Highlights
-
•
Clonal spread of New Delhi metallo-β-lactamase 1 Pseudomonas aerurinosa ST773 of Ukrainian origin in European countries.
-
•
Genomic analysis revealed highly related isolates, distant from non-Ukrainian strains.
-
•
The prophage content shows genetic relatedness of New Delhi metallo-β-lactamase 1 P. aerurinosa ST773 isolates.
-
•
Collaborative efforts are needed to combat antimicrobial resistance on a global scale.
Keywords: NDM-1-ST773 P. aeruginosa, Ukrainian patients, Clonal dissemination, Spain, The Netherlands, Bacteriophages
Abstract
Objectives
We describe the clonal spread of New Delhi metallo-β-lactamase (NDM) 1–producing Pseudomonas aeruginosa isolates belonging to the ST773 clone in Spain and the Netherlands, associated with the transfer of Ukrainian patients during the war.
Methods
Between March and December 2022, nine NDM-1–producing P. aeruginosa ST773 isolates were recovered from nine Ukrainian patients evacuated to two Spanish (n = 3) and five Dutch (n = 6) hospitals. Antimicrobial susceptibility testing was studied (Sensititre, Microscan, EUCAST-2023). Whole genome sequencing (Illumina, Oxford-Nanopore) was used to analyze the genetic relatedness, the resistome, and the prophage content.
Results
All NDM-1–producing P. aeruginosa ST773 isolates exhibited resistance to all tested antimicrobials except colistin, aztreonam, and cefiderocol. Genomic analysis revealed that all isolates had an identical resistome and a chromosomally encoded integrative conjugative element carrying the blaNDM-1 gene. The core genome multilocus sequence typing and core genome single nucleotide polymorphisms analysis showed highly related isolates, irrespective of country of isolation, distant from other NDM-1-ST773 P. aeruginosa not collected in Ukraine. Both analysis revealed two closely related clusters, spanning the Spanish and Dutch isolates. In addition, a high content of prophages was identified in all strains, most of them in more than one isolate simultaneously, regardless of their origin country. Moreover, an identical phage tail-like bacteriocin cluster was identified in all NDM-1-ST773 P. aeruginosa.
Conclusions
We report a clonal dissemination of NDM-producing P. aeruginosa ST773 to the Netherlands and Spain associated with patients from Ukraine. Our work highlights the importance of genomic surveillance and to understand the dynamics of resistance in multidrug-resistant bacteria after the transfer of patients from conflict zones. International collaboration is crucial to address global antimicrobial resistance.
Introduction
Between 2014 and 2021, Ukraine reported higher rates of antimicrobial resistance in regional and military hospitals compared to other European Union/European Economic Area countries, particularly, concerning gram-negative bacteria [[1], [2], [3]]. Given the ongoing conflict in Ukraine and the anticipated movement of migrants, the European Centre for Disease Prevention and Control issued a guideline in March 2022, recommending the screening for carriage of multidrug-resistant organisms and pre-emptive isolation of patients displaced and injured, transferred from hospitals in Ukraine, or with a history of hospital admission in Ukraine in the last 12 months [4]. Despite these proactive measures, some countries have notified cases of infection/colonization attributed to certain multidrug-resistant clones of Acinetobacter baumannii, Enterobacterales, and Pseudomonas aeruginosa, which are typically associated with health care settings in Ukraine [[5], [6], [7], [8]].
P. aeruginosa is an opportunistic pathogen that causes severe infections, particularly, in the hospital setting and in immunocompromised patients. P. aeruginosa high-risk clones are disseminated worldwide and often harbor acquired resistance genes that confer them a multidrug or extensively drug-resistant (MDR/XDR) profile, which facilitates their spread in the hospital setting and gives them the opportunity to obtain more resistance determinants from other gram-negative pathogens [9,10]. In addition, P. aeruginosa has shown a great ability to integrate other genetic structures such as temperate bacteriophages, named prophages, that can confer the parasitized bacterium or lysogen evolutionary advantages [11].
The Netherlands and Spain were the first countries to report cases of New Delhi metallo-β-lactamase (NDM) 1–producing P. aeruginosa isolates belonging to the ST773 clone imported from Ukraine during 2022 after the initiation of the war [6,8]. In this work, we characterized by whole genome sequencing (WGS) a collection of NDM-1-ST773 P. aeruginosa isolates recovered in these two countries in 2022, focusing on their genetic relationship, the characterization of mobile genetic elements involved in the dissemination of blaNDM, and the prophage content.
Methods
Bacterial isolates and patient's background
Between March and December 2022, nine P. aeruginosa isolates compatible with the production of a metallo-β-lactamase were isolated in clinical or surveillance samples from nine Ukrainian patients transferred to general hospitals in Madrid, Spain (Hospital Universitario 12 de Octubre y Hospital Universitario Ramón y Cajal) and in five provinces at the Netherlands (Overijssel, Utrecht, Gelderland, Noord-Holland, and Limburg) (Table 1). In Spain, phenotypic characterization and WGS was performed at each of the hospitals where the isolates were recovered. The Dutch samples were phenotypically characterized in each of the centers, and then were sent to the National Institute for Public Health and the Environment for WGS. Clinical, epidemiologic, and demographic data were retrospectively reviewed. All sequences were sent to the Hospital Universitario Ramón y Cajal (Madrid, Spain) for the subsequent genomic characterization. The study was approved by the ethical committee (Ref. 104/23).
Table 1.
Epidemiological data of Ukrainian patients infected/colonized by NDM-1-ST773 P. aeruginosa isolates evacuated to Spain and the Netherlands during 2022. Antimicrobial susceptibility data are also included.
| Isolate | Gender | Admission diagnosis | Source of infection (+) NDM-Pa | Month (2022) | Week (2022) | Province (Country) | Remarks | Other microorganisms detected during the admission | Resistance profilea,b |
|---|---|---|---|---|---|---|---|---|---|
| Isolate_ESP_1 | M | Septic pseudoarthritis in left femur | Abscess (Wound) | March | 13 | Madrid (Spain) | Previous admission to a Kyiv hospital (left femur septic pseudoarthritis) | OXA-48+NDM-1 K. pneumoniae | P/T4, FEP, CZA, C/T, IMI, IMI/REL, MER, MEV, TOB, AMK |
| Isolate_ESP_2 | F | Mediastinitis | Bone sample (surgical site) | May | 20 | Madrid (Spain) | Refugee with previous heart surgery in November 2021 in Ukraine. | VIM-1 K. oxytoca, Aspergillus fumigatus, extended spectrum beta-lactamase-Escherichia coli and Corynebacterium sp. | P/T4, FEP, CZA, C/T, IMI, IMI/REL, MER, MEV, TOB, AMK |
| Isolate_ESP_3 | M | Surgical infection in the right jaw | Abscess (Wound) | June | 26 | Madrid (Spain) | Ukrainian soldier with shotgun in the jaw (previous admissions to military hospitals in Ukraine, in Dnipro and Vynnitsya) | OXA-48+NDM-1 K. pneumoniae, OXA-48 K. pneumoniae | P/T4, FEP, CZA, C/T, IMI, IMI/REL, MER, MEV, TOB, AMK |
| Isolate_NLD_1 | M | Unknown | Tissue | July | 28 | Overijssel (The Netherlands) | Ukrainian person | Not recovered | MER |
| Isolate_NLD_2 | F | SSTI | Wound | August | 31 | Utrecht (The Netherlands) | Ukrainian person | NDM-1 K. pneumoniae, OXA-23-Acinetobacter baumannii, OXA-66-A. baumannii | MER |
| Isolate_NLD_3 | M | SSTI | Wound | August | 33 | Overijssel (The Netherlands) | Ukrainian soldier | OXA-72-A. baumannii, NDM-1 K. pneumoniae, OXA-48 K. pneumoniae | MER |
| Isolate_NLD_4 | M | Carrier screening | Rectal swab | August | 34 | Gelderland (The Netherlands) | Ukrainian soldier with shot wound direct transfer from military hospital in Poland | Not recovered | MER |
| Isolate_NLD_5 | M | SSTI | Tissue biopsy | September | 39 | Noord-Holland (The Netherlands) | Soldier with trauma from Ukraine | NDM-1-Proteus stuartii | MER |
| Isolate_NLD_6 | M | Carrier screening | Rectal swab | December | 51 | Limburg (The Netherlands) | Ukrainian person | NDM-5-E. coli, KPC-3-E. coli, KPC-2 K. pneumoniae, NDM-1 K. pneumoniae, OXA-48 K. pneumoniae | MER |
AMK, amikacin; CZA, ceftazidime-avibactam; C/T, ceftolozane-tazobactam; F, female; FEP, cefepime; IMI, imipenem; IMI/REL, imipenem-relebactam; M, male; MER, meropenem; MEV, meropenem-vaborbactam; P/T4, piperacillin-tazobactam; SSTI, skin and soft tissue infection; TOB, tobramycin.
Antimicrobials for which NDM-1-S773-P. aeruginosa isolates showed resistant minimum inhibitory concentration values (according to EUCAST).
In isolates recovered in The Netherlands, only meropenem susceptibility was reported.
Phenotypic characterization
In the isolates detected in Spain, antimicrobial susceptibility was determined by broth microdilution using a semiautomated microdilution system (MicroScan, Beckman Coulter Diagnostics, USA) and the SENSITITRE EUMDROXF panel (ThermoFisher, USA). Minimum inhibitory concentrations (MICs) were interpreted using EUCAST-2023 clinical break points (http://www.eucast.org/clinical_breakpoints/). Cefiderocol susceptibility was assessed by gradient strips and disk diffusion (Liofilchem, Roseto degli Abruzzi, Italy). In the isolates collected in the Netherlands, meropenem susceptibility was determined using gradient strips (Liofilchem). Carbapenemase production was confirmed using the KPC/MBL/OXA-48 Confirm Kit (Rosco Diagnostica, Taastrup, Denmark), the immunochromatography test O.K.N.V.I. RESIST 5 (CORIS BioConcept, Gembloux, Belgium), the eazyplex Superbug CRE system (Amplex-Biosystems, Delaware), the NG-Test CARBA 5 (NG-Biotech, France), the Allplex Entero-DR Assay kit (Seegene Inc., Bogotá, Colombia), or the carbapenem inactivation method [12].
WGS and bioinformatic analysis
Short-read sequencing was performed using the Illumina NovaSeq 6000 platform (OGC, Oxford, UK) (Spain) and Illumina NextSeq550 (the Netherlands), with 2- × 150-bp paired-end reads. Nanopore long-read technology was also carried out using a MinION flow cell (R9.4.1) in accordance with SQK-LSK110 sequencing procedures. The Unicycler tool (v0.4.8) was used to obtain the complete hybrid genome assemblies from the combination of short- and long-reads, as previously reported [8]. All consensus assemblies were annotated using RAST Server (v2.0) (https://rast.nmpdr.org/). Assemblies were deposited at DDBJ/ENA/GenBank under the project number PRJNA949836.
Molecular characterization and resistome analysis
Antimicrobial resistance genes were screened using ABRicate (v1.0.1) (ResFinder and CARD databases, threshold, 90% coverage; 98% identity). In silico multilocus sequence typing (MLST) assignment was carried out using MLST (v2.16.1) (https://github.com/tseemann/mlst). Variant calling was also performed for the analysis of chromosomal determinants involved in antibiotic resistance, as described previously [13]. Core genome single nucleotide polymorphisms (cgSNPs) maximum likelihood phylogenetic trees were reconstructed and visualized using IQ-TREE (v2.0.7) software and iTOL, respectively. The core genome MLST (cgMLST) of all genomes was also created and validated using the pipeline chewBBACA [14]. In all these analysis, P. aeruginosa PAO1 (GenBank accession no. NC_002516.2) was used as the reference genome.
Mobile genetic elements
The complete genomes and the mobile genetic elements implicated in the dissemination of β-lactamase genes were characterized, reconstructed, and visualized using ICEfinder (https://bioinfo-mml.sjtu.edu.cn/ICEfinder/ICEfinder.html), Blast webtool (blast.ncbi.nlm.gov), Kablammo (http://kablammo.wasmuthlab.org), and Proksee (https://proksee.ca/).
Bacteriophages and pyocins
Prophage analysis was performed using the command line software VIBRANT and Blast [15]. Phage taxonomy was stablished by creating a phage proteomic tree using ViPTree version 3.7 with these prophages and phages on the virus-host database (https://www.genome.jp/viptree/) [16]. Phages clustered together were analyzed to be of the same genus using VIRIDIC (https://rhea.icbm.uni-oldenburg.de/viridic/). These results were confirmed by BLAST analysis of the terminase large subunit, whenever this gene was present in the genomes, accepting query cover and identity values above 95% as the same species. Pyocin clusters were classified by BLASTp analysis of the tail fiber proteins against the NCBI database, considering query cover and identity values above 90% as the same pyocin subtype [17].
Prophage and pyocins were annotated by RAST and Pharokka [18]. Unassigned Open Reading Frames (ORFs) were manually annotated by HMMER (http://hmmer.org/) and HHPRED, as previously described [17]. Anti-CRISPR proteins were analyzed by BLASTp against the Anti-CRISPRdb database [19]. Regions encoding prophages and pyocins were visualized using Proksee.
Results
Bacterial isolates and clinical data
Nine unrelated patients of Ukrainian origin (median age, 42 years [range: 23-74 years]; seven of nine male patients) colonized (two of nine) or infected (seven of nine) by NDM-producing P. aeruginosa isolates were detected in two Spanish (three patients) and five Dutch (six patients) hospitals between March and December 2022. All patients were transferred or evacuated from Ukraine as a direct consequence of the war. Previous hospital admission in Ukraine in the previous 12 months was documented in at least six patients (five injured soldiers and one older woman undergoing heart surgery). In addition, colonization or infection by other carbapenemase-producing Enterobacterales was also detected in seven patients (Table 1).
All NDM-producing P. aeruginosa isolates recovered in Spain were resistant to all tested antimicrobials, except for colistin (MIC ≤2 mg/L), aztreonam (MIC: 8-12 mg/L), and cefiderocol (MIC range 0.75-2 mg/L; disk diffusion method: 20-23 mm).
Resistance gene content and mutational resistome
Screening for antibiotic resistance genes revealed that all isolates carried an identical content of resistance genes affecting different antimicrobial groups: aminoglycosides [aac(3), aadA11, aph(3′)-IIb, rmtB4], β-lactams (blaNDM-1, blaOXA-395, blaPDC-16), chloramphenicol (catB7), fosfomycin (fosA), sulphonamides (two copies of sul1), and tetracyclines (tetG). One antiseptic-resistant gene (qacE) and one gene affecting the quaternary ammonium compound efflux SMR transporter (qnrVC1) were also found in all strains. One Dutch isolate (Isolate_NLD_2) carried two copies of the blaNDM-1 gene (Table 2).
Table 2.
Results of the genomic characterization of all NDM-1–producing P. aeruginosa isolates recovered from Ukrainian patients in Spain and the Netherlands during 2022.
| Isolate | ST | Clustera | Chromosomeb (pb) | blaNDM-1-ICE size(pb) | Plasmidc (pb) | Antibiotic-resistant genes content | Accession number |
|---|---|---|---|---|---|---|---|
| Isolate_ESP_1 | 773 | I | 6,852,907 | 116,999 | pH12O (46,720) | aac(3), aadA11, aph(3′)-IIb, blaNDM-1, blaOXA-395, blaPDC-16, catB7, fosA, qacE, qnrVC1, rmtB4, sul1, sul1, tet(G) | CP142443b, CP142444c |
| Isolate_ESP_2 | 773 | I | 6,922,344 | 116,997 | pB81 (51,081) | aac(3), aadA11, aph(3′)-IIb, blaNDM-1, blaOXA-395, blaPDC-16, catB7, fosA, qacE, qnrVC1, rmtB4, sul1, sul1, tet(G) | CP142449b, CP142450c |
| Isolate_ESP_3 | 773 | II | 6,850,251 | 116,998 | - | aac(3), aadA11, aph(3′)-IIb, blaNDM-1, blaOXA-395, blaPDC-16, catB7, fosA, qacE, qnrVC1, rmtB4, sul1, sul1, tet(G) | CP142448b |
| Isolate_NLD_1 | 773 | II | 6,836,351 | 116,995 | - | aac(3), aadA11, aph(3′)-IIb, blaNDM-1, blaOXA-395, blaPDC-16, catB7, fosA, qacE, qnrVC1, rmtB4, sul1, sul1, tet(G) | JBAWKG000000000b |
| Isolate_NLD_2 | 773 | I | 6,862,835 | 120,224d | - | aac(3), aadA11, aph(3′)-IIb, blaNDM-1, blaNDM-1, blaOXA-395, blaPDC-16, catB7, fosA, qacE, qnrVC1, rmtB4, sul1, sul1, tet(G) | JBAWKH000000000b |
| Isolate_NLD_3 | 773 | I | 6,921,153 | 116,989 | pS75 (50,326) | aac(3), aadA11, aph(3′)-IIb, blaNDM-1, blaOXA-395, blaPDC-16, catB7, fosA, qacE, qnrVC1, rmtB4, sul1, sul1, tet(G) | JBAWKF000000000b |
| Isolate_NLD_4 | 773 | II | 6,908,226 | 116,997 | - | aac(3), aadA11, aph(3′)-IIb, blaNDM-1, blaOXA-395, blaPDC-16, catB7, fosA, qacE, qnrVC1, rmtB4, sul1, sul1, tet(G) | CP142447b |
| Isolate_NLD_5 | 773 | I | 6,911,612 | 116,993 | - | aac(3), aadA11, aph(3′)-IIb, blaNDM-1, blaOXA-395, blaPDC-16, catB7, fosA, qacE, qnrVC1, rmtB4, sul1, sul1, tet(G) | CP142446b |
| Isolate_NLD_6 | 773 | I | 6,853,133 | 116,997 | - | aac(3), aadA11, aph(3′)-IIb, blaNDM-1, blaOXA-395, blaPDC-16, catB7, fosA, qacE, qnrVC1, rmtB4, sul1, sul1, tet(G) | CP142445b |
Clusters detected with the cgMLST and cgSNPs analysis using P. aeruginosa PAO1 (NC_002516.2) as reference genome.
Accession number of chromosomes of NDM-1–producing P. aeruginosa isolates.
Accession number of plasmids detected in NDM-1–producing P. aeruginosa isolates.
Two copies of blaNDM-1 in the ICE, that is flanked by attL and attR (gtctcgtttcccgctccaaacat) but inverted in the chromosome with respect to the other isolates.
The variant calling analysis of chromosomal genes also showed an identical mutational resistome, except for four genes, in which exclusive mutations were found (Table S1). These genes were involved in the transcriptional activation of P. aeruginosa virulence genes (lasR), the regulation of MexXY (mexY) and MexAB-OprM (nalC) effux pumps, and the lipid A modification (pagL). T543A-mexY (eight of nine) and G71E-nalC (eight of nine) mutations were encountered in several of the strains (Table S1).
Molecular typing and mobile genetic elements
All NDM-producing P. aeruginosa isolates belonged to the same clone (ST773) and shared a high percentage of coverage (98-99%) and identity (99.96-100%). All ST773 P. aeruginosa isolates carried a chromosomally encoded blaNDM-1 gene along with a bleomycin resistance gene between two IS91 (IS91-blaNDM-1-bleMBL-IS91), contained on an identical ∼117 Kb integrative conjugative element (ICE) flanked by 23-pb attL and attR sequences (gtctcgtttcccgctccaaacat) (Figure S1, Table 2). The Dutch isolate (Isolate_NLD_2) that carried two copies of the blaNDM-1 gene (IS91-bleMBL-blaNDM-1-bleMBL-blaNDM-1-IS91) had a larger ICE (120.224 bp) that was located inverted in the chromosome but with identical attR and attL regions (Figure S2). xerD and parA genes, involved in the mobilization and integration of pathogenicity genomic islands structures in P. aeruginosa, were identified flanking all blaNDM-1-ICEs. Other common regions involved in mobilization and maintenance of the pathogenicity genomic islands such as the pil operon were also found. In all isolates, the blaNDM-1-ICE contained five additional resistance genes affecting other antimicrobial groups: sulfonamides (sul1), aminoglycosides (acc[3] and rmtB4), chloramphenicol (floR), and tetracyclines [tet(G)] (Figure S1).
Plasmid reconstruction showed that two Spanish (ESP_1 and ESP_2, each from one center) and one Dutch (NDL_3) NDM-1-ST773 P. aeruginosa isolates contained a closely related plasmid (∼47-51 Kb) without any resistance genes. Genes encoding membrane proteins belonged to the secretion system type IV transport proteins (VirB1, VirB4, VirB6, VirB8, VirB9, VirB10, VirB11, and VirD4) were detected, along with genes encoding an RNA metabolism protein (retron-type RNA-directed DNA polymerase) and proteins involved in regulation and cell signaling (HigA toxin and HigB antitoxin) (Table 2, Figure S3).
cgMLST and cgSNP analysis
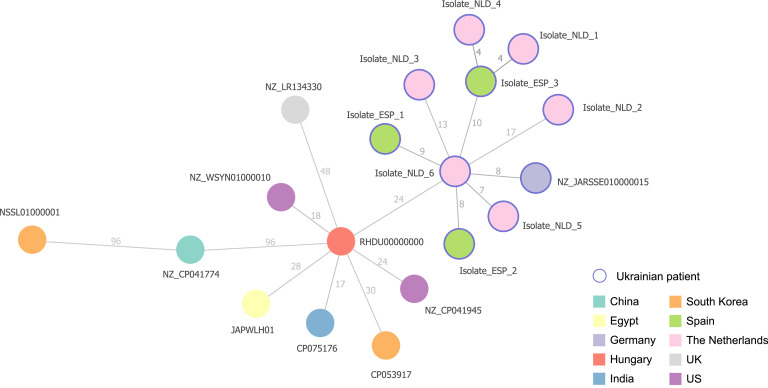
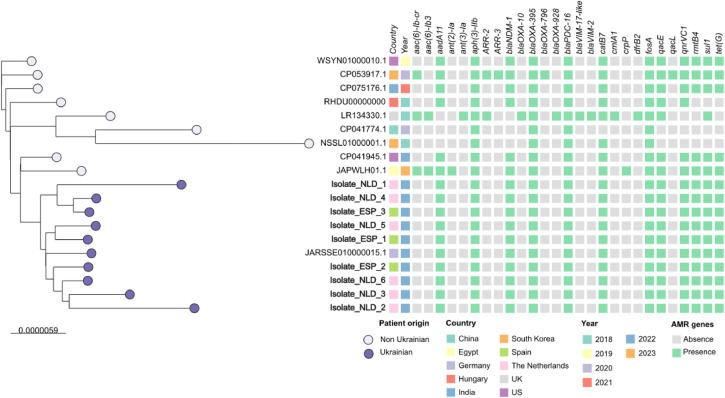
A cgMLST phylogenetic tree of the Spanish and Dutch isolates in context of ten related ST773-P. aeruginosa isolates downloaded from Pathogenwatch (https://pathogen.watch) was constructed. Of a total of 5,694 loci searched, 5,250 loci were present in the genome of all isolates. This analysis showed that NDM-1-ST773-P. aeruginosa from patients of Ukrainian origin were grouped in the same cluster and more distant from the strains collected in other countries and in other years. Overall, NDM-1-ST773-P. aeruginosa strains diverged in distance from 4 to 17 loci and from 4 to 96 loci from strains collected in other geographical regions (Figure 1). Using P. aeruginosa PAO1 as reference genome, the core genome of all NDM-1-ST773 P. aeruginosa ranged between 5.882.658 and 5.887.317 bp, with an evolutionary distance of 54.168-54.576 SNPs (9.2-9.3 SNPs/Mb). The maximum likelihood phylogenetic tree based on the cgSNP analysis of all genomes gave highly similar results that the neighbor Joining tree from the cgMLST schema (Figure 2). Both methods clustered all Ukrainian ST773 P. aeruginosa strains in two clusters: cluster I, including two Spanish isolates (ESP_1 and ESP_2), and four Dutch strains (NLD_2, NLD_3, NLD_5, and NLD_6), all of them from different centers and recovered between March and December, and cluster II, formed by one Spanish (ESP_3) and two Dutch isolates (NLD_1 and NLD_4), also from different centers and from different months (June, July, and August, respectively) (Figures 1 and 2).
Figure 1.
cgMLST based phylogenetic tree constructed with all NDM-1-ST773-P. aeruginosa isolates recovered in this study (purple surrounding circle; ESP: Spain; NLD: the Netherlands) and ten closely related ST773-P. aeruginosa isolates from other locations. P. aeruginosa PAO1 was used as reference genome (NC_002516.2). cgMLST is a minimum spanning tree based on the cgMLST allelic profile of P. aeruginosa PAO1, determined by the pipeline chewBACCA and visualized using Phyloviz Online tool.
Figure 2.
cgSNP based phylogenetic tree constructed with all NDM-1-ST773 P. aeruginosa isolates (bold letters) recovered in this study and 10 closely related ST773 P. aeruginosa isolates from other locations. Patient origin (Ukrainian, purple; non-Ukrainian, grey), country, year of collection, and presence (green) or absence (grey) of antimicrobial resistance genes detected using Resfinder database are indicated. cgSNP is a maximum likelihood tree based on core genome SNP analysis, reconstructed and visualized using IQ-TREE (v2.0.7) and iTOL, respectively.
AMR, antimicrobial resistance.
Prophages and pyocins analysis
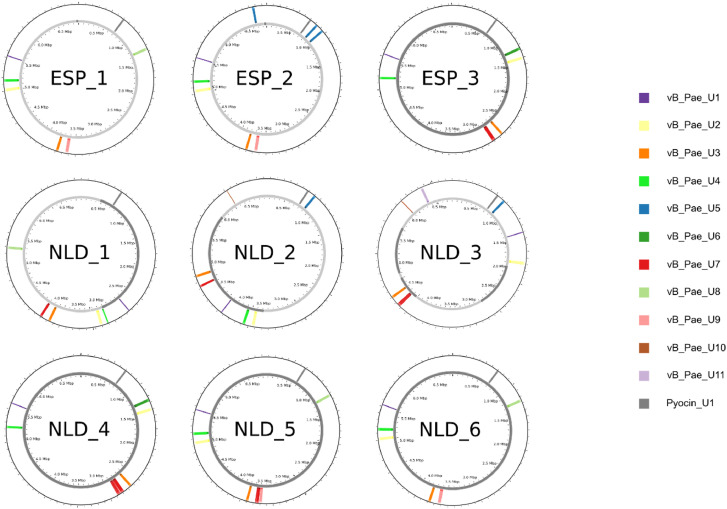
A total of 11 different prophages were identified in all P. aeruginosa strains, and 10 of them were present in more than one strain simultaneously. A phage tail-like bacteriocin (PTLB) cluster was also identified in all isolates, at first misidentified as a prophage (Figure 3, Table S2). The prophage and PTLB cluster characteristics are summarized in Table 3.
Figure 3.
Map of all NDM-1-ST773 P. aeruginosa strains and the relative position of prophages and phage tail-like bacteriocin clusters detected within them. ESP, Spain; NLD, the Netherlands.
Table 3.
Characteristics of the eleven prophages and the phage tail-like bacteriocin cluster (pyocin cluster) identified in the NDM-1-ST773 P. aeruginosa strains from Ukrainian patients.
| Prophage /phage tail-like bacteriocin cluster name | Family /Pyocin subtype | Length (Kb) | GC content (%) | ORF (n°) | Gene density (gene/Kb) | Host strains | Hypothetical proteins (%) | Upstream gene | Downstream gene | Accession number |
|---|---|---|---|---|---|---|---|---|---|---|
| vB_Pae_U1 | Unclassified Caudoviricetes | 16.2 | 59.3 | 26 | 1.60 | 9/9 | 65.4 | Quinolinatephosphoribosyltransferase (EC 2.4.2.19) | tRNA-Thr-CGT | PP003913 |
| vB_Pae_U2 | Unclassified Caudoviricetes (Siphovirus tail morphology group) | 45.1 | 61.3 | 67 | 1.49 | 9/9 | 58.8 | tRNA-Pseudo-GAG | Antitoxin HigA | PP003914 |
| vB_Pae_U3 | Unclassified Caudoviricetes (Siphovirus tail morphology group) | 38.6 | 62.1 | 53 | 1.37 | 9/9 | 50.9 | Type I secretion system, outer membrane component LapEb | PP003915 | |
| vB_Pae_U4a | Casadabanvirus | 36.4 | 64.4 | 53 | 1.46 | 3/9 | 39.6 | FAD-dependent monooxygenase PhzS | Pyridoxamine 5′-phosphate oxidase PhzG (EC 1.4.3.5) | PP003916 |
| Aminotransferase, class IIIb | ||||||||||
| Bacteriocin/lantibiotic efflux ABC transporter, permease/ATP-binding proteinb | ||||||||||
| vB_Pae_U5 | Unclassified Caudoviricetes | 40.4 | 63.3 | 52 | 1.29 | 8/9 | 60.4 | hypothetical protein | tRNA-Leu-CAA | PP003917 |
| tRNA-Gly-CCC | ||||||||||
| vB_Pae_U6 | Unclassified Caudoviricetes | 43.6 | 62.2 | 66 | 1.51 | 2/9 | 84.8 | Rossmann fold nucleotide-binding protein Smf possibly involved in DNA uptake | tRNA-Gly-CCC | PP003918 |
| vB_Pae_U7a | Detrevirus | 57.8 | 58.9 | 103 | 1.78 | 6/9 | 64.1 | Bis-ABC ATPase YbiT | Probable 5-carboxymethyl-2-hydroxymuconate delta isomerase | PP003919 |
| vB_Pae_U8 | Unclassified Caudoviricetes | 41.1 | 62.6 | 60 | 1.46 | 4/9 | 61.7 | Rossmann fold nucleotide-binding protein Smf possibly involved in DNA uptake | tRNA-Leu-CAA | PP003920 |
| vB_Pae_U9 | Unclassified Caudoviricetes | 49.7 | 59.4 | 85 | 1.71 | 4/9 | 72.1 | Probable 5-carboxymethyl-2-hydroxymuconate delta isomerase | Bis-ABC ATPaseYbiT | PP003921 |
| vB_Pae_U10 | Unclassified Caudoviricetes | 10.5 | 60.8 | 13 | 1.24 | 2/9 | 38.5 | Glycosyltransferaseb | PP003922 | |
| vB_Pae_U11 | Beetrevirus | 39.0 | 63.1 | 56 | 1.44 | 1/9 | 51.8 | tRNA-dihydrouridine(20/20a) synthase (EC 1.3.1.91) | hypotheticalprotein | PP003923 |
| Pyocin_U1 | R5-F cluster | 28.8 | 64.5 | 38 | 1.32 | 9/9 | 21.1 | Anthranilate synthase, amidotransferase component (EC 4.1.3.27) | Anthranilate synthase, aminase component (EC 4.1.3.27) | PP003924 |
aThree copies of vB_Pae_U4 and two copies of vB_Pae_U7 were detected in isolates ESP_2 and NLD_4, respectively.
ORF truncated by the insertion of the prophage.
The PTLB cluster presented a conserved structure: a PrtN/PrtR activator/repressor tandem, a zinc finger transcription factor, a phage antitermination Q-like protein, an R5-type (Rigid, with a structure similar to a Myovirus tail) pyocin, a lytic cassette, and an F-type (Flexible, analog to a Siphovirus tail) pyocin (Figure S4).
Prophage length ranged from 10.5 Kb (vB_Pae_U10) to 57.8 Kb (vB_Pae_U7), with a GC content ranging from 58.9 % (vB_Pae_U7) to 64.4 % (vB_Pae_U4). The number of ORF was also very variable, with 13 ORF the shortest (vB_Pae_U10) and 103 the longest (vB_Pae_U7). The ORF function of each prophage genome is summarized in Figure S5. A considerable number of ORF remained with an unknown function after annotation, ranging from 38.5% of hypothetical proteins (vB_Pae_U10) to 84.8% (vB_Pae_U6). The PTLB cluster had 21.1% of hypothetical proteins. Prophage vB_Pae_U4 was also found to code for an anti-CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) proteins with homology with AcrIF3 (score 8.85 × 10-104).
Regarding insertion sites, the PTLB cluster was localized within the tryptophan operon, between the anthranilate synthase components I (trpE) and II (trpG). Half of the prophages (6/12) were inserted next to transfer RNA coding genes. Prophages vB_Pae_U2, vB_Pae_U4 (in strain ESP_2) and vB_Pae_U10 were inserted inside other genes, truncating them and possibly resulting in impaired gene expression. Prophage vB_Pae_U2 was inserted inside the lapE gene in all strains, which codes for an outer membrane pore forming protein as a component of the type I secretion system. In ESP_2, prophage vB_Pae_U4 was inserted inside a class III aminotransferase gene and inside an ABC-type bacteriocin/lantibiotic exporter. This strain was found to have three copies of the same phage, the third one being between an flavin adenine dinucleotide–dependent mooxygenase (phzS) and a pyridoxamine 5’-phosphate oxidase (phzG), as in two Dutch strains (NLD_2 and NLD_3). Finally, prophage vB_Pae_U10 was found integrated into the glycosyltransferase gene (Table 3).
Discussion
In this work, we report the clonal spread in 2022 of NDM-1 P. aeruginosa isolates belonging to clone ST773 in two European countries (Spain and the Netherlands) due to the transfer of Ukrainian patients because of the war.
In P. aeruginosa, resistance to carbapenems is usually due to mutations in chromosomal genes and, to a lesser extent, to horizontal acquisition of carbapenemase-encoding genes [9,10,20]. In Europe, P. aeruginosa is usually associated with the production of Verona integron-encoded MBL enzymes, and the report of NDM carbapenemases is limited to nosocomial outbreaks or imported cases from endemic areas [13,[21], [22], [23]]. NDM enzymes can hydrolyze all β-lactams, except for monobactams, conferring an almost pan-β-lactam-resistant phenotype that usually leaves few treatment options [24]. In our collection, all NDM-1-ST773 P. aeruginosa isolates were resistant to all tested antimicrobials, except for colistin, aztreonam, and cefiderocol. This high-level of resistance coincides with the alarming rates previously reported in Ukrainian health care settings [2,3].
The ST773 P. aeruginosa clone has been previously described associated with the production of NDM-1 in some countries such as the United States, Nepal, and South Korea [23,25,26]. However, the NDM-1-ST773 P. aeruginosa clone had not been described in Europe before until the arrival of Ukrainian patients [[6], [7], [8]]. Our analysis showed that isolates of Ukrainian origin were more genetically related to each other than to other NDM-1-ST773 P. aeruginosa strains recovered from other locations. Note that one NDM-1-ST773 P. aeruginosa recovered from a Ukrainian soldier admitted to a US military hospital in Germany in 2022 was found to be part of the same cluster as the Spanish and Dutch strains, suggesting clonal spread to Germany [7]. The cgMLST and cgSNP analysis also revealed the clustering of our strains into distinct but close subgroups, spanning the Spanish and Dutch isolates, indicating that the strains were closely related despite belonging to patients who had been transferred to different countries. The introduction of these isolates in European countries poses a risk of potential local transmission events within and between health care facilities in the country of detection.
In addition, the blaNDM-1 genetic environment in our isolates (blaNDM-1-ICE) was closely related to that found in other ST773 P. aeruginosa isolates [23,25,26]. The identical resistome and shared genetic elements also suggest a clonal expansion of the NDM-1-ST773 P. aeruginosa clone. The presence of additional resistance genes within the ICE underscores the potential for further acquisition of resistance determinants, posing a continual threat to antimicrobial efficacy. Furthermore, the closely related plasmid identified in Spanish and Dutch isolates, devoid of resistance genes, suggests that plasmids could also be a potential vehicle for gene exchange in P. aeruginosa. Understanding the genetic platforms that facilitates the spread of virulence and resistance genes is crucial for developing targeted intervention strategies.
Recent articles have shown that lysogeny is a common trait among P. aeruginosa clinical isolates and that prophage role and characteristics remain understudied when compared to lytic phages [27]. In our NDM-1-ST773 P. aeruginosa strains, we found a high content of prophages than in other studies [27]. Most of them were found in more than one strain simultaneously, irrespective of the origin country. Moreover, an identical pyocin (PTLB) cluster was also found in all strains, again showing a high genetic relatedness among the isolates. The pyocin cluster showed a similar pattern as R-F-type bacteriocins previously described in P. aeruginosa [28]. In our isolates, differences were only found in those regions of the ST773 genome where prophages were inserted, suggesting a dynamic interaction between the bacteriophage and the host bacterium. Consistent with other studies, up to three of these prophages were located truncating other genes and presumably resulting in non-functional proteins [29,30]. Prophages inserted within essential genes further emphasize their potential impact on bacterial physiology, most likely influencing virulence and antibiotic resistance.
On the other hand, it should be noted that in at least seven of these patients other MDR bacteria were isolated in addition to the NDM-1-ST773 P. aeruginosa. Among others, NDM-producing Enterobaterales isolates were detected, including ST147 K. pneumoniae producing OXA-48 or co-producing OXA-48+NDM-1 [8]. The ST147 K. pneumoniae clone has already been linked to the movement of Ukrainian patients to other central European countries in 2022 [5]. Co-colonization or co-infection with other MDR also carrying carbapenemases and/or extended-spectrum β-lactamases genes, such as ST78-OXA-23 Acinetobacter baumannii, ST395-OXA-48+NDM-1 K. pneumoniae, ST1047-IMP-1 P. aeruginosa, and ST357-VEB-9 P. aeruginosa, has also been reported in Ukrainian patients in Germany [7].
The timeline of cases after the conflict and the association with patients who had previous hospitalization in Ukraine suggests a direct link to the conflict-driven migration. This underlines the importance of collaborative efforts in surveillance, early detection, and the establishment of protocols for managing patients with a history of health care exposure in conflict zones. The combination of WGS and epidemiologic data allows a better understanding of the evolution of resistance mechanisms and the transmission dynamics, which helps to apply adapted infection control measures.
In conclusion, our study provides a comprehensive genomic perspective on the emergence and dissemination of NDM-1–producing P. aeruginosa ST773 clone in Europe after the conflict in Ukraine. The interconnectedness of health care systems and the adaptability of these strains underscore the need for international collaboration in combating the global challenge of antimicrobial resistance. Continued genomic surveillance, along with the reinforcement of infection control measures, is imperative to mitigate the impact of these highly resistant clones on public health.
Declarations of competing interest
The authors have no competing interests to declare.
Acknowledgments
Funding
This study was supported by Plan Nacional de I + D + i 2013–2016 and Instituto de Salud Carlos III, Subdirección General de Redes y Centros de Investigación Cooperativa, Ministerio de Economía, Industria y Competitividad, Spanish Network for Research in Infectious Diseases (REIPI RD16/0016/0011), co-financed by the European Development Regional Fund ‘A way to achieve Europe’ (ERDF), Operative program Intelligent Growth 2014–2020, research grant (PI22/01283), (PI19/00571) and CIBER de Enfermedades Infecciosas (CIBERINFEC) (CB21/13/00084), Instituto de Salud Carlos III, Madrid, Spain. MH-G is supported by a postdoctoral contract by CIBERINFEC (CB21/13/00084). MGA is supported by the Río Hortega program (CM22/00159, ISCIII).
Ethical approval
This study was approved by the Ramón y Cajal University Hospital Ethics Committee (Reference 104/23).
Author contributions
All authors have made substantial contributions to this work. MHG and MGA were involved in the design of the study, acquisition, analysis and interpretation of data, as well as in the drafting and revising the manuscript. MPC, BGB, EV, JV, MT, APAH have contributed to the acquisition/ data analysis and revising the manuscript. PRG and RC are responsible for the conception, design and supervision of the study, as well as drafting and revising the article critically for important intellectual content. All authors have reviewed and given their approval for the publication of the final manuscript.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ijregi.2024.100415.
Appendix. Supplementary materials
References
- 1.European Centre for Disease Prevention and Control . European Centre for Disease Prevention and Control; Boulevard: 2022. Antimicrobial resistance surveillance in Europe, 2022–2020 data. [Google Scholar]
- 2.Kondratiuk V, Jones BT, Kovalchuk V, Kovalenko I, Ganiuk V, Kondratiuk O, et al. Phenotypic and genotypic characterization of antibiotic resistance in military hospital-associated bacteria from war injuries in the Eastern Ukraine conflict between 2014 and 2020. J Hosp Infect. 2021;112:69–76. doi: 10.1016/j.jhin.2021.03.020. [DOI] [PubMed] [Google Scholar]
- 3.Salmanov A, Shchehlov D, Svyrydiuk O, Bortnik I, Mamonova M, Korniyenko S, et al. Epidemiology of healthcare-associated infections and mechanisms of antimicrobial resistance of responsible pathogens in Ukraine: a multicentre study. J Hosp Infect. 2023;131:129–138. doi: 10.1016/j.jhin.2022.10.007. [DOI] [PubMed] [Google Scholar]
- 4.European Centre for Disease Prevention and Control . European Centre for Disease Prevention and Control; Boulevard: 2022. Operational considerations for the prevention and control of infectious diseases - Russia's aggression towards Ukraine. [Google Scholar]
- 5.Sandfort M, Hans JB, Fischer MA, Reichert F, Cremanns M, Eisfeld J, et al. Increase in NDM-1 and NDM-1/OXA-48-producing Klebsiella pneumoniae in Germany associated with the war in Ukraine, 2022. Euro Surveill. 2022;27 doi: 10.2807/1560-7917.ES.2022.27.50.2200926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zwittink RD, Wielders CCH, Notermans DW, Verkaik NJ, Schoffelen AF, Witteveen S, et al. Multidrug-resistant organisms in patients from Ukraine in the Netherlands, March to August 2022. Euro Surveill. 2022;27 doi: 10.2807/1560-7917.ES.2022.27.50.2200896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mc Gann PT, Lebreton F, Jones BT, Dao HD, Nelson MJ, Luo T, Wyatt AC, et al. Six extensively drug- resistant bacteria in an injured soldier, Ukraine. Emerginf Infect Dis. 2013;8:1692–1695. doi: 10.1007/s00268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hernández-García M, Cabello M, Ponce-Alonso M, Herrador-Gómez PM, Gioia F, Cobo J, et al. First detection in Spain of NDM-1-producing Pseudomonas aeruginosa in two patients transferred from Ukraine to a university hospital. J Glob Antimicrob Resist. 2024;36:105–111. doi: 10.1016/j.jgar.2023.12.022. [DOI] [PubMed] [Google Scholar]
- 9.Del Barrio-Tofiño E, López-Causapé C, Oliver A. Pseudomonas aeruginosa epidemic high-risk clones and their association with horizontally-acquired β-lactamases: 2020 update. Int J Antimicrob Agents. 2020;56 doi: 10.1016/j.ijantimicag.2020.106196. [DOI] [PubMed] [Google Scholar]
- 10.Horcajada JP, Montero M, Oliver A, Sorlí L, Luque S, Gómez-Zorrilla S, et al. Epidemiology and treatment of multidrug-resistant and extensively drug-resistant Pseudomonas aeruginosa infections. Clin Microbiol Rev. 2019;32:1–52. doi: 10.1128/CMR.00031-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson G, Banerjee S, Putonti C. Diversity of Pseudomonas aeruginosa temperate phages. mSphere. 2022;1 doi: 10.5281/msphere.5072377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Der Zwaluw K, De Haan A, Pluister GN, Bootsma HJ, De Neeling AJ, Schouls LM. The Carbapenem Inactivation Method (CIM), a simple and low-cost alternative for the carba NP test to assess phenotypic carbapenemase activity in Gram-negative rods. PLoS One. 2015;10 doi: 10.1371/journal.pone.0123690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hernández-García M, García-Castillo M, García-Fernández S, Melo-Cristino J, Pinto MF, Goncalves E, et al. Distinct epidemiology and resistance mechanisms affecting ceftolozane /tazobactam in Pseudomonas aeruginosa isolates recovered from ICU patients in Spain and Portugal depicted by WGS. J Antimicrob Chemother. 2021;76:370–379. doi: 10.1093/jac/dkaa430. [DOI] [PubMed] [Google Scholar]
- 14.Silva M, Machado MP, Silva DN, Rossi M, Moran-Gilad J, Santos S, et al. chewBBACA: a complete suite for gene-by-gene schema creation and strain identification. Microb Genom. 2018;4 doi: 10.1099/mgen.0.000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kieft K, Zhou Z, Anantharaman K. VIBRANT: automated recovery, annotation and curation of microbial viruses, and evaluation of viral community function from genomic sequences. Microbiome. 2020;8:90. doi: 10.1186/s40168-020-00867-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mihara T, Nishimura Y, Shimizu Y, Nishiyama H, Yoshikawa G, Uehara H, et al. Linking virus genomes with host taxonomy. Viruses. 2016;8:66. doi: 10.3390/v8030066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blasco L, de Aledo MG, Ortiz-Cartagena C, Blériot I, Pacios O, López M, et al. Study of 32 new phage tail-like bacteriocins (pyocins) from a clinical collection of Pseudomonas aeruginosa and of their potential use as typing markers and antimicrobial agents. Sci Rep. 2023;13:117. doi: 10.1038/s41598-022-27341-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bouras G, Nepal R, Houtak G, Psaltis AJ, Wormald PJ, Vreugde S. Pharokka: a fast scalable bacteriophage annotation tool. Bioinformatics. 2023;39:btac776. doi: 10.1093/bioinformatics/btac776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dong C, Hao GF, Hua HL, Liu S, Labena AA, Chai G, et al. Anti-CRISPRdb: a comprehensive online resource for anti-CRISPR proteins. Nucleic Acids Res. 2018;46:D393–D398. doi: 10.1093/nar/gkx835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tenover FC, Nicolau DP, Gill CM. Carbapenemase-producing Pseudomonas aeruginosa –an emerging challenge. Emerg Microbes Infect. 2022;11:811–814. doi: 10.1080/22221751.2022.2048972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wendel AF, Malecki M, Mattner F, Xanthopoulou K, Wille J, Seifert H, et al. Genomic-based transmission analysis of carbapenem-resistant Pseudomonas aeruginosa at a tertiary care centre in Cologne (Germany) from 2015 to 2020. JAC Antimicrob Resist. 2022;4:dlac057. doi: 10.1093/jacamr/dlac057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hammoudi Halat D, Ayoub Moubareck C. The intriguing carbapenemases of Pseudomonas aeruginosa: current status, genetic profile, and global epidemiology. Yale J Biol Med. 2022;95:507–515. [PMC free article] [PubMed] [Google Scholar]
- 23.Khan A, Shropshire WC, Hanson B, Dinh AQ, Wanger A, Ostrosky-Zeichner L, et al. Simultaneous infection with Enterobacteriaceae and Pseudomonas aeruginosa harboring multiple carbapenemases in a returning traveler colonized with Candida auris. Antimicrob Agents Chemother. 2020;64 doi: 10.1128/AAC.01466-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R, et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis. 2010;10:597–602. doi: 10.1016/S1473-3099(10)70143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi YJ, Kim YA, Junglim K, Jeong SH, Shin JH, Shin KS, et al. Emergence of NDM-1-producing Pseudomonas aeruginosa sequence Type 773 clone: shift of carbapenemase molecular epidemiology and spread of 16S rRNA methylase genes in Korea. Ann Lab Med. 2023;43:196–199. doi: 10.3343/alm.2023.43.2.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takahashi T, Tada T, Shrestha S, Hishinuma T, Sherchan JB, Tohya M, et al. Molecular characterisation of carbapenem-resistant Pseudomonas aeruginosa clinical isolates in Nepal. J Glob Antimicrob Resist. 2021;26:279–284. doi: 10.1016/j.jgar.2021.07.003. [DOI] [PubMed] [Google Scholar]
- 27.González de Aledo M, Blasco L, Lopez M, Ortiz-Cartagena C, Bleriot I, Pacios O, et al. Prophage identification and molecular analysis in the genomes of Pseudomonas aeruginosa strains isolated from critical care patients. mSphere. 2023;8 doi: 10.1128/msphere.00128-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saha S, Ojobor CD, Li ASC, Mackinnon E, North OI, Bondy-Denomy J, et al. F-type pyocins are diverse noncontractile phage tail-like weapons for killing Pseudomonas aeruginosa. J Bacteriol. 2023;205 doi: 10.1128/jb.00029-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith TJ, Sondermann H, O'Toole GA. Type 1 does the two-step: Type 1 secretion substrates with a functional periplasmic intermediate. J Bacteriol. 2018;200:e00168. doi: 10.1128/JB.00168-18. e00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campbell A. Prophage insertion sites. Res Microbiol. 2003;154:277–282. doi: 10.1016/S0923-2508(03)00071-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.