Y., Yang C., Liu Y. Over-expression of PTEN on proliferation and apoptosis in canine mammary tumors cells. Animal Cells Syst. (Seoul). 2016;20:325–334. [Google Scholar]
- Ueno T., Sato W., Horie Y., Komatsu M., Tanida I., Yoshida M., Ohshima S., Mak T.W., Watanabe S., Kominami E. Loss of Pten, a tumor suppressor, causes the strong inhibition of autophagy without affecting LC3 lipidation. Autophagy. 2008;4:692–700. doi: 10.4161/auto.6085. [DOI] [PubMed] [Google Scholar]
- Vucicevic L., Misirkic-Marjanovic M., Paunovic V., Kravic-Stevovic T., Martinovic T., Ciric D., Maric N., Petricevic S., Harhaji-Trajkovic L., Bumbasirevic V. Autophagy inhibition uncovers the neurotoxic action of the antipsychotic drug olanzapine. Autophagy. 2014;10:2362–2378. doi: 10.4161/15548627.2014.984270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wang X., Wang H., Bao J., Jia N., Huang H., Li A. PTEN protects kidney against acute kidney injury by alleviating apoptosis and promoting autophagy via regulating HIF1-α and mTOR through PI3K/Akt pathway. Exp. Cell Res. 2021;406 doi: 10.1016/j.yexcr.2021.112729. [DOI] [PubMed] [Google Scholar]
- Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L. SWISS-MODEL: homology modelling of protein structures and complexes. Nucl. Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T., Li D., Liu F., Qi L., Yan G., Wang M. Regulation on Beclin-1 expression by mTOR in CoCl2-induced HT22 cell ischemia-reperfusion injury. Brain Res. 2015;1614:60–66. doi: 10.1016/j.brainres.2015.04.016. [DOI] [PubMed] [Google Scholar]
- Yang Y., Lu Y., Zhou Y., Sun H., Ma Y., Tan J., Li N., Li H. Identification and characterization of microRNAs, especially gga-miR-181b-5p, in chicken macrophages associated with avian pathogenic E. coli infection. Avian Pathol. 2023;52:185–198. doi: 10.1080/03079457.2023.2181146. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Hoell P., Ahlemeyer B., Krieglstein J. PTEN: a crucial mediator of mitochondria-dependent apoptosis. Apoptosis. 2006;11:197–207. doi: 10.1007/s10495-006-3714-5. [DOI] [PubMed] [Google Scholar]
- Zhu S., Ying Y., Ye L., Ying W., Ye J., Wu Q., Chen M., Zhu H., Li X., Dou H. Systemic administration of fibroblast growth factor 21 improves the recovery of spinal cord injury (SCI) in rats and attenuates SCI-induced autophagy. Front. Pharmacol. 2021;11 doi: 10.3389/fphar.2020.628369. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.