Abstract
Background
In recent years, germline gain-of-function (GOF) mutations in signal transducer and activator of transcription 3 (STAT3) have been identified as a cause of early-onset multiorgan autoimmune diseases with the widespread use of next-generation sequencing, and targeted therapies such as tocilizumab have been reported to be effective.
Objective
We sought to assess whether a novel STAT3 mutation detected by whole-exome sequencing is pathogenic and examine the efficacy of targeted therapy.
Methods
A pediatric patient with idiopathic pulmonary hemosiderosis, autoimmune thyroiditis, inflammatory bowel disease unclassified, leukocytosis, thrombocytosis, and severe growth failure was examined.
Results
This 7-year-old boy had idiopathic pulmonary hemosiderosis at the age of 6 months. Despite high-dose steroid therapy, pulmonary fibrosis progressed. Furthermore, he presented with severe growth failure, autoimmune thyroiditis, leukocytosis, thrombocytosis, and inflammation bowel disease unclassified. Given the presence of multiple autoimmune diseases, whole-exome sequencing was performed, which detected germline de novo heterozygous STAT3 mutation (NM_139276.2; c.2144C>A, p.(P715Q)). Dual-luciferase reporter assay revealed this novel STAT3 mutation as GOF. After starting tocilizumab therapy at the age of 6, hospital stays decreased, and the progression of pulmonary fibrosis was decelerated without increasing the steroid dose. New autoimmune diseases did not develop, and no apparent adverse effects on growth have been observed.
Conclusions
Tocilizumab may be effective for patients with STAT3 GOF mutation, including those requiring long-term management of idiopathic pulmonary hemosiderosis. Diagnosis of patients with early-onset multiorgan autoimmune diseases in which STAT3 GOF is suspected should be confirmed by genetic testing and functional analysis to consider the introduction of targeted therapies.
Key words: STAT3, GOF, whole-exome sequencing, idiopathic pulmonary hemosiderosis, tocilizumab, IL-6, KL-6
Signal transducer and activator of transcription (STAT) 3 is a transcription factor and acts downstream of inflammatory cytokines such as IL-6. Activated STAT3 phosphorylated by the Janus kinase (JAK)-STAT pathway forms a dimer, translocated into the nucleus, and promotes gene expression.1, 2, 3 STAT3-knockout mice are lethal embryonically, and the protein is essential for survival. Its function has been analyzed using conditional knockout mice with tissue-specific STAT3 deficiency.4
One of STAT3’s involvement is in the expression of acute-phase proteins in inflammation. When IL-6 acts on hepatocytes, C-reactive protein (CRP) is produced via the JAK-STAT pathway, and this is widely used as an inflammation marker. IL-6, a major stimulator of STAT3, has become a therapeutic target for autoimmune diseases. Tocilizumab (TCZ) is an antibody preparation that blocks the IL-6 receptor and is used to treat diseases such as rheumatoid arthritis.
A loss of function (LOF) of STAT3 is known as hyper-IgE syndrome, which causes immunodeficiency.5,6 Recently, owing to the prevalence of next-generation sequencing, germline gain-of-function (GOF) mutations of STAT3 are identified to cause early-onset multiorgan autoimmunity and growth failure.7, 8, 9 Some studies have reported that targeted therapies such as TCZ and JAK inhibitors were effective for disease management of patients with STAT3 GOF.10,11
We detected a novel STAT3 mutation using whole-exome sequencing, concluded it to be a GOF mutation using dual-luciferase reporter assay, and administered TCZ therapy.
Methods
Whole-exome sequencing
The patient was recruited through the “Initiative on Rare and Undiagnosed Diseases.”12 The molecular research protocol was approved by the local institutional board. The genomic DNA of both the patient and his parents was extracted from peripheral blood leukocytes using a standard phenol extraction protocol. All exons were captured using the SureSelect XT Human All Exon V6 (Agilent Technologies, Santa Clara, Calif), and exome analyses were performed using the HiSeq platform (Illumina, San Diego, Calif). Subsequently, bioinformatic analysis was performed as previously reported.13 Briefly, sequence reads were mapped to the human reference genome (GRCh37) according to Burrows–Wheeler Aligner and the Genome Analysis Tool Kit best-practice guidelines, using packages in the integrated analysis suite variant tools. After excluding variants found in the general population on the databases, the called variants were annotated with SnpEff.14
Cloning of the human STAT3 gene
PBMCs were isolated from EDTA blood sample of a patient with STAT3 mutation using Lymphoprep (Abbott Diagnostics Technologies AS, Oslo, Norway). Total RNA was isolated from 1.0 × 107 cells using QIAshredder and RNeasy Mini Kit (Qiagen, Hilden, Germany). cDNA was synthesized with the QuantiTect Reverse Transcription Kit (Qiagen). PCR was performed using KOD FX (TOYOBO Co Ltd, Osaka, Japan) and specific primers (Table I). Full-length human STAT3 cDNA added deoxyadenosine to the 3ʹ end using Takara Ex Taq (Takara Bio, Inc, Shiga, Japan) was inserted into the pcDNA3.1/V5-His-TOPO vector (Invitrogen, Carlsbad, Calif) to generate pcDNA3.1-STAT3-V5-His vectors. DH5α (TOYOBO) was mixed with plasmid vectors, spread on LB plates added with ampicillin, and incubated overnight at 37°C. Plasmid vectors obtained from single colonies were purified using QIAprep Spin Miniprep Kit (Qiagen). Purified vectors were sequenced with BigDye Terminator v3.1/1.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, Calif) using the sequencing primers (Table Ⅰ). Sequencing was performed on an Applied Biosystems 3130 Genetic Analyzer.
Table I.
STAT3 primers for PCR, Sanger sequencing, and site-directed mutagenesis
| PCR for full-length STAT3 cDNA | |
| Name | Sequences (5'–3') |
| STAT3 forward | GCCGCCGCCATGGCCCAATGGAATCAGC |
| STAT3 reverse | CATGGGGGAGGTAGCGCACTCCGAGGTC |
| Sanger sequencing | |
| Name | Sequences (5'–3') |
| T7 | TAATACGACTCACTATAGGG |
| STAT3 seq 1 forward | AAACTGGATAACGTCATTAGCAGAATCTCA |
| STAT3 seq 1 reverse | TGAGATTCTGCTAATGACGTTATCCAGTTT |
| STAT3 seq 2 forward | CTGGCAGAGAAACTCTTGGGACCTGGTGTG |
| STAT3 seq 2 reverse | CACACCAGGTCCCAAGAGTTTCTCTGCCAG |
| BGH reverse | TAGAAGGCACAGTCGAGG |
| Site-directed mutagenesis | |
| Name | Sequences (5'–3') |
| c.2144C>T (p.P715L) forward | GTGACACTAACGACCTGCAGCAATAC |
| c.2144C>T (p.P715L) reverse | GGTCGTTAGTGTCACACAGATAAACT |
| c.2144C>G (p.P715R) forward | GTGACACGAACGACCTGCAGCAATAC |
| c.2144C>G (p.P715R) reverse | GGTCGTTCGTGTCACACAGATAAACT |
| c.2143C>T (p.P715S) forward | TGTGACATCAACGACCTGCAGCAATA |
| c.2143C>T (p.P715S) reverse | GTCGTTGATGTCACACAGATAAACTT |
| c.2143C>A (p.P715T) forward | TGTGACAACAACGACCTGCAGCAATA |
| c.2143C>A (p.P715T) reverse | GTCGTTGTTGTCACACAGATAAACTT |
| c.2143C>G (p.P715A) forward | TGTGACAGCAACGACCTGCAGCAATA |
| c.2143C>G (p.P715A) reverse | GTCGTTGCTGTCACACAGATAAACTT |
| c.2147C>T (p.T716M) forward | ACACCAATGACCTGCAGCAATACCAT |
| c.2147C>T (p.T716M) reverse | GCAGGTCATTGGTGTCACACAGATAA |
| c.1909G>A (p.V637M) forward | CCAGTCCATGGAACCATACACAAAGC |
| c.1909G>A (p.V637M) reverse | GGTTCCATGGACTGGATCTGGGTCTT |
Site-directed mutagenesis
Mutations in human STAT3 (p.P715L, p.P715R, p.P715S, p.P715T, p.P715A, p.T716M, and p.V637M), except for p.P715Q, were generated from pcDNA3.1-STAT3 wild-type (WT)-V5-His using primeSTAR Mutagenesis Basal Kit (Takara Bio, Inc, Shiga, Japan) and specific primers (Table I), following the manufacturer’s guidelines.
Cell culture
Human hepatoma Hep3B cells were obtained from the Cell Resource Center for Biomedical Research (Tohoku University, Sendai, Japan). Cells were maintained in high-glucose Dulbecco modified Eagle medium (FUJIFILM Wako Pure Chemical Corporation, Osaka, Japan) supplemented with 10% FBS (Gibco, Grand Island, NY) and 1% penicillin-streptomycin (Nacalai Tesque, Kyoto, Japan) at 37°C under a humidified atmosphere of 5% CO2.
Dual-luciferase reporter assay
Hep3B cells were seeded in a 96-well plate at a density of 1 × 104 cells/well suspended in 100 μL of high-glucose Dulbecco modified Eagle medium supplemented with 10% FBS and were transfected after 24 hours with 200 ng pcDNA3.1-STAT3-V5-His, 200 ng IRF1 promoter-luc vector,15 and 10 ng pRL-TK vector using Lipofectamine 2000 and Opti-MEM I Reduced Serum Medium (Invitrogen). Cells were incubated in the transfection mix for 24 hours.
Cells were washed with PBS(−) (FUJIFILM Wako Pure Chemical Corporation), added to 100 μL of Dulbecco modified Eagle medium supplemented with 0.5% FBS, and incubated for 36 hours, and 10 ng/mL recombinant human IL-6 (PeproTech, Rocky Hill, NJ) was also included for the final 12 hours. Cells were washed with PBS(−) and lysed for the luciferase assay using Dual-Luciferase Reporter Assay System (Promega, Madison, Wis).
DNA-STAT3 binding assay
Nuclear extracts of Hep3B cells were obtained using Nuclear Extract Kit (Active Motif, Carlsbad, Calif). The binding activity of STAT3 to DNA was evaluated using TransAM STAT3 Transcription Factor Assay Kit (Active Motif). Eight micrograms of nuclear extracts was used in each reaction. The OD was measured at a wavelength of 450 nm by a microplate reader (Bio-Rad Laboratories, Hercules, Calif).
Western blotting
Hep3B cells were lysed in RIPA buffer (10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Nonidet P-40, 0.1% sodium deoxycholate, 0.1% SDS) supplemented with 1% protease inhibitor and phosphatase inhibitor cocktails (Nacalai Tesque, Kyoto, Japan).
Extracted proteins were subjected to SDS-PAGE and Western blotting. Samples transferred onto PVDF membranes were treated with 1:1000 diluted anti–human phospho-Stat3 (Tyr705) antibody (D3A7, Cell Signaling Technology, Danvers, Mass), anti–human Stat3 antibody (124H6, Cell Signaling Technology), anti–V5 antibody (V5005, Nacalai Tesque), anti–GAPDH antibody (FL-335, Santa Cruz Biotechnology, Santa Cruz, Calif), and anti–Lamin B antibody (M-20, Santa Cruz Biotechnology). These were followed by treatment with 1:2000 or 1:5000 diluted anti–rabbit IgG horseradish peroxidase–linked antibody (#7074, Cell Signaling Technology) or anti–mouse IgG HRP-linked antibody (#7076, Cell Signaling Technology) or anti–goat IgG-HRP antibody (sc-2020, Santa Cruz Biotechnology).
Flow cytometry
Samples for flow cytometric analysis were prepared according to the manufacturer’s protocol. In brief, heparinized whole blood (100 μL) was incubated with fluorochrome-conjugated antibodies for 20 minutes on ice. The antibodies used were anti–CD3-FITC (#300406), anti–CD4-PE/Cy7 (#344612), and anti–CD8-BV510 (#301048; all from Biolegend, San Diego, Calif). To lyse red blood cells, 1× FACS lysing solution (BD Biosciences, San Jose, Calif) was added to each sample and incubated in the dark at room temperature for 15 minutes. After centrifugation, the pelleted blood cells were washed with D-PBS(−) containing 0.5% (wt/vol) BSA and 2 mM EDTA and the resuspended cells were analyzed on a FACSAria Fusion flow cytometer (BD Biosciences). FACS data were analyzed using FlowJo Software (BD Biosciences).
Statistical analysis
In the dual-luciferase reporter assay, multiple comparisons with 1-way ANOVA followed by the Dunnett multiple comparison test were performed. Compared with WT, a P value of less than .05 was considered statistically significant. If not indicated otherwise, data are represented as mean and SD.
Ethical considerations
Parental informed consent for all studies involving human subjects was obtained because the patient was a child. The Ethical Review Boards of Kochi Medical School approved the cyclophosphamide and hydroxychloroquine administration for idiopathic pulmonary hemosiderosis (IPH) and TCZ administration in the patient with STAT3 GOF mutation.
Results
The patient was a 7-year-old boy who was born full term, and his birth weight was appropriate for gestational age.
Endocrine manifestations: Congenital hypothyroidism and autoimmune thyroiditis
Immediately after birth, he was diagnosed with hypothyroidism by neonatal mass screening and was taking thyroid hormone preparation orally from age 9 days. In the later inspection, he was diagnosed with autoimmune thyroiditis because of high serum thyroglobulin antibodies and thyroid peroxidase antibodies.
Lung manifestations: IPH and interstitial lung disease
At the age of 6 months, he had a norovirus infection. At that time, he presented with anemia (hemoglobin 7.0 g/dL) and was given iron preparations. After treatment initiation, the anemia did not improve, and he was referred to our hospital at age 7 months.
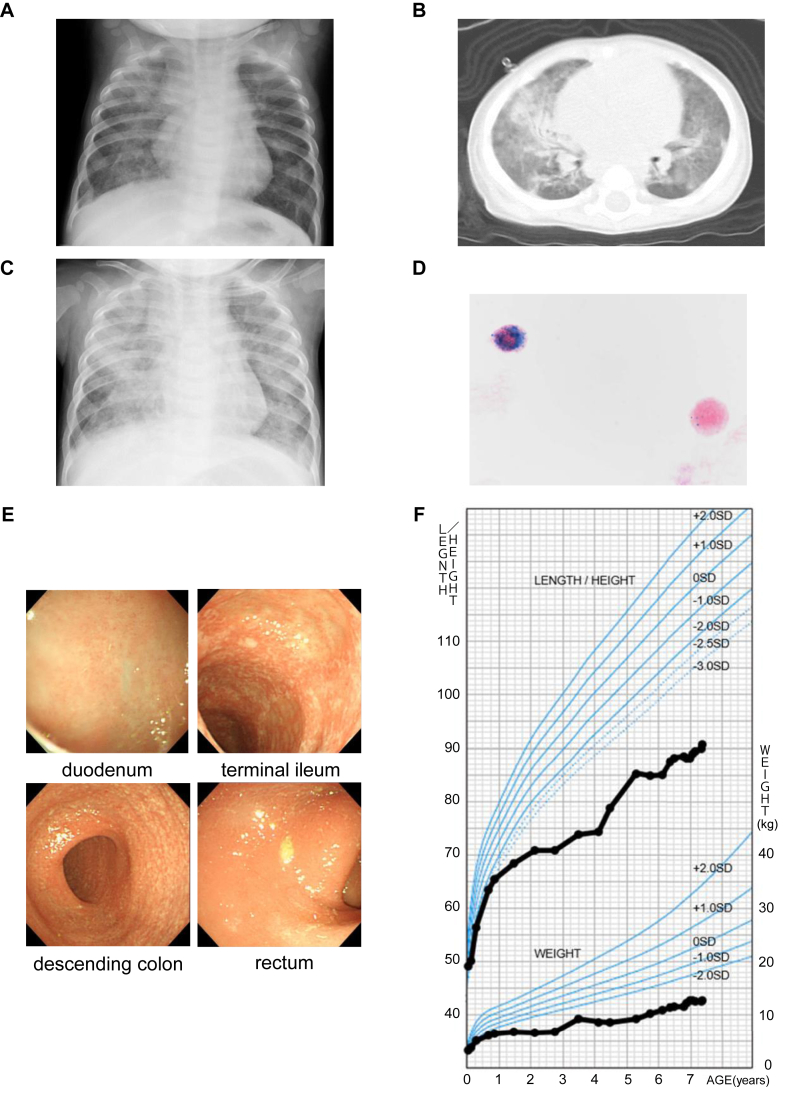
Initial investigations on admission revealed anemia (red blood cell count, 35.6 × 105/μL, hemoglobin 7.0 g/dL, and hematocrit 28.9%) with elevated reticulocyte count (3.0 × 105/μL), leukocytosis (white blood cell count, 28,600 /μL) with lymphocytosis (17,400/μL), and elevated platelet count (6.3 × 105/μL). Inflammatory markers (CRP, 3.8 mg/L; CH50, 54.9 U/mL; C3, 83.0 mg/dL; C4, 27.0 mg/dL; and ferritin, 41.2 ng/mL) and immunoglobulin (IgG, 662 mg/dL; IgA, 89 mg/dL; IgM, 42 mg/dL; and IgE, 0.3 U/mL) were normal. Pulmonary fibrosis markers (KL-6, 1000 U/mL; SP-A, 62.4 ng/mL; SP-D, 224.0 ng/mL) were markedly elevated. Chest X-ray imaging and chest computed tomography showed ground-glass opacity in both lungs (Fig 1, A and B). Atypical pneumonia was suspected, and so clarithromycin was started.
Fig 1.
Multiorgan autoimmune diseases and severe growth failure that developed by early childhood. A and B, Chest X-ray imaging and computed tomography at 6 months showed ground-glass opacity in both lungs. C, Chest X-ray on day 4 of hospitalization showed ground-glass opacity in both lungs, more strongly than on day 1. D, Gastric fluid testing with hematoxylin-eosin and Prussian blue staining demonstrated hemosiderin-laden macrophages. E, Gastrointestinal endoscopy at 2 years 10 months. Mucosal inflammation was observed from the ileum to the entire large intestine. F, Growth chart up to age 7. From approximately 6 months when IPH developed, both height and weight fell below −2.0 SD. Furthermore, growth failure became more prominent approximately from 2 to 4 years when gastrointestinal symptoms due to IBDU were present. TCZ was administered from 6 years and 2 months. IBDU, IBD unclassified. Modified from Cross-sectional Growth Chart for Boys.16
On admission, he did not present with tachypnea (respiratory rate, 30 cycles/min), and his SpO2 was normal (98% on room air). However, after admission, his respiratory rate increased, and SpO2 decreased, requiring oxygen administration. A chest X-ray image showed worsening of the ground-glass opacity (Fig 1, C). Gastric fluid testing with hematoxylin-eosin and Prussian blue staining demonstrated hemosiderin-laden macrophages (Fig 1, D), indicating alveolar hemorrhage. He was diagnosed with pulmonary hemosiderosis and received high-dose prednisolone therapy. Subsequently, respiratory failure improved promptly.
Autoantibodies (ANA, RF, anti-DNA antibody, P-ANCA, C-ANCA, anti–smooth muscle antibody, and anti–GBM antibody) were not elevated. To differentiate from Heiner syndrome, Western blotting, Ouchterlony double immunodiffusion, and immunoprecipitation were performed, and antibodies to cow milk were not detected in the serum. As a diagnostic treatment, he started dairy elimination; however, the pulmonary hemorrhage did not improve. Based on these results, IPH was considered.
He had multiple relapses of IPH, with reducing doses of prednisolone. To reduce the adverse effect of steroids, liposteroid therapy was initiated.17,18 Immunosuppressants (mizoribine, azathioprine, and cyclophosphamide) were also used concomitantly.
At age 2 years and 10 months, bronchoalveolar lavage was performed during intubation and mechanical ventilation due to the exacerbated respiratory status, and hemosiderin-laden macrophages were also detected in the bronchoalveolar lavage fluid.
At age 5 years, hydroxychloroquine treatment was started to prevent recurrent pulmonary hemorrhages. Since then, episodes of dyspnea with progressive anemia decreased; however, serum KL-6 levels gradually increased, which indicated pulmonary fibrosis progression.
Gastrointestinal manifestations: Inflammatory bowel disease unclassified
He had a tendency toward constipation since the age of 1 year. When he was 2 years old, he presented with severe abdominal distension and diarrhea. Fecal calprotectin level was high (3800 mg/kg). An upper gastrointestinal endoscopy revealed mild duodenal villous atrophy. Colonoscopy did not find erythema and ulceration. The pathological finding of mucosal biopsy revealed crypt abscess in a part of the large intestine, and no lesions in the stomach and duodenum were found. He was diagnosed with inflammatory bowel disease (IBD) unclassified and took mesalazine and probiotics. However, mesalazine was discontinued because of the worsening fecal incontinence.
Because of treatment resistance, anti–TNF antibody infliximab was introduced. After the second infliximab dose (15 days after the first dose), his general condition was worsened by severe diarrhea and dehydration. He underwent a second endoscopy 3 months after the first dose. No major problems in stomach and duodenum (Fig 1, E) were found; however, pathologically, mild inflammation was observed in the duodenal mucosa. In the lower gastrointestinal tract, mucosal inflammation was observed from the ileum to the entire large intestine, and pathological findings showed villous atrophy at the end of the ileal mucosa and crypt abscess from the ileum to the entire large intestinal mucosa. Cytomegalovirus staining was negative. Infliximab was discontinued after a total of 2 doses. He was put under fasting control for a while. After the diarrhea subsided, he resumed enteral feeding with elemental diet.
Two months later, endoscopic and pathological findings in both the upper and lower gastrointestinal tracts were improved. Since then, gastrointestinal symptoms remained clinically inconspicuous under treatment for IPH (oral prednisolone and intravenous liposteroid).
In the later inspection, serum tissue transglutaminase IgA was not elevated (<4 U/mL). Together with previous mucosal biopsy findings, celiac disease was considered very unlikely.
Leukocytosis (lymphocytosis, monocytosis, and/or neutrophilia) and thrombocytosis
At age 9 days, thrombocytosis was observed. At 5 months, he had leukocytosis with lymphocytosis. At 6 months, he also had monocytosis. The maximum white blood cell, lymphocyte, and platelet counts before steroid introduction at age 7 months for IPH were 32,700 /μL, 21,900 /μL, and 8.8 × 105 /μL, respectively. He had no lymphadenopathy or splenomegaly. As for thrombocytosis, serum erythropoietin levels (10.4 mU/mL) were not elevated, his bone marrow findings were normal, and JAK2 p.V617F mutation was not detected in the PBMCs. At approximately the age of 1.5 years, which was 1 year after the start of steroid therapy, the lymphocytosis gradually improved to approximately 5000 /μL, which was within the reference range. On the contrary, he had neutrophilia (∼15,000-20,000/μL) and continued to have leukocytosis (∼20,000-30,000 /μL) and monocytosis (∼1000-3000 /μL). Thrombocytosis (∼6-12 × 105 /μL) also persisted; however, no clear thromboembolic symptoms were observed.
Severe growth failure: Short stature and poor weight gain after birth without developmental delay
At birth, height and weight were within the standard range (Fig 1, F).16 He had fallen below the standard range from approximately 3 to 4 months after birth. By the time, he was diagnosed with pulmonary hemosiderosis, at age 6 months, his height and weight were −2.4 SD. His bone age was 3 to 6 months at the actual age of 1 year and 2 months, and less than 1 year and 6 months at the actual age of 4 years and 10 months, showing a marked delay. The growth hormone stimulation tests (arginine and insulin tests) were performed twice; however, the patterns were normal, and he was not diagnosed with growth hormone deficiency. His mental development at the age of 6 was normal.
A novel STAT3 GOF mutation detected by whole-exome sequencing and diagnosed by dual-luciferase reporter assay
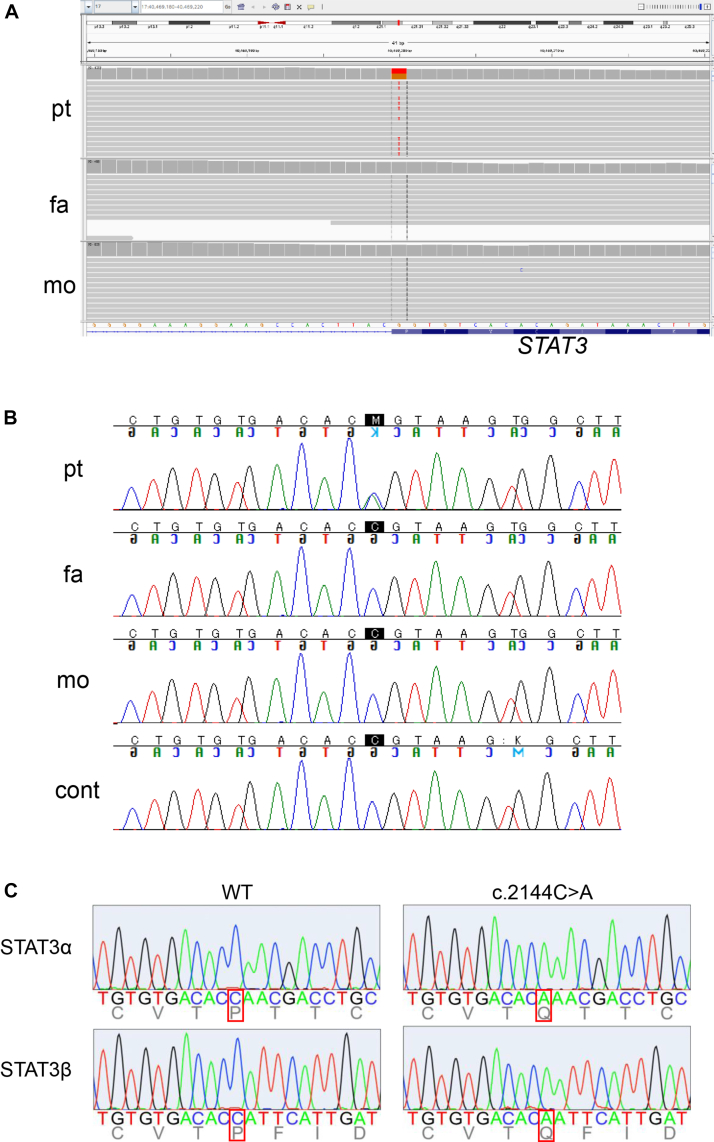
Given the early onset of an autoimmune disease that progressively involves multiple organ systems, inborn error of immunity was suspected. G-banding was normal (46, XY), and fluorescence in situ hybridization of chromosome 21 was negative. His whole-exome sequencing and those of his parents were performed, to identify 9 candidate variants, based on the allele frequency in the general population such as gnomAD and hereditary form. Among these, however, only STAT3 was likely accountable for the phenotype of this patient. This mutation was a germline heterozygous de novo STAT3 mutation (NM_139276.2; c.2144C>A, p.(P715Q)), which was undetectable in his parents (Fig 2, A and B), and had not been reported in the literature.
Fig 2.
The de novo STAT3 mutation. A, The WES data of de novo mutation in STAT3 displayed in IGV viewer. B, Sanger sequencing of the STAT3 variant in genomic DNA. C, Sanger validation of the STAT3 cDNA from the constructed vectors. The c.2144C>A variant was confirmed in both STAT3α and STAT3β, which are transcription variants of STAT3. cont, Control; fa, father; mo, mother; pt, patient; WES, whole-exome sequencing.
To investigate the pathological significance of this STAT3 mutation, WT and mutant STAT3 cDNA were obtained by reverse transcription of mRNA from the patient’s PBMCs, amplified by PCR, and inserted into plasmid vectors. Two vectors containing STAT3 (WT or c.2144C>A) were selected using Sanger validation (Fig 2, C). Previously known mutations of STAT3 (GOF mutations: p.P715L and p.P716M, LOF mutation: p.V637M) were obtained by site-directed mutagenesis based on the vector-inserted STAT3 WT.
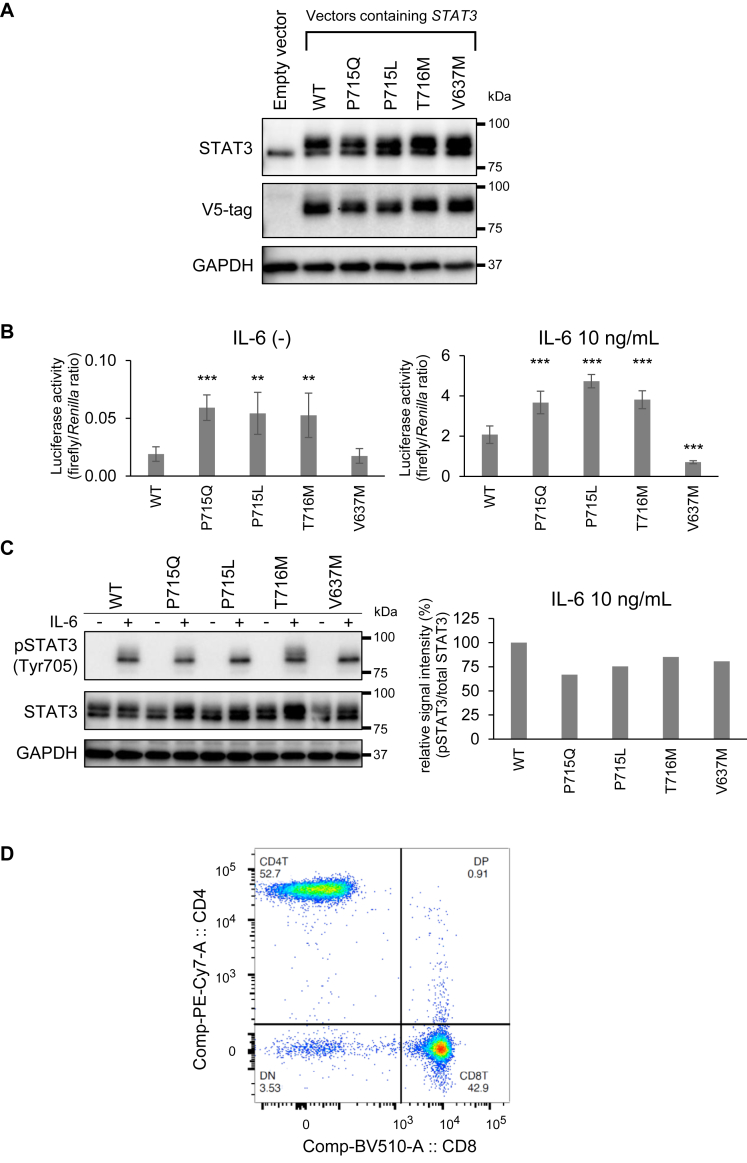
After confirming STAT3 overexpression in Hep3B cells transfected with STAT3-containing vectors (Fig 3, A), dual-luciferase reporter assay was performed to examine transcriptional activities of these variants (Fig 3, B). With or without IL-6 stimulation, high transcriptional activities were observed in cells that overexpressed STAT3 p.P715Q as well as known STAT3 GOF variants7,9 compared with STAT3 WT. In contrast, STAT3 p.V637M overexpression attenuated luciferase activity under IL-6 stimulation compared with WT, illustrating a dominant-negative function of this variant. From these results, we concluded that a novel STAT3 p.P715Q mutation is GOF. In Western blotting, STAT3 phosphorylation was not different between WT and variants overexpressed in Hep3B cells, with or without IL-6 stimulation (Fig 3, C).
Fig 3.
A novel STAT3 p.P715Q mutation was diagnosed as GOF. A, Western blotting of STAT3 protein and V5-tag expressed in Hep3B cells after transient transfection of indicated plasmid vectors. The STAT3 p.P715Q expression was examined alongside that of the WT, 2 previously described GOF mutations (p.P715L and p.T716M), and a previously described LOF mutation (p.V637M). B, STAT3 activity under nonstimulated and IL-6 (10 ng/mL)-stimulated conditions following transient transfection of indicated plasmid vectors into Hep3B cells. Data are presented as an average (n = 6) under each experimental condition ± SEM. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001. C, Western blotting of STAT3 protein and its phosphorylation in Hep3B cells overexpressing STAT3α. Each half of the samples was stimulated with IL-6 (10 ng/mL) for 20 minutes for the analysis. D, Flow cytometric analysis of whole blood leukocytes. The proportion of double-negative (CD3+CD4−CD8−) T cells in CD3+ T cells was 3.5%. GAPDH, Glyceraldehyde 3-phosphate dehydrogenase.
Analysis of peripheral blood lymphocyte subsets: Autoimmune lymphoproliferative syndrome–like immune phenotype
In the flow cytometry of whole blood (Fig 3, D), the proportion of double-negative (CD4−CD8−) T cells in CD3+ T cells was 3.5%.
Analysis of signal enhancement mechanism by STAT3 GOF variants and the effect of the amino acid replacement at position 715 in STAT3
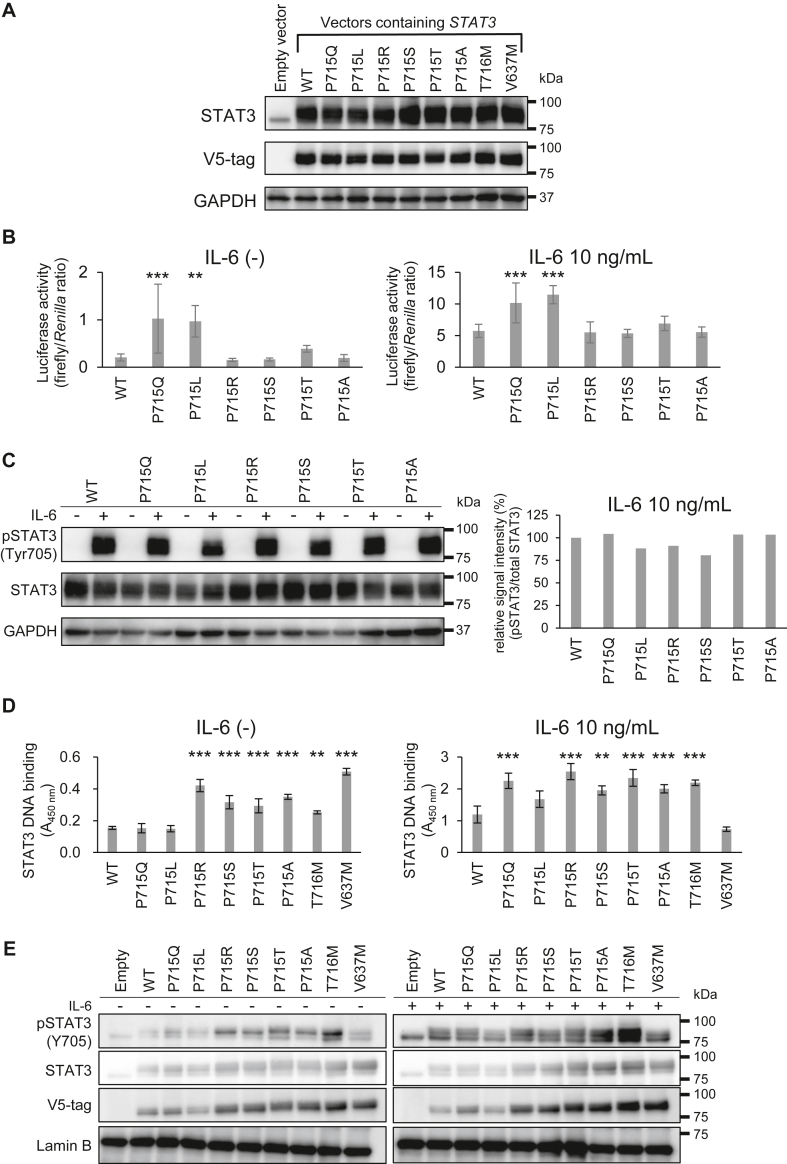
To investigate the effect of amino acid mutations at position 715, variants with amino acid mutations at position 715 (p.P715R, p.P715S, p.P715T, and p.P715A) were generated by introducing single nucleotide substitutions and expressed in Hep3B cells (Fig 4, A).
Fig 4.
Analysis of signal enhancement mechanism by STAT3 GOF variants and the effect of the amino acid replacement at position 715 in STAT3. A, Western blotting of STAT3 protein and V5-tag expressed in Hep3B cells after transient transfection of indicated plasmid vectors. B, STAT3 activity under nonstimulated and IL-6 (10 ng/mL)-stimulated conditions following transient transfection of indicated plasmid vectors into Hep3B cells. Data are presented as an average (n = 6) under each experimental condition ± SEM. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001. C, Western blotting of STAT3 protein and its phosphorylation in Hep3B cells overexpressing STAT3α. Each half of the samples was stimulated with IL-6 (10 ng/mL) for 20 minutes for the analysis. D, DNA-STAT3 binding assay under nonstimulated and IL-6 (10 ng/mL)-stimulated conditions. E, Western blotting of nuclear extracts of Hep3B cells. GAPDH, Glyceraldehyde 3-phosphate dehydrogenase.
In cells overexpressing these new STAT3 variants, transcriptional activities in dual-luciferase reporter assay as well as Tyr705 phosphorylation intensities in Western blotting were not different compared with WT, regardless of the presence or absence of IL-6 stimulation (Fig 4, B and C). However, unexpectedly, these new variants exhibited enhanced DNA-binding activities accompanied with increased nuclear accumulation determined by V5-tag (Fig 4, D and E). In p.P715Q, but not in p.P715L, both DNA-binding activity and nuclear accumulation under IL-6–stimulated conditions were enhanced compared with WT. From these complicated results, we conclude that not all mutants at position 715 of STAT3 are GOF, and the potential mechanism of GOF is discussed in the Discussion section later.
TCZ, targeted therapy for STAT3 GOF mutation
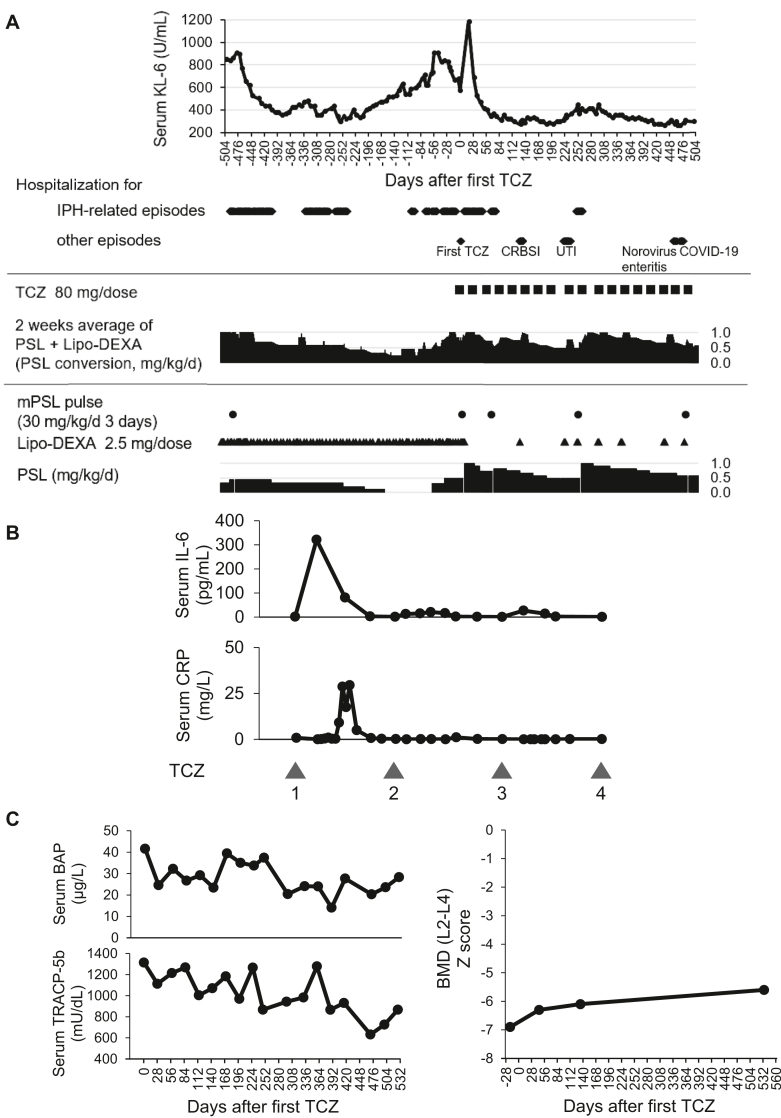
At the age of 6, disease control of IPH had become difficult. After diagnosis of STAT3 GOF, TCZ was administered at a dosage of 8 mg/kg intravenous infusion every 4 weeks (Fig 5, A). Six days after the initial administration, pulmonary hemorrhage was observed, but improved with 1 course of methylprednisolone pulse treatment. The rapid increase in serum IL-6 level after the first TCZ administration (Fig 5, B) was observed and attenuated with repeated dosing. Regarding IBD unclassified, there were no gastrointestinal symptoms. Fecal calprotectin level was not elevated (89.6 mg/kg) 1 month after introducing TCZ. A mild increase in bone mineral density Z score was observed from −6.9 (baseline) to −5.6 (1.5 years after TCZ treatment) with a decrease in bone turnover markers (Fig 5, C).
Fig 5.
TCZ may be effective for patients with STAT3 GOF mutation, including those requiring long-term management of IPH. A, Hospitalizations, main treatments, and serum KL-6 concentrations at approximately 72 weeks after starting TCZ therapy. B, Serum IL-6 and CRP concentration after starting TCZ therapy. TCZ was administered every 4 weeks. C, Serum concentrations of bone turnover markers and BMDs of the lumber spine (L2-L4) around starting TCZ therapy at the age of 6. BAP, Bone-specific alkaline phosphatase; BMD, bone mineral density; COVID-19, coronavirus disease 2019; CRBSI, catheter-related blood stream infection; DEXA, dexamethasone; mPSL, methylprednisolone; PSL, prednisolone; TRACP-5b, tartrate-resistant acid phosphatase 5b; UTI, urinary tract infection.
Despite a few episodes of suspected pulmonary hemorrhage, approximately 72 weeks after TCZ introduction, the monthly average of hospital stays decreased (associated with exacerbation of IPH: 14.6 vs 4.5 days, all episodes: 14.6 vs 4.5 days) without increasing the steroid dose (Table II).
Table II.
Monthly average of hospital stays before and after 72 wk of starting TCZ therapy
| Hospital stays (/30 d) | Pre-TCZ | Post-TCZ |
|---|---|---|
| All episodes | 14.6 | 7.1 |
| IPH-related episodes only | 14.6 | 4.5 |
Discussion
No study has reported germline STAT3 p.P715Q mutation and described IPH development in patients with STAT3 GOF. IPH is a rare disease,19 and its onset in this patient was thought to be triggered by STAT3 GOF.
Lymphoproliferative disease, observed in 73% of patients with STAT3 GOF, is reported to be the most common manifestation, and diffuse lymphadenopathy and splenomegaly are often present. Autoimmune cytopenia is reported to be the second most common clinical manifestation.20 Our patient exhibited no such findings but presented with marked lymphocytosis before steroid introduction and thrombocytosis. These findings together with the increase in peripheral blood double-negative T cells may be signs of lymphoproliferative disease related to STAT3 GOF,21 although it is not typical autoimmune lymphoproliferative syndrome–like phenotype.
TCZ was selected as a targeted therapy for STAT3 GOF owing to its established safety in children. After introducing TCZ, no new autoimmune diseases developed. Although leukocytosis and thrombocytosis remained, this patient might be protected from autoimmune cytopenia, which is often seen in patients with STAT3 GOF. The absence of fecal calprotectin elevation and abdominal symptoms suggests that IBD unclassified did not progress to definitive IBD. Although TCZ did not reverse growth retardation, mild increase in bone mineral density Z score accompanied by a decrease in bone turnover markers, especially bone resorption marker tartrate-resistant acid phosphatase 5b,22 was observed. This may be ascribed to an inhibitory effect of TCZ on osteoclast differentiation, which is enhanced by IL-6 and STAT3.23,24 It appears that the risk of contracting infectious diseases has increased, possibly due to immunosuppression by TCZ. However, it is possible that decreased activity of IPH allowed him to start school, thereby increasing opportunities of infection.
As for IPH, serum KL-6 levels and hospital stays associated with IPH exacerbation decreased without increasing the steroid dose. Based on the treatment course of this patient, TCZ might be partially effective for IPH, particularly decelerating the progression of pulmonary fibrosis. Studies have reported that the IL-6 pathway affects pulmonary fibrosis,25,26 and interstitial lung disease developed in nearly half the patients with STAT3 GOF,10,20 suggesting that not only IPH but also STAT3 overactivation contributes to pulmonary fibrosis.
In this patient, the serum CRP levels were low even before TCZ therapy, suggesting that the involvement of IL-6 in the pathology was not so strong. However, IPH exacerbation and the rapid increase in serum IL-6 levels with elevated CRP after the first TCZ administration (Fig 5, B) suggest that locally produced IL-6 may have been and might affect the pathology of IPH. Serum IL-6 elevation after TCZ administration disappeared with the continuation of TCZ therapy, as in the previous report for rheumatoid arthritis and Castleman disease.27 These results suggest that TCZ is effective not only for STAT3 GOF but also for long-term management of IPH. Because many cytokines other than IL-6 are involved in the STAT3 pathway, treatment with JAK inhibitors that broadly suppress their signals might be another therapeutic option. Some studies have reported that the addition of a JAK inhibitor was effective to STAT3 GOF.10,28, 29, 30, 31
STAT3 activation is characterized by the Tyr705 phosphorylation located in the transactivation domain (TAD), where the p.P715Q, p.P715L, and p.T716M mutations are also located. However, similar to previous reports,9,32, 33, 34, 35, 36 phosphorylation intensity of Tyr705 was not affected in these variants, suggesting that different mechanisms account for GOF. TAD of STAT3 contributes to homodimerization via binding to SH2. Increased nuclear accumulation of the p.P715Q compared with WT (Fig 4, E; visualized by V5-tag) suggests that enhanced dimerization and/or nuclear translocation contributes to the function of this variant. Nevertheless, both the GOF p.T716M and LOF p.V637M similarly exhibited enhanced nuclear accumulation. In addition, nuclear accumulation of p.P715L, another GOF, was not enhanced compared with WT, although this mutant showed increased transcriptional activity comparable to those of p.P715Q and p.T716M. Furthermore, newly synthesized variants at amino acid position 715 in STAT3 (p.P715R, p.P715S, p.P715T, and p.P715A) showed increased nuclear accumulation and DNA-binding activity (Fig 4, D and E), but their transcriptional activities were not enhanced compared with WT (Fig 4, B). These results suggest that neither nuclear accumulation nor DNA-binding activity determines the final function of STAT3 variants at Pro715 and that there remains another mechanism that leads to GOF.
Proline is an amino acid that confers unique conformational constraints on the peptide chain in that the side chain is cyclized back onto the backbone amide position,37 replacing Pro715 with another amino acid may have a significant impact on the structure of TAD. STAT3 TAD contributes not only to homodimerization but also to transactivation, like other TADs that interact with components of the preinitiation complex to enhance recruitment and stabilization of the general factors at target promoters.2,3,38 Therefore, STAT3 variants with TAD mutations may have alterations in the transactivation of the target gene. We speculate that p.P715L and probably p.P715Q may be more efficient in recruiting the transcriptional machinery, whereas 4 new variants of STAT3 (p.P715R, p.P715S, p.P715T, and p.P715A) that showed enhanced nuclear accumulation and DNA-binding activities may have decreased efficiency in the transcriptional machinery recruitment, resulting in enhanced or unaltered transcriptional activities, respectively, in the reporter assay. Thus, mutations of Pro715 in STAT3 may have various effects on the function of TAD, possibly resulting in altered dimerization, nuclear accumulation, and/or transcription. We presume that the final transcriptional activities of STAT3 variants are determined by the sum of the efficiencies of these multiple processes. Further investigation is needed to determine the mechanism of altered functions due to mutations at Pro715 in STAT3.
In our case, whole-exome sequencing revealed a STAT3 GOF mutation, opening up new treatment options. Early diagnosis based on identification of STAT3 mutations is desirable to select better treatment and improve prognosis. However, increasing number of case reports have revealed that STAT3 GOF has very diverse manifestations, and the STAT3 mutation site does not appear to determine the manifestations20; thus, suspecting and diagnosing this disease are very difficult at its early stage. Further accumulation of cases is expected to increase our understanding of this disease. Furthermore, not limited to STAT3 GOF, accumulation of more knowledge such as on manifestations of other genetic diseases is warranted in the future. When it is unknown whether the identified gene mutation is pathogenic, further functional analysis of the mutant protein is recommended.39, 40, 41
Disclosure statement
This work was supported by the Initiative on Rare and Undiagnosed Diseases (grant no. JP23ek0109549) from the Japan Agency for Medical Research and Development.
Disclosure of potential conflict of interest: The authors declare that they have no relevant conflicts of interest.
Acknowledgments
We thank Akira Shiraishi, Masataka Ishimura, Shouichi Ohga, and the staff (Department of Pediatrics, Graduate School of Medical Sciences, Kyushu University) for taking the consultation on managing IPH. We thank Takatomo Maeyama, Yuri Etani, and the staff (Department of Pediatric Gastroenterology, Nutrition, and Endocrinology, Osaka Women’s and Children’s Hospital) for their detailed examination, including gastrointestinal endoscopy and bronchoscopy.
References
- 1.Hillmer E.J., Zhang H., Li H.S., Watowich S.S. STAT3 signaling in immunity. Cytokine Growth Factor Rev. 2016;31:1–15. doi: 10.1016/j.cytogfr.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Horvath C.M. STAT proteins and transcriptional responses to extracellular signals. Trends Biochem Sci. 2000;25:496–502. doi: 10.1016/s0968-0004(00)01624-8. [DOI] [PubMed] [Google Scholar]
- 3.Levy D.E., Darnell J.E., Jr. Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 4.Takeda K., Kaisho T., Yoshida N., Takeda J., Kishimoto T., Akira S. Stat3 activation is responsible for IL-6-dependent T cell proliferation through preventing apoptosis: generation and characterization of T cell-specific Stat3-deficient mice. J Immunol. 1998;161:4652–4660. [PubMed] [Google Scholar]
- 5.Holland S.M., DeLeo F.R., Elloumi H.Z., Hsu A.P., Uzel G., Brodsky N., et al. STAT3 mutations in the hyper-IgE syndrome. N Engl J Med. 2007;357:1608–1619. doi: 10.1056/NEJMoa073687. [DOI] [PubMed] [Google Scholar]
- 6.Woellner C., Gertz E.M., Schaffer A.A., Lagos M., Perro M., Glocker E.O., et al. Mutations in STAT3 and diagnostic guidelines for hyper-IgE syndrome. J Allergy Clin Immunol. 2010;125:424–432.e8. doi: 10.1016/j.jaci.2009.10.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flanagan S.E., Haapaniemi E., Russell M.A., Caswell R., Allen H.L., De Franco E., et al. Activating germline mutations in STAT3 cause early-onset multi-organ autoimmune disease. Nat Genet. 2014;46:812–814. doi: 10.1038/ng.3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haapaniemi E.M., Kaustio M., Rajala H.L., van Adrichem A.J., Kainulainen L., Glumoff V., et al. Autoimmunity, hypogammaglobulinemia, lymphoproliferation, and mycobacterial disease in patients with activating mutations in STAT3. Blood. 2015;125:639–648. doi: 10.1182/blood-2014-04-570101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Milner J.D., Vogel T.P., Forbes L., Ma C.A., Stray-Pedersen A., Niemela J.E., et al. Early-onset lymphoproliferation and autoimmunity caused by germline STAT3 gain-of-function mutations. Blood. 2015;125:591–599. doi: 10.1182/blood-2014-09-602763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fabre A., Marchal S., Barlogis V., Mari B., Barbry P., Rohrlich P.S., et al. Clinical aspects of STAT3 gain-of-function germline mutations: a systematic review. J Allergy Clin Immunol Pract. 2019;7:1958–1969.e9. doi: 10.1016/j.jaip.2019.02.018. [DOI] [PubMed] [Google Scholar]
- 11.Wang W., Liu L., Hui X., Wang Y., Ying W., Zhou Q., et al. Efficacy of tocilizumab therapy in a patient with severe pancytopenia associated with a STAT3 gain-of-function mutation. BMC Immunol. 2021;22:19. doi: 10.1186/s12865-021-00411-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takahashi Y., Date H., Oi H., Adachi T., Imanishi N., Kimura E., et al. Six years’ accomplishment of the Initiative on Rare and Undiagnosed Diseases: nationwide project in Japan to discover causes, mechanisms, and cures. J Hum Genet. 2022;67:505–513. doi: 10.1038/s10038-022-01025-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamada M., Suzuki H., Watanabe A., Uehara T., Takenouchi T., Mizuno S., et al. Role of chimeric transcript formation in the pathogenesis of birth defects. Congenit Anom (Kyoto) 2021;61:76–81. doi: 10.1111/cga.12400. [DOI] [PubMed] [Google Scholar]
- 14.Cingolani P., Platts A., Wang le L., Coon M., Nguyen T., Wang L., et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Narazaki M., Fujimoto M., Matsumoto T., Morita Y., Saito H., Kajita T., et al. Three distinct domains of SSI-1/SOCS-1/JAB protein are required for its suppression of interleukin 6 signaling. Proc Natl Acad Sci U S A. 1998;95:13130–13134. doi: 10.1073/pnas.95.22.13130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Isojima T., Kato N., Ito Y., Kanzaki S., Murata M. Growth standard charts for Japanese children with mean and standard deviation (SD) values based on the year 2000 national survey. Clin Pediatr Endocrinol. 2016;25:71–76. doi: 10.1297/cpe.25.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doi T., Ohga S., Ishimura M., Takada H., Ishii K., Ihara K., et al. Long-term liposteroid therapy for idiopathic pulmonary hemosiderosis. Eur J Pediatr. 2013;172:1475–1481. doi: 10.1007/s00431-013-2065-9. [DOI] [PubMed] [Google Scholar]
- 18.Doi T., Ohga S., Ishimura M., Takada H., Ishii K., Ihara K., et al. Erratum to: Long-term liposteroid therapy for idiopathic pulmonary hemosiderosis. Eur J Pediatr. 2015;174:1701. doi: 10.1007/s00431-015-2638-x. [DOI] [PubMed] [Google Scholar]
- 19.Taytard J., Nathan N., de Blic J., Fayon M., Epaud R., Deschildre A., et al. New insights into pediatric idiopathic pulmonary hemosiderosis: the French RespiRare((R)) cohort. Orphanet J Rare Dis. 2013;8:161. doi: 10.1186/1750-1172-8-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leiding J.W., Vogel T.P., Santarlas V.G.J., Mhaskar R., Smith M.R., Carisey A., et al. Monogenic early-onset lymphoproliferation and autoimmunity: natural history of STAT3 gain-of-function syndrome. J Allergy Clin Immunol. 2023;151:1081–1095. doi: 10.1016/j.jaci.2022.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vogel T.P., Leiding J.W., Cooper M.A., Forbes Satter L.R. STAT3 gain-of-function syndrome. Front Pediatr. 2022;10 doi: 10.3389/fped.2022.770077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Halleen J.M., Tiitinen S.L., Ylipahkala H., Fagerlund K.M., Vaananen H.K. Tartrate-resistant acid phosphatase 5b (TRACP 5b) as a marker of bone resorption. Clin Lab. 2006;52:499–510. [PubMed] [Google Scholar]
- 23.Takayanagi H. Osteoimmunology and the effects of the immune system on bone. Nat Rev Rheumatol. 2009;5:667–676. doi: 10.1038/nrrheum.2009.217. [DOI] [PubMed] [Google Scholar]
- 24.Yokota K. Osteoclast differentiation in rheumatoid arthritis. Immunol Med. 2024;47:6–11. doi: 10.1080/25785826.2023.2220931. [DOI] [PubMed] [Google Scholar]
- 25.Pechkovsky D.V., Prele C.M., Wong J., Hogaboam C.M., McAnulty R.J., Laurent G.J., et al. STAT3-mediated signaling dysregulates lung fibroblast-myofibroblast activation and differentiation in UIP/IPF. Am J Pathol. 2012;180:1398–1412. doi: 10.1016/j.ajpath.2011.12.022. [DOI] [PubMed] [Google Scholar]
- 26.Epstein Shochet G., Brook E., Bardenstein-Wald B., Shitrit D. TGF-beta pathway activation by idiopathic pulmonary fibrosis (IPF) fibroblast derived soluble factors is mediated by IL-6 trans-signaling. Respir Res. 2020;21:56. doi: 10.1186/s12931-020-1319-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nishimoto N., Terao K., Mima T., Nakahara H., Takagi N., Kakehi T. Mechanisms and pathologic significances in increase in serum interleukin-6 (IL-6) and soluble IL-6 receptor after administration of an anti-IL-6 receptor antibody, tocilizumab, in patients with rheumatoid arthritis and Castleman disease. Blood. 2008;112:3959–3964. doi: 10.1182/blood-2008-05-155846. [DOI] [PubMed] [Google Scholar]
- 28.Parlato M., Charbit-Henrion F., Abi Nader E., Begue B., Guegan N., Bruneau J., et al. Efficacy of ruxolitinib therapy in a patient with severe enterocolitis associated with a STAT3 gain-of-function mutation. Gastroenterology. 2019;156:1206–1210.e1. doi: 10.1053/j.gastro.2018.11.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Forbes L.R., Vogel T.P., Cooper M.A., Castro-Wagner J., Schussler E., Weinacht K.G., et al. Jakinibs for the treatment of immune dysregulation in patients with gain-of-function signal transducer and activator of transcription 1 (STAT1) or STAT3 mutations. J Allergy Clin Immunol. 2018;142:1665–1669. doi: 10.1016/j.jaci.2018.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Silva-Carmona M., Vogel T.P., Marchal S., Guesmi M., Dubus J.C., Leroy S., et al. Successful treatment of interstitial lung disease in STAT3 gain-of-function using JAK inhibitors. Am J Respir Crit Care Med. 2020;202:893–897. doi: 10.1164/rccm.201906-1204LE. [DOI] [PubMed] [Google Scholar]
- 31.Wegehaupt O., Muckenhaupt T., Johnson M.B., Schwab K.O., Speckmann C. Ruxolitinib controls lymphoproliferation and diabetes in a STAT3-GOF patient. J Clin Immunol. 2020;40:1207–1210. doi: 10.1007/s10875-020-00864-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jagle S., Heeg M., Grun S., Rensing-Ehl A., Maccari M.E., Klemann C., et al. Distinct molecular response patterns of activating STAT3 mutations associate with penetrance of lymphoproliferation and autoimmunity. Clin Immunol. 2020;210 doi: 10.1016/j.clim.2019.108316. [DOI] [PubMed] [Google Scholar]
- 33.Mauracher A.A., Eekels J.J.M., Woytschak J., van Drogen A., Bosch A., Prader S., et al. Erythropoiesis defect observed in STAT3 GOF patients with severe anemia. J Allergy Clin Immunol. 2020;145:1297–1301. doi: 10.1016/j.jaci.2019.11.042. [DOI] [PubMed] [Google Scholar]
- 34.Saarimaki-Vire J., Balboa D., Russell M.A., Saarikettu J., Kinnunen M., Keskitalo S., et al. An activating STAT3 mutation causes neonatal diabetes through premature induction of pancreatic differentiation. Cell Rep. 2017;19:281–294. doi: 10.1016/j.celrep.2017.03.055. [DOI] [PubMed] [Google Scholar]
- 35.Sarfati E., Hadjadj J., Fusaro M., Klifa R., Grimaud M., Berteloot L., et al. Life-saving, dose-adjusted, targeted therapy in a patient with a STAT3 gain-of-function mutation. J Clin Immunol. 2021;41:807–810. doi: 10.1007/s10875-020-00914-3. [DOI] [PubMed] [Google Scholar]
- 36.Wienke J., Janssen W., Scholman R., Spits H., van Gijn M., Boes M., et al. A novel human STAT3 mutation presents with autoimmunity involving Th17 hyperactivation. Oncotarget. 2015;6:20037–20042. doi: 10.18632/oncotarget.5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vanhoof G., Goossens F., De Meester I., Hendriks D., Scharpé S. Proline motifs in peptides and their biological processing. FASEB J. 1995;9:736–744. [PubMed] [Google Scholar]
- 38.Frietze S., Farnham P.J. Transcription factor effector domains. Subcell Biochem. 2011;52:261–277. doi: 10.1007/978-90-481-9069-0_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abraham R.S., Butte M.J. The new “wholly trinity” in the diagnosis and management of inborn errors of immunity. J Allergy Clin Immunol Pract. 2021;9:613–625. doi: 10.1016/j.jaip.2020.11.044. [DOI] [PubMed] [Google Scholar]
- 40.Heimall J. Now is the time to use molecular gene testing for the diagnosis of primary immune deficiencies. J Allergy Clin Immunol Pract. 2019;7:833–838. doi: 10.1016/j.jaip.2018.12.022. [DOI] [PubMed] [Google Scholar]
- 41.Leiding J.W., Forbes L.R. Mechanism-based precision therapy for the treatment of primary immunodeficiency and primary immunodysregulatory diseases. J Allergy Clin Immunol Pract. 2019;7:761–773. doi: 10.1016/j.jaip.2018.12.017. [DOI] [PubMed] [Google Scholar]