Summary
Glioblastoma (GB) is the most common primary malignant brain tumor, characterized by resistance to therapy. Despite aggressive treatment options, GB remains an incurable disease. Invasiveness and heterogeneity are key GB features that cannot be studied in preclinical in vitro models. In this study, we investigated the effects of standard therapy using patient-derived GB organoids (GBOs). GBOs reflect the complexity and heterogeneity of the original tumor tissue. No significant effect on GBO viability or invasion was observed after irradiation and temozolomide treatment. E3 ubiquitin-protein ligase (MDM2), cyclin-dependent kinase inhibitor 1A (CDKN1A), and the serine/threonine kinases ATM and ATR were upregulated at the gene and protein levels after treatment. Our results show that the p53 pathway and DNA-damage response mechanisms were triggered, suggesting that GBOs recapitulate GB therapy resistance. GBOs thus provide a highly efficient platform to assess the specific responses of GB patients to therapy and to further explore therapy resistance.
Subject areas: cellular physiology, cellular toxicology, in vitro toxicology including 3D culture, technical aspects of cell biology, cancer
Graphical abstract

Highlights
-
•
GBOs retain cellular diversity, heterogeneity, and transcriptional profile of GBs
-
•
GBOs recapitulate GB therapy resistance
-
•
Expression of ATM, ATR, MDM2, and CDKN1A is increased in GBOs
Cellular physiology; Cellular toxicology; In vitro toxicology including D culture; Technical aspects of cell biology; Cancer
Introduction
Glioblastoma (GB) is the most malignant primary brain tumor. The mean of 5-year overall survival of GB patients after the diagnosis is only 6.8%, varying with age, tumor location, and gender, age being the major risk factor.1 According to the World Health Organization (WHO) 2021 classification of central nervous system tumors, GB is defined as diffuse astrocytic glioma grade IV without mutations in isocitrate dehydrogenase (IDH) genes and with features of microvascular proliferation, necrosis, and/or unique molecular fingerprint.2 Our understanding of the pathology of these tumors increased steadily, but still there are no significant improvements in treatment or patient outcome. Standard-of-care therapy as of 2005 includes maximal surgical resection with concomitant radiotherapy (60 Gy in 30 fractions) alone or in combination with daily treatment with the alkylating agent temozolomide (TMZ) followed by adjuvant six cycles of TMZ, resulted in prolong survival of GB patients to 16.0 months. The recent introduction of TTFields (tumor treating fields) with TMZ increased median overall survival to 20.9 months 3 Treatment resistance is mainly caused by the heterogeneity of the tumor, the presence of therapy resilient glioblastoma stem cells (GSCs),4 and the diffuse infiltration of GB cells into the healthy brain parenchyma, which makes complete surgical resection difficult and leads to tumor recurrence.
GBs are characterized by their extensive inter-tumor and intra-tumor heterogeneity at various levels, such as the genetic, epigenomic, metabolic, and cellular levels.5 GBs are stratified by different molecular properties into various subtypes. The Cancer Genome Atlas (TCGA) categorizes GBs on the base of the bulk tumor analyses of DNA copy number, gene expression, and methylation status into three subtypes: proneural (PN), classical (CL), and mesenchymal (MES).6,7 These subtypes differ in their prognostic and therapeutic implications, with the MES subtype being the most aggressive and associated with the worst prognosis. 8 Regardless of classification, GB cells and GSCs exhibit inherent phenotypic plasticity and transition between different states,9 which is determined by environmental factors, i.e., an interaction with surrounding cells in the tumor microenvironment (TME), which modulate the response of GB to therapy.10,11
Malignant brain tumors are complex ecosystems composed of various neoplastic and stromal components.12 In addition to tumor cells, they contain a mixture of various brain-resident and brain-infiltrating cells, including immune cells, endothelial cells, astrocytes, and neurons, as well as extracellular matrix (ECM).13 The role of TME may have been underestimated in GB, as it can also modulate response to therapy and be targeted along with tumor cells.14
The development of clinically relevant models that accurately represent the complexity of tumors is necessary to improve our understanding of the properties of GB, mimic their different therapeutic responses, and thus enable the development of personalized therapy. Three-dimensional (3D) culture systems, such as patient-derived organoids15,16 and explants17 represent advanced in vitro models that preserve a complex TME and thus provide a preclinical model that better mimics tumor responses to therapy in vivo. This model is a valuable tool for exploring tumor heterogeneity and interactions between cancer cells and TME, which may lead to the discovery of new potential therapeutic targets, as well as to monitoring the effects of standard and novel therapeutic regimens.
The aim of the present study was to investigate the response of patient-derived GB organoids (GBOs) to standard-of-care therapy and to explore the treatment resistance of GB in association with complex TME. Here, we generated GBOs from tumor tissues from 22 patients, of whom 17 GBOs were exposed to irradiation (IR) and chemotherapy with TMZ. Cellular composition and gene expression signatures of GB subtypes, epithelial-to-mesenchymal transition (EMT) markers, GSCs, immunosuppression markers, cytokine signaling, immune cells, and DNA-damage response/cell cycle genes were compared between GBOs and parental tumors. The effect of IR and TMZ on GBOs was assessed by cell viability, invasion, and apoptosis assays and monitored by scanning electron microscopy (SEM). We investigated changes in the expression of therapy, TMZ and IR target genes in GBOs complemented by analyses in patient-derived primary GB cells and GSCs.
Results
GBOs retain TME cellular composition of the parental tumor
We studied the effects of GB standard therapy, IR and TMZ, in the context of the TME using the GBO model. The success rate of established GBOs derived from GB tissues in our study was 69% and was highly dependent on the tissue preservation and characteristics of the original tumor (Figures 1A and 1B). Immunofluorescence labeling of cell-type-specific markers confirmed that TME associated stromal and immune cells are preserved within GBOs in culture (Figures 1C and S2). Macrophages and microglia (CD68 and Iba1-positive cells) and endothelial cells (CD31+, CD105+) were detected, besides astrocytes and differentiated GB cells (GFAP+) and GSCs (SOX2+, CD9+, CD44+, and OLIG2+). In addition, lymphocytes (CD3+) and the PD-1/PD-L1 axis were detected in the GBO samples (Figure S2). In general, the abundance of TME cells in GBOs reflected their abundance in patient tumor tissue, but significant differences were also observed between individual GBOs and tumor tissue sections of the same patient, reflecting intra-tumoural heterogeneity. GBOs derived from GB tissue samples resemble the features of high-grade gliomas such as the cellular as well nuclear atypia of GB tumors. They do contain mitotic figures and pleomorphic nuclei, gemistocytic cells as well as multinucleated giant cells (Figure S3). As non-tumour cells were expected to become gradually lost in the GBOs with time in culture,15 GBOs were analyzed within 3–4 weeks in cultures. We also compared basal cytokine secretion from GBOs and GB cells derived from the same patient. Only a small number of secreted proteins from GBOs were more or less equally secreted from GB cells, indicating that these were secreted by GB cells. Similar amounts of fractalkine IL-8, MCP-1/CCL2, RANTES/CCL5, and SDF1α+β, were secreted by both cultures. GROα was more secreted by GB cells. In contrast cytokines, such as IL-6, M-CSF, and MCP-2 were secreted in larger amounts by GBO than by GB cells, indicating that their secretion was induced in GB cells by the TME present in GBOs, whereas other cytokines were secreted only by GBO stromal, vasculature and immune cells, e.g., VEGF and the cytokines such as IL-22, MCP-3, MIP-1α, and MIP-1β, respectively (Figure 1D).
Figure 1.
Overview of the procedures used to generate GBOs from resected tumor tissue, cell-type-specific immunostaining of the GBOs and parental tumors and cytokine secretion from GBOs
(A) Schematic presentation of main steps for generating GBOs from resected tumor tissue, created with bioRender.com.
(B) Growth rate as the quantification of relative area of viable GBOs for five weeks. Data represent mean values ±S.E.M. (n = 3 GBOs samples per patient (P)).
(C) Immunofluorescence analyses of the GBO TME. GBOs express markers of GSCs (SOX2, CD9, and CD44) and TME cells, such as markers of cells of vasculature (CD31 and CD105), as well as macrophages and microglia (CD68 and Iba1). Representative images from a tissue sample from one patient are shown. Scale bar: 100 μm.
(D) Selected cytokines and growth factors are secreted by GBOs in comparison to cultured GB cells from the same patient.
GBOs reflect the gene expression profile of the parental tumor
To compare gene expression patterns between tissues and GBOs from the same tumor, we analyzed the expression of genes associated with a GSC signature, genes involved in EMT, immunosuppression, DNA-damage responses and cell cycle, proliferation and invasion, cytokine signaling, GB subtype related genes, and genes associated with immune cell populations within the TME (Table 1; Table S3).
Table 1.
List of selective genes for GB subtype and markers of other processes analyzed by RT-qPCR
| Groups | Marker genes |
|---|---|
| PN Subtype | P2RX7, STMN4, SOX10, ERBB3, OLIG2, and NOTCH |
| CL Subtype | ACSBG1, KCNF1, S100A4, and NFKB1 |
| MES Subtype | COL1A2, COL1A1, TGFB1, THBS1, and DAB2 |
| GSCs | CD9, FUT4, ID1, PROM1, SOX2, OLIG-2, and CD44 |
| EMT | STAT3, CDH1, CHI3L1, CD44, and SNAI1 |
| GB immunosuppression | IDO1 and IL6 |
| Immune cells in TME | AIF1, FCGR3A, CD68, FOXP3, and NCAM1 |
| Cytokines | CCL2, CXCL12, CXCR4, and IL6 |
| DNA-damage response | ATM, ATR, MDM2, and CHEK1 |
| Cell cycle | CDKN1A and CDKN2A |
| Proliferation and invasion | MTOR and PIK3CA |
A subset of 22 GBOs from the corresponding tumor tissues of 17 patients were screened for the expression profile of the gene groups mentioned above, by quantitative reverse-transcription PCR (RT-qPCR). We performed a statistical comparison of the gene expression profiles among GBOs and corresponding tissue samples. The Pearson correlation coefficient was used to determine the associations in mRNA expression. A correlation heatmap of GBOs (Figure 2A) and GB tissues (Figure 2B) identified four distinct clusters of correlated genes described in Table 1. In both GBOs and tissues, cluster 1 consists of genes related to GSCs, DNA-damage responses, cell-cycle progression, and proliferation/invasion. There is also a subset of genes belonging to the group of genes involved in EMT, PN, and CL subtype. Cluster 2 consists of genes that determine the PN subtype; cluster 3 consists of genes associated with the MES subtype, and a number of genes associated with GSCs and cytokine signaling; cluster 4 consists of genes associated with cytokine signaling and immune cell related genes of the TME. We found similar correlation patterns in GBOs and the corresponding tumors. For example, PN-related genes (marked with ochre squares in Figures 2A and 2B) are grouped together and form a cluster in the heatmap. These results suggest that patient-derived GBOs reflect the transcriptional profile of the original tumors.
Figure 2.
Gene expressions in GBOs vs. tumor tissues
The heatmaps represent correlation matrixes of mRNA expressions in (A) GBOs and (B) tissues. Pearson correlations between gene expressions in (A) GBO samples (nO = 22) and (B) corresponding tissues (nT = 22) are displayed. These correlation coefficients are represented in the heatmap: blue means a positive correlation and red a negative correlation. For a more structured visualization of the correlation data within the mRNA expression, clustering of the genes, i.e., grouping of data based on the relationships among variables was carried out. All the features can be divided into four main groups (clusters). In both GBOs and GB tissues, cluster 1 mostly consists of genes related to GSCs (violet squares) and genes involved in DNA-damage responses, the cell cycle (pink squares) and proliferation/invasion (crossed pink squares); cluster 2 consists of genes determining the PN subtype (ochre squares); cluster 3 consists of genes determining the MES subtype (blue squares) and genes involved in cytokine signaling (orange squares); and cluster 4 consists of genes involved in cytokine signaling (orange squares) and immune cells of the TME (yellow squares). Pearson correlation coefficients p values are presented in Tables S4 and S6.
(C) Difference of means in the expression of genes AIF1, FCGR3A (CD16), CD68, FOXP3, and NCAM1 are presented. The paired mean differences between GBO and GB tissues are shown in the estimation plots. Both groups are plotted on the left axes as a scatter graph showing individual values: each paired set of the individual patient is connected by a line. Statistical analyses were performed with GraphPad Prism software using paired t test. The paired mean difference is plotted on the right axis, and the 95% confidence interval is indicated by the error bars. GB samples and the corresponding clinical data are listed in Tables S1 and S2.
When immune cell genes were compared between individual patient’ GBOs and the corresponding GB tissues, the estimation plots show that gene expression is consistent but not significantly different in most cases. Major changes were observed in the expression of TME-related genes, e.g., the AIF1, FCGR3A (CD16), and NCAM1 genes which is consistent with the fact that the TME becomes underrepresented in GBO cultures over time (Figure 2C). In the other groups of genes, only some of the genes were significantly dysregulated (Figure S4).
IR and TMZ do not affect the viability and invasion of GBOs but trigger apoptosis in surface cells
Glioblastoma organoids were treated with IR and TMZ. GBOs were irradiated with a single dose (10 Gy) and exposed with 50 μM TMZ every 48 h for 1 week. Response to therapy as a potential decrease in cell viability was observed in GBOs from two out of six patients; however, when combining results for all patients, GBO viability was not significantly affected by this protocol of IR or/and TMZ (Figure 3A). Scatterplots for viability of individual patient GBOs are shown in Figure S5.
Figure 3.
Treatment with IR and TMZ does not significantly affect GBO viability or invasion but does affect apoptosis
(A) The effect of IR (10 Gy, single dose), TMZ (50 μM 1 week), and their combination on GBO viability, which was measured with Cell-Titer Glo 3D assay. Data are presented as means ± S.E.M. of six patient samples (nO = 6).
(B) The effect of IR (10 Gy, single dose), TMZ (50 μM), and their combination on GBO invasion in Matrigel, which was quantified 48 h after the treatment. Data are presented as mean values ± S.E.M. of six patient samples (nO = 6). Statistical analyses were performed with GraphPad Prism software using the ordinary one-way ANOVA test. Legend: NIBXXX, patient ID; C, core; R, rim; REC, recurrent GB.
(C) Representative images of GBO invasion for one condition (TMZ treatment) after 24 and 48 h are shown. Scale bar: 500 μm.
(D and E) Scanning electron microscopy (SEM) images of non-treated (D-a, D-b, and D-c) and TMZ- and IR-treated (D-d, D-e, and D-f) GBOs at low (D- and D-d), medium (D-b and D-e), and high magnification (D-c and D-f). Both non-treated and treated GBOs are round and compact, whereas the granular aspect at high magnification appears to be caused by protruding cells at the GBO surface that possess high numbers of long protrusions (arrows in D-c). Treatment with TMZ and IR resulted in apoptotic cells that lost their membrane integrity (arrowheads in D-f) and most of their protrusions (D-c). Scale bars: (D-a,d) 100 μm; (D-b, c, e, and f) 10 μm (E) GBOs from 2 patients (nO = 2) were irradiated with a single dose of IR (10 Gy) and exposed to 50 μM TMZ for 1 week. The percentage of cells in early and late apoptosis after TMZ and IR treatment are shown (mean ± S.E.M.). After GBO dissociation, cells were labeled with annexin-V-FITC (x axis) and propidium iodide (PI) (y axis) and analyzed by flow cytometry. Dot blots represent the results from GBOs from one patient. Statistical analyses were performed using GraphPad Prism software, using ordinary one-way ANOVA test (∗p < 0.05, ∗∗p < 0.01). GB samples and the corresponding clinical data are listed in Tables S1 and S2.
We set up an invasion assay of GBOs embedded in Matrigel and monitored the invasion of GBOs into the 3D matrix after 48 h. Decreased cell invasion of GBOs from two patients was observed following IR, TMZ, or their combination. Combining results for all patients for single or combined treatment had no significant effect on GBO invasion (Figure 3B). Scatterplots for relative invasion of individual patient GBOs are shown in Figure S6. Combination of standard therapy had also no effect on proliferation of cancer cells analyzed by Ki-67 immunofluorescence (Figure S7).
The morphology of GBOs and the effects of standard therapy on GBO surfaces were studied with SEM. Representative low-magnification SEM images of GBO show a compact round structure with a granular surface (Figures 3D-a). At a higher magnification, the granular structure is shown to be caused by protruding cells (Figures 3D-b) that possess numerous astrocyte-like long protrusions (arrows; Figures 3D-c). Treatment with 50 μM TMZ and 10 Gy IR did not alter the overall shape of GBOs (Figures 3D-d), but less protruding cells (Figures 3D-e) with shorter protrusions (Figures 3D–3F) were observed at GBO surfaces at higher magnification. Moreover, numerous cells lost their membrane integrity likely due to apoptosis or necrosis (Figures 3D–3F). We further tested whether standard therapy induces cell death via early and late apoptosis in GBOs derived from two different patient samples (NIB232 R and NIB279 C). GBOs were treated with a single dose of IR (10 Gy) and exposed with 50 μM TMZ every 48 h for 1 week. Significant changes in the percentage of cells in early and late apoptosis were observed after the treatment. In the two patient GBO samples, IR, TMZ, and their combination increased the percentage of apoptotic cells by 7%, 6%, and 5%, respectively. This suggests that the observed effect on apoptosis was attributed to a small percentage of affected cells on the GBO surfaces (Figure 3E).
Standard therapy increases the expression of DNA-damage response genes and proteins in GBOs
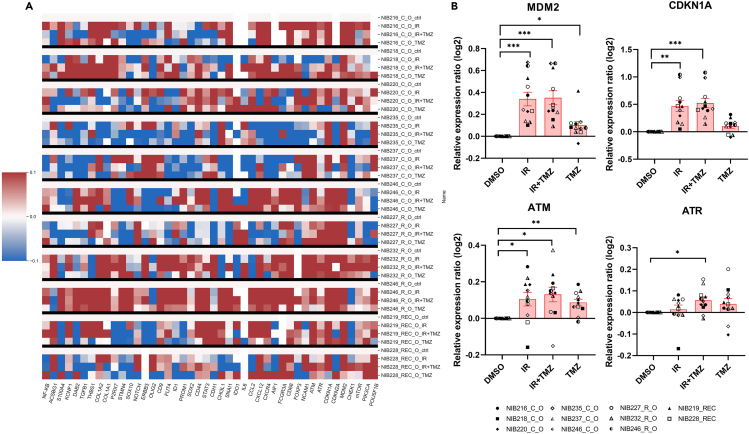
To evaluate the effect of standard therapy on GBOs at the level of gene expression, we analyzed the expression patterns of the genes listed in Table 1. We exposed GBOs from 11 patients to a single dose of IR (10 Gy) and treatment with 50 μM TMZ every 48 h for a week. In total, 44 genes were assessed, and changes in their expression after IR treatment or the combination of IR with TMZ treatment, compared to TMZ alone, were observed (Figure 4A). When combining results for all patients, four genes, ATM and ATR, CDKN1A and MDM2, involved in the cascade of DNA-damage response p53 signaling, were significantly upregulated in GBOs after treatment (Figure 4B). Furthermore, we evaluated CDKN1A (p21) and MDM2 expression in two patients derived GBOs at the protein level, using immunofluorescence and western blot which showed higher expression of both proteins after the treatment with IR and TMZ (Figure 5).
Figure 4.
A heatmap of relative gene expression ratios in treated GBOs and upregulation of DNA-damage response and cell cycle
(A) In total, 11 patient GBO samples (nO = 11) were used. Relative gene expression ratios in GBOs treated with IR (10 Gy, single dose), TMZ (50 μM, 1 week), or the combination of IR and TMZ are presented as downregulation in blue and upregulation in red color. The baseline expression is represented in white. The tested genes are shown below, and the GBOs and treatments are given at the right side of the heatmap.
(B) Selected genes ATM, ATR, CDKN1A, and MDM2 genes were significantly upregulated in GBOs treated with IR and TMZ standard therapy. Data are presented as mean values ±S.E.M. of GBO samples (nO = 11). Significance of gene expressions among treatment groups vs. control (DMSO) were evaluated on log2 transformed values (log2(treatment/control[DMSO])). Statistical analyses were performed with GraphPad Prism software using the one-way repeated measures paired ANOVA test. Data are presented as mean values ±S.E.M. (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001). Legend: NIBXXX, patient ID; C, core; R, rim; REC, recurrent GB. GB samples and the corresponding clinical data are listed in Tables S1 and S2.
Figure 5.
Upregulation of MDM2 and p21 proteins in treated GBOs
(A and B) MDM2 and P21 immunostaining in non-treated and treated GBOs. (A) MDM2 (red), GFAP (glial fibrillary acidic protein) (green), Hoechst (blue) and (B) p21 (green), GFAP (red) and Hoechst (blue) labeling in two patient GBO sections (NIB246 and NIB218) in control GBO samples (DMSO) and after treatment with IR and TMZ. MDM2 and p21 are upregulated in GBOs after treatment. NIB246 responded with higher upregulation of proteins after treatment compared to NIB218. Scale bars from 250 μm to 500 μm and 100 μm (enlarged images).
(C) Western blot analyses of MDM2 and p21 protein expression in non-treated and TMZ+IR treated GBOs. Results are expressed as fold-change relative to DMSO control. Data were normalized to β-tubulin control. Legend: NIBXXX, patient ID; C, core; R, rim.
To reveal changes in correlation between expression of upregulated genes and other genes in GBOs after treatment, we performed a combined cluster and correlation analysis in the non-treated versus treated GBOs (Figure 6). The Pearson correlation coefficient was used to determine the associations in mRNA expression levels of the genes. Changes in the correlation between identified upregulated genes (MDM2, CDKN1A, ATM, and ATR) and other genes tested were observed after treatment. CDKN1A correlated with PN gene (NOTCH), CL gene (KCNF1), EMT genes (STAT3 and CDH1), DNA-damage response gene (ATR), proliferation/invasion gene (MTOR), GSC marker gene (OLIG2), and immune cell related gene (FOXP3) after treatment (based on p values computed with Benjamini-Hochberg correction) (Table S4. Pearson correlation coefficients p-values for non-treated GBOs correlation matrix, related to Figures 2 and 6, Table S5. Pearson correlation coefficients p-values for treated GBOs correlation matrix, related to Figures 2 and 6, Table S6. Pearson correlation coefficients p-values for tissues correlation matrix, related to Figures 2 and 6). ATM correlated with PN gene (P2RX7), CL gene (ACSBG1), proliferation/invasion gene (PIK3CA), and GSC marker gene (OLIG2); novel positive correlations between ATR and PN gene (NOTCH), CL genes (KCNF1, NFKB), GSC marker genes (SOX2, FUT4), immune cell related gene (NCAM1), cell-cycle gene (CDKN1A), and cytokine signaling (CXCR4 and CCL2) were observed after treatment (Figure 6C). These results show novel correlations of upregulated genes MDM2, CDKN1A, ATM, and ATR with increased GSC, EMT, immune cell, and subtypes genes in GBOs after treatment.
Figure 6.
Correlation matrix heatmaps and Venn diagrams of correlated genes of non-treated and treated GBOs
(A and B) The heatmaps represent correlation matrixes between mRNA expression in (A) non-treated GBOs and (B) treated GBOs. Pearson correlations between gene expressions in non-treated GBO samples (nO = 22) and treated GBO samples (nTO = 22) are displayed. Pearson correlation coefficients p values are presented in Tables S4 and S5. These correlation coefficients are represented in the heatmap: blue means a positive correlation and red a negative correlation.
(C) Venn diagrams showing common genes correlated with ATM and ATR genes between non-treated and treated GBOs and novel significant positive correlations between CDKN1A/ATM/ATR and other genes in treated GBOs.
Differential responses of GB cells and GSCs to therapy at the gene expression level
To compare the effect of standard therapy on GB cells, GSCs, and GBOs at the gene level, patient-derived GB cells and GSCs (both n = 5) were subjected to a single dose of IR (2 Gy) and a 48 h treatment with 50 μM TMZ alone and both combined. In contrast to GBOs, GSCs showed significant upregulation of expression of CD44, MTOR, THBS1, and ATM genes, whereas OLIG2 gene expression was downregulated. In differentiated GB cells, ATR expression was upregulated and TGFB1 expression was downregulated after the treatment (Figure 7), indicating different response compared with GBOs.
Figure 7.
Treatment-related changes in the expression of specific genes in differentiated GB cells and GSCs
Relative gene expression ratios in treated cells with IR (2 Gy, single dose), TMZ (50 μM, single dose for 48 h), and combination (IR + TMZ) are presented as downregulation in blue and upregulation in red color. The baseline expression is represented in white. The tested genes are shown below, and cell and treatment types are shown on the right. GSCs (nGSCs = 5) and differentiated GB cells (DIFF) (nDIFF = 5) are shown in gray and blue, respectively. In GSCs (n = 5), standard treatments increased CD44, MTOR, THBS1, and ATM and decreased OLIG2 gene expression (bar charts in gray). In differentiated GB cells (n = 5), same treatment decreased TGFB1 and/or increased ATR gene expression. Significance of relative gene expression among groups were evaluated on log2 transformed values (log2(treatment/control (DMSO)). Statistical analyses were performed with GraphPad Prism software using the one-way repeated measures paired ANOVA test. Data are presented as mean values ±S.E.M. ∗p < 0.05, ∗∗p < 0.01. Legend: NIBXXX, patient ID; C, core.
Discussion
Patient-derived cancer cells are valuable tools for basic research and pre-clinical studies. Differentiated GB cells and GSCs have long been used to study GB and have provided important insights into the biology of the disease. However, tumors are not only composed of cancer cells but also represent a complex ecosystem of diverse cells and other elements of the TME.12 Recently, 3D organoid cultures of human tumor tissue have emerged as a representative platform for in vitro modeling of cancer heterogeneity and interactions with the TME.18
GBOs were generated within 2–4 weeks after the surgery, and upon the molecular analyses of the tumors, targeted therapies can be tested in vitro before translating them to patients. The key aspect of this method is to prevent the dissociation of tumor tissue into single cells and to maintain the natural cell-cell contacts that allow the formation and development of GBOs in the absence of exogenous growth factors, serum, and ECM. Thus, GBOs retain the cellular diversity, structural heterogeneity, and transcriptional profiles of their parental tumors. This enables studying dynamic interactions of cancer stem cells and differentiated cells within their TME, which consists of immune cells, stromal cells and ECM. The only restriction is the varying, limited division number of the TME cells, changing the original cellular composition of the GBO after prolonged culture.
We have successfully generated GBOs following the protocol by Jacob et al.15,16 and were the first to established GBOs from either the tumor’s rim or the core of the primary and recurrent tumors. Several studies using patient-derived models have focused mainly on tumor cells obtained from the tumor core, as this is the region that the neurosurgeon can most safely remove to avoid damage to healthy brain parenchyma. In contrast, unremoved GB cells at the tumor edges are invasive and may have a more stem-like resistant phenotype of GSCs or their progenitors,4,12 both types being most responsible for GB recurrences.19 Therefore, establishing GBOs from the invasive edge of the tumor offers a promising approach to reveal the novel GSC-associated targets.
GBOs retained the cellular composition and diversity of the TME observed in primary tumors using immunochemical markers of GSCs (SOX2, CD9, and CD44), differentiated GB cells and astrocytes GFAP, macrophages (CD68), microglia (Iba), lymphocytes (CD3), and endothelial cells (CD31). This was confirmed by a broader panel of specific gene expression markers (Figures 2 and 6). To confirm the presence of the TME and cellular communication within the TME, the secretion of growth factors and cytokines from GBOs and GB cells was analyzed. We were able to confirm that GBOs secrete a variety of different growth factors and cytokines that were not secreted by cultured GB cells and are thus caused by the TME of GBOs.
Similar gene expression profiles of GBOs compared to their parental tissues were confirmed by the presence of four distinct gene clusters that were identified in both groups, GBOs and tumor tissues (Figure 2). Cluster 1 consist of the largest group of genes encoding known GSCs stemness genes,4,12 genes involved in the EMT process that induces cell invasion,20 cell cycle, and DNA-damage responses. This is in agreement with several studies4,20,21,22 showing that GSCs have higher expression and activity of DNA-damage response proteins, that contribute to higher resistance to radiotherapy. Clusters 2 and 3 were related to GB subtypes classification6,7 of genes determining the PN and MES subtypes, respectively and genes encoding for GSCs. Cluster 3 also contained genes involved in cytokine signaling. Cluster 4 contained mainly genes encoding for specific TME immune cells and some genes involved in the EMT process and cytokine signaling. Overall, selective gene expression comparisons clearly showed a representative pattern, especially of GSC marker genes and EMT-related genes that do not differ significantly among GBOs and compared to the corresponding tissues. Major differences were observed in the expression of genes encoding for immune cells, e.g., AIF1, FCGR3A, and NCAM1, most likely due to a reduction of immune cells in GBOs during prolonged cultivation.15
The major aim of this study was to uncover the cellular and molecular mechanism underlying the resistance of GBOs to TMZ and IR alone or their combination, which to some extent mimics standard therapy in patients. The study was complemented by studies on primary GB and GSC cell lines derived from patient tumors. Reduced cell viability was observed in two out of the six GBOs, but on average, the viability of the GBOs was not significantly impaired (Figure 3). Invasion of the two patient-derived GBOs decreased with standard therapy, but again, no significant overall effect of IR and TMZ was observed. Jacob et al.15 also found that similar therapeutic responses were more efficient in some but not other patients. By applying electron microscopy, we observed changes of GBO surfaces after being exposed to standard therapy. Specifically, standard therapy did not alter the shape of GBOs, although numerous cells on the GBO surface lost their membrane integrity, shown as a small but significant change in the proportion of cells in early and late apoptosis in GBOs from two patients.
We followed the changes in expression of selected genes in GBOs that responded to treatment with IR and IR in combination with TMZ and compared the observation of these effects in patient-derived primary GBs and GSCs lines (Figure 7). In general, the expression of some genes was similarly changed in GSCs and differentiated GB cells, while others were found only in GBOs, reflecting the influence of TME impact on treatment efficacy. Among stemness related genes, upregulated in GSCs, but not in GBOs, was CD44, a known marker of the MES GSCs/GB subtype, suggested the PN to MES subtype transition upon treatment, also observed in GSCs/and GB tumors in irradiated patients.19 The cells that escape radiation induced stress by changing their phenotype/subtype, become more proliferative23 with exhibit higher expression of MTOR (mammalian target of rapamycin) and ATM (ATM serine/threonine kinase), both of which are involved in the expression of the DNA-damage response and represent a phenotypically robust sub-population that should be targeted.24 Overall, we show here that differentiated GB cells and GSCs, in contrast to GBOs, respond differently to therapy, with a different combination of genes than GBOs, indicating an important influence of the TME on treatment. Therefore, the interpretation of the data on isolated GB/GSC cells in vitro can only be translated with caution to the expected clinical effects of the therapy.
The most coherent data from this study is the DNA-damage response associated with significant upregulation of ATM and ATR kinases, MDM2 and CDKN1A, of 44 different genes in GBOs after treatment with IR, TMZ, or in combination (Figure 4). These data show that with standard GBO therapy, the DNA damage response was triggered by the activation of cell membrane-associated serine/threonine kinases such as ATM and ATR. ATM and ATR can phosphorylate p53,25 which is then translocated to the cell nucleus. Its transcriptional activation induces the expression of various genes that interrupt cell division, such as cell cycle inhibitor p21/CDKN1A. This is likely for activation of DNA-damage repair enzymes or to induce either senescence, apoptosis, or proliferation of repaired cells, as well as for its own inhibition by upregulating the synthesis of its inhibitor MDM2 as has been shown previously.26 The p53 inhibitor MDM2, being E3 ubiquitin-protein ligase,25 leads the p53/MDM2 complex to proteasomal degradation.26 Overexpression of MDM2 has been found in a variety of cancers, where it further suppresses p53 in p53 non-mutated tumors, and is associated with chemo- and radio-resistance in human malignancies.25
Cyclin-dependent kinase inhibitor p21/Cip1 (CDKN1A), plays an important role in cell cycle regulation by ensuring genomic stability, causing p53-dependent mediation of cell-cycle arrest in the G1 phase. Overexpression of CDKN1A/p21 causes cells to adopt a more aggressive phenotype capable of overcoming cell cycle blockade, senescence, and apoptosis.27 Here, we showed that patient NIB246 responded with higher expression of MDM2 and CDKN1A genes after treatment with more abundant levels of both proteins, as did patient NIB218. Taken together, standard therapy did not significantly affect cellular processes in GBOs, such as viability and invasion, but induced relatively small effects on apoptosis. This may be due to three causes. First using the TMZ, IR, or both in different doses and frequency, as used in patients. Using fractionated IR will mimic the effects of standard clinical settings better, so our further analysis includes this. Secondly, including different samples regarding MGMT methylation and p53 expression in the GBO cohort. Third, due to sufficient upregulation of expression of genes involved in DNA-damage repair, leading to acquired radioresistance. The latter is also a cause of the relatively poor effect of these therapies in GB patients.
In summary, we have shown that compared to 2D GB and 3D GSC cell models, which represent uniform tumor cell populations grown under artificial conditions and promote clonal selection across multiple passages, the 3D GBO model retains the cellular diversity, heterogeneity, and transcriptional profiles of GB and GSCs. The GBO model also preserves the TME of its parental tumors, albeit for a limited time. After treatment with IR and TMZ, which to some extent mimics patients’ standard therapy, the expression of ATM and ATR, MDM2 and CDKN1A was significantly increased in GBOs, confirming that the p53 signaling pathway and DNA damage response mechanisms were triggered. However, the poor response of GBOs to treatment, which resulted in only a small proportion of GB cells undergoing apoptosis, suggests that GBOs indeed recapitulate the therapy resistance of GB. The GBO model proved to be very useful for screening and identifying new mechanisms of therapy resistance that may lead to better treatment of GB in the future.
Limitations of the study
In this study, we highlight the ability of patient-derived organoids (GBOs) to recapitulate inter-patient variability and reflect the cellular composition and transcriptomic profiles of corresponding tumor tissues, providing a valuable platform for GB therapy response and resistance studies. However, the study has some limitations that should be considered. GBOs were generated from core or rim tissue samples as well as recurrent samples. Not enough rim samples were collected to compare whether the region-specific transcriptome profiles were preserved in their GBOs. The study was also limited by the matched primary and recurrent samples, which were insufficient to allow longitudinal studies and shed light on the molecular basis of recurrent GB tumors. To recapitulate the standard treatment, GBOs were given a single dose of 10 Gy, which is a limitation of this study as fractionated radiotherapy is more clinically relevant.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal to SOX2 | Abcam | Cat#ab171380; RRID:AB_2732072 |
| Rabbit polyclonal to GFAP | Abcam | Cat#ab211271; RRID: N/A |
| Mouse monoclonal to Iba1 | Abcam | Cat#ab15690; RRID:AB_2224403 |
| Rabbit polyclonal to CD68 | Atlas antibodies | Cat#HPA048982; RRID:AB_2680587 |
| Mouse monoclonal to CD44 | BioRad | Cat#MCA2504; RRID:AB_808430 |
| Rabbit monoclonal to CD9 | Cell signaling technology | Cat#13403; RRID:AB_2732848 |
| Mouse monoclonal to CD31 (PECAM-1) | Cell signaling technology | Cat#3528; RRID:AB_2160882 |
| Rabbit polyclonal to CD105 (ENG) | Atlas antibodies | Cat#HPA067440; RRID:AB_2685840 |
| Rabbit Monoclonal to CD3E | Abcam | Cat#ab16669; RRID:AB_443425 |
| Mouse monoclonal to PD-1 | Cell signaling technology | Cat#43248S; RRID:AB_2728836 |
| Rabbit Monoclonal to PD-L1 | Cell signaling technology | Cat#13684T; RRID:AB_2687655 |
| Mouse monoclonal to MDM2 [2A10] | Abcam | Cat#ab16895; RRID:AB_2143534 |
| Rabbit monoclonal to p21 [EPR362] | Abcam | Cat#ab109520; RRID:AB_10860537 |
| Rabbit monoclonal to MDM2 (D1V27) | Cell signaling technology | Cat#86934; RRID:AB_2784534 |
| Goat anti-Rabbit IgG (H + L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Invitrogen | Cat#A11008; RRID:AB_143165 |
| Goat anti-Mouse IgG (H + L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 546 | Invitrogen | Cat#A11003; RRID:AB_141370 |
| Goat anti-mouse IgG (H + L) IRDye(R) 800CW | LI-COR | Cat#926-32210; RRID:AB_621842 |
| Chemicals, peptides, and recombinant proteins | ||
| 2-mercaptoethanol | Sigma Aldrich | Cat#M6250-100ML |
| 4% formaldehyde solution | Merck | Cat#1004960700 |
| 4–12% Bis-Tris NuPAGE® gels | Thermo Fisher Scientific | Cat#NP0321BOX |
| Annexin-V-FITC | Miltenyi Biotech | Cat#130-097-928 |
| aqueous osmium tetroxide | SPI Supplies | Cat#2598 |
| B-27 | Invitrogen | Cat#12587010 |
| bFGF | Invitrogen | Cat#13256029 |
| BSA | Sigma-Aldrich | Cat#A9418-100g |
| collagenase I | Gibco | Cat#17018-029 |
| collagenase II | Gibco | Cat#17101015 |
| collagenase IV | Gibco | Cat#17104019 |
| DMEM | Hyclone | Cat#52100-039 |
| DMEM F-12 | Gibco | Cat#11320033 |
| EGF | Invitrogen | Cat#PHG0311 |
| GlutaMax | Gibco | Cat#35050061 |
| heparin | Sigma-Aldrich | Cat#H3149-10KU |
| hexamethyldisilazane | Sigma-Aldrich | Cat#804324 |
| Hoechst 33258 | Sigma-Aldrich | Cat#94403 |
| human insulin | Sigma-Aldrich | Cat#I9278 |
| Immobilon®-FL PVDF Membrane | Merck | Cat#IPFL00010 |
| L-glutamine | Sigma-Aldrich | Cat#G7513 |
| N-2 | Gibco | Cat#17502048 |
| NEAA | Sigma-Aldrich | Cat#M7145 |
| Neurobasal Medium | Invitrogen | Cat#21103-049 |
| Odyssey® blocking buffer | Thermo Fisher Scientific | Cat#15590545 |
| penicillin/streptomycin | Sigma-Aldrich | Cat#SI-P0781 |
| ProLong Gold AntiFade reagent | Invitrogen | Cat#P36980 |
| propidium iodide | Miltenyi Biotech | Cat#130-093-233 |
| PhosSTOP™ | Roche | Cat#04 906 845 001 |
| Red Blood Cell (RBC) lysis buffer | Thermo Fisher Scientific | Cat#00-4333-57 |
| RIPA buffer | Thermo Fisher Scientific | Cat#89900 |
| TMZ | Sigma-Aldrich | Cat#T2577-25MG |
| TrypLE Express | Gibco | Cat#12064-013 |
| trypsin–EDTA solution | Gibco | Cat#25200056 |
| xylene | Chem-Lab | Cat#CL00.2402.2500 |
| Critical commercial assays | ||
| 48.48 Dynamic Arrays IFC | Fluidigm | Cat#BMK-M-48.48 |
| AllPrep DNA/RNA/Protein Mini Kit | Qiagen | Cat#80004 |
| Cell Titer Glo 3D cell viability assay | Promega | Cat#G9681 |
| High-Capacity cDNA Reverse Transcription Kit | Thermo Fisher Scientific | Cat#4368814 |
| Human Cytokine/Chemokine 71-Plex Discovery Assay® Array | Eve Technologies | https://www.evetechnologies.com/product/clinical-cytokine-chemokine-growth-factor-71-plex-panel-hd71-clin/#1 |
| MycoAlert Mycoplasma Detection Kit | Lonza | Cat#LT07-318 |
| Experimental models: Cell lines | ||
| NCH421k | Cell Lines Service GMBH | Cat#300118 (male);RRID:CVCL_X910 |
| NCH644 | Cell Lines Service GMBH | Cat#300124 (female); RRID:CVCL_X914 |
| Patient derived cell lines | This paper | N/A |
| Patient derived GB organoids (GBOs) | This paper | N/A |
| Oligonucleotides | ||
| List of TaqMan gene expression assays (Thermo Fisher Scientific) used for RT-qPCR. | Available in Supplementary file (Table S7) | N/A |
| Software and algorithms | ||
| Adobe PhotoShop | Adobe | https://www.adobe.com/products/photoshop.html; RRID:SCR_014199 |
| BioRender | Science Suite Inc. DBA BioRender | Agreement number: IG271U22G4; RRID:SCR_018361 |
| Empiria Studio Software | LI-COR | https://www.licor.com/bio/empiria-studio/; RRID:SCR_022512 |
| FlowJo™ v10.8 Software | BD Life Sciences | N/A |
| Fluidigm® BioMark™ HD Data Collection Software v3 | Standard Biotools (Fluidigm) | N/A |
| GraphPad Prism | 2024 GraphPad Software | 8.4.3 (https://www.graphpad.com/features); RRID:SCR_002798 |
| ImageJ | Schneider et al. 28 | 1.53a; RRID:SCR_003070 |
| ImagePro 10 | Media Cybernetics | V10 https://mediacy.com/tag/image-pro-v10/ |
| Luminex™ 100 system | Eve Technologies Corp | N/A |
| Nikon software NIS-Elements | Nikon | https://www.microscope.healthcare.nikon.com/products/software/nis-elements |
| Python 3.9.0 | Van Rossum and Drake 29 | version 3.9; RRID:SCR_008394 |
| QuantGenious | Baebler et al. 30 | http://quantgenius.nib.si/user/login |
| Other | ||
| Orbital shaker (CO2 resistant) | Thermo Fischer Scientific | Cat#88881102 |
| Gulmay Medical 225 X-ray Generator | Gulmay | N/A |
| Fine Science Tools Dowell Scissors | Fine Science Tools | Cat#15040-11 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Metka Novak (metka.novak@nib.si).
Materials availability statement
There are restrictions to the availability of GBOs due to limited amounts of patient biopsies.
Data and code availability
-
•
De-identified patient data is available in this paper’s supplemental information. Any additional data reported in this paper will be shared by the lead contact upon request.
-
•
Original code can be shared by the lead contact upon request.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and study participant details
Established cell lines
The established GSC cell lines NCH421k (male) and NCH644 (female) were obtained from CLS (Cell Lines Service GMBH, Eppelheim, Germany). Cells were grown as floating spheres in complete Neurobasal Medium (Invitrogen, Life Technologies, Carlsbad, CA, USA) containing 2 mM L-glutamine, 1× penicillin/streptomycin (P/S) (both: Sigma-Aldrich, Misuri, USA), 1× B-27 (Invitrogen, Massachusetts, USA), 1 U/mL heparin (Sigma-Aldrich, Misuri, USA), 20 ng/mL bFGF and EGF (both: Invitrogen, Life Technologies, Carlsbad, CA, USA). Cell lines were maintained at 37°C with 5% CO2 and 95% humidity. Once GSC spheres reached 200 μm in diameter, they were dissociated using TrypLE Express (Gibco, Thermo Fisher Scientific, Massachusetts, USA).
Patient samples
Human tumor samples were obtained from patients with GB (WHO grade IV) operated at the Department of Neurosurgery, University Medical Center Ljubljana, Slovenia. Each sample was first taken from the core region of the tumor (named core), according to the enhancement area on an image guidance (MRI) navigation system. The second sample was taken from the invasive edge or margin (named rim) of the initial sample and was defined by the 5-aminolevulinicacid (5ALA) fluorescence positive area beyond the enhancement, according to the image guidance navigation system.31 Tumor diagnoses were determined using the standard histopathology and molecular protocols at the Institute of Pathology of the Medical Faculty, University of Ljubljana based on the latest WHO classification 2021.2 The clinical data and tumor characteristics (histopathological and molecular data) were provided by the Department of Neurosurgery and the Institute of Pathology of the Faculty of Medicine, University of Ljubljana, Slovenia. The study was approved by the National Medical Ethics Committee of the Republic of Slovenia (approval numbers 92/06/12, 0120–190/2018/4, and 0120–190/2018/26). Written informed consent was obtained from the patients and/or their authorized representatives in accordance with the Declaration of Helsinki.32
Primary GB cell lines
For the isolation of primary GB cells, fresh GB tumor tissue biopsies were dissected with a scalpel in high-glucose Dulbecco’s Modified Eagle’s Medium (DMEM) (Hyclone, GE Healthcare, Chicago, IL, USA) supplemented with 10% fetal bovine serum (FBS; Gibco, Thermo Fisher Scientific, Waltham, MA, USA), 2 mM L-glutamine (Sigma-Aldrich, Misuri, USA), and 1× P/S (Sigma-Aldrich, St. Louis, MO, USA) and were seeded in six-well cell culture plates (Corning, New York, USA). Growing cells were detached with a 0.25% trypsin–EDTA solution (Gibco, Thermo Fisher Scientific, Waltham, MA, USA) and transferred to T25 or T75 cell culture flasks (Corning, New York, USA). For subsequent analyses, cells were grown after at least three passages. GB cells were defined by their selective expression of the GFAP marker by qPCR (Figure S1).
For the isolation of primary GSCs, tumor tissue pieces were dissected by a scalpel and digested in a digestion buffer (200 U/mL, containing collagenase II and collagenase IV (Gibco, Thermo Fisher Scientific, Waltham, MA, USA) in basic Neurobasal Medium (Invitrogen, Life Technologies, Carlsbad, CA, USA)). Cell suspensions were filtered using a cell strainer with 100 μm pores (BD Falcon, Corning, NY, USA). Single cells were collected and resuspended in complete Neurobasal Medium containing 2 mM L-glutamine (Sigma-Aldrich, Misuri, USA), 1× P/S (both: Sigma-Aldrich), 1× B-27 w/o vitamin A (Invitrogen, Life Technologies, Carlsbad, CA, USA), 1 U/mL heparin (Sigma-Aldrich, St. Louis, MO, USA), 20 ng/mL bFGF, and EGF (both: Invitrogen). GSCs were cultured as floating spheres in a non-treated cell culture flask (Sarstedt Inc., Nümbrecht, Germany). Once GSC spheres reached a diameter of 200 μm, they were dissociated using TrypLE Express (Gibco, Thermo Fisher Scientific, Waltham, MA, USA). The GSCs were analyzed for GB and GSCs markers by qPCR (Figure S1).
GBOs
GBOs were established from fresh primary and recurrent (REC) GB biopsies of the tumor core (C) or rim (R) tissue samples, according to the protocol of Jacob et al.15 After transport in the processing medium (Hibernate A (Gibco, Thermo Fisher Scientific, Massachusetts, USA), 1x GlutaMax (Gibco, Thermo Fisher Scientific, Waltham, MA, USA), 1x P/S (Gibco, Thermo Fisher Scientific, Waltham, MA, USA) on ice, patient-derived tumor tissue was washed with 1x PBS (Gibco, Thermo Fisher Scientific, Waltham, MA, USA) to remove blood and debris. Fresh processing medium was added, and the tumor tissue was cut into small pieces of approximately 0.5–1 mm using fine dissection scissors (Fine science tools, Foster City, USA). The tumor pieces were washed twice with 1× PBS (Gibco, Thermo Fisher Scientific, Waltham, MA, USA) and incubated in 1× Red Blood Cell (RBC) lysis buffer (Thermo Fisher Scientific, Waltham, MA, USA) for 10 min with gentle shaking at room temperature. After two washes in DMEM F-12 medium (Gibco, Thermo Fisher Scientific, Waltham, MA, USA), tumor pieces were distributed into ultra-low attachment 6-well culture plates (Corning, New York, USA) with 4 mL GBO medium containing 50% DMEM F-12 (Gibco, Thermo Fisher Scientific, Waltham, MA, USA), 50% Neurobasal (Gibco, Thermo Fisher Scientific, Waltham, MA, USA), 1× GlutaMax (Gibco, Thermo Fisher Scientific, Waltham, MA, USA), 1× NEAAs (Sigma-Aldrich, St. Louis, MO, USA), 1× P/S (Gibco, Thermo Fisher Scientific, Waltham, MA, USA), 1× N-2 supplement (Gibco, Thermo Fisher Scientific, Waltham, MA, USA), 1× B-27 supplement w/o vitamin A (Gibco, Thermo Fisher Scientific, Waltham, MA, USA), 1× 2-mercaptoethanol (Gibco, Thermo Fisher Scientific, Waltham, MA, USA), and 2.5 μg/mL human insulin (Sigma-Aldrich, St. Louis, MO, USA). The 6-well culture plates were placed on a CO2-resistant orbital shaker (Thermo Fisher Scientific, Waltham, MA, USA) in a sterile incubator at 37°C, 5% CO2, and 90% humidity. 75% of the GBO medium was replaced every 48 h. Depending on tissue quality and characteristics, tumor pieces rounded within 1–2 weeks of culture. GBOs were generally used for experiments within 3–4 weeks. For longer cultivation, GBOs were cut into pieces of 200–500 μm diameter with fine scissors to prevent necrosis in the center. GBOs and cell cultures were monitored for Mycoplasma using MycoAlert Mycoplasma Detection Kit (Lonza, Basel, Switzerland).
To monitor the growth of GBOs over 5 weeks, GBOs of similar size (0.5–1 mm diameter) were placed in individual wells of a 48-well tissue culture plate containing 250 μL GBO medium per well. Images of each GBO were taken each week at the same timepoint over 5 weeks using a bright-field microscope. The area of each GBO was quantified using Nikon software NIS-Elements by selecting the area with automatic threshold setting. The area at each time point was divided by the area at time point 0 to calculate a growth ratio for each time point. Three individual GBOs were measured for each patient sample (n = 3).
Method details
Immunostaining
GBOs were washed with 1× PBS (Gibco, Thermo Fisher Scientific, Waltham, MA, USA) and fixed in 4% formaldehyde solution (Merck, Darmstadt, Germany) for 72 h at 4°C. Then, the fixed GBOs were dehydrated and embedded in paraffin. Paraffin blocks were cut into 4 μm thick slices. Formalin-fixed paraffin-embedded sections on slides were deparaffinised in xylene (Chem-Lab, Zedelgem, Belgium) and rehydrated in ethanol (Merck, Darmstadt, Germany). Antigens were retrieved by heating in 10 mM sodium citrate buffer (pH 6.0) at 95°C for 20 min. After cooling, GBO sections were blocked for 1 h at room temperature in 10% (v/v) FBS (Gibco, Thermo Fisher Scientific, Waltham, MA, USA) or normal goat serum (Sigma-Aldrich, St. Louis, MO, USA), 0.1% Triton X-100 (v/v) and 1% BSA (w/v) (both Sigma-Aldrich, St. Louis, MO, USA) in 1× PBS. Primary antibodies diluted in 1% BSA (w/v) in 1× PBS (key resources table) were added to the sections and incubated overnight at 4°C in a humidity chamber. The dilutions of the antibodies were: anti-SOX2 (1:50); anti-GFAP (1:1000); anti-Iba1 (1:200); anti-CD68 1:2500; anti-CD44 (1:100); anti-CD9 (1:200); anti-CD31 (1:1000); anti-CD105 (1:1000); anti-CD3 (1:1000); anti-PD1(1:200); anti-PD-L1 (1:200); anti-MDM2 (1:200); and anti-p21 (1:200). The slides were then washed twice in 0.5% BSA (w/v) in 1× PBS. Secondary antibodies diluted in 0.5% BSA (w/v) in 1× PBS (key resources table) (1:200) were added to the slides and incubated for 1 h in a humidity chamber at room temperature. After washing with 1× PBS, nuclei were stained with Hoechst 33258 solution (Sigma-Aldrich; diluted 1:1000 in 1× PBS) for 5 min at room temperature. Slides were washed with 1× PBS and mounted in ProLong Gold AntiFade reagent (Invitrogen, Life Technologies, Carlsbad, CA, USA), cover slipped, and sealed with nail polish. Inverted fluorescent microscope Eclipse Ti (Nikon, Tokyo, Japan) and NIS-Elements, Nikon software were used to image fluorescence. The images were processed with Image-Pro v10 software (Media Cybernetics) and cropped and prepared with Adobe Photoshop CS6 (Adobe).
Western blot analysis
Western blot analysis of GBOs was performed as described previously. 33 Briefly, GBO extracts were prepared in RIPA buffer (Thermo Fisher Scientific, Waltham, MA, US), supplemented with protease and phosphatase inhibitors (complete Mini; Phos STOP, Sigma-Aldrich, St. Louis, MO, USA), incubated on ice for 15 min cleared by centrifugation (13,000 rpm, 15 min). Protein concentration was measured on DeNovix DS-11 FX (DeNovix, USA). 50 μg of total protein was loaded on 4–12% Bis-Tris NuPAGE gels (Thermo Fisher Scientific, Waltham, MA, USA) for 60 min with 180 V iBlot 2 system (Thermo Fisher Scientific, Waltham, MA, USA) was used to transfer proteins on a PVDF membrane (Immobilon-FL PVDF Membrane, Merck, NJ, USA). Odyssey blocking buffer probed with primary antibodies overnight (at +4 C) against MDM2 and p21 was used. Differences in loading were controlled by antibody beta-tubulin. IRDye(R) 800CW goat anti-mouse IgG (H + L) and IRDye(R) 680RD goat anti-rabbit IgG (H + L) secondary antibodies were used. The image was taken by using an Odyssey DLx imager (LI-COR, Nebraska, USA). The signal fold change was analyzed by Empiria Studio Software (LI-COR, Nebraska, USA).
Real-time quantitative PCR
Tumor samples, GB cells, and GBOs were snap frozen and stored in liquid nitrogen for further analysis. Total RNA was isolated using an AllPrep DNA/RNA/Protein Mini Kit (Qiagen, Germantown, MD, USA) according to the manufacturer’s instructions; cDNA was generated from 1 μg of total RNA from each sample using a High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific, Waltham, MA, USA). To evaluate the expression level of genes, RT-qPCR was performed using Fluidigm BioMark HD System RT-PCR (Fluidigm Corporation, San Francisco, CA, USA) and 48.48 Dynamic Arrays IFC. cDNA of 42 samples and 24 TaqMan Gene Expression assays (ThermoFisher Scientific, Table S7) were mixed pairwise in nanoliter chambers to enable parallel analysis of 2304 reactions. Visualization and analysis of the RT-qPCR results were performed using the Biomark Data Collection software, the Fluidigm RT-qPCR analysis software (both: Fluidigm Corporation), and the quantGenius software.30 Relative copy numbers of cDNA were normalized to housekeeping genes HPRT1 and GAPDH.
For RT-qPCR analysis, 22 GBOs and the corresponding parental tissues were analyzed. For the studies of therapy effects on GBOs, 11 treated and non-treated GBOs were analyzed. In addition, NCH421k and NCH644 GSC cell lines, primary GSCs (n = 3) and primary GB cells (n = 5) were analyzed for relative mRNA expression of specific markers. These included selective markers for the GB subtypes, markers for GSCs, genes involved in EMT and GB immunosuppression, genes associated with immune cell populations within the TME, cytokine signaling, and genes involved in DNA-damage response, the cell cycle (Table 1). GB samples and the corresponding clinical data are listed in Tables S1 and S2. Selection of genes is supported with references in Table S3, related to Table 1.
Cytokine secretion assay
The cytokines, chemokines and growth factors in the culture media were detected using a Human Cytokine/Chemokine 71-Plex Discovery Assay Array (HD71). Human Cytokine/Chemokine 71-Plex Discovery Assay Array (HD71) includes: 6CKine, BCA-1, CTACK, EGF, ENA-78, Eotaxin, Eotaxin-2, Eotaxin-3, FGF-2, Flt3L, Fractalkine, G-CSF, GM-CSF, GROα, I-309, IFNα2, IFNγ, IL-1α, IL-1β, IL-1RA, IL-2, IL-3, IL-4, IL-5, IL-6, IL-7, IL-8, IL-9, IL-10, IL-12p40, IL-12p70, IL-13, IL-15, IL-16, IL-17A, IL-17E/IL-25, IL-17F, IL-18, IL-20, IL-21, IL-22, IL-23, IL-27, IL-28, IL-33, IP-10, LIF, MCP-1, MCP-2, MCP-3, MCP-4, M-CSF, MDC, MIG, MIP-1α, MIP-1β, MIP-1δ, PDGF-AA, PDGF-AB/BB, RANTES, sCD40L, SCF, SDF-1α+β, TARC, TGFα, TNFα, TNFβ, TPO, TRAIL, TSLP, VEGF-A. A multiplexing analysis was performed using the Luminex 100 system by Eve Technologies Corp. (Calgary, AB, Canada) on GB cells and GBOs from the same patient.
GBO viability assay
Non-irradiated GBOs or GBOs irradiated with Gulmay Medical 225 X-ray Generator with 0.55 mm Cu and 1.8 mm Al filtering instrument with a single dose of 10 Gy were added to individual wells of a 24-well plate (Corning, New York, USA). GBOs were then cultured for one week on an orbital shaker rotating at 120 rpm within a 37°C, 5% CO2, and 90% humidity sterile incubator in 500 μL GBO medium containing either 50 μM TMZ (Sigma-Aldrich, Misuri, USA), or 0.1% DMSO (Sigma-Aldrich, Misuri, USA) vehicle control. The medium containing fresh drug was replaced every 48 h. For a given treatment, GBOs were treated in triplicate. The Cell Titer Glo 3D cell viability assay (Promega, Madison, WI, USA) was used to assess cell viability according to manufacturer’s instructions.
Invasion assay
Non-irradiated GBOs or GBOs irradiated with Gulmay Medical 225 X-ray Generator with 0.55 mm Cu and 1.8 mm Al filtering instrument with a single dose of 10 Gy were placed in individual wells of the U-bottom 96-well plate (VWR). GBO medium was removed and 100 μL of 30% Matrigel (Corning, NY, USA) in GBO medium with or without 50 μM TMZ (Sigma-Aldrich, St. Louis, MO, USA) was added per well. GBO invasion was imaged after 24 h or 48 h on inverted microscope (Nikon Eclipse Ti, Tokyo, Japan) and NIS-Elements, Nikon software. Images were analyzed in ImageJ. 28 Invasion was quantified either by counting the number of invasive cells (for GBOs exhibiting single-cell invasion) or by measuring the invasion area (for GBOs exhibiting collective invasion). The number of invasive cells or the invasion area were normalized to average GBO diameter to account for differences in sizes of GBOs.
Scanning electron microscopy
For scanning electron microscopy (SEM), GBOs were fixed in 2.5% glutaraldehyde in 1x PBS for 1 h at 37°C and postfixed with 1% aqueous osmium tetroxide (OsO4) for 3 h at room temperature. After washing with deionized water, GBOs were dehydrated in an ascending concentration series of ethanol and air-dried in hexamethyldisilazane (HMDS). Dry samples were attached to aluminum holders using conductive silver paste and sputter coated with platinum. Imaging was performed using a JSM-7500F field emission scanning electron microscope (JEOL, Akishima, Japan).
Apoptosis assay
Assessment of apoptosis induced by standard therapy (IR, TMZ, or combination) in GBOs was performed by flow cytometry. Six GBOs per condition were irradiated with Gulmay Medical 225 X-ray Generator with 0.55 mm Cu and 1.8 mm Al filtering instrument with a single dose of irradiation (IR; 10 Gy) or/and subjected to daily treatment with 50 μM TMZ for one week. GBOs were then enzymatically dissociated with TrypLE and Collagenase I solution in a 2:1 ratio and washed with 1× PBS. Early/late apoptotic cells were detected by a 15 min labeling with Annexin-V-FITC (Miltenyi Biotech, Bergisch Gladbach, Germany) in the dark at room T and subsequent 5 min labeling with 1 μg/mL propidium iodide (Miltenyi Biotech, Bergisch Gladbach, Germany). Stained cells were analyzed with a flow cytometer MACSQuant Analyzer (Miltenyi Biotec Bergisch Gladbach, Germany) and FlowJo v10.8 Software (BD Life Sciences, New Jersey, USA). Solvent control 0.1% DMSO was used as a negative control. Live cells are both annexin V and propidium iodide negative. At early stage of apoptosis the cells bind annexin V while still excluding propidium iodide. At late stage of apoptosis they bind annexin V-FITC and stain brightly with PI.
Quantification and statistical analysis
Statistical analyses
All statistical tests and sample sizes as well as the software used to calculate the statistics (GraphPad Prism) are included in the legends to the figures and in the text. All data are presented as mean ± SEM. The p-values were indicated in the figures as follows: ∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001 and not statistically significant if p > 0.05. The "n" value given refers to either cells, parental tumor pieces or GBOs from an n-number of patients or to the number of GBOs from one patient used in the method.
Pearson correlation and clustering
The Pearson correlation coefficient was used to determine the associations within mRNA expression in GBOs (non-treated and treated) and corresponding tissues. These correlation coefficients were represented in a heatmap, which is a two-way display of a data matrix where the individual cells are displayed as colored rectangles. The color of an individual cell is proportional to its position along a color gradient. The dependence between mRNA expressions was evaluated using the Pearson correlation test, and the resulting p-values were adjusted for multiple comparisons using the Benjamini-Hochberg procedure. The latter provides a more reliable approach to identifying significant correlations while reducing the likelihood of false positives. The values below 0.05 were considered statistically significant. The p-value annotation legend is the same as the one previously reported, i.e., ∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001, and it is shown in Table S4. Pearson correlation coefficients p-values for non-treated GBOs correlation matrix, related to Figures 2 and 6, Table S5. Pearson correlation coefficients p-values for treated GBOs correlation matrix, related to Figures 2 and 6, Table S6. Pearson correlation coefficients p-values for tissues correlation matrix, related to Figures 2 and 6.
Additionally, for a more structured visualisation of the correlation data within the mRNA expression, a clustering procedure was carried out. In particular, an agglomerative clustering was performed using the Euclidean distance as distance metric and the average method as a linkage criterion. The latter defines the distance between groups as the average distance between each of the members. The resulting graph is the dendrogram, which is plotted on top of Figure 2. The analyses are computed using Python packages.29
Acknowledgments
This work was supported by the Slovenian Research Agency (Program and research grants P1-0245, J3-4504, J3-2526, NC-0023, Young researcher grants 10040137 to B.M. and 10040147 to A.H.) and by the European Program of Cross-Border Cooperation for Slovenia-Italy Interreg TRANS-GLIOMA. The research was funded by HE project Twinning for excellence to strategically advance research in carcinogenesis and cancer (CutCancer; 101079113). M.M. has received funding from the European Union’s Horizon 2020 research and innovation program under the Marie Sklodowska-Curie - Innovative Training Network 2020, grant agreement no 956394 (PARENT project). A.S. has received funding from the Slovenian Research Agency under the Research Program P2-0209. We thank Fadi Jacob for advice in the preparation of GBOs. We also thank the Institute of Pathology (Ajla Hajrlahović, Miša Omerzel) for the preparation of tissue and GBO sections. We thank prof. Cornelis van Noorden at Amsterdam UMC for critically proofreading the manuscript.
Author contributions
Conceptualization, B.M., A.H., M.N., and B.B; methodology and analysis, B.M., A.H., M.M., and M.V.; writing-original draft, B.M., A.H., M.N., and B.B.; writing – review & editing, M.N., B.M., B.B., M.M., M.V., A.P., R.B., J.M., M.S.V., T.L.T, and A.S.; samples provision, A.P., R.B., and J.M.; data provision and analysis, J.M., A.M., A.Z., and M.S.V.
Declaration of interests
The authors report no competing interests.
Published: July 31, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.110604.
Supplemental information
References
- 1.Wen P.Y., Weller M., Lee E.Q., Alexander B.M., Barnholtz-Sloan J.S., Barthel F.P., Batchelor T.T., Bindra R.S., Chang S.M., Chiocca E.A., et al. Glioblastoma in adults: a Society for Neuro-Oncology (SNO) and European Society of Neuro-Oncology (EANO) consensus review on current management and future directions. Neuro Oncol. 2020;22:1073–1113. doi: 10.1093/neuonc/noaa106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Louis D.N., Perry A., Wesseling P., Brat D.J., Cree I.A., Figarella-Branger D., Hawkins C., Ng H.K., Pfister S.M., Reifenberger G., et al. The 2021 WHO Classification of Tumors of the Central Nervous System: a summary. Neuro Oncol. 2021;23:1231–1251. doi: 10.1093/NEUONC/NOAB106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stupp R., Taillibert S., Kanner A., Read W., Steinberg D., Lhermitte B., Toms S., Idbaih A., Ahluwalia M.S., Fink K., et al. Effect of tumor-treating fields plus maintenance temozolomide vs maintenance temozolomide alone on survival in patients with glioblastoma: a randomized clinical trial. J. Am. Med. Assoc. 2017;318:2306–2316. doi: 10.1001/jama.2017.18718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lathia J.D., Mack S.C., Mulkearns-hubert E.E., Valentim C.L.L., Rich J.N. Cancer stem cells in glioblastoma. Genes Dev. 2015;29:1203–1217. doi: 10.1101/gad.261982.115.tumors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Inda M.D.M., Bonavia R., Seoane J. Glioblastoma Multiforme: A Look Inside Its Heterogeneous. Nature Cancers (Basel) 2014;6:226–239. doi: 10.3390/CANCERS6010226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verhaak R.G.W., Hoadley K.A., Purdom E., Wang V., Qi Y., Wilkerson M.D., Miller C.R., Ding L., Golub T., Mesirov J.P., et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Behnan J., Stangeland B., Hosainey S.A.M., Joel M., Olsen T.K., Micci F., Glover J.C., Isakson P., Brinchmann J.E. Differential propagation of stroma and cancer stem cells dictates tumorigenesis and multipotency. Oncogene. 2016;36:570–584. doi: 10.1038/onc.2016.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ehman E.C., Johnson G.B., Villanueva-meyer J.E., Cha S., Leynes A.P., Larson P.E.Z., Hope T.A., Hope T.A. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. J. Magn. Reson. Imaging. 2017;46:1247–1262. doi: 10.1002/jmri.25711.PET/MRI. [DOI] [Google Scholar]
- 9.Neftel C., Laffy J., Filbin M.G., Hara T., Shore M.E., Rahme G.J., Richman A.R., Silverbush D., Shaw M.L., Hebert C.M., et al. An Integrative Model of Cellular States, Plasticity, and Genetics for Glioblastoma. Cell. 2019;178:835–849.e21. doi: 10.1016/J.CELL.2019.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hara T., Chanoch-Myers R., Mathewson N.D., Myskiw C., Atta L., Bussema L., Eichhorn S.W., Greenwald A.C., Kinker G.S., Rodman C., et al. Interactions between cancer cells and immune cells drive transitions to mesenchymal-like states in glioblastoma. Cancer Cell. 2021;39:779–792.e11. doi: 10.1016/J.CCELL.2021.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmitt M.J., Company C., Dramaretska Y., Barozzi I., Göhrig A., Kertalli S., Großmann M., Naumann H., Sanchez-Bailon M.P., Hulsman D., et al. Phenotypic mapping of pathologic cross-talk between glioblastoma and innate immune cells by synthetic genetic tracing. Cancer Discov. 2021;11:754–777. doi: 10.1158/2159-8290.CD-20-0219/333477/AM/PHENOTYPIC-MAPPING-OF-PATHOLOGICAL-CROSSTALK. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gimple R.C., Yang K., Halbert M.E., Agnihotri S., Rich J.N. Brain cancer stem cells: resilience through adaptive plasticity and hierarchical heterogeneity. Nat. Rev. Cancer. 2022;22:497–514. doi: 10.1038/s41568-022-00486-x. [DOI] [PubMed] [Google Scholar]
- 13.Khaddour K., Johanns T.M., Ansstas G. The landscape of novel therapeutics and challenges in glioblastoma multiforme: Contemporary state and future directions. Pharmaceuticals. 2020;13:389–426. doi: 10.3390/ph13110389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bikfalvi A., da Costa C.A., Avril T., Barnier J.-V., Bauchet L., Brisson L., Cartron P.F., Castel H., Chevet E., Chneiweiss H., et al. Challenges in glioblastoma research: focus on the tumor microenvironment. Trends Cancer. 2023;9:9–27. doi: 10.1016/J.TRECAN.2022.09.005. [DOI] [PubMed] [Google Scholar]
- 15.Jacob F., Salinas R.D., Zhang D.Y., Nguyen P.T.T., Schnoll J.G., Wong S.Z.H., Thokala R., Sheikh S., Saxena D., Prokop S., et al. A Patient-Derived Glioblastoma Organoid Model and Biobank Recapitulates Inter- and Intra-tumoral Heterogeneity. Cell. 2020;180:188–204.e22. doi: 10.1016/j.cell.2019.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacob F., Ming G.L., Song H. Generation and biobanking of patient-derived glioblastoma organoids and their application in CAR T cell testing. Nat. Protoc. 2020;15:4000–4033. doi: 10.1038/s41596-020-0402-9. [DOI] [PubMed] [Google Scholar]
- 17.LeBlanc V.G., Trinh D.L., Aslanpour S., Hughes M., Livingstone D., Jin D., Ahn B.Y., Blough M.D., Cairncross J.G., Chan J.A., et al. Single-cell landscapes of primary glioblastomas and matched explants and cell lines show variable retention of inter- and intratumor heterogeneity. Cancer Cell. 2022;40:379–392.e9. doi: 10.1016/j.ccell.2022.02.016. [DOI] [PubMed] [Google Scholar]
- 18.Ratliff M., Kim H., Qi H., Kim M., Ku B., Azorin D.D., Hausmann D., Khajuria R.K., Patel A., Maier E., et al. Patient-Derived Tumor Organoids for Guidance of Personalized Drug Therapies in Recurrent Glioblastoma. Int. J. Mol. Sci. 2022;23:6572. doi: 10.3390/IJMS23126572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Minata M., Audia A., Shi J., Lu S., Bernstock J., Pavlyukov M.S., Das A., Kim S.H., Shin Y.J., Lee Y., et al. Phenotypic Plasticity of Invasive Edge Glioma Stem-like Cells in Response to Ionizing Radiation. Cell Rep. 2019;26:1893–1905.e7. doi: 10.1016/j.celrep.2019.01.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Majc B., Sever T., Zarić M., Breznik B., Turk B., Lah T.T. BBA - Molecular Cell Research Epithelial-to-mesenchymal transition as the driver of changing carcinoma. Biochim. Biophys. Acta. Mol. Cell Res. 2020;1867:118782. doi: 10.1016/j.bbamcr.2020.118782. [DOI] [PubMed] [Google Scholar]
- 21.Wang X., Prager B.C., Wu Q., Kim L.J.Y., Gimple R.C., Shi Y., Yang K., Morton A.R., Zhou W., Zhu Z., et al. Reciprocal Signaling between Glioblastoma Stem Cells and Differentiated Tumor Cells Promotes Malignant Progression. Cell Stem Cell. 2018;22:514–528.e5. doi: 10.1016/j.stem.2018.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skvortsova I., Debbage P., Kumar V., Skvortsov S. Radiation resistance: Cancer stem cells (CSCs) and their enigmatic pro-survival signaling. Semin. Cancer Biol. 2015;35:39–44. doi: 10.1016/j.semcancer.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 23.Steponaitis G., Tamasauskas A. Mesenchymal and Proneural Subtypes of Glioblastoma Disclose Branching Based on GSC Associated Signature. Int. J. Mol. Sci. 2021;22:4964. doi: 10.3390/IJMS22094964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhat K., Balasubramaniyan V., Vaillant B., Ezhilarasan R., Hummelink K., Hollingsworth F., Wani K., Heathcock L., James J., Goodman L., et al. Mesenchymal Differentiation Mediated by NF-κB Promotes Radiation Resistance in Glioblastoma. Cancer Cell. 2013;24:331–346. doi: 10.1016/j.ccr.2013.08.001.Mesenchymal. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hou H., Sun D., Zhang X. The role of MDM2 amplification and overexpression in therapeutic resistance of malignant tumors. Cancer Cell Int. 2019;19:216–218. doi: 10.1186/S12935-019-0937-4/TABLES/1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bourdon J.C., Fernandes K., Murray-Zmijewski F., Liu G., Diot A., Xirodimas D.P., Saville M.K., Lane D.P. p53 isoforms can regulate p53 transcriptional activity. Genes Dev. 2005;19:2122–2137. doi: 10.1101/gad.1339905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zamagni A., Pasini A., Pirini F., Ravaioli S., Giordano E., Tesei A., Calistri D., Ulivi P., Fabbri F., Foca F., et al. CDKN1A upregulation and cisplatin-pemetrexed resistance in non-small cell lung cancer cells. Int. J. Oncol. 2020;56:1574–1584. doi: 10.3892/IJO.2020.5024/DOWNLOAD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Rossum G., Drake F.L. Python 3 Reference Manual. CreateSpace; Scotts Valley, CA: 2009. https://books.google.si/books/about/Introduction_to_Python_3.html?id=51LMQQAACAAJ&redir_esc=y [Google Scholar]
- 30.Baebler Š., Svalina M., Petek M., Stare K., Rotter A., Pompe-Novak M., Gruden K. quantGenius: implementation of a decision support system for qPCR-based gene quantification. BMC Bioinformatics. 2017;18:276. doi: 10.1186/s12859-017-1688-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Porčnik A., Novak M., Breznik B., Majc B., Hrastar B., Šamec N., Zottel A., Jovčevska I., Vittori M., Rotter A., et al. TRIM28 Selective Nanobody Reduces Glioblastoma Stem Cell Invasion. Molecules. 2021;26:5141. doi: 10.3390/MOLECULES26175141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.2008. Preamble WMA Declaration of Helsinki-Ethical Principles for Medical Research Involving Human Subjects. [PubMed] [Google Scholar]
- 33.Kaminskyy V.O., Hååg P., Novak M., Végvári Á., Arapi V., Lewensohn R., Viktorsson K. EPHA2 Interacts with DNA-PKcs in Cell Nucleus and Controls Ionizing Radiation Responses in Non-Small Cell Lung Cancer Cells. Cancers. 2021;13:1010–1013. doi: 10.3390/CANCERS13051010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
De-identified patient data is available in this paper’s supplemental information. Any additional data reported in this paper will be shared by the lead contact upon request.
-
•
Original code can be shared by the lead contact upon request.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.