Abstract
Merging traditional Chinese medicine's (TCM) principles of medicine-food homology with modern flavor chemistry, this research unveils PungentDB (http://www.pungentdb.org.cn/home), a database documenting 205 unique pungent flavor compounds from 231 TCMs. It provides detailed insights into their chemical attributes, biological targets (including IC50/EC50 values), and molecular structures (2D/3D), enriched with visualizations of target organ distribution and protein structures, exploring the pungent flavor space with the help of a feature-rich visual interface. This collection, derived from over 3249 sources and highlighting 9129 targets, delves into the compounds' unique pungent flavors—taste, aroma, and thermal sensations—and their interaction with taste and olfactory receptors. PungentDB bridges ancient wisdom and culinary innovation, offering a nuanced exploration of pungent flavors' role in enhancing food quality, safety, and sensory experiences. This initiative propels flavor chemistry forward, serving as a pivotal resource for food science advancement and the innovative application of pungent flavors.
Keywords: PungentDB, Flavor chemistry, Traditional Chinese medicine, Medicine food homology, Sensory science, Food flavor enhancement, Chemical analysis of food
Hightlights
-
•
PungentDB: A bridge between traditional Chinese medicine and contemporary food flavor chemistry.
-
•
Comprehensive profiling of 205+ pungent compounds from 231 traditional Chinese medicines.
-
•
Advanced visualization tools for exploring molecular structures and receptor interactions.
-
•
Facilitates interdisciplinary research by integrating sensory science with chemical analysis.
-
•
Paves the way for novel flavor enhancement strategies in food technology.
1. Introduction
The exploration of traditional Chinese medicine (TCM), particularly the concept of medicine-food homology, offers a unique perspective on the therapeutic and dietary value of natural products (Gong, Ji, Xu, Zhang, & Li, 2020; Zhang et al., 2023). This ancient wisdom posits that food and medicine share common sources and effects, enhancing both health and sensory attributes in culinary practices. Integrating these traditional insights with modern scientific approaches, especially in flavor chemistry, opens new avenues for understanding the complex interplay between diet and health. This convergence is reshaping dietary practices and steering the development of functional foods that cater to both health efficacy and sensory appeal (Yin et al., 2021; Yin et al., 2023).
Advancements in analytical techniques, such as high-performance liquid chromatography (HPLC), gas chromatography–mass spectrometry (GC–MS), and liquid chromatography-mass spectrometry (LC-MS), have revolutionized our ability to identify and quantify the complex array of flavor compounds in foods (Weng et al., 2021). These methodologies provide a detailed understanding of the chemical foundations of flavor and its implications for food safety, quality, and sensory properties. The advent of ‘omics’ technologies—metabolomics, proteomics, and genomics—further enhances our capability to dissect the molecular profiles of foods, offering insights into how these compounds interact with human biology to influence taste, aroma, and health outcomes (Datir & Regan, 2022; Li et al., 2021).
Despite significant strides, the food industry faces ongoing challenges in maintaining the balance between flavor enhancement and nutritional integrity. Processing techniques often alter the flavor profile of food, sometimes at the expense of nutritional value (Utpott, Rodrigues, de Oliveira Rios, Mercali, & Flôres, 2022). Additionally, scaling biotechnological methods such as microbial fermentation and enzymatic modification presents challenges in consistency and acceptability. However, these methods hold promise for developing novel flavor compounds that can enhance health benefits without compromising taste or safety (Soto-Vaca, Gutierrez, Losso, Xu, & Finley, 2012).
Sensory science plays a crucial role in translating chemical profiles into perceptible sensory experiences, effectively bridging the gap between the biochemical properties of food compounds and their perceived flavors. This field has evolved to encompass not only basic tastes but also the intricate sensations of aroma, texture, and mouthfeel (Koehler et al., 2024). Sensory evaluations are essential for understanding how processing methods, storage conditions, and culinary preparations impact the overall flavor profile of foods. This knowledge guides the optimization of processing techniques to preserve or enhance the sensory quality of food products (Colbert, Triplett, & Maier, 2022).
In our study, we introduce PungentDB, a comprehensive database that integrates the principles of TCM with modern scientific methodologies to explore the pungent flavors of foods and their medicinal correlations. PungentDB includes detailed chemical, biological, and sensory information on pungent compounds identified from TCM sources, providing an innovative tool for researchers and practitioners alike to understand and leverage the health-promoting properties of these compounds. Our research addresses the gap in systematic documentation and provides a platform for the scientific community to study the complex interactions between pungent flavors and their sensory and therapeutic effects. This approach supports the advancement of flavor science and contributes to the development of functional foods that are both nutritious and appealing.
2. Material and methods
2.1. Data acquisition
We collected compounds from pungent TCMs of medicine food homology and identified pungent compounds from various databases including FlavorDB (Garg et al., 2018), FooDB (http://foodb.ca), and Flavornet (Arn & Acree, 1998), FEMA(Adams et al., 2004). Information of pungent TCMs of medicine food homology were extracted from pharmacopoeia of the People's Republic of China (2015), which consists of 5608 different species of drugs, including herbs, plant oils and extracts. The pungent TCMs' compounds were sourced from TCMSP (Ru et al., 2014), TCMID (Huang et al., 2018), BATMAN-TCM (Kong et al., 2024), TCM-Mesh (Zhang, Yu, Bai, & Ning, 2017), TCM Database@Taiwan (Chen, 2011). The 2D/3D chemical structures, physicochemical properties, and SDF/XML files for most pungent flavor molecules were obtained from PubChem (Wang et al., 2009). Users can download these files in either 2D or 3D formats.
We compiled a set of physical descriptors and identifiers for these compounds, including PubChem CID, FEMA number, InChI, InChI Key, CAS registry number, molecular weight, logP, XlogP (XlogP-AA), Smiles molecular solubility, water solubility, H-bond donors, H-bond acceptors and Rotatable bonds, flavor profile, taste and odors, drawing from PubChem (Wang et al., 2009), FlavorDB (Garg et al., 2018), FooDB (http://foodb.ca), FEMA (Adams et al., 2004). The water solubility descriptors for some pungent compounds were computed using the ALOGPS 2.1 program (free resources available at http://www.vcclab.org/lab/alogps/).
The targets information of pungent compounds were extracted from publications using Pubtator (Wei, Kao, & Lu, 2013) and several disease-gene databases including DrugBank (http://www.drugbank.ca/), STITCH (http://stitch.embl.de/), ChEMBL (https://www.ebi.ac.uk/chembl/), BindingDB (http://www.bindingdb.org/bind/index.jsp). Therapeutic Target Database (http://bidd.nus.edu.sg/ group/ttd/), and Comparative Toxicogenomics Database (CTD, http://ctdbase.org/).
Relevant pungent sensory taste receptors, including TRPs and ORs families, and their IC50 and EC50 values were derived from literatures and ChEMBL (Gaulton et al., 2012) (https://www.ebi.ac.uk/chembl/). These sources span five major publishers: Elsevier ScienceDirect (https://www.sciencedirect.com/), Nature (https://www.nature.com/), Science Online (http://www.sciencemag.org/), Springer (https://link.springer.com/), Wiley (http://onlinelibrary.wily.com). Notably, 20 % of these sources were published in Cell, Nature, Science journals and their sub-journals.
2.2. Identification of the pungent flavor compounds
To identify the pungent compounds in TCMs, this study employed two main approaches. The first approach involved cross-referencing compounds from pungent TCMs with known databases of pungent compounds. These databases included the GB2760–2014 National Food Safety Standard and Food Additives Use Standard (China), FlavorDB (Garg et al., 2018), FooDB (http://foodb.ca), Flavornet (Arn & Acree, 1998), and FEMA (Adams et al., 2004). By intersecting these databases, a comprehensive list of pungent compounds was compiled.
The second approach focused on identifying relevant compounds through extensive literature review. This involved gathering data on the interactions of compounds with sensory receptors, specifically the transient receptor potential (TRP) and olfactory receptor (OR) families. The pungency is attributable to chemicals that activate a specific transient receptor potential (TRP) cation channel family and olfactory receptors (ORs). Moreover, the ligands of TRPs are for the most part spices and flavor molecules. Data collection was performed from journal articles published by major publishers such as Elsevier ScienceDirect, Nature, Science Online, Springer, and Wiley. This thorough literature review ensured a robust dataset of pungent compounds linked to sensory receptors.
Through these methods, we ensure the comprehensive identification of pungent compounds in TCMs, leveraging both established databases and extensive literature to provide a thorough understanding of these compounds and their sensory impacts.
2.3. Organ target location mapping and system distribution with gene organizer
In Traditional Chinese Medicine (TCM), meridian theory suggests that a TCM's therapeutic efficacy is directed towards specific organs or meridians, indicating its target within the body. Due to the limitation of most expression datasets, there is a strong bias towards certain organs and tissues (e.g. brain, blood and skin), and many other body parts are rare or completely absent. TCM theory focuses on the interaction of TCMs with human body with a holistic view. To map these targets accurately, we utilized Gene ORGANizer (Gokhman et al., 2017), a comprehensive and fully curated database containing over 150,000 gene-body part associations, covering more than 7000 human genes. Gene ORGANizer provides robust evidence for understanding the organ target locations and system distribution of pungent compounds from TCMs, thus elucidating the concept of meridian tropism at the molecular level.
The methodology involved mapping the targets of pungent TCM compounds to organ-specific locations and human systems using Gene ORGANizer. This was achieved by analyzing the correlation between genes and their associated organs and systems based on phenotypic data. The research aimed to establish a foundational understanding of the relationship between TCM meridian theory and organ-specific targeting, focusing on phenotype-genotype relationships. This process is crucial for accurate clinical diagnosis and treatment, as symptom phenotypes are diligently observed by physicians.
2.4. Assay of pungent compounds
The assay data for pungent compounds in PungentDB were sourced from ChEMBL and peer-reviewed scientific publications. Data extraction involved a meticulous manual curation process to ensure accuracy and relevance. ChEMBL, a comprehensive bioactivity database, provided detailed bioassay information, including IC50 and EC50 values, target names, assay organisms, and activity types (Gaulton et al., 2012). Additionally, relevant data were manually extracted from various peer-reviewed journals, capturing detailed bioactivity profiles of pungent compounds.
Each compound's assay data were annotated with its name, PubChem Identifier (PID), structure (2D and 3D), IC50 value, EC50 value, assay organism, and target names. The data were organized to link each compound with its respective assays and targets, creating a comprehensive list that includes the PID, substance name, target name, and assay references. This process ensured that each entry was cross-referenced with relevant literature sources and ChEMBL entries, enabling users to access detailed bioactivity records and additional resources through the ‘Target Report Card’ and ‘Document Report Card’ in ChEMBL.
2.5. Molecular structure search
To facilitate the identification of pungent compounds, we implemented a 2D structure similarity search feature within PungentDB. This feature allows users to retrieve compounds similar to a given chemical structure query. The JSME Molecule Editor (Bienfait & Ertl, 2013) (http://peter-ertl.com/jsme/), an advanced tool for drawing chemical structures, was integrated into the database as an optional built-in editor. This interface enables users to sketch structures or substructures of interest for their queries.
For the actual similarity search, we utilized RDKit (Bento et al., 2020). The 2D structures of small molecules were converted into SMILES format, facilitating efficient chemical structure similarity searches. The similarity between the query molecule and database entries is measured using the Tanimoto coefficient, a standard 2D similarity metric. The Tanimoto coefficient ranges from 0 to 1, with 1 indicating maximum similarity. The results of the similarity search depend on the user's specified threshold settings.
2.6. Advanced search
To enable refined searches, the ‘Advanced Search’ feature in PungentDB was developed to allow users to query the database based on a wide array of molecular properties and identifiers. The design process involved integrating multiple search parameters to facilitate complex queries. Key molecular properties included in the search criteria are molecular weight, hydrogen bond donors and acceptors, rotatable bonds, LogP, water solubility, and XLogP3 (or XLogP3-AA). Additionally, the search functionality supports queries by various compound identifiers such as PungentDB ID, PubChem CID, SMILES, FEMA number, InChI, InChI Key, CAS registry number, IUPAC systematic name, and molecular formula.
To construct this feature, the database schema was enhanced to index these properties efficiently. An intuitive user interface was designed to allow users to specify search criteria easily, either by entering textual data or using a chemical structure drawing tool. The backend processing uses advanced algorithms to match the search criteria with the database entries, ensuring quick and accurate retrieval of relevant compounds. Moreover, the system supports the inclusion of receptor-specific searches, allowing users to filter compounds based on their interaction with specific pungent taste receptors (TRP family) and olfactory receptors (ORs). This was achieved by linking the molecular properties of compounds with known receptor interactions, compiled from extensive literature and database sources.
2.7. Visualization tools/features
To facilitate the exploration of pungent flavor molecules and their interactions, PungentDB employs a suite of advanced visualization tools and features. These tools enable users to search for structurally similar molecules within the database and provide interactive data visualizations for quick access to desired information. PungentDB uses Cytoscape.js (v3.5.0, http://js.cytoscape.org/) to construct dynamic, browser-based networks that display multilevel interactions among pungent TCMs, pungent compounds, and their targets (TRPs and ORs). Additionally, D3.js (https://d3js.org/) is utilized to visualize the network of pungent flavor compound-pharmacological actions, illustrating multiple pharmacological effects exerted by each pungent compound.
For 3D structure visualization, LiteMol Viewer (Sehnal et al., 2017) (https://litemol.org) provides efficient visualization of chemical structures, including biological context annotations from resources such as UniProt. 3Dmol.js (Rego & Koes, 2015) (http://3dmol.csb.pitt.edu) offers a high-performance interactive viewer for 3D molecular data, while NGL Viewer (Rose & Hildebrand, 2015) (http://proteinformatics.charite.de/) uses WebGL to display molecules with various representations. For 2D/3D structure display, users can interactively sketch in a structure or substructure of interest using JSME Molecule Editor (Bienfait & Ertl, 2013) (http://peter-ertl.com/jsme/), and perform structural similarity searches with RDKit (Bento et al., 2020), using the Tanimoto coefficient to assess the degree of similarity between the query molecule and database entries.
The D3.js JavaScript library is also used to implement organ location maps and system distributions for pungent compounds' targets based on gene phenotype, aiding in the visualization of how these compounds interact with different parts of the human body.
2.8. Visualization and interaction of pungent compounds with TRP and OR receptor structures
To support the visualization and interactive exploration of TRP and olfactory receptor (OR) family macromolecules, we implemented several advanced tools within PungentDB. We created a custom database consisting of protein sequences from 71 TRP receptors and 33 ORs (both human and other species). The sequences are searchable using PungentDB's local BLAST service, powered by the NCBI BLAST package/Webblast and ViroBLAST (Chen, Li, Hou, Zhang, & Qiao, 2021; Johnson et al., 2008). This service facilitates the search for sequence similarities across multiple databases, aiding in the study of TCM genomics (Deng, Nickle, Learn, Maust, & Mullins, 2007).
We integrated high-performance interactive viewers such as 3Dmol.js, LiteMol Viewer, and NGL. These tools enable users to view, manipulate, and analyze 3D molecular data directly within their web browsers. 3Dmol.js provides a full-featured API and a straightforward interface for embedding molecular data (Rego & Koes, 2015). LiteMol Viewer offers rapid 3D visualization and uses annotations from UniProt to map protein functions and regulatory activities (Sehnal et al., 2017). NGL Viewer displays large molecular complexes using WebGL, supporting various representations like cartoon, spacefill, and licorice (Rose & Hildebrand, 2015).
2.9. Webserver tech stack
PungentDB has been designed as a Relational Database using SQLite (https://www.sqlite.org/index.html). The webserver has been built using the Python web development framework, flask (http://flask.pocoo.org/). Flask has no database abstraction layer, form validation, or any other components where pre-existing third-party libraries provide common functions (https://en.wikipedia.org/wiki/Flask(web_framework)). However, Flask supports extensions that can add application features as if they were implemented in Flask itself. More specifically, this is done by means of a Flask micro-framework over a Nginx reverse server and a Gunicorn server executing Flask (Mufid, Basofi, Al Rasyid, & Rochimansyah, 2019). The front-end has been built using HTML5, CSS and JavaScript. The jQuery, Bootstrap, D3.js and cytoscape.js were used to add to the functionality of PungentDB. Ningx (http://nginx.org/en/) is an HTTP and reverse proxy server, a mail proxy server, and a generic TCP/UDP proxy server. The basic HTTP server features are serving static and index files, accelerating reverse proxying with caching and loading balancing and fault tolerance. Moreover, Ningx can limit the number of simultaneous connections or requests coming from one address. The site is best viewed in latest versions of Firefox, Google Chrome, Opera, Internet Explorer, Safari and Microsoft Edge.
3. Results
3.1. Database overview
The database offers interactive tools for browsing, searching, and visualizing pungent flavor compounds and their associated receptors, facilitating research on the chemical and sensory attributes of these compounds.
PungentDB combines different dimensions of pungent flavor constituting the ‘Protein/DNA Blasting Layer’, ‘Entity Layer’, and ‘Visualization Layer’ (Fig. 1). The former incorporates facets of ingredients which are entities from plant sources often used in TCMs, whereas the latter represents molecules responsible for pungent flavor sensation associated receptors and their descriptors. By bringing relevant information from pungent TCMs of medicine food homology, PungentDB provides a comprehensive dataset backed by a user-friendly interface, creative visualizations, and pungent flavor compounds sources for exploring features that contribute to the potential medical efficacy. Thus, PungentDB paves the way for an improved understanding of pungent flavor perception from pungent TCMs of medicine food homology arising out of complex interplay of pungent flavor compounds with biological systems and medical applications in pungent TCMs of medicine food homology.
Fig. 1.
PungentDB is a seamless amalgamation of ‘Protein/DNA Blasting Layer’, ‘Entity Layer’, and ‘Visualization Layer’. The resource provides a comprehensive dataset along with a userfriendly interface, visualization and interlinked search engines for exploring the pungent flavor universe from pungent TCMs of medicine food homology.
3.2. Compounds data acquisition results
We collected and identified more than 20,000 compounds from 231 pungent TCMs of medicine food homology. The collected data include 2D/3D chemical structures, physicochemical properties, and identifiers for each pungent compound, such as PubChem CID, FEMA number, InChI, InChI Key, CAS registry number, molecular weight, logP, and XlogP (XlogP-AA). The water solubility for some pungent compounds was computed using the ALOGPS 2.1 program. The targets information of 205 pungent compounds was extracted from publications and several disease-gene databases.
3.3. Identification of pungent flavor compounds
We identified a comprehensive list of pungent compounds present in Traditional Chinese Medicines (TCMs) by integrating established databases and extensive literature. This approach provided a thorough understanding of these compounds and their sensory impacts. The identified pungent compounds were found to activate specific transient receptor potential (TRP) cation channels and olfactory receptors. This finding highlights the interaction of flavor molecules with biological sensory mechanisms, a phenomenon historically recognized by ancient Chinese medical practitioners through long-term clinical practice.
PungentDB (http://www.pungentdb.org.cn/home/) compiles data on over 205 pungent compounds from 231 TCMs, including molecular properties, biological targets, pharmacological actions, and compound identifiers. The dataset for further analysis includes identified pungent flavor compounds from FlavorDB, FooDB, Flavornet, FEMA, and the pharmacopoeia of the People's Republic of China. The GB2760–2014 national standard of spice served as a reference for identifying these pungent flavor compounds. So the spices flavor molecules were considered as the basis of pungent flavor. These compounds were consistently found to activate specific TRP cation channels and olfactory receptors, thus validating their roles as the fundamental elements of pungent flavor.
3.4. Organ target location mapping
Using Gene ORGANizer (Gokhman et al., 2017), we mapped the targets of pungent TCMs' pungent flavor compounds into an organ location map/human system level. The correlation of genes associated with organs and human systems was analyzed based on phenotype. This mapping provides strong evidence for elucidating the connotation of meridian tropism at the target level.
The utilization of Gene ORGANizer allowed for the detailed mapping of pungent TCM compounds' targets into specific organs and human systems. This mapping revealed significant correlations between genes associated with particular organs and their phenotypic expressions, providing strong evidence for the concept of meridian tropism at the target level.
The results demonstrated that certain organs and tissues (e.g., brain, blood, and skin) are more frequently represented in the expression datasets, while many other body parts are underrepresented or absent. This highlights the need for comprehensive datasets to achieve a holistic view of TCM interactions with the human body. By mapping these targets, we were able to provide a more nuanced understanding of how pungent compounds interact with various body systems, which is essential for elucidating the therapeutic mechanisms of TCM.
Overall, this research significantly contributes to the understanding of organ-specific targeting and the practical application of TCM meridian theory in modern medical practice. It lays the groundwork for future studies aimed at exploring the intricate relationships between TCM theories and contemporary biological evidence.
3.5. Assay results
The activity data for the pungent compounds, including IC50 and EC50 values, were extracted from ChEMBL and 4329 full-text peer-reviewed publications. The data includes structures, annotations, assay organism, target names, and activity types. The assay list, with links to relevant pungent compounds and their targets, is accessible via PungentDB. The list of assays for pungent compounds is displayed with the links to the PID, Name (substances that have been tested for their bioactivity), Target Name (the proteins or systems being monitored by an assay), Assay References (literature source for the assay) (Supplementary Fig. S1). The ‘Target Name’ and ‘Assay References’ in this assay list link to the ‘Target Report Card’ and ‘Document Report Card’ in ChEMBL, providing users with comprehensive details and facilitating deeper exploration of pungent compounds and their bioactivities.
3.6. Molecular structure search
The implementation of the 2D structure similarity search in PungentDB provides users with a powerful tool to identify compounds with similar chemical structures. Users can easily sketch a structure or substructure of interest using the JSME Molecule Editor. The RDKit-powered search then converts these structures into SMILES format and calculates the Tanimoto coefficient to determine the similarity between the query molecule and database entries. This process allows for precise identification of structurally similar pungent compounds, aiding in the exploration and analysis of the molecular landscape of pungent TCMs. The feature supports a wide range of research applications, from basic research to more advanced pharmacological studies.
3.7. Advanced search
The advanced search feature allows for refined queries based on molecular properties, compound identifiers, and associated receptors. Users can browse a table with all PungentDB compounds and their properties, providing detailed information on pungent flavor molecules.
The implementation of the ‘Advanced Search’ feature in PungentDB significantly enhances the database's functionality. Advanced Search provides an option for refined search by querying PungentDB data on the basis of a variety of molecular properties (Mol. Weight, Hydrogen Bond Donor, Hydrogen Bond Acceptor, Rotatable Bonds, LogP, Water solubility, XLogP3 or XLogP3-AA), compound identifiers (PungentDB ID, PubChem CID, Smiles, FEMA number, InChI, InChI Key, CAS registry number, IUPAC systematic name, Mol. Formula), associated pungent taste receptors (TRPs family), and olfactory receptors (ORs), etc. Users can also perform detailed searches using a combination of molecular properties and identifiers, facilitating the identification of compounds that meet specific research criteria. For example, users can filter compounds by molecular weight ranges, hydrogen bonding characteristics, or specific identifiers like PubChem CID and SMILES (see Possible usage 1 and Fig. S1 A (III)). This advanced querying capability allows researchers to pinpoint compounds with desired properties quickly. For numeric fields either a range or discrete values can be provided as a query and subsequently obtain details of the pungent flavor molecules.
Furthermore, the ability to search based on receptor interactions enables targeted studies on pungent taste and olfactory receptors. This feature provides comprehensive data on the association between compounds and their receptor targets, aiding in the exploration of structure-activity relationships. The search results are presented in a user-friendly format, with a comprehensive table listing all matching PungentDB compounds and their properties. Users can easily browse and analyze the detailed information on each compound, including its molecular structure, physical properties, and bioactivity data.
Overall, the ‘Advanced Search’ functionality in PungentDB offers a powerful tool for researchers, enhancing the ability to perform complex queries and retrieve detailed information on pungent flavor compounds efficiently. This feature supports a wide range of research applications, from basic science to applied studies in flavor chemistry and traditional medicine.
3.8. Visualization tools/features on pungent flavor compounds and their profiles
PungentDB provides a range of visualization tools, including 2D/3D chemical structure displays generated using LiteMol Viewer, Cytoscape.js, D3.js, 3Dmol.js, and NGL. These tools offer interactive ways to explore the pungent compounds from TCMs, visualize their networks, biological activities, organ location map and system distribution feature and inspect features of associated receptors.
The visualization tools and features in PungentDB provide users with a comprehensive and interactive platform to explore the pungent flavor universe from TCMs. The Cytoscape.js-based network module allows for the exploration of multilevel interactions among pungent TCMs, pungent compounds, and their targets. Users can click on entities to view their associations with TCMs and related pungent flavor compounds, facilitating an in-depth understanding of these relationships (Fig. 2A). The D3.js visualizations illustrate the multiple pharmacological effects exerted by each pungent compound, providing insights into their diverse biological activities (Fig. 2B).
Fig. 2.
Visualization tools/features in PungentDB.
For 3D structure visualization, LiteMol Viewer enables users to efficiently view and manipulate the 3D structures of pungent compounds, including rotating the structures 360 degrees and capturing screenshots. It also provides biological context annotations from UniProt, enhancing the user's understanding of the molecular basis of pungency (Fig. 2C).
To make it easier to determine pungent compounds act on human body parts, organ location map/system distribution for pungent compounds' targets based on gene phenotype was implemented with the D3.js JavaScript library (https://d3js.org) (Fig. 2D).
Overall, these visualization tools and features enhance the usability of PungentDB, enabling researchers to conduct detailed and interactive explorations of pungent flavor compounds from TCMs, thereby supporting their scientific inquiries and advancing the field of flavor chemistry.
3.9. Visualization and interactive analysis of TRP and OR receptor structures
The entity details of particular human associated pungent taste receptors (TRPs family), olfactory receptors allows users to interactively inspect the features of receptors listed in the Receptors associated to pungent tab.
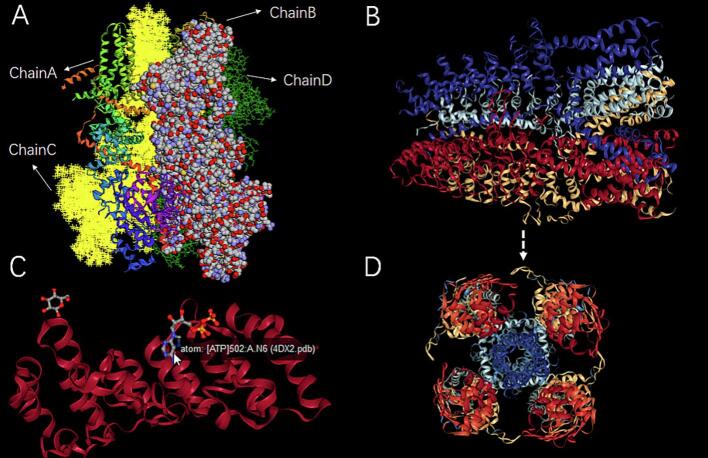
3Dmol.js is an high-performance interactive viewer for 3D molecular data that requires no plugins to work in web browsers and it provides a full featured API for developers as well as a straightforward declarative interface that lets users easily share and embed molecular data in websites (Rego & Koes, 2015). This biomacromolecular consists of four chains, which are shown in different representation and color modes (Fig. 3A).
Fig. 3.
Gallery of 3D molecular structures of TRPs family proteins visualizations showing various representations with three different molecular viewers. (A) The structure of a TRPs ion channel (PDB ID: 6BQV). This molecule consists of four chains with 3Dmol.js, which are shown in different representation and color modes. Chain A is presented as cartoon mode colored by spectrum; Chain B is presented as sphere mode colored by spectrum; Chain C is presented as cross mode colored by yellow; Chain D is presented as line mode colored by green. (B) and (D) are generated biomacromolecular structure of TRPA1 (PDB ID: 3J9P) with NGL from different perspectives. (C) The structure of a TRPs ion channel (PDB ID: 4DX2) is implemented with the molecular viewer LiteMol. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The web page enables visualization and manipulation with the 3D structure using LiteMol Viewer (Sehnal et al., 2017), which is powerful and blazing-fast tool for handling 3D macromolecular data in the browser (https://litemol.org). LiteMol uses annotations from UniProt (e.g. a function of protein as well as activity regulation) and maps them on the protein annotations section (Fig. 3C).
Moreover, there were another optional Viewers NGL (http://proteinformatics.charite.de/), 3Dmol (http://3dmol.csb.pitt.edu). NGL Viewer is a web application for molecular visualization. WebGL is employed to display molecules like proteins and DNA/RNA with a variety of representations. NGL can interactively display large molecular complexes and is also unaffected by the retirement of third-party plug-ins like Flash and Java Applets (Rose & Hildebrand, 2015). It supports common structural fileformats (e.g. PDB, mmCIF) and a variety of molecular representations (e.g. ‘cartoon, spacefill, licorice’) (Fig. 3B, D).
Users can visualize the 3D structures of these receptors, which are essential for understanding the molecular basis of their interactions with pungent flavor compounds. The interface supports various representations, providing a detailed view of the molecular architecture.
The integration of LiteMol Viewer allows for efficient manipulation of 3D structures, facilitating detailed analysis of ligand-receptor interactions. Users can rotate structures, zoom in on specific areas, and capture screenshots for further study. NGL Viewer supports interactive display of large molecular complexes, unaffected by the limitations of traditional plug-ins, thus enhancing the user experience (Fig. 3B, D).
PungentDB's local BLAST service, TCM-Blat (Chen et al., 2021), powered by the NCBI BLAST package/Webblast and ViroBLAST, allows for searching TRP and olfactory receptor protein sequences. This service supports TCM genomics research by providing sequence similarity searches against multiple databases.
Overall, the visualization tools in PungentDB provide a comprehensive and interactive platform for exploring the structural features of TRP and OR family receptors, significantly contributing to the understanding of their roles in sensory perception and pharmacological effects.
4. Discussion
The food industry faces ongoing challenges in balancing flavor enhancement with nutritional integrity. Processing techniques often modify the flavor profile of food, sometimes at the expense of nutritional value. Additionally, scaling up biotechnological methods like microbial fermentation and enzymatic modification presents challenges in consistency and acceptability (Nielsen, Tillegreen, & Petranovic, 2022). Yet, these methods offer potential for developing novel flavor compounds that enhance health benefits without compromising taste or safety (Hu et al., 2023; Yamamoto & Inui-Yamamoto, 2023).
The simple perception of pungent TCMs of medicine food homology arises from the interaction of flavor molecules with the biological machinery by ancient medical practitioners' human sensory system. Flavors derived from TCMs have shaped the pungent flavor throughout ancient long-term clinical practice in China.
Pungent flavor is an expression of pungent olfactory and gustatory/taste sensations experienced through a multitude of chemical processes triggered by pungent molecules. Pungent compounds widely exist in pungent Traditional Chinese Medicines (TCMs) of medicine food homology. Pungent flavor in TCM term (sour, salty, sweet, bitter and pungent) serve specific functions in identifying materia medica compounds found in long term clinical practice of traditional Chinese medicine (TCM), and are recognized by some specific pungent taste receptors (transient receptor potential cation channel subfamily, TRPs) or olfactory receptors (ORs) in the oral cavity and nasal cavity. Interestingly, pungent flavor in TCMs indicate not only pungent taste but also pungent odor. Due to the limitation of biomedical level in ancient times, ancient Chinese physicians could recognize and discover medicinal herbs through human basic perception (taste, smell, touch, etc.) (An et al., 2020). They defined pungent taste, spicy smell or aromatic odor herbs as pungent flavor TCMs. The pungent flavor in TCM term was the objective expression of sensory taste (taste, olfaction, etc.) by Chinese ancient physicians. However, they didn't know material basis and the pungent flavor's molecular mechanism of pungent flavor TCMs (Z. Chen, Cao, Zhang, & Qiao, 2019). PungentDB was created with the aim of integrating multidimensional aspects of pungent flavor molecules and representing their molecular features, flavor profiles and details of related pungent flavor TCMs.
To summarize, taste recognition is complexity and import for pungent TCMs of medicine food homology sensing and odors. The pungent recognition is possibly plays additional physiological roles and medical value. Characterizing molecules as pungent and assigning them to a specific receptor represents a crucial step towards prediction and rational modulation of spiciness. Creating a database of known pungent compounds, which, in addition to associations between pungent molecules and their receptors, also contains the chemicophysical properties of the ligands and organ location/system distribution data relevant for pungent molecules' targets, is an essential step to discover the potential efficacy in this direction.
4.1. Integration of TCM and flavor chemistry
Our database not only aggregates chemical data but also aligns it with TCM principles, offering a unique synthesis not extensively covered in current literature. Previous studies, such as those by Gong et al.(Gong et al., 2020) and Zhang et al. (K. Zhang et al., 2023), have separately discussed the roles of TCM in food science and the chemical analysis of flavors. PungentDB bridges these aspects by providing a platform that correlates pungent compounds with their historical and therapeutic contexts in TCM, offering a holistic view that was previously fragmented.
The methodologies employed for data collection and analysis in PungentDB improve upon traditional techniques by incorporating advanced computational tools that enhance the accuracy and granularity of data on pungent flavors. Our database utilizes state-of-the-art bioinformatics tools to systematically categorize and analyze pungent compounds, thus providing more detailed insights into their potential applications.
The comprehensive data compilation in PungentDB, particularly regarding the interaction of pungent compounds with TRP and OR receptors as observed in our results, paves the way for future experimental and clinical research. By providing a detailed biochemical context, our database supports the development of new hypotheses regarding the sensory and therapeutic roles of pungent compounds. This aspect is slightly touched upon in existing databases but is extensively detailed in PungentDB, offering a robust foundation for advancing research in food chemistry and pharmacology.
4.2. Comparison of database content and methodology
Our methods of data collection, particularly the integration of 2D/3D chemical structures, physicochemical properties, and extensive metadata for each compound, follow rigorous standards that surpass many existing databases. For instance, while databases such as PubChem (Kim et al., 2023) and FooDB (http://foodb.ca) provide similar types of data, PungentDB incorporates specific TCM-related attributes that are often overlooked, such as associations with pungent flavor and therapeutic roles as highlighted by sources like the pharmacopoeia of the People's Republic of China and TCMID (Huang et al., 2018).
The use of ALOGPS 2.1 (Tetko & Tanchuk, 2002) for computing water solubility and gathering target information from multiple disease-gene databases demonstrates an advanced approach not typically employed in traditional databases. This methodological sophistication allows for a more accurate prediction of compound behavior in biological systems, which is crucial for the practical application of these compounds in food science and medicine.
By collating an extensive range of identifiers and properties (e.g., PubChem CID, FEMA number, molecular weight, logP), PungentDB provides a more comprehensive tool for researchers. The database's focus on pungent TCMs fills a significant gap in existing literature by linking these compounds' sensory and chemical properties to their historical and medicinal use in TCM, which is rarely addressed together in other databases.
4.3. Identification techniques and database integration
Our methodology for identifying pungent flavor compounds through the integration of established databases (e.g., FlavorDB, FooDB, Flavornet, FEMA) and literature enhances the depth of our analysis. This approach aligns with the practices discussed in the pharmacopoeia of the People's Republic of China and is further supported by our use of the GB2760–2014 national standard.
The compounds we identified were consistently found to activate specific transient receptor potential (TRP) cation channels and olfactory receptors, validating their roles in pungent flavor perception. This finding is critical as it underscores the biochemical basis of sensory perceptions discussed in studies like those by Chen et al. (Chen et al., 2019), which explore the molecular mechanisms underlying the sensory properties of pungent compounds.
4.4. Organ-specific target mapping
Our approach with Gene ORGANizer provided a novel insight into the organ-specific effects of pungent compounds, which aligns with and expands upon foundational studies in pharmacology and TCM, such as those by Gokhman et al. (Gokhman et al., 2017) who developed the Gene ORGANizer tool. Our findings demonstrate a detailed correlation between gene expressions and organ-specific effects, supporting the theory of meridian tropism in TCM at a molecular level.
By analyzing the correlations between genes and phenotypic expressions associated with different organs, our study provides empirical evidence supporting traditional claims of TCM. These insights have important implications for the development of targeted therapeutic strategies using TCM principles. Future research should focus on expanding these organ-target mappings with more comprehensive genomic data.
4.5. Bioactivity and assay data
The assay results from ChEMBL and various peer-reviewed publications provided detailed bioactivity data, including IC50 and EC50 values for pungent compounds. This comprehensive collection mirrors the approach seen in large-scale bioactivity databases and studies such as those by Gaulton et al. (Gaulton et al., 2012) and is in line with the methodologies employed in high-throughput screening in pharmaceutical research. Our integration of this data into PungentDB enhances its utility for exploring pungent compounds' pharmacological potentials.
Our approach to making these assay results accessible through PungentDB with links to PID, substance names, target names, and assay references allows users to explore the bioactivities of compounds in a user-friendly manner. This method of data presentation and integration supports the emerging trend of combining cheminformatics tools with traditional databases to improve data accessibility and utility, as highlighted in the work by Johnson et al. (Johnson et al., 2008).
4.6. 2D/3D structure similarity search
Our implementation of the 2D structure similarity search in PungentDB leverages the RDKit toolkit to compute Tanimoto coefficients for molecular similarity assessments, aligning with the practices described in contemporary cheminformatics research (Bento et al., 2020).This approach allows for robust identification of structurally similar pungent compounds, enhancing the database's utility for pharmacological and flavor science research.
Unlike conventional databases that may offer limited search functionalities, PungentDB integrates the JSME Molecule Editor, enabling users to intuitively sketch structures or substructures (Bienfait & Ertl, 2013). This feature reflects advancements discussed by Zdrazil et al. (Zdrazil et al., 2024) in ChEMBL, which similarly aims to provide comprehensive access to chemical data for broader scientific applications.
Building on this foundation, future enhancements to PungentDB will include the integration of 3D structure search capabilities and improved algorithms for molecular similarity to accommodate more complex queries and diverse research needs. These developments are anticipated to align with emerging trends in database utility and accessibility, driving forward the fields of flavor chemistry and pharmacology.
4.7. Integration of advanced search capabilities
PungentDB's advanced search feature offers significant advancements over traditional database searches by allowing highly refined queries based on a wide range of molecular properties and identifiers such as molecular weight, LogP, and receptor associations. This feature is crucial for researchers focusing on the specificity of interactions between pungent compounds and sensory receptors, which is less emphasized in databases like ChEMBL or PubChem. By integrating these functionalities, PungentDB provides a tailored tool for exploring pungent compounds in Traditional Chinese Medicines (TCMs), enhancing the research efficiency in pharmacological and flavor chemistry studies.
Unlike general chemical databases, PungentDB is specifically designed to address the unique aspects of pungent compounds in TCMs. For instance, while tools like the RDKit and ChemSpider (Pence & Williams, 2010) offer chemical search capabilities, they do not provide integration with TCM-specific receptor data. PungentDB fills this gap by combining chemical searches with detailed receptor information, particularly the TRP and OR families relevant to pungency perception. This integration is supported by extensive literature on receptor-ligand interactions that influence the sensory perception of pungency, adding a layer of specificity that is crucial for the application of TCMs in both medicinal and culinary contexts.
Several studies underscore the importance of integrating chemical properties with receptor data to enhance the understanding of flavor and pharmacological profiles. For instance, research by Humbeck & Koch (Humbeck & Koch, 2017) on bioactivity databases highlights the need for specialized tools that cater to specific scientific inquiries such as those provided by PungentDB. Moreover, the work by Gong et al. (Gong et al., 2020) on the sensory and health impacts of bioactive compounds aligns with our approach to enhance the usability of data for TCM applications.
4.8. Visualization tools in PungentDB
PungentDB utilizes a variety of advanced visualization tools such as LiteMol Viewer, Cytoscape.js, D3.js, 3Dmol.js, and NGL. These tools facilitate an interactive exploration of the chemical structures and biological interactions of pungent compounds derived from TCMs. The detailed visualization capabilities enable researchers to examine the molecular interactions and network dynamics of these compounds, providing a deeper understanding of their pharmacological and sensory properties.
The application of visualization tools in PungentDB mirrors advancements in computational chemistry and bioinformatics, where such technologies have been pivotal. For instance, studies like those by Sehnal et al. (Sehnal et al., 2017) and Rego et al. (Rego & Koes, 2015) have highlighted the utility of LiteMol Viewer and 3Dmol.js in enhancing the accessibility and understanding of complex molecular data. Similarly, the network visualization capabilities of Cytoscape.js align with the findings of Shannon et al. (Shannon et al., 2003), who demonstrated its effectiveness in elucidating biological pathways and protein interactions.
The incorporation of these visualization tools in PungentDB not only corroborates the methodologies employed in existing research but also extends their application to the study of pungent flavors in TCMs. By providing a platform for detailed molecular and network analysis, PungentDB enhances the existing knowledge base and supports the discovery of novel insights into the sensory and biological mechanisms of pungent compounds.
4.9. Enhancing flavor research with advanced visualization tools
The integration of advanced visualization tools such as 3Dmol.js and LiteMol Viewer in PungentDB significantly enhances our understanding of the molecular mechanisms underlying pungent flavors. These tools enable intricate examinations of the TRP and OR receptors, which play a critical role in sensory perception by interacting with pungent compounds (Rego & Koes, 2015; Sehnal et al., 2017). By providing interactive 3D visualizations, researchers can better understand the structural basis of receptor-ligand interactions, which is crucial for designing compounds with optimized sensory profiles.
Furthermore, the use of these tools allows for a detailed analysis of the interactions between pungent molecules and their target receptors, offering insights into the diverse pharmacological effects of these compounds. The interactive tools and visualization capabilities of PungentDB has set a new benchmark in the exploration of pungent flavors. Our approach enhances user interaction and data accessibility. However, PungentDB introduces specific enhancements in the visualization of flavor compound interactions with biological receptors, providing a more intuitive understanding of chemical-sensory relationships. This advancement over existing databases is crucial for both academic researchers and industry practitioners seeking deeper insights into flavor dynamics.
Looking forward, the integration of TCM with modern flavor science and food technology suggests a promising avenue for the development of next-generation functional foods. By leveraging the vast potential of food-medicine homology and advanced analytical tools, the food science community can innovate new product formulations that support both health and pleasure. This endeavor will require multidisciplinary collaborations spanning TCM, modern science, culinary arts, and technology to fully realize the potential of flavor chemistry in enhancing human health and well-being.
Despite our best efforts, PungentDB is not an exhaustive repository of all pungent molecules and ingredients from pungent TCMs of medicine food homology. Our data on pungent molecules of an ingredient is limited by the availability of information about them from literature survey and relevant databases. Similarly, the pungent compounds represented in the database are not exhaustive in themselves as their choice is limited by another pungent TCMs' compounds from magnumopus named Chinese Materia Medica.
In future, we intend to increase the coverage of pungent TCMs of medicine food homology and extract the pungent compounds from these sources. Our endeavor is to integrate aspects of pungent flavor profiles with those of entities and molecules that has hitherto been unexplored based on TRPs and ORs family receptors from newly increased pungent TCMs' chemical constituents. Fundamental questions as well as potential therapeutic applications of the modulation of pungent associated sensory taste and olfactory receptors continue to motivate further development of the PungentDB database. The number of TRPs and ORs family receptors in a given species may relate to the receptors' receptive range. The ability to understand and predict these effects relies on the knowledge of what is ‘pungent’ (sensory taste and odor) to different species, and on the ability to cluster ligands based on their chemical similarity from more pungent TCMs of medicine food homology.
PungentDB will be extended in the future to include additional species, and thus contributing to elucidating evolutionary aspects of pungent sensory taste and olfactory perception. By leveraging the vast potential of food-medicine homology and advanced analytical tools, the food science community can innovate new product formulations that support both health and pleasure. This endeavor will require multidisciplinary collaborations spanning TCM, modern science, culinary arts, and technology to fully realize the potential of flavor chemistry in enhancing human health and well-being.
As we proceed, it is crucial that the research community continues to engage with these advanced methodologies to refine our understanding of flavor chemistry and its impact on food quality and human health. The insights garnered from combining traditional knowledge with modern science will not only enrich our dietary patterns but also enhance the therapeutic potential of our meals, adhering to the ethos of medicine-food homology that has been recognized for centuries in TCM. The ongoing research efforts documented in this special issue are testimony to the dynamic and evolving field of flavor science, which remains at the forefront of scientific and culinary innovation.
5. Conclusion
PungentDB emerges as a pioneering resource, elucidating the intricate relationships between more than 205 pungent flavor compounds extracted from 231 traditional Chinese medicines (TCMs) and their interactions with taste and olfactory receptors. This database not only sheds light on the chemical essence of pungency but also highlights the potential of these compounds to significantly improve food quality, safety, and sensory experiences. It provides a comprehensive platform that combines detailed compound information, including bioactive profiles, pharmacological actions, and molecular structures, thereby enabling robust scientific inquiry and fostering innovation in the realm of food flavor chemistry.
The fusion of traditional insights with modern scientific practices through PungentDB exemplifies the significance of adopting a multidisciplinary approach in food science research. By offering sophisticated visualization tools and extensive datasets, PungentDB enhances our capacity to navigate the molecular landscape of flavors. This advancement opens new avenues for the development and refinement of food flavors, leveraging both natural and biotechnological methodologies. The database stands at the forefront, propelling the food science field towards groundbreaking discoveries in flavor enhancement and sensory analysis, underscoring the imperative of interdisciplinary collaboration to tackle the complexities of flavor chemistry.
Looking ahead, the anticipated expansion of PungentDB to include a broader spectrum of pungent compounds from varied sources is set to widen the research domain in flavor chemistry further. Future iterations aim to integrate cutting-edge ‘omics’ analyses and sensory evaluation studies, deepening our comprehension of flavor's molecular foundations and their practical implications in food science and technology. In essence, PungentDB represents a critical resource for both researchers and practitioners, facilitating significant advancements in flavor science that promise to have a lasting impact on both traditional and contemporary culinary practices.
Availability
PungentDB is available at http://www.pungentdb.org.cn/home.
Authors' contributions
ZC, YZ and YQ conceived the study. ZC drafted the manuscript. ZC, YC, XL, and JL collected the data. ZC, XS, BX and ZW analyzed the data.ZC YC YZ and YQ wrote the codes, evaluated the software, and provided scientific and conceptual interpretations.
Funding
This work was supported by the National Natural Science Foundation of China (No.81430094), Outstanding Youth Program of China Academy of Chinese Medical Sciences (Z0882), Young Talents Support Project of China Association of Chinese Medicine (2022–2024) - Level Matching Funds for Institute of Basic Research in Clinical Medicine, China Academy of Chinese Medical Sciences (Z0866).
CRediT authorship contribution statement
Zhao Chen: Writing – original draft, Visualization, Validation, Supervision, Software, Resources, Project administration, Methodology, Investigation, Data curation. Zhixin Wang: Writing – review & editing, Supervision, Investigation, Formal analysis. Yanfeng Cao: Validation, Resources, Formal analysis, Data curation. Xinyuan Shi: Visualization, Supervision, Resources, Investigation, Conceptualization. Bing Xu: Writing – review & editing, Software, Methodology, Formal analysis. Xi Li: Writing – review & editing, Validation, Methodology, Investigation, Formal analysis, Data curation. Jing Li: Visualization, Resources, Project administration, Formal analysis, Data curation. Yanling Zhang: Writing – review & editing, Supervision, Methodology, Investigation. Yanjiang Qiao: Writing – review & editing, Supervision, Methodology, Funding acquisition, Conceptualization.
Declaration of competing interest
In the preparation of our manuscript titled “PungentDB: Bridging Traditional Chinese Medicine of medicine food homology and Modern Food Flavor Chemistry,” submitted to the special issue on Flavor Chemistry of Food in Food Chemistry: X, we have meticulously adhered to the ethical standards of scientific research and publishing. We hereby declare that there are no conflicts of interest, including financial, consultative, institutional, or other relationships that might lead to bias or a conflict of interest.
Our research is purely academic in nature, aiming to contribute to the body of knowledge within the realms of food science, traditional Chinese medicine, and flavor chemistry. All data were collected and processed with the utmost integrity, and all contributors to this work have been duly acknowledged. This work has not received funding or support from any organization that could have influenced its outcome.
Furthermore, this manuscript is original, has not been published elsewhere, and is not currently under consideration by any other journal. We have cited all references appropriately to acknowledge the contributions of previous research in our work. Through this submission, we aim to contribute valuable insights into the interplay between traditional Chinese medicine and modern food flavor chemistry, with a particular focus on the chemistry of pungent flavors and their potential applications in enhancing food quality, safety, and sensory properties.
Conflict of interest statement: None declared.
Acknowledgements
We would like to thank Xingde Ren, Xiaosa Shi, Zexin Xie, Siyang Dong, Xinyue Yang, Yalan Yang, Lan Yang, Shijing Yang, Sixian Lu for data collection and valuable suggestions. We thank the National Natural Science Foundation of China (No.81430094) for its support for this work.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.fochx.2024.101742.
Contributor Information
Zhao Chen, Email: zhaochen223713@126.com.
Yanling Zhang, Email: zhangyanling@bucm.edu.cn.
Yanjiang Qiao, Email: yjqiao@bucm.edu.cn.
Appendix A. Supplementary data
Overview of the PungentDB database: This supplementary material includes a detailed presentation of the PungentDB database, highlighting its comprehensive nature and diverse applications. Additionally, we provide case studies that illustrate the utility of PungentDB across various contexts. A series of video demonstrations recorded from the PungentDB website showcases these examples, accessible via the 'Examples' link within the database interface.
Data availability
Data will be made available on request.
References
- Adams T.B., Cohen S.M., Doull J., Feron V.J., Goodman J.I., Marnett L.J., Waddell W.J. The FEMA GRAS assessment of cinnamyl derivatives used as flavor ingredients. Food and Chemical Toxicology. 2004;42(2):157–185. doi: 10.1016/j.fct.2003.08.021. [DOI] [PubMed] [Google Scholar]
- An F., Li M., Zhao Y., Zhang Y., Mu D., Hu X., Wu R. Metatranscriptome-based investigation of flavor-producing core microbiota in different fermentation stages of dajiang, a traditional fermented soybean paste of Northeast China. Food Chemistry. 2020;128509 doi: 10.1016/j.foodchem.2020.128509. [DOI] [PubMed] [Google Scholar]
- Arn H., Acree T. Flavornet: A database of aroma compounds based on odor potency in natural products. Developments in Food Science. 1998;40:27–28. doi: 10.1016/S0167-4501(98)80029-0. [DOI] [Google Scholar]
- Bento A.P., Hersey A., Félix E., Landrum G., Gaulton A., Atkinson F., Leach A.R. An open source chemical structure curation pipeline using RDKit. Journal of Cheminformatics. 2020;12:1–16. doi: 10.1186/s13321-020-00456-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienfait B., Ertl P. JSME: A free molecule editor in JavaScript. Journal of Cheminformatics. 2013;5:1–6. doi: 10.1186/1758-2946-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.Y. TCM database@Taiwan: The world’s largest traditional Chinese medicine database for drug screening in silico. PLoS One. 2011;6(1) doi: 10.1371/journal.pone.0015939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Cao Y., Zhang Y., Qiao Y. A novel discovery: Holistic efficacy at the special organ level of pungent flavored compounds from pungent traditional Chinese medicine. International Journal of Molecular Sciences. 2019;20(3):752. doi: 10.3390/ijms20030752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Li J., Hou N., Zhang Y., Qiao Y. TCM-Blast for traditional Chinese medicine genome alignment with integrated resources. BMC Plant Biology. 2021;21:1–7. doi: 10.1186/s12870-021-03096-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbert S.E., Triplett C.S., Maier J.X. The role of viscosity in flavor preference: Plasticity and interactions with taste. Chemical Senses. 2022;47:bjac018. doi: 10.1093/chemse/bjac018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datir S., Regan S. Advances in physiological, transcriptomic, proteomic, metabolomic, and molecular genetic approaches for enhancing mango fruit quality. Journal of Agricultural and Food Chemistry. 2022;71(1):20–34. doi: 10.1021/acs.jafc.2c05958. [DOI] [PubMed] [Google Scholar]
- Deng W., Nickle D.C., Learn G.H., Maust B., Mullins J.I. ViroBLAST: A stand-alone BLAST web server for flexible queries of multiple databases and user’s datasets. Bioinformatics. 2007;23(17):2334–2336. doi: 10.1093/bioinformatics/btm331. [DOI] [PubMed] [Google Scholar]
- Garg N., Sethupathy A., Tuwani R., Nk R., Dokania S., Iyer A., Kathuria K. FlavorDB: A database of flavor molecules. Nucleic Acids Research. 2018;46(D1):7. doi: 10.1093/nar/gkx957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaulton A., Bellis L.J., Bento A.P., Chambers J., Davies M., Hersey A., Al-Lazikani B. ChEMBL: A large-scale bioactivity database for drug discovery. Nucleic Acids Research. 2012;40(D1):D1100–D1107. doi: 10.1093/nar/gkr777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhman D., Kelman G., Amartely A., Gershon G., Tsur S., Carmel L. Gene ORGANizer: Linking genes to the organs they affect. Nucleic Acids Research. 2017;45(W1):W138–W145. doi: 10.1093/nar/gkx302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong X., Ji M., Xu J., Zhang C., Li M. Hypoglycemic effects of bioactive ingredients from medicine food homology and medicinal health food species used in China. Critical Reviews in Food Science and Nutrition. 2020;60(14):2303–2326. doi: 10.1080/10408398.2019.1634517. [DOI] [PubMed] [Google Scholar]
- Hu J., Chen Z., Huang X., Yan Z., Li Y., Zhu Y., Zhou P. Hyaluronic acid applied as a natural flavor enhancer and its mechanism exploration. Food Bioscience. 2023;55 doi: 10.1016/j.fbio.2023.102969. [DOI] [Google Scholar]
- Huang L., Xie D., Yu Y., Liu H., Shi Y., Shi T., Wen C. TCMID 2.0: A comprehensive resource for TCM. Nucleic Acids Research. 2018;46(D1):D1117–D1120. doi: 10.1093/nar/gkx1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbeck L., Koch O. What can we learn from bioactivity data? Chemoinformatics tools and applications in chemical biology research. ACS Chemical Biology. 2017;12(1):23–35. doi: 10.1021/acschembio.6b00706. [DOI] [PubMed] [Google Scholar]
- Johnson M., Zaretskaya I., Raytselis Y., Merezhuk Y., McGinnis S., Madden T.L. NCBI BLAST: A better web interface. Nucleic Acids Research. 2008;36(suppl_2):W5–W9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Chen J., Cheng T., Gindulyte A., He J., He S., Yu B. PubChem 2023 update. Nucleic Acids Research. 2023;51(D1):D1373–D1380. doi: 10.1093/nar/gkac956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler M., Benthin J., Karanth S., Wiesenfarth M., Sebald K., Somoza V. Biophysical investigations using atomic force microscopy can elucidate the link between mouthfeel and flavour perception. Nature Food. 2024;1-7 doi: 10.1038/s43016-024-00958-3. [DOI] [PubMed] [Google Scholar]
- Kong X., Liu C., Zhang Z., Cheng M., Mei Z., Li X., Jiang P. BATMAN-TCM 2.0: An enhanced integrative database for known and predicted interactions between traditional Chinese medicine ingredients and target proteins. Nucleic Acids Research. 2024;52(D1):D1110–D1120. doi: 10.1093/nar/gkad926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Tian Y., Jiang P., Lin Y., Liu X., Yang H. Recent advances in the application of metabolomics for food safety control and food quality analyses. Critical Reviews in Food Science and Nutrition. 2021;61(9):1448–1469. doi: 10.1080/10408398.2020.1761287. [DOI] [PubMed] [Google Scholar]
- Mufid M.R., Basofi A., Al Rasyid M.U.H., Rochimansyah I.F. 2019 international electronics symposium (IES) IEEE; 2019. Design an mvc model using python for flask framework development; pp. 214–219. [Google Scholar]
- Nielsen J., Tillegreen C.B., Petranovic D. Innovation trends in industrial biotechnology. Trends in Biotechnology. 2022;40(10):1160–1172. doi: 10.1016/j.tibtech.2022.03.007. [DOI] [PubMed] [Google Scholar]
- Pence H.E., Williams A. ChemSpider: An online chemical information resource. Journal of Chemical Education. 2010;87(11):1123–1124. doi: 10.1021/ed100697w. [DOI] [Google Scholar]
- Rego N., Koes D. 3Dmol. Js: Molecular visualization with WebGL. Bioinformatics. 2015;31(8):1322–1324. doi: 10.1093/bioinformatics/btu829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose A.S., Hildebrand P.W. NGL viewer: A web application for molecular visualization. Nucleic Acids Research. 2015;43(W1):W576–W579. doi: 10.1093/nar/gkv402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ru J., Li P., Wang J., Zhou W., Li B., Huang C., Yang L. TCMSP: A database of systems pharmacology for drug discovery from herbal medicines. Journal of Cheminformatics. 2014;6:13. doi: 10.1186/1758-2946-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehnal D., Deshpande M., Vařeková R.S., Mir S., Berka K., Midlik A., Koča J. LiteMol suite: Interactive web-based visualization of large-scale macromolecular structure data. Nature Methods. 2017;14(12):1121–1122. doi: 10.1038/nmeth.4499. [DOI] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Research. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto-Vaca A., Gutierrez A., Losso J.N., Xu Z., Finley J.W. Evolution of phenolic compounds from color and flavor problems to health benefits. Journal of Agricultural and Food Chemistry. 2012;60(27):6658–6677. doi: 10.1021/jf300861c. [DOI] [PubMed] [Google Scholar]
- Tetko I.V., Tanchuk V.Y. Application of associative neural networks for prediction of lipophilicity in ALOGPS 2.1 program. Journal of Chemical Information and Computer Sciences. 2002;42(5):1136–1145. doi: 10.1021/ci025515j. [DOI] [PubMed] [Google Scholar]
- Utpott M., Rodrigues E., de Oliveira Rios A., Mercali G.D., Flôres S.H. Metabolomics: An analytical technique for food processing evaluation. Food Chemistry. 2022;366 doi: 10.1016/j.foodchem.2021.130685. [DOI] [PubMed] [Google Scholar]
- Wang Y., Xiao J., Suzek T.O., Zhang J., Wang J., Bryant S.H. PubChem: A public information system for analyzing bioactivities of small molecules. Nucleic Acids Research. 2009;37(Web Server issue), W623-633 doi: 10.1093/nar/gkp456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C.H., Kao H.Y., Lu Z. PubTator: A web-based text mining tool for assisting biocuration. Nucleic Acids Research. 2013;41(Web Server issue), W518-522 doi: 10.1093/nar/gkt441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Z., Sun L., Wang F., Sui X., Fang Y., Tang X., Shen X. Assessment the flavor of soybean meal hydrolyzed with Alcalase enzyme under different hydrolysis conditions by E-nose, E-tongue and HS-SPME-GC–MS. Food Chemistry: X. 2021;12 doi: 10.1016/j.fochx.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T., Inui-Yamamoto C. The flavor-enhancing action of glutamate and its mechanism involving the notion of kokumi. npj Science of Food. 2023;7(1):3. doi: 10.1038/s41538-023-00178-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J., Wu M., Lin R., Li X., Ding H., Han L., Qu H. Application and development trends of gas chromatography–ion mobility spectrometry for traditional Chinese medicine, clinical, food and environmental analysis. Microchemical Journal. 2021;168 doi: 10.1016/j.microc.2021.106527. [DOI] [Google Scholar]
- Yin W., Shang M., Li X., Sang S., Chen L., Long J., Qiu C. Recent developments in sources, chemical constituents, health benefits and food applications of essential oils extracted from medicine food homology plants. Food Bioscience. 2023;102997 doi: 10.1016/j.fbio.2023.102997. [DOI] [Google Scholar]
- Zdrazil B., Felix E., Hunter F., Manners E.J., Blackshaw J., Corbett S., Mosquera J.F. The ChEMBL database in 2023: A drug discovery platform spanning multiple bioactivity data types and time periods. Nucleic Acids Research. 2024;52(D1):D1180–D1192. doi: 10.1093/nar/gkad1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Zhang T.-T., Guo R.-R., Ye Q., Zhao H.-L., Huang X.-H. The regulation of key flavor of traditional fermented food by microbial metabolism: A review. Food Chemistry: X. 2023;100871 doi: 10.1016/j.fochx.2023.100871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R.-Z., Yu S.-J., Bai H., Ning K. TCM-mesh: The database and analytical system for network pharmacology analysis for TCM preparations. Scientific Reports. 2017;7(1):2821. doi: 10.1038/s41598-017-03039-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Overview of the PungentDB database: This supplementary material includes a detailed presentation of the PungentDB database, highlighting its comprehensive nature and diverse applications. Additionally, we provide case studies that illustrate the utility of PungentDB across various contexts. A series of video demonstrations recorded from the PungentDB website showcases these examples, accessible via the 'Examples' link within the database interface.
Data Availability Statement
Data will be made available on request.