Summary
As the most abundant glial cells in the central nervous system (CNS), astrocytes dynamically respond to neurotoxic stress, however, the key molecular regulators controlling the inflammatory status of these sentinels during neurotoxic stress are many and complex. Herein, we demonstrate that the m6A epitranscriptomic mRNA modification tightly regulates the pro-inflammatory functions of astrocytes. Specifically, the astrocytic neurotoxic stressor, manganese (Mn), downregulated the m6A reader YTHDF2 in human and mouse astrocyte cultures and in the mouse brain. Functionally, YTHDF2 knockdown augmented, while its overexpression dampened, the neurotoxic stress-induced proinflammatory response, suggesting YTHDF2 serves as a key upstream regulator of inflammatory responses in astrocytes. Mechanistically, YTHDF2 RIP-sequencing identified MAP2K4 (MKK4; SEK1) mRNA as a YTHDF2 target influencing inflammatory signaling. Our target validation revealed that Mn-exposed astrocytes mediate proinflammatory responses by activating the phosphorylation of SEK1, JNK, and cJUN signaling. Collectively, YTHDF2 serves as a key upstream ‘molecular switch’ controlling SEK1(MAP2K4)-JNK-cJUN proinflammatory signaling in astrocytes.
Subject areas: Molecular mechanism of gene regulation, Molecular network, Neurotoxicology, Cell biology, Transcriptomics
Graphical abstract

Highlights
-
•
Mn exposure induces proinflammatory response in astrocytes by reducing YTHDF2
-
•
YTHDF2 targets MAP2K4 mRNA for decay in astrocytes
-
•
Mn-induced YTHDF2 downregulation upregulates the proinflammatory SEK1 pathway
-
•
Selective depletion of YTHDF2 in astrocytes induces astrogliosis in mice
Molecular mechanism of gene regulation; Molecular network; Neurotoxicology; Cell biology; Transcriptomics
Introduction
As the most abundant non-neuronal cells of the central nervous system (CNS), astrocytes are vital for brain and neuronal homeostasis. Their supportive functions include antioxidant defense, glutamate and water-ion-pH homeostasis, blood-brain barrier maintenance, synapse formation and maturation, and neurotrophic factor and cytokine production.1,2,3,4 Astrocytes respond dynamically to neurotoxic stressors to protect and support neuronal health. When exposed to harmful substances or environments, astrocytes can undergo changes in their morphology and function, becoming reactive astrocytes. While this reactive response aims to limit damage and maintain homeostasis, chronic or sustained activation of astrocytes can also lead to uncontrolled inflammatory responses and contribute to various neurodegenerative conditions. Nevertheless, the precise molecular factors that govern the pro-inflammatory state of these guardian cells under chronic neurotoxic stress conditions remain enigmatic.
A substantial body of evidence has established that neurotoxic concentrations of the metal Mn significantly target and adversely affect astrocyte physiology.5,6,7,8,9 Indeed, astrocytes exhibit a significantly higher affinity for Mn than do neurons given their higher divalent metal transporter content.10,11 Mn then accumulates through sequestration in mitochondria via the calcium uniporter,5,11,12 exerting neurotoxic stress. Homeostatic dysregulation by Mn can induce inflammation in astrocytes, and chronic cellular alterations can transform quiescent astrocytes into reactive astrocytes, leading to neuronal toxicity.5,13,14,15,16 In response to Mn, astrocytes release various chemokines, cytokines, and other neurotoxic factors and sustain these responses potentially through the NF-κB and MAPK cascades.17,18 Furthermore, chronic Mn exposure can induce Parkinsonian conditions by primarily inducing astrocytic dysfunction.19,20 Therefore, we adopted Mn-treated astrocytes as a model to understand the role of epitranscriptomic changes underlying chronic astrocyte activation.
N6-methyladenosine (m6A) is the most prevalent epitranscriptomic modification21 regulating mRNA translation and decay. The DRACH (D = A/G/U, R = A/G, H = A/C/U) consensus sequences on mRNAs are targeted by the m6A writer complex, composed of METTL3 and METTL14 (methyltransferase-like), and accessory regulatory proteins such as WTAP, during which a methyl group is added to the sixth nitrogen position of adenosine.22,23,24 Additionally, m6A modifications are reversible and can be removed by m6A demethylases such as ALKBH5 and FTO.25,26 Recent studies reveal that m6A-modified mRNAs have reduced stability, yielding shorter half-lives that result in decreased time spent in ribosomal translation pools.27 The m6A reader, YTHDF2 (YT521-B homology domain family), is known to promote the initiation of RNA decay, while YTHDF1 has been defined to promote m6A mRNA translation, and whereas YTHDF3 may facilitate both processes.28 Understanding why such mechanisms would be requisite for cellular physiology and pathology varies across many biological disciplines, but the m6A readers which execute the fate of m6A mRNAs remain as fundamental elements of the ensemble. Since m6A modifications can dictate the fate of mRNA and translational rate, understanding its role in inflammatory processes would provide insights into upstream regulation of cytokine and chemokine mRNA production. In this regard, YTHDF2 has been highlighted in certain cellular contexts, including hypoxia, survival/apoptosis, proliferation, and inflammation, and has been shown to directly target secretory mRNAs such as IL-11,29 transcription factor RELA, and MAP kinases,30,31,32 revealing that YTHDF2’s reader function may be highly significant in inflammation. Pro-inflammatory astrocytic responses present with the upregulation of numerous chemokines and cytokines, which are regulated by multiple kinases and transcription factors.5,33,34 However, the upstream epitranscriptomic regulatory events governing the neuroinflammatory signaling have yet to be delineated, hindering our ability to understand the molecular mechanism by which cytokine and chemokine mRNA dynamics are controlled during aberrant astrocyte activation. To address this, herein, we adopted an approach using Mn as an astrocyte-specific neurotoxic stressor to determine whether m6A epitranscriptomic regulators modulate the neuroinflammatory response during astrocyte activation. Interestingly, our results reveal that the epitranscriptomic reader YTHDF2 acts as a negative epitranscriptomic regulator of the SEK1(MAP2K4)-JNK-cJUN proinflammatory signaling cascade in astrocytes, suggesting that YTHDF2 may be an exploitable target for controlling astrocytic inflammation in neuroinflammatory conditions.
Results
The astrocytic neurotoxic stressor Mn downregulates YTHDF2 expression in cell culture
As the major function of YTHDF2 in accelerating the decay of m6A mRNAs was recently demonstrated,27 we reasoned this epitranscriptomic reader may influence inflammatory signaling, which relays rapid onset and decay depending on the insults. Only a few studies have determined the expression levels of YTHDF2 under different stress conditions. Limited observations in cancer cells revealed heat-shock stress upregulated YTHDF2,35 whereas hypoxia induced by oxygen deprivation or by cobalt chloride treatment downregulated YTHDF2.36 Cellular hypoxia has been shown to precede oxidative stress responses,37 and we recently reported that the astrocytic neurotoxic stressor Mn induced mitochondrial dysfunction and oxidative stress in astrocytes to augment neuroinflammation.5 Previously, we showed that 100 μM Mn can evoke pro-inflammatory gene expression and the release of chemokines/cytokines in both primary mouse astrocytes and the human U251 astrocyte cells,5 thus, we used 100 μM for all in vitro Mn treatment experiments. Primary mouse astrocytes isolated from 1 to 2-day-old post-natal pups were treated with 100 μM Mn and revealed a time-dependent decrease in YTHDF2 protein levels beginning after 3 h and persisting across the entire 24-h period (Figure 1A). Based on these results and previous studies showing the presence and sustainability of astrocytic inflammation, we continued using the 24-h Mn treatment timepoint.5 Immunocytochemistry was also performed with primary mouse astrocytes, revealing primarily cytoplasmic perinuclear localization of YTHDF2 with notable decreases after Mn treatment (Figure 1B). To test the specificity of Mn-induced downregulation of YTHDF2, we treated human U251 astrocytes with individual metal chlorides (PbCl2, CuCl2, FeCl2) at 100 μM as well for 24 h and observed specific downregulation of YTHDF2 by Mn (Figure 1C) but not with other metals. YTHDF2 mRNA in U251 astrocytes showed small but significant decreases post Mn exposure, while YTHDF1 and YTHDF3 were not significantly affected by Mn (Figure 1D). Next, we investigated global m6A expression using LCMS/MS in U251 astrocytes. Interestingly, we observed a general decrease (Figure 1E). This decrease in global m6A expression may be the result of changes in m6A writers and erasers. Indeed, when probing for additional m6A modifiers, we observed that both METTL3 and METTL14 decreased after Mn treatment (Figure S1A). Conversely, the demethylase ALKBH5 increased after Mn treatment (Figure S1B and S1C). In efforts to corroborate key findings, we utilized human induced pluripotent stem cells (iPSC) and differentiated them into astrocytes. Afterward, we exposed them to Mn and observed similar expression patterns as in the U251 astrocytes, with decreased YTHDF2 and increased ALKBH5 (Figure S1D). Lastly, considering the substantial downregulation of the YTHDF2 protein instead of its gene following Mn exposure, we proposed that Mn-induced YTHDF2 downregulation may be primarily attributed to the degradation of YTHDF2 protein. To understand how Mn may promote YTHDF2 degradation, we performed cotreatments of Mn and MG-132 (proteasome inhibitor) in U251 astrocytes and revealed inhibition of the proteasome prevents drastic decreases in YTHDF2 by Mn, as indicated by increased amounts of the high molecular weight ubiquitinated proteins (Figure S1E), suggesting that Mn may promote YTHDF2 degradation via a proteasomal pathway such as the recently investigated SKP2-associated E3 ubiquitin ligase complex pathway.38 Collectively, these results demonstrate that Mn-induced neurotoxic stress can modulate key epitranscriptomic regulators such as YTHDF2 in astrocytic cells.
Figure 1.
Mn treatment decreases YTHDF2 in cell culture
(A) Primary mouse astrocytes were exposed to 100 μM for 1–24 h and YTHDF2 levels were determined by Western blot. The bottom panel shows the results of densitometric analysis of YTHDF2 bands normalized by β-actin (n = 4–5). YTHDF2 decreases time-dependently beginning after 3 h in primary mouse astrocytes.
(B) ICC representation at 40x depicting YTHDF2 decreases in Mn-exposed primary mouse astrocytes at 24 h (100 μm scale).
(C) 24-h treatment of human U251 astrocytes with different heavy metals, all at 100 μM (n = 3).
(D) Mn decreases YTHDF2 mRNA at 24 h in U251 astrocytes (n = 5).
(E) Mn decreases global m6A levels at 24 h in U251 astrocytes, as measured by LC-MS/MS (n = 6). Data are means ± SEM. Two-group comparisons performed using unpaired t-test. p-values < 0.05 considered significant evidence.
YTHDF2 levels can affect pro-inflammatory chemokine/cytokine responses in neurotoxic stress-exposed astrocytes
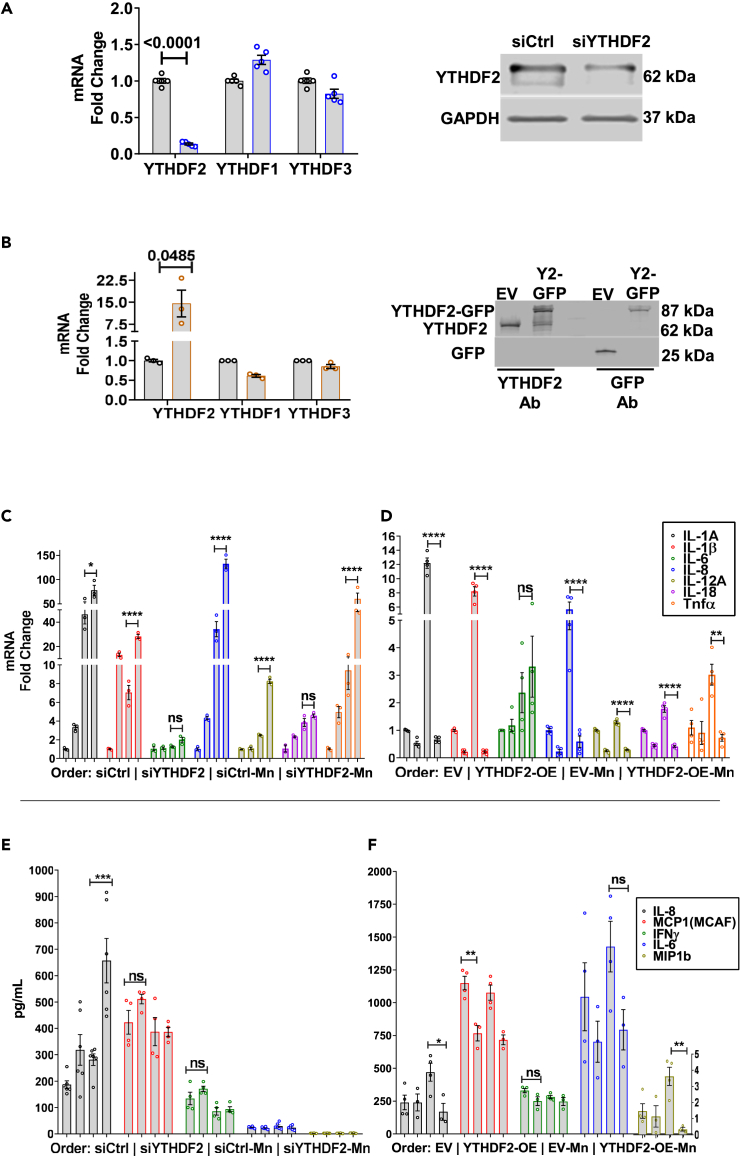
In response to neurotoxic stressors such as Mn, astrocytes activate pro-inflammatory signaling pathways that elicit the production of pro-inflammatory chemokines/cytokines.39 The production of these pro-inflammatory mRNA transcripts could be increased because of prolonged half-lives in the ribosomal translating pools. Using SRAMP m6A site predictor,40 we analyzed various proinflammatory transcripts for m6A DRACH sequences and found that most of them contain DRACH sequences within their coding sequences, suggesting m6A marks may be added by METTL3/METTL14 complex and read by the m6A reader proteins like YTHDF2. YTHDF2 can decrease mRNA stability through recruitment of CCR4-NOT41 or HRSP12–RNase P/MRP42 complexes, thus, we hypothesized the reduction of YTHDF2 may lead to an upregulated pro-inflammatory state by increased stability of pro-inflammatory transcripts, or indirectly by the stabilization of their upstream signaling factors. To test this hypothesis, we knocked down YTHDF2 transiently in U251 astrocytes using the YTHDF2-specific siRNA (siYTHDF2) for 48 h, followed by 24 h of 100 μM Mn treatment. Additionally, we also generated stably overexpressing YTHDF2-GFP U251 astrocytes to jointly determine whether Mn-induced YTHDF2 downregulation contributes to Mn-stimulated pro-inflammatory chemokine/cytokine responses. The specific knockdown and overexpression of YTHDF2, but not YTHDF2’s paralogs were validated by RT-qPCR and Western blot analyses (Figures 2A and 2B). qPCR analysis revealed that siYTHDF2-transfected U251s had an overall pro-inflammatory basal state, which was exacerbated by Mn exposure, as indicated by IL-1α, IL-1β, IL-8, IL-12α, and TNFα (Figure 2C). Conversely, overexpression of YTHDF2 showed no basal changes or reduced proinflammatory basal states, depending upon the chemokine/cytokine. Stable overexpression of YTHDF2 in U251 astrocytes markedly attenuated Mn-stimulated gene upregulation of the investigated chemokine/cytokine, except IL-6 (Figure 2D). Interestingly, IL-6 showed similar patterns of gene expression when comparing YTHDF2 knockdown and overexpression states (Figures 2C and 2D), suggesting YTHDF2 may have some degree of specificity, either directly upon certain chemokine/cytokines or indirectly upon certain upstream signaling factors, or maybe sensitive to exogenously driven genetic changes.
Figure 2.
YTHDF2 levels can affect pro-inflammatory chemokine/cytokine responses in Mn-exposed astrocytes
(A) Validation of YTHDF2 knockdown using siRNA, both qPCR and immunoblotting (n = 5–6).
(B) Validation of YTHDF2 overexpression, both qPCR and immunoblotting (n = 3).
(C) siYTHDF2 increased basal pro-inflammatory gene expression and exacerbated after Mn exposure (n = 3).
(D) Overexpression of YTHDF2 suppressed basal pro-inflammatory gene expression and prevented upregulation after Mn exposure (n = 4).
(E and F) Multiplex ELISA analysis of the treatment media of siYTHDF2 and YTHDF2 overexpression experiments for cytokines/chemokines (n = 4–6). IL-8 was most consistently affected by YTHDF2 levels, showing exacerbated release in siYTHDF2 experiments, while its release was prevented in YTHDF2 overexpression experiments. MCP1 (MCAF), IFNγ, IL-6, and MIP1b showed similar trends but with less consistency overall. Data are means ± SEM. Two-group comparisons performed using unpaired t-test. p-values < 0.05 considered significant evidence. Two-way ANOVA with FDR Two-stage step-up method of Benjamini, Krieger, and Yekutieli for multi-group comparison. Q-values < 0.05 considered significant evidence.
Observations of differential gene expression of pro-inflammatory chemokine/cytokines upon knockdown and overexpression of YTHDF2 prompted us to assess if these same chemokine/cytokines were upregulated and released at the protein level. The treatment media from the gene expression experiments discussed above were assayed by the Luminex Bio-Plex ELISA system. Of the analyzed chemokine/cytokines, Mn exposure of U251 astrocytes had the greatest consistency on the secretion of IL-8. The knockdown of YTHDF2 elevated the basal level of IL-8 secretion. Upon Mn exposure, this response was significantly exacerbated as compared to siCtrl (siRNA Control) group exposed to Mn (Figure 2E), corroborating the gene expression results for IL-8 (Figure 2C). MCP1, IFNγ, IL-6, and MIP1b remained largely unaffected or marginally variable after YTHDF2 knockdown and/or Mn treatment (Figure 2E). Conversely, Mn-exposed EV (Empty Vector) astrocytes upregulated and secreted high levels of IL-8, while Mn-exposed YTHDF2-overexpressing cells did not upregulate and secrete IL-8 protein levels (Figure 2F), corroborating the gene expression results (Figure 2D). IL-6 and MIP1b responded similarly to IL-8, increasing in Mn-exposed EV cells but not in Mn-exposed YTHDF2-OE cells (Figure 2F). MCP1 was significantly lower basally in YTHDF2-overexpressing cells, but Mn treatment did not affect MCP1 release in both stable cells (Figure 2F). Corroborating the knockdown results (Figure 2E), IFNγ release was not affected by YTHDF2 overexpression and/or Mn treatment (Figure 2F). Additionally, we compared Mn-induced chemokine/cytokine levels to TNFα (100 ng/mL)-induced chemokine/cytokine levels, as TNFα is a potent pro-inflammatory cytokine (Figure S2A). Overall, we observed largely similar trends, especially when assessing YTHDF2-overexpressing cells. Notably, YTHDF2 overexpression significantly attenuated TNFα-induced levels of IL-1β, IL-8, IFNγ, and MCP1, while only MCP1 approached statistical significance in YTHDF2 knockdown experiments (Figure S2A). These differences between Mn and TNFα in chemokine/cytokine induction may be the result of different signaling pathways or a difference in the potency of the two exogenous stimuli.
Based on our corroborated gene and protein results, we proceeded to perform an mRNA stability experiment using actinomycin D to determine if YTHDF2 directly regulates the mRNA decay of certain pro-inflammatory chemokine/cytokine genes. For this, we selected IL-8 (based on Figures 2C and 2D) and determined its mRNA stability in both YTHDF2 knockdown and overexpression U251 astrocytes (Figures S2B and S2C). Since basal levels of IL-8 and other chemokine/cytokines were upregulated in siYTHDF2 cells, the stability of IL-8 mRNA in YTHDF2 knockdown cells was evaluated under non-stimulatory conditions. siYTHDF2 cells showed no increased half-life of IL-8 over 2 and 4-h time points (Figure S2B). On the other hand, EV and YTHDF2 overexpression cells were evaluated under Mn exposure when the relative abundance of chemokines/cytokines is increased for ease of comparison yet showed no sign of decreased half-life for IL-8 mRNA (Figure S2C). These functional experiments suggest YTHDF2 has an anti-inflammatory function in astrocytes, as YTHDF2 overexpression attenuates astrocytic inflammation elicited by Mn. Since YTHDF2 does not appear to regulate IL-8 directly, it may be executing its effects upon the upstream regulators of pro-inflammatory pathways.
YTHDF2 negatively regulates SEK1(MAP2K4)-JNK-cJUN pathway
YTHDF2 has been shown to target many mRNAs of signaling proteins involved in stress responses of varying conditions and cell types.30,31,32,43 Since the actinomycin D mRNA stability assay showed no direct influence of YTHDF2 upon IL-8 mRNA, we postulated that the pro-inflammatory response observed under the Mn exposure model may lie in the abundance of certain mRNAs that translate into signaling proteins or transcription factors whose downstream effects are to upregulate pro-inflammatory gene transcription. For that reason, we performed RNA-sequencing of Mn-exposed U251 astrocytes to evaluate the overall transcriptome in pursuit of corroborating the functional experiments of Figure 2. GO analysis of the regulated transcription factor targets revealed an overrepresentation of transcription factor families (bolded) including the AP-1 components (JUN and FOS) and HIF components (HIF1α and EPAS1) (Figure 3A). To support this, a network of significant differentially expressed genes (DEGs) was generated to provide a visual representation of direct and indirect gene regulation through transcription factors or protein-protein interactions (Figure S3A). In this DEG network, IL-1α and IL-8 were significantly upregulated genes by Mn exposure, supporting our functional results from Figure 2, and further indicating their upstream regulation by the JNK pathway. Functional annotations of the main connected network of genes revolve around cell proliferation, apoptosis, cell adhesion, wound healing, angiogenesis, and responses to hypoxia. Overall, these RNA-sequencing results suggest Mn exposure can highly stress astrocytes leading to altered survival/apoptotic signaling, hypoxic signaling, and overall inflammatory reactivity.
Figure 3.
RNA- and RIP-sequencing of Mn-exposed U251 astrocytes reveals YTHDF2 targeting of MAP2K4 (SEK1)
(A and B) (A) Z-scores of Mn/Ctrl GO for Transcriptional Factor Targets, indicating important roles for cJUN and HIF1α (B) RIP-sequencing graphical representation of statistically significant YTHDF2 targets in Ctrl that were affected by Mn exposure. YTHDF2 targets are identified as having ≥+1 log2(RIP/input) ratio. MAP2K4 was identified as a lost YTHDF2 target under Mn exposure, suggesting regulation of the SEK1(MAP2K4)-JNK-cJUN pathway.
(C) siYTHDF2 astrocytes present with longer MAP2K4 mRNA half-life (n = 3).
(D) ICC representation at 40x depicting siYTHDF2 astrocytes have increased SEK1 protein levels (100 μm scale).
(E) YTHDF2 overexpressing astrocytes present with shorter MAP2K4 half-life under Mn exposure (n = 4–5).
(F) ICC representation at 40x depicting YTHDF2-overexpressing astrocytes have decreased SEK1 protein levels (100 μm scale).
(G and H) Immunoblotting and quantification revealing cJUN phosphorylation is increased in Mn-exposed astrocytes and sustained in siYTHDF2 Mn-exposed astrocytes. SEK1 protein levels are basally increased in siYTHDF2 astrocytes (n = 3–4).
(I and J) Immunoblotting and quantification revealing cJUN phosphorylation is increased in Mn-exposed astrocytes and prevented in YTHDF2-overexpressing Mn-exposed astrocytes. SEK1 protein levels are basally decreased in YTHDF2-overexpressing astrocytes (n = 4). Data are means ± SEM. Two-group comparisons performed using unpaired t-test, with 2-fold gene threshold and 1.96 Z-score threshold using Altanalyze. Adjusted p-values < 0.05 considered significant evidence. For mRNA half-life comparisons, One-phase decay non-linear regression analysis was performed. p-values < 0.05 considered significant evidence. two-way ANOVA with FDR Two-stage step-up method of Benjamini, Krieger, and Yekutieli for multi-group comparison. Q-values < 0.05 considered significant evidence.
Given the strong hypoxic signature induced by Mn exposure in the RNA-sequencing data, we functionally validated it using siHIF1α in combination with Mn exposure. After siRNA transfection and Mn treatment, nuclear and cytosolic lysates were prepared to analyze HIF1α levels. Mn induced a strong upregulation of nuclear HIF1α, while siHIF1α transfection completely abolished HIF1α levels. Cytosolic HIF1α levels were undetectable likely because of rapid degradation and slow lysate processing methodology (Figure S4A). Interestingly, previous studies showed hypoxic states whether induced by O2 depletion29 or by cobalt chloride36 decrease YTHDF2 levels. In Zhong et al., YTHDF2 overexpression under hypoxia had a small effect in preventing HIF1α upregulation, suggesting YTHDF2 may target HIF1α mRNA,36 providing one possible anti-inflammatory mechanism of controlling chemo/cytokine release, as HIF1α knockdown attenuated Mn-induced gene expression of IL-1α and IL-1β (Figure S4B). Nuclear and cytosolic lysates of both siYTHDF2 and YTHDF2-OE cells revealed no observable increases in HIF1α in siYTHDF2-transfected cells, but there was an observable reduction in HIF1α levels in YTHDF2-OE cells (Figure S4C). Actinomycin D mRNA stability experiments did not support HIF1α mRNA as targets of YTHDF2 (data not shown). These results suggested YTHDF2 overexpression indirectly affects HIF1α protein levels, and HIF1α may only be partially responsible for the induction of certain chemokines/cytokines.
Next, we performed YTHDF2-RIP-sequencing assays on both control and Mn-exposed U251 astrocytes to seek out plausible mRNA targets of YTHDF2 that, when degraded, would alleviate the pro-inflammatory response. Using the log2(RIP/input) calculation on our normalized TPM sequencing data, we denoted YTHDF2 targets as ≥+1. In brief, we identified a total of 1,992 YTHDF2 transcript targets based on the control group. After performing a statistical comparison between control and Mn groups, we identified only 92 statistically significant (pvalue< 0.005) YTHDF2 targets, of which 90% were lost targets in the Mn group. Lost targets are defined as positive YTHDF2 targets in control conditions but are no longer significant YTHDF2 targets in the Mn group, putatively because of a decrease in YTHDF2 levels. Firstly, in agreement with our IL-8 mRNA stability experiments, IL-8 was not an observed target of YTHDF2, nor were any other studied chemokines/cytokines. Secondly, HIF1α, ARNT, and EPAS1 were not observed as YTHDF2 targets either, eliminating the HIF components as candidates for YTHDF2 regulation of inflammation. And thirdly, as NF-κB transcriptional activation has been shown to be important in regulating Mn toxicity and inflammation in astrocytes17,44 and as others have shown YTHDF2 to potentially regulate NF-κB signaling,45 we did seek out if NF-κB or any other related components like RELA would be targets of YTHDF2. However, NF-κB and its related components were not targets of YTHDF2 in our analysis. Nevertheless, our RIP-sequencing results did show that the mRNA of an upstream component of the MAPK pro-inflammatory signaling cascade, dual specificity mitogen-activated protein kinase 4 (SEK1 (protein), MAP2K4 or MKK4 (gene)), was positively bound by YTHDF2 in the controls but presented as a lost target in Mn, suggesting MAP2K4 mRNA is bound and directed toward decay by YTHDF2 (Figure 3B). In response to various environmental stressors such as Mn, SEK1 may be activated through phosphorylation of its serine and threonine residues at positions 257 and 261. Activated SEK1 leads to phosphorylation of JNK, which further phosphorylates its main cellular substrate, cJUN. Our preliminary analysis of cJUN S63 revealed increased phosphorylation post-Mn treatment and sustained phosphorylation in siYTHDF2 astrocytes treated with Mn. Conversely, YTHDF2 overexpression suppressed cJUN S63 phosphorylation (Figure S4D). The activated cJUN interacts with JunB, JunD, c-Fos, or ATF to constitute the AP-1 transcription factor, which regulates gene expression that includes pro-inflammatory genes such as IL-8 and IL-1α.46,47,48,49 A supplemental STRING analysis of SEK1, JNKs, JUN and its associated AP-1 partners, along with IL-8, demonstrated MAP2K4-JNK-cJUN signaling can lead to the transcriptional upregulation of IL-8 and IL-1α (Figure S4E), supporting YTHDF2’s indirect control of chemokine/cytokine mRNA and protein, which represents one mechanism of anti-inflammatory signaling in astrocytes.
To validate the YTHDF2 RIP-sequencing target MAP2K4, we first analyzed its mRNA sequence using the m6A prediction server, SRAMP,40 and found a total of 12 DRACH consensus sites, 4 of which were very high confidence for m6A deposition (Figure S4F). Then, we performed functional mRNA stability experiments using actinomycin D to assess MAP2K4’s half-life in both YTHDF2 knockdown and overexpression cells. YTHDF2 knockdown led to a doubling of MAP2K4’s half-life over the time span of 4 h (Figure 3C). Since MAP2K4’s half-life was doubled, we performed immunocytochemistry to gauge whether MAP2K4 protein (SEK1) was affected by the overall change in mRNA, and indeed, the abundance of SEK1 was also elevated (Figure 3D). Conversely, in Mn-exposed, YTHDF2-overexpressing cells, we observed a significant decrease in the half-life of MAP2K4 (Figure 3E), which was also accompanied by a decrease of its protein SEK1’s immunoreactivity (Figure 3F). These results support that MAP2K4 transcripts can be destabilized by YTHDF2, which can overall influence the protein abundance of SEK1.
Since YTHDF2’s regulation of MAP2K4 transcripts was potent enough to affect the abundance of its protein SEK1, we wanted to quantitatively determine if this mechanism affected the downstream phosphorylation and activation of cJUN, the principal component of the AP-1 transcription factor, which was an overrepresented transcription factor target within our RNA-sequencing dataset (Figure 3A). SEK1 is the principal kinase responsible for promoting cJUN activation by first phosphorylating the JNK kinases, which then subsequently phosphorylate cJUN to trigger AP-1 transcription factor complex formation and activation that promote pro-inflammatory gene expression.50,51,52,53 As such, we first assessed the signaling cascade under YTHDF2 knockdown conditions along with Mn exposure in U251 astrocytes. Under YTHDF2 knockdown only, SEK1 protein was upregulated, as observed previously by actinomycin D assay (Figure 3C) and immunocytochemistry (Figure 3D), however, there were no detectable changes in phosphorylation of SEK1. Under Mn exposure, phosphorylation of SEK1 was significantly upregulated and sustained in Mn-exposed YTHDF2 knockdown cells. Likewise, downstream partners JNK and cJUN were highly phosphorylated under Mn exposure and sustained in Mn-exposed YTHDF2 knockdown cells (Figures 3G and 3H). Interestingly, YTHDF2 knockdown-only cells had a notable elevation of cJUN phosphorylation observed by immunoblotting and immunocytochemistry (Figures 3G, 3H, and S4D), suggesting that there may be some leaky basal elevation of the SEK1(MAP2K4)-JNK-cJUN cascade. Furthermore, total JNK protein levels behaved similarly to SEK1 protein levels, however, none of the JNK mRNAs (MAPK8, MAPK9, and MAPK10) were observed by YTHDF2-RIP-sequencing. Because cJUN phosphorylation was highly upregulated by Mn exposure and has been linked to biological processes such as inflammation, cell survival, and apoptosis, these results support that MAP2K4 mRNA is a target of YTHDF2, and under YTHDF2 downregulation, cJUN phosphorylation is sustained and saturated upon Mn exposure in astrocytes, leading to sustained pro-inflammatory signaling.
Lastly, we performed similar signaling experiments using YTHDF2-overexpressing cells to determine if Mn-induced SEK1, JNK, and cJUN phosphorylation can be prevented by YTHDF2 overexpression. Under unstimulated conditions, SEK1 protein was downregulated by YTHDF2 overexpression (Figures 3I and 3J), supporting the findings from the mRNA stability assay and immunocytochemistry functional experiments (Figures 3E and 3F). Again, there were no notable changes in phosphorylation of SEK1. Under Mn exposure, phosphorylation of SEK1 was significantly upregulated, but it was prevented by YTHDF2 overexpression. Likewise, JNK and cJUN were highly phosphorylated in EV cells under Mn exposure, but the phosphorylation was significantly reduced in Mn-exposed YTHDF2-overexpressing cells (Figures 3I and 3J). Interestingly, YTHDF2-overexpressing cells displayed a similar basal elevation of cJUN phosphorylation as observed in the YTHDF2 knockdown cells by immunoblotting (Figures 3I and 3J) and immunocytochemistry (Figure S4D), suggesting that cJUN phosphorylation is sensitive upon disturbances in the abundance of YTHDF2. Collectively, these functional signaling studies support that astrocytes exposed to the neurotoxic stressor Mn upregulate the SEK1(MAP2K4)-JNK-cJUN signaling cascade by decreasing YTHDF2 levels to sustain proliferative, survival, and inflammatory signaling.
YTHDF2 is decreased in Mn-gavaged mice, and a conditional knockout of YTHDF2 in the astrocytes of mice leads to increased astrocyte reactivity in substantia nigra pars compacta/reticulata and globus pallidus
We examined YTHDF2 levels in mice gavaged with 30 mg/kg of Mn to verify our in vitro results. We evaluated the brain regions relevant to Mn neurotoxicity including the globus pallidus, the principal region affected in humans,54,55,56,57 and the substantia nigra pars compacta/reticulata (SN), an additional site of interest where neuroinflammatory and neurotoxic effects have been observed.56,57,58,59,60,61 In the globus pallidus, increased GFAP reactivity was prevalent in Mn-exposed mice (Figure 4A). YTHDF2 immunoreactivity in GFAP-positive cells was generally observed to be decreased (Figure 4A), despite the overall YTHDF2 immunoreactivity in GFAP-positive cells being low. Surprisingly, the predominant localization of YTHDF2 was in larger cytoplasmic cells, presumably neurons (Figure 4A).62 We also immunoblotted whole tissue lysates from the substantia nigra and observed a significant decrease in YTHDF2 levels, approximately ≤20% (Figure 4B).
Figure 4.
YTHDF2 is decreased in Mn-gavaged mice, and a conditional knockout of YTHDF2 in the astrocytes of mice leads to increased astrocyte reactivity in substantia nigra pars compacta/reticulata and globus pallidus
(A) IHC representation at 40x depicting YTHDF2 decreases and colocalization in GFAP+ cells in the globus pallidus of Mn-gavaged mice (100 μm scale).
(B) Immunoblotting of the substantia nigra showing decreases of YTHDF2 in mice gavaged with Mn (n = 9).
(C) Schematic of Y2cKO mice generation. Tamoxifen (tiY2cKO) induces Cre recombination of YTHDF2 exon 4 to produce a band at 748 bp in astrocytes isolated from striatal/hippocampal brain tissue using biotinylated-EAAT1/GLAST-1 antibody.
(D) Globus pallidus validation of YTHDF2 protein reduction and increased GFAP immunoreactivity, and also increased m6A staining upon YTHDF2 deletion. YTHDF2 was quantified by parent-child extraction using GFAP as the parent (n = 3–4) (100 μm scale).
(E) Globus pallidus representative images of increased C3d immunoreactivity in GFAP-positive cells in the tiY2cKO mice (100 μm scale).
(F) Immunoblotting and quantification of substantia nigra- showing tiY2cKO mice with increased GFAP immunoreactivity similar to Mn-gavaged mice, indicating specific loss of YTHDF2 in astrocytes increases their reactivity (n = 4–6).
(G) Substantia nigra representative images of increased C3d immunoreactivity in GFAP-positive cells in the tiY2cKO mice (100 μm scale). Data are means ± SEM. Student’s t test or two-way ANOVA with FDR Two-stage step-up method of Benjamini, Krieger, and Yekutieli for multi-group comparison. Q-values < 0.05 considered significant evidence, with q-values between 0.05 and 0.01 considered as weaker evidence taking into consideration variations in data and trends.
Our in vitro studies showed that targeting SEK1 by YTHDF2 had profound effects in astrocytes exposed to Mn. We next sought to determine if such effects could be observed in vivo by generating an inducible conditional YTHDF2 knockout mouse line under the GFAP promoter, abbreviated as Y2cKO (Figure 4C). Y2cKO mice were administered 100 mg/kg tamoxifen in Miglyol-812N (tiY2cKO) or just Miglyol-812N (vehicle (viY2cKO) intraperitoneally daily for 5 days. DNA recombination was validated by immunoprecipitating astrocytes from brain tissues through positive selection using a biotinylated-EAAT1 antibody, and then performing PCR/genotyping (Figure 4C). We further validated the YTHDF2 protein expression in GFAP+ cells of the globus pallidus, showing decreased astrocytic YTHDF2 immunoreactivity in tiY2cKO mice. Interestingly, GFAP immunoreactivity was significantly upregulated in tiY2cKO mice, and likewise, so was global m6A (Figure 4D). Since observations of increased GFAP reactivity are not alone sufficient to define the physiological states of astrocytes,63 thus we co-stained for complement 3d (C3d) and GFAP, as C3d has been noted to be a functional co-indicator of astrocytes that can mediate neurotoxicity.64,65 There was an increase in the number of C3d immunoreactive GFAP+ cells (Figure 4E), suggesting that loss of YTHDF2 protein indeed elicits astrocytic inflammation in vivo.
After 5 days of tamoxifen induction, mice were gavaged with 30 mg/kg of Mn daily for 30 days. YTHDF2 levels were significantly decreased in pooled tiY2cKO groups as a factor using two-way ANOVA. Mn-treated viY2cKO mice had a ≤20% decrease of YTHDF2 (Figure 4F), similar to the above-mentioned findings (Figure 4B). Strikingly, tiY2cKO control mice had increased GFAP levels, comparable to that of viY2cKO Mn mice, however, there was no exacerbation of GFAP reactivity following Mn exposure (Figure 4F). These results indicate that genetic deletion of astrocytic Ythdf2 was sufficient to perturb astrocyte function and increase GFAP reactivity. We again stained for C3d, and tiY2cKO, viY2cKO Mn, and tiY2cKO Mn groups all had higher numbers of GFAP/C3d-positive astrocytes in the substantia nigra (Figure 4G) compared to the control viY2cKO group. Collectively, these findings support the overall in vitro findings that loss of YTHDF2 in astrocytes can induce sustained inflammation.
Discussion
The overall destabilization effect that m6A modifications confer upon RNAs, especially mRNAs that code for proteins, has promoted a wave of intense investigation in many disciplines such as cancer, virology, developmental biology, and immunology to name a few. Initial findings demonstrating specific m6A reader proteins are requisite for determining and executing the fate of m6A-modified mRNAs provided this impetus.66 YTHDF2 was shown to destabilize and promote the decay of m6A mRNAs,27 whereas YTHDF1 could promote translation,67 and YTHDF3 could potentially facilitate translation by association with YTHDF1 and afterward associate with YTHDF2 to facilitate decay.28 This putative mechanistic framework overall decreases the half-life of m6A mRNAs, specifically, those modified within coding sequences or near 3′ UTR regions, suggesting m6A modifications may be necessary for critical cellular responses requiring large transcriptional bursts.21 Under this framework, we postulated that YTHDF2 may be the most critical m6A reader as it principally promotes m6A mRNA decay. Herein, we show that YTHDF2 levels are decreased in astrocytes exposed to the astrocytic neurotoxic stressor Mn in vitro and in mice. Functional evaluation of YTHDF2 levels demonstrates that YTHDF2 negatively regulates MAP2K4 (SEK1), thereby affecting the direct downstream JNK-cJUN signaling pathway, which can control survival, proliferation, and inflammation. For the first time to our knowledge, decreased YTHDF2 levels allow astrocytes to mount and sustain a proper response to the environmental toxicant Mn (Figure 5).
Figure 5.
Integrated working hypothesis of YTHDF2’s role on the SEK1(MAP2K4)-JNK-cJUN pathway
Upon Mn exposure, ALKBH5 levels increase while YTHDF2 decreases, leading to increased half-life of MAP2K4 mRNA. Increased abundance of MAP2K4 leads to more SEK1 protein, allowing for the sustained activation (increased phosphorylation) of the downstream pathway targets JNK and cJUN by Mn, which promotes the pro-inflammatory response in astrocytes. When YTHDF2 is overexpressed, MAP2K4 mRNA is degraded. This leads to lower SEK1 protein levels, reducing the pathway activation of JNK and cJUN (decreased phosphorylation) by Mn, resulting in an anti-inflammatory response.
The neurotoxic stressor Mn is a heavy metal known to perturb metabolic functions and redox reactions in all neural cells in vitro.5,58,68 Additionally, it is known to activate signaling pathways such as NF-κB that can promote pro-inflammatory responses in astrocytes and microglia, which can inevitably damage neurons.17 These observations support the importance of investigating neuroinflammatory responses as being major drivers of neurodegeneration in Mn neurotoxicity. Astrocytes are generally seen as maintainers of neural homeostasis, but in recent years, more findings have surfaced demonstrating the importance of astrocytes in neuroinflammation, revealing their abilities to transform into neurotoxic cells.14,64 These pro-inflammatory inductions require a large transcriptional burst of mRNAs necessary to elicit and sustain the response to combat the insults. As such, having mechanisms that could regulate mRNA turnover quickly becomes important in altering cellular states in response to environmental changes. In this study, we observed a time-dependent decrease in m6A reader YTHDF2 upon exposing astrocytes to Mn, in which levels declined after 3 h post Mn, being lowest at 12 and 24 h (Figure 1A). Because Mn can elicit pro-inflammatory responses in astrocytes, this decrease in YTHDF2 levels would suggest that astrocytes need to decrease YTHDF2 to allow a sustained pro-inflammatory response. The mechanism by which Mn could decrease YTHDF2 levels is uncertain. Here, we do observe small decreases in YTHDF2 transcripts, but the largest decrease appears to be post-translational, possibly through the proteasome as MG-132 prevented Mn-induced decreases of YTHDF2 (Figure S1E). Recently, it has been shown CDK1 can maintain the protein stability of YTHDF2, where inhibition of CDK1 led to high polyubiquitination of YTHDF2 by the SKP2 E3 ubiquitin ligase complex38, however, it is unclear what effects Mn confers upon CDK1. Additionally, our results support the findings that hypoxia, whether induced by oxygen deprivation or cobalt chloride (CoCl2), can reduce YTHDF2 levels (Figures 3A and S4B) via an unidentified HIF1α mechanism,36 or via HIF2α transcriptional repression,29 both of which are increased in Mn-exposed astrocytes. With the decrease of YTHDF2 levels, we hypothesized an elevation in global m6A levels, however, the levels of m6A had decreased instead (Figure 1E), signifying a more complex cellular response regarding m6A modifications and their associated regulatory tiers. High doses of arsenite have been shown to upregulate the m6A demethylases, while low doses did not.39 Our dosage of Mn is considered a high dose5,17 and is in support of this finding. The demethylase ALKBH5 was upregulated by Mn treatment as shown by our results in U251 and furthermore in iPSC astrocytes (Figures S1B–S1D). Such results present an interesting hypothesis concerning pro-inflammatory responses, whereby astrocytes and other cell types may elevate m6A demethylases and downregulate YTHDF2 concomitantly to ensure an increased half-life of mRNAs necessary to be translated for a given insult. Moreover, we also show that METTL3 and METTL14 writers are decreased by Mn exposure, further suggesting a cooperative role between the m6A regulatory tiers to enhance m6A-driven responses following neurotoxic stressor Mn exposure. On the other hand, tiY2cKO mice showed increased m6A as initially expected (Figure 4D), suggesting in vitro and in vivo contexts exert significantly different influences on m6A methylation. These avenues remain to be fully elucidated, but what is certain is that YTHDF2 overexpression can suppress pro-inflammatory gene expression and the production of specific chemokines/cytokines, of which IL-8 was most significantly affected in our experiments (Figures 2C–2F). This anti-inflammatory response of YTHDF2 is in agreement with other recent cancer studies showing suppression of IL-11 and IL-8.29,45 Furthermore, although Mn failed to induce the production of MCP1 (also known as MCAF/CCL2) in our experiments, manipulation of YTHDF2 levels appreciably affected its basal levels, where the knockdown of YTHDF2 increased MCP1, and the overexpression of YTHD2 reduced MCP1 (Figures 2E and 2F). Interestingly, TNFα significantly increased MCP1 levels that were exacerbated by YTHDF2 knockdown and suppressed by YTHDF2 overexpression (Figure S2A). This is a significant finding in that MCP1 has been shown to be a principal chemokine produced by astrocytes, even those stimulated by Mn.17,69
Our initial hypothesis was that YTHDF2 could bind various m6A mRNAs of secretory factors such as chemokines/cytokines, as in the case of IL-11 targeting in hepatocellular carcinoma.29 YTHDF2-RIP-sequencing did not reveal any lost chemokine/cytokine targets by our methods. Others have previously shown that YTHDF2 can bind upstream signaling factor mRNAs that lead to the production of pro-inflammatory chemokines/cytokines.30,32,45 These signaling factors include RELA, MAP2K4, and MAP4K4. Our results pinpointed MAP2K4 mRNA as a YTHDF2 target, lost under Mn exposure (Figures 3B–3J). MAP2K4 or MKK4 (SEK1) is necessary for hepatocyte and fibroblast proliferation51,70 and for the protection against apoptosis in thymocytes.70 Additionally, the SEK1(MAP2K4)-JNK-cJUN pathway has been shown to be upregulated in astrocytes upon infection with mutant retrovirus MoMuLV-ts1 leading to COX-2 upregulation,53 and is upregulated in models of ischemia.71 JNK phosphorylation is upregulated in in vitro astrocytes treated with lipopolysaccharide or cotreated with IFNγ,52,72 and JNK expression is high in GFAP-positive astrocytes in mechanical allodynia.73 Indeed, in many stress models the SEK1(MAP2K4)-JNK-cJUN pathway is activated, as also observed in our Mn-exposed astrocytes (Figures 3G–3J and S4D). YTHDF2 overexpression drastically prevented activation of the pathway by decreasing MAP2K4 mRNA (Figures 3C–3F), thereby reducing total SEK1 levels and reducing phosphorylation of JNK and cJUN by Mn exposure. SEK1, JNK, and cJUN have all been found to be necessary for survival and proliferation.70,74,75 Additionally, IL-8 and MCP1 are known targets of cJUN and the AP-1 transcription factor family, especially in astrocytes,76,77,78 lending further explanatory power that would suggest these particular chemokines may be necessary for basal proliferation capabilities. Interestingly, GFAP levels were also reduced in YTHDF2-overexpressing cells (Figures 3G and 3I), and GFAP is a known transcriptional target of cJUN,79 providing another integrated explanation for the connection between the SEK1(MAP2K4)-JNK-cJUN pathway and motility.80 Overall, YTHDF2 appears to significantly and negatively regulate the SEK-JNK-cJUN pathway that converges upon multiple cellular responses, which include survival/apoptosis, proliferation, migration, and inflammation.
Our observations of the negative correlation between YTHDF2 levels and Mn-induction of pro-inflammatory genes/proteins in astrocytes suggested YTHDF2 may act as an anti-inflammatory m6A reader by negatively regulating the SEK1(MAP2K4)-JNK-cJUN pathway. In our examination of mice gavaged daily with Mn at 30 mg/kg for 30 days, YTHDF2 levels showed evidence of decreases in the substantia nigra and in astrocytes of the globus pallidus (Figures 4A and 4B). To determine if there was physiological evidence of our in vitro studies, we mimicked them in vivo by generating an inducible conditional knockout of YTHDF2 under the GFAP promoter (Y2cKO) (Figure 4C). In this study, we observed a decreasing trend in YTHDF2 levels in Mn-treated mice as in our preliminary study, approximately ≤20% (Figure 4F). Whether higher doses of Mn and/or longer timepoints in Mn gavage studies are more effective in reducing YTHDF2 remain to be tested. Alternatively, other Mn administration models should be tried to establish consistency and efficacy for disease modeling, especially inhalation models. Toxicity from inhaled Mn was first observed in Mn ore crushers.81,82 It is observably more potent based on a recent study that nasally administered 30 mg/kg for 21 days, in which dopaminergic cell (tyrosine hydroxylase +) loss was evident in the substantia nigra83, however, the globus pallidus was not investigated. Nevertheless, significant GFAP increases were observed in tiY2cKO mice, comparable to Mn-treated viY2cKO mice (Figure 4F). As astrocytes increase GFAP immunoreactivity when exhibiting detrimental and protective functions,63 we asked whether any co-indicators were also present to provide a clue about the astrocyte function. GFAP/C3d+ astrocytes were more abundant in tiY2cKO and Mn-treated viY2cKO mice in both the substantia nigra and the rostral globus pallidus (Figures 4E and 4G). Collectively, this YTHDF2 conditional knockout study would suggest YTHDF2 reduction or deletion in astrocytes perturbs their function, revealing their cellular states may be pro-inflammatory in Mn exposure models.
In summary, our studies indicate that YTHDF2 targets MAP2K4 mRNA to negatively regulate the SEK1(MAP2K4)-JNK-cJUN pathway in astrocytes, which is known to converge upon survival/apoptotic, proliferative, migratory, and inflammatory responses. Furthermore, m6A perturbation and decreases in YTHDF2 in our animal model of Mn neurotoxicity support the relevance of m6A epitranscriptomics in other neuroinflammatory and neurodegenerative conditions.84 Overall, YTHDF2’s m6A reader roles may be exploited for therapeutic benefits in controlling processes underlying chronic neurodegenerative diseases.
Limitations of the study
Our studies have a few limitations. Our mechanistic in vitro studies were performed in human U251-MG glioblastoma, a cancer cell line. Although this is a potential caveat in relation to Mn-induced Parkinsonism, our supportive in vivo results largely mitigate this. Moreover, as it is a cancer cell line, the elucidated mechanism herein will provide aid to the cancer research community and those studying m6A epitranscriptomics. Next, because we did not observe an exacerbation of astrocytic inflammation in tiY2cKO mice under our experimental conditions, we did not fully recapitulate our in vitro siYTHDF2 experiments. We suspect the confounding factor may be the lower YTHDF2 levels in astrocytes as compared to cells with bigger somas such as neurons (Figure 4A). This would indicate overexpression of YTHDF2 in vivo may be a more suitable alternative to ascertaining YTHDF2’s effects on astrocyte functionality and inflammation, which we will pursue in future studies.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit YTHDF2 | Proteintech | Cat# 24744-1-AP; RRID: AB_2687435 |
| Mouse GFAP | EMD Bioscience | Cat# MAB360; RRID: AB_11212597 |
| Rabbit GFAP | EMD Bioscience | Cat# AB5804; RRID: AB_2109645 |
| Mouse HIF1α | BD Biosciences | Cat# 610959; RRID: AB_398272 |
| Rabbit pSEK1 | Cell Signaling Technology | Cat# 9156; RRID: AB_2297420 |
| Rabbit SEK1 | Cell Signaling Technology | Cat# 9152; RRID: AB_330905 |
| Rabbit cJUN S63 | Cell Signaling Technology | Cat# 2361; RRID: AB_490908 |
| Mouse cJUN | Santa Cruz | Cat# sc-74543; RRID: AB_1121646 |
| Rabbit pJNK | Cell Signaling Technology | Cat# 4671; RRID: AB_331338 |
| Rabbit JNK | Cell Signaling Technology | Cat# 9258; RRID: AB_2141027 |
| Mouse β-Actin | Sigma | Cat# A5441; RRID: AB_476744 |
| Rabbit β-Actin | Sigma | Cat# A2103; RRID: AB_476694 |
| Mouse GAPDH | Sigma | Cat# G8795; RRID: AB_1078991 |
| Goat C3d | R&D Systems | Cat# AF2655; RRID: AB_2066622 |
| Goat GFP | Abcam | Cat# ab6673; RRID: AB_305643 |
| Rabbit Lamin B1 | Abcam | Cat# ab16048; RRID: AB_443298 |
| Biotinylated EAAT1/GLAST-1 | Novus Biologicals | Cat# NB100-1869B; RRID: AB_3150018 |
| Rabbit METTL3 | Abcam | Cat# ab195352; RRID: AB_2721254 |
| Rabbit METTL14 | Proteintech | Cat# 26158-1-AP; RRID: AB_2800447 |
| Rabbit ALKBH5 | Novus Biologicals | Cat# NBP1-82188; RRID: AB_11037354 |
| Chemicals, peptides, and recombinant proteins | ||
| MG-132 | Tocris Bioscience | Cat# 1748 |
| Lipofectamine 2000 | Invitrogen | Cat# 11668019 |
| Actinomycin D | Invitrogen | Cat# 11805017 |
| Miglyol 812N | IOI Oleochemical | Cat# NA |
| Critical commercial assays | ||
| NE-PER Nuclear and Cytoplasmic Extraction Kit | Thermo Fisher Scientific | Cat# 78833 |
| CD11b+ Selection Kit 2 | Stem Cell Technologies | Cat# 18970 |
| Biotin Positive Selection Kit 2 | Stem Cell Technologies | Cat# 17683 |
| Bio-plex Pro Human Cytokine 17-plex Kit | Bio-rad | Cat# M5000031YV |
| GenElute mRNA Kit | Sigma | Cat# MRN70 |
| Deposited data | ||
| RNA and YTHDF2 RIP-sequencing | This paper | NCBI GEO Accession GSE254720 |
| Experimental models: Cell lines | ||
| U251-MG | ATCC | Cat# HTB-17 |
| Human Induced Pluripotent Stem Cells | Allen Institute | ID# AICS-0087-031 cL.31 |
| Experimental models: Organisms/strains | ||
| C57BL/6NCrl | Charles River | Cat# NA |
| B6.Cg-Tg(GFAP-cre/ERT2)505Fmv/J mice | Jackson Laboratory | Cat# 012849 |
| Ythdf2(loxP/loxP) | Chuan He Laboratory | Cat# NA |
| Tamoxifen-inducible YTHDF2 conditional under Gfap promoter (Y2cKO) | This manuscript | Cat# NA |
| Oligonucleotides | ||
| YTHDF2 siRNA 5′-AAGGACGTTCCCAATAGCCAA-3′ | Qiagen | Cat# 1027424 |
| HIF1α siRNA 5′-GCTGATTTGTGAACCCAT-3′ | Ambion | Cat# NA |
| AllStars negative control siRNA | Qiagen | 1027281 |
| YTHDF2 loxP recombination F = GACCTGGATTAGCAGGAGT | IDT | Cat# NA |
| YTHDF2 loxP recombination R = CAGACCACTCCAACACAGAA | IDT | Cat# NA |
| YTHDF1 (F: ACACAACCTCCATCTTCGAC & R: ACTGGTTCGCCCTCATTG) | IDT | Cat# NA |
| YTHDF2 (F: TAGCCAACTGCGACACATTC & R: CACGACCTTGACGTTCCTTT) | IDT | Cat# NA |
| YTHDF3 (F: TGACAACAAACCGGTTACCA & TGTTTCTATTTCTCTCCCTACGC) | IDT | Cat# NA |
| MAP2K4 (F: TCCCAATCCTACAGGAGTTCAA & R: CCAGTGTTGTTCAGGGGAGA) | IDT | Cat# NA |
| Recombinant DNA | ||
| YTHDF2 pLenti-C-mGFP-P2A-Puro | OriGene | Cat# RC230306L4 |
| Software and algorithms | ||
| GraphPad Prism 8.0 | GraphPad | Cat# NA |
| AltAnalyze | Altanalyze.org | Cat# NA |
Resource availability
Lead contact
Inquiries and requests for resources should be directed to and will be fulfilled by Anumantha G. Kanthasamy (Anumantha.kanthasamy@uga.edu).
Materials availability
YTHDF2 conditional (Gfap) knockout mice generated in this manuscript will be available upon request within a reasonable time frame depending upon the status of the colony. The mice have yet to be deposited in a repository, however, this may change in the future. Please send your inquiries to the designated lead contact.
Data code and availability
-
•
Sequencing data have been deposited at NCBI GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Quantitative PCR, immunoblotting, and immunofluorescence will be shared upon a reasonable request to the lead contact.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon reasonable request.
Experimental model and study participant details
Mouse model
All animal procedures were approved by Iowa State University’s Institutional Animal Care and Use Committee (IACUC). Mice were housed under standard conditions for constant temperature (22 ± 1°C), relative humidity (30%), and a 12-h light cycle with food and water available ad libitum. Eight-week-old C57BL/6NCrl male mice purchased from Charles River were orally gavaged with Mn (30 mg/kg) dissolved in ultrapure water daily for 30 days. This dose was chosen based on previous studies on mouse models of Mn neurotoxicity, which generally use the Mn doses between 5 and 30 mg/kg/day for periods of 21–120 days.85 The control animals received daily injections of the same volume of water.
Tamoxifen-inducible conditional YTHDF2-knockout mouse (Y2cKO)
To create the tamoxifen-inducible conditional YTHDF2-knockout mouse (Y2cKO), the Ythdf2(loxP/loxP) mice acquired from Dr. Chuan He at the University of Chicago Medicine were crossed with the B6.Cg-Tg(GFAP-cre/ERT2)505Fmv/J mice, which were purchased from the Jackson Laboratory (Stock# 012849). The F1 generation was backcrossed to the Ythdf2(loxP/loxP) mice to generate the Y2cKO mice. Both male and female Y2cKO mice were given intraperitoneal injections of tamoxifen (100 mg/kg/d in Miglyol 812N oil) or vehicle for 5 days and then gavaged daily with 30 mg/kg Mn or water for 30 days as described above.
Primary mouse astrocytes
For primary mouse astrocytes, 1-2-day-old mouse pups (C57BL/6) were decapitated, and their brains harvested. Subsequently, the meninges were removed, and the brains were incubated in 0.25% Trypsin-EDTA for 15 min in a 37°C water bath and mixed every 5 min. Afterward, the brains were washed in DMEM/F12 growth media supplemented with 10% FBS, 1% penicillin/streptomycin, 1% L-glutamine, 1% sodium pyruvate, and 1% non-essential amino acids, homogenized by pipetting, and filtered through a 70-μm filter. The resulting cell suspension was then plated in flasks to attach and grow for 16 days. The growth media was aspirated and replaced with fresh growth media on the 6th d. After 16 days, the cells were collected and subjected to CD11b-positive selection to remove microglia. The negative fraction containing astrocytes was maintained in DMEM media supplemented with 10% FBS, 1% glutamine, and 1% penicillin/streptomycin, and treated with metals in DMEM containing 2% FBS.
Cell line
The human U251-MG astrocyte cell line obtained from ATCC (HTB-17) was maintained in MEM supplemented with 10% FBS and 1% penicillin/streptomycin and treated in MEM with 2% FBS. Cells were cultured at 37°C with 5% CO2. The source sex of the cells is male and was authenticated by short tandem repeat PCR profiling.
Human induced pluripotent stem cells
The human induced pluripotent stem cells (iPSC) were maintained in mTeSR Plus medium (STEMCELL Tech, Vancouver, Canada) supplemented with 1X penicillin/streptomycin (Gibco) in 5% CO2 at 37°C. The medium was changed every other day. We modified the protocol for neural induction from STEMCELL Tech and astrocyte differentiation from the previously established protocol.86 To generate neural progenitor cells (NPCs) from iPSC, we used STEMdiff SMADi Neural Induction Kit (STEMCELL) following the manufacturer’s protocol with minor changes. Briefly, human iPSCs were seeded on AggreWell800 24-well plate and cultured with STEMdiff Neural Induction Medium + SMADi for 5 days to make embryoid bodies (EB). After 5 days, the EBs were harvested from a single well of an AggreWell800 plate and plated onto a Matrigel-coated well of a tissue culture-treated 6-well plate. They were cultured in the same medium for another 5 days, and neural rosettes were mechanically selected using glass Pasteur pipettes. NPCs were cultured in NPC expansion medium: DMEM/F-12 + Glutamax supplemented with N2 supplement (Gibco), B27 supplement without vitamin A (Gibco), 10 ng/mL EGF (R&D Systems, Minneapolis, MN), 10 ng/mL bFGF (R&D Systems), and 1X penicillin/streptomycin. For astrocytic differentiation, NPCs were seeded on a Matrigel-coated 6-well plate. The next day, the medium was switched to astrocyte induction medium: DMEM/F-12 + Glutamax supplemented with N2, B27 without vitamin A, 10 ng/mL EGF, 10 ng/mL human LIF (R&D Systems), and 1X penicillin/streptomycin. The medium was changed every two days for 2 weeks and cells were split when they reached ∼90% confluence. After that, the cells were treated with astrocyte medium (DMEM/F-12 + Glutamax supplemented with B27 without vitamin A and 1X penicillin/streptomycin) containing 20 ng/mL CNTF (R&D systems) for 4 weeks. After 4 weeks, mature astrocytes were obtained for downstream experiments. These astroyctes were treated with 500 μM of MnCl2.
Method details
Chemicals & reagents
Cell culture media, supplements, and transfection reagents (lipofectamine 2000 in conjunction with Opti-MEM) were purchased from Life Technologies (Waltham, MA). SAFC fetal bovine serum (FBS), manganese chloride tetrahydrate (MnCl2), and puromycin were obtained from Sigma (St. Louis, MO), while lead chloride (PbCl2), copper chloride (CuCl2), and iron chloride (FeCl2) were purchased from Fisher Scientific (Waltham, MA). The proteasome inhibitor MG-132 was purchased from Tocris Bioscience (Minneapolis, MN). Antibodies used are as follows: rabbit YTHDF2 (Proteintech, Rosemont, IL: #24744-1-AP), mouse GFAP (EMD Bioscience, Burlington, MA: #MAB360), rabbit GFAP (EMD Bioscience: #AB5804), mouse HIF1α (BD Biosciences, Becton Drive Franklin Lakes, NJ: #610959), rabbit Lamin B1 (Abcam, Cambridge, UK: #ab16048), rabbit pSEK1 (Cell Signaling Technology (CST), Danvers, MA: #9156), rabbit SEK1 (CST: #9152), rabbit cJUN S63 (CST: #2361), mouse cJUN (Santa Cruz, Dallas, TX: #sc-74543), rabbit pJNK (CST: #4671), rabbit JNK (CST: #9258), mouse β-actin (Sigma: #A5441), rabbit β-actin (Sigma: #A2103), mouse GAPDH (Sigma: #G8795), goat complement component C3d (R&D Systems, Minneapolis, MN: #AF2655), goat GFP (Abcam: ab6673), rabbit METTL14 (Proteintech, Rosemont, IL: #26158-1-AP), rabbit METTL3 (Abcam: #ab195352), and rabbit ALKBH5 (Novus Biologicals, Centennial, CO: #NBP1-82188).
Overexpression and siRNA transfection
To generate a stable cell line expressing the human YTHDF2, U251-MG cells were stably transfected with a GFP-tagged YTHDF2 expression plasmid pLenti-C-mGFP-P2A-Puro (OriGene, #RC230306L4) or GFP empty vector by lipofectamine 2000 according to the procedure recommended by the manufacturer. The stable transfectants were selected and maintained in 3 μg/mL of puromycin added to the growth media.
The siRNA transfection of U251 astrocytes was performed by using lipofectamine 2000 following the manufacturer’s instructions and scaled as necessary with a default of 100 pmol siRNA for a 10 cm2 area. The YTHDF2-specific siRNA and AllStars negative control siRNA (#1027281) were purchased from Qiagen, while the HIF1α-specific siRNA was obtained from Ambion. The siRNA sequence for YTHDF2 is 5′-AAGGACGTTCCCAATAGCCAA-3′ and for HIF1α is 5′-GCTGATTTGTGAACCCAT-3’ (Ambion). After 48 h from the initial transfection, the cells were subjected to Mn treatment experiments. For all cell experiments, MnCl2 treatments were performed at 100 μM, and likewise, so were PbCl2, CuCl2, and FeCl2 treatments. Treatment with the proteasome inhibitor MG-132 was performed at 5 μM. Actinomycin D treatments were performed at 10 μg/mL.
Immunocytochemistry (ICC) and Immunohistochemistry (IHC)
For ICC, 13-mm coverslips were placed into 24-well plates and coated with poly-D-lysine. After experimental treatments, the media was aspirated, and 4% paraformaldehyde (PFA) was added to the wells, which were then incubated for 30 min at room temperature. Afterward, the coverslips were rinsed 5 times with PBS and then blocked using 5% donkey serum with 0.25% Triton X-100 in PBS for 1 h at room temperature. Primary antibodies were incubated overnight at room temperature, while secondary AlexaFluor antibodies, all from Invitrogen, were incubated for 1 h at room temperature. Washing was performed using PBS. Coverslips were then imaged using a Keyence All-in-one microscope (Keyence, Osaka, Japan).
For IHC, mice were perfused with 4% PFA, after which the brains were placed in 4% PFA for at least 24 h. For paraffin-embedded sectioning, the brains were placed onto a matrix and cut into 2-mm coronal pieces and then embedded into cassettes. Paraffin-embedded sections were cut at 5 μm. For cryosectioning, brains were transferred into a 30% sucrose solution and allowed to sink. Afterward, the brains were embedded and frozen in the Optimal Cutting Temperature compound. Cryosections were cut at 30 μM and allowed to free-float in cryosolution (30% sucrose and 30% ethylene glycol in PBS). The sections were rinsed in PBS and then subjected to antigen retrieval solution (10 mM sodium citrate, pH 7.6). The sections were blocked using 5% donkey serum with 0.25% Triton X-100 in PBS for 1 h at room temperature. Primary antibodies were incubated overnight, while secondary antibodies were incubated for 1 h at room temperature. The sections were imaged using the same Keyence All-in-one microscope.
Immunoblotting
Cells or brain tissues were homogenized in modified RIPA buffer containing Halt protease and phosphatase inhibitors, 1 mM sodium orthovanadate, and 1 mM phenylmethanesulfonyl fluoride. Afterward, the lysate was sonicated, cleared by centrifugation at 4°C, and the supernatant was collected. The protein concentrations were determined by a Bradford assay. Ten to forty μg of protein was loaded for SDS-PAGE gel electrophoresis and transferred onto nitrocellulose membranes. Membranes were blocked using Li-cor Intercept blocking buffer for 1 h at room temperature. Primary antibodies were incubated overnight at room temperature, while secondary antibodies were incubated for 1 h at room temperature. Washing was performed using PBS-Tween. All membranes were imaged using a Li-Cor Odyssey infrared imaging system (Li-cor, Lincoln, NE), and data were analyzed using ImageJ software or Odyssey software 2.0 (Li-Cor). β-actin or GAPDH served as an internal control for loading.
Nuclear and cytoplasmic fractionation
Cells were collected by scraping and washing with PBS 1 time. Twenty μL of the packed cell volume was used with CER I (200 μL), CER II (11 μL), and NER (100 μL) buffers according to manufacturer’s protocol of the NE-PER Nuclear and Cytoplasmic Extraction Reagent kit (Thermo Fisher Scientific). Afterward, the immunoblotting procedure given above was followed, with 10 μg of protein from both the nuclear and cytosolic fractions loaded onto the same gel.
Biotinylated EAAT1/GLAST-1 Immunoprecipitation
Striatal and hippocampal brain tissues from tamoxifen-induced Y2cKO (tiY2cKO) and vehicle Y2cKO (viY2cKO) mice were isolated and incubated in 0.25% trypsin-EDTA (TE) at 37°C for 15 min. Then, the TE was removed, and the brain tissue was rinsed in Hibernate A (minus calcium) media (BrainBits LLC, Springfield, IL). Next, the tissue was mechanically dissociated through intermittent pulsing using a micro-tube homogenizer. The resulting cell suspension was filtered using a 70-μm filter and centrifuged for 5 min at 1,500 rpm room temperature. The cell pellet was resuspended in Ca2+- and Mg2+-free Dulbecco’s phosphate buffered saline (DPBS) supplemented with 2% FBS and 1 mM EDTA and subjected to positive biotin selection according to manufacturer’s protocol (StemCell Technologies). A biotinylated EAAT1/GLAST-1 antibody (Novus Biologicals) was used in conjunction with this kit to positively select astrocytes. Purified astrocytes were lysed, and their DNA was collected via the alkaline extraction method, followed by PCR and gel electrophoresis for DNA recombination. The primer sequences (5′-3′) are: Forward = GACCTGGATTAGCAGGAGT; Reverse = CAGACCACTCCAACACAGAA. The expected sizes of the amplicons are as follows: Ythdf2 wild-type, 2882 bp; Ythdf2 LacZ modified, 9932 bp; Ythdf2 LacZ removed (floxed), 3019 bp; and Ythdf2 Cre recombined (post tamoxifen), 748 bp.
Luminex ELISA assays
For chemokine/cytokine analysis, the treatment media was collected and analyzed using the Bio-Plex Pro Human Cytokine 17-plex Assay kit (#M5000031YV) following the established kit protocol. The analytes were detected using the Luminex Bio-Plex 200 system and quantified against a known standard curve, which is represented in picograms/milliliter (pg/mL).
Quantitative real-time RT-PCR (qRT-PCR)
To extract RNA, cells were lysed using 1 mL of TRIzol reagent. Chloroform (200 μL) was added, and samples were vigorously mixed for 10 s, allowed to settle for 3 min at room temperature, and then centrifuged for 15 min at 12,000 rcf 4°C to obtain phase separation. The top aqueous layer was collected, mixed with isopropanol (at least 1:1), incubated for 10 min at room temperature, and then centrifuged for 15 min at 12,000 rcf 4°C. Isopropanol was removed, while the RNA pellets were washed with 75% ethanol and then centrifuged for 15 min at 20,627 rcf 4°C. After air-drying for 10 min at room temperature, the RNA pellets were dissolved in RNase/DNase free ultrapure water and quantified using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific). One microgram of RNA was converted into cDNA using a High-Capacity cDNA Reverse Transcription kit (Applied Biosystems). qPCR was performed using PowerUp Sybr Green reagents (Applied Biosystems) and run on the QuantStudio 3 system (Applied Biosystems, Waltham, Massachusetts). Human 18S rRNA was used as the housekeeping gene for normalization. The primer sequences for the YTHDF paralogs and MAP2K4 are as follows (5′-3′): YTHDF1 (F: ACACAACCTCCATCTTCGAC & R: ACTGGTTCGCCCTCATTG), YTHDF2 (F: TAGCCAACTGCGACACATTC & R: CACGACCTTGACGTTCCTTT), YTHDF3 (F: TGACAACAAACCGGTTACCA & TGTTTCTATTTCTCTCCCTACGC), and MAP2K4 (F: TCCCAATCCTACAGGAGTTCAA & R: CCAGTGTTGTTCAGGGGAGA). Validated QuantiTect primer sets obtained from (Qiagen) for IL-1α (QT00001127), IL-1β (QT00021385), IL-8 or CXCL8 (QT00000322), IL-12α (QT00000357), IL-18 (QT00014560), TNFα (QT00029162) were also used. The data were analyzed using the ΔΔCt method.
Actinomycin D mRNA stability assay
After transfection and/or treatments, cells were treated with 10 μg/mL of actinomycin D (Invitrogen) for 2 and 4 h, with 0 h as the initial condition. Cells were then collected by scraping and washing with PBS once. RNA was isolated and quantified as described in the qRT-PCR section.
RNA- and RIP-sequencing and analysis
The RNA- and RIP-sequencing analysis was adapted from previous studies.27,41 Briefly, cells were lysed in 2 volumes of modified TNE buffer containing 1% NP-40, Halt protease and phosphatase inhibitors, 1 mM sodium orthovanadate, 1 mM phenylmethanesulfonyl fluoride, and SUPERase RNase inhibitor (Invitrogen, 1:100). Lysates were cleared twice through centrifugation, and 2.5% of the total cleared lysate was collected and mixed with 1 mL of TRIzol for the RNA input fraction. RNA input was poly (A) purified using the GenElute mRNA kit (Sigma, #MRN 70). The remaining cleared lysate was subjected to immunoprecipitation with 10 μg of YTHDF2 antibody under constant rotation at 4°C for 4 h. The Protein A/G Magnetic Beads from Thermo Scientific Pierce (#88803) were then added to incubate under constant rotation at 4°C for 1 h. Then the YTHDF2 mRNP complexes were magnetically isolated, washed with TNE buffer, and placed in 1 mL of TRIzol for RNA isolation for the immunoprecipitated fraction. Sequencing was performed using a 50-cycle (single-end) with a HiSeq 3000 system (Illumina) with a total of 100 ng of RNA from both input and immunoprecipitated fractions. Fastq files were processed with the default settings of Altanalyze (embedded with Kallisto), and the expression files were subsequently processed through the embedded gene ontology software GO-Elite.87,88,89 For RIP-seq targets, we used the log2(RIP/input) calculation on our TPM sequencing data, with values ≥+1 as denoted as targets of YTHDF2, while values <+1 were considered non-targets of YTHDF2.90 Fastq and processed data have been uploaded to NCBI GEO (accession GSE254720).
Global m6A LC-MS/MS
Global m6A was quantified as previously described.27 Briefly, RNA was isolated by TRIzol and subjected to 2 rounds of poly (A) purification using the Gen-Elute mRNA kit (MRN 70). Thirty nanograms of mRNA were digested by nuclease P1 at 37 °C for 3 h, followed by FastAP alkaline phosphatase digestion at 37 °C for 6 h. The samples were diluted and filtered through a 0.22-μm filter, and 5 μL of each sample was injected into the LC-MS/MS. Reverse phase UPLC using a C18 column separated the nucleosides, which were detected by an Agilent 6410 QQQ LC mass spectrometer in positive electrospray ionization mode. Quantification was performed using a standard curve after taking the nucleoside to base ion mass transitions of 282 to 150 (m6A), and 268 to 136 (A). The ratio of m6A to A was calculated and expressed as a percentage of the control.
Quantification and statistical analysis
GraphPad 8.0 was utilized for data visualization and statistical analysis. For two group comparisons, we applied the unpaired t-test. Welch correction was applied for comparisons with unequal variance and Mann-Whitney for nonnormally distributed data. For multiple group comparisons, two-way ANOVA was applied, with corrections for multiple comparisons controlled using FDR two-stage step-up method of Benjamini, Krieger and Yekutieli. For mRNA half-life comparisons, one-phase decay non-linear regression analysis was performed. Exact p-values are reported. p-values less than 0.05 were considered as strong evidence; however, p-values between 0.05 and 0.01 were considered as weak evidence taking into consideration the variations in data and trends.91
Acknowledgments
This work was supported in part by the National Institutes of Health R01 Grants ES027245, ES026892, NS121692 (A.G.K.) and NS124226 (A.K.). In addition, A.G.K. was also supported by the W. Eugene and Linda Lloyd Endowed Chair, the Scott and Nancy Armbrust Biomedical Science Endowment, the Johnny Isakson Endowed Chair, the Georgia Research Alliance Eminent Scholar, and the Coach Mark Richt Neurological Disease Research funds.
Author contributions
E.M. and A.E. contributed equally in conceiving, performing experiments, and writing and revising the manuscript as necessary. P.J.H., C.M., A.J., and S.S. provided insights and performed experiments. D.R., C.G., A.K.H., A.Z., and B.P. performed experiments. G.Z., H.J., V.A., and A.K. provided critical feedback for the work and performed manuscript revisions. C.H. provided resources for performing RNA- and RIP-sequencing and the initial Ythdf2-floxed mice. A.G.K. conceived the study and provided intellectual and financial resources, critical feedback, manuscript preparation, and general guidance.
Declaration of interests
A.G.K. and V.A. have an equity interest in PK Biosciences Corporation and Probiome Therapeutics, located at the University of Georgia, GA. The terms of this arrangement have been reviewed and approved by the University of Georgia in accordance with their conflict-of-interest policies. No other authors have conflicts of interest.
Published: July 31, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.110619.
Supplemental information
References
- 1.Volterra A., Meldolesi J. Astrocytes, from brain glue to communication elements: the revolution continues. Nat. Rev. Neurosci. 2005;6:626–640. doi: 10.1038/nrn1722. [DOI] [PubMed] [Google Scholar]
- 2.Hamby M.E., Sofroniew M.V. Reactive astrocytes as therapeutic targets for CNS disorders. Neurotherapeutics. 2010;7:494–506. doi: 10.1016/j.nurt.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barreto G.E., Gonzalez J., Torres Y., Morales L. Astrocytic-neuronal crosstalk: Implications for neuroprotection from brain injury. Neurosci. Res. 2011;71:107–113. doi: 10.1016/j.neures.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 4.Deitmer J.W., Rose C.R. Ion changes and signalling in perisynaptic glia. Brain Res. Rev. 2010;63:113–129. doi: 10.1016/j.brainresrev.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 5.Sarkar S., Malovic E., Harischandra D.S., Ngwa H.A., Ghosh A., Hogan C., Rokad D., Zenitsky G., Jin H., Anantharam V., et al. Manganese exposure induces neuroinflammation by impairing mitochondrial dynamics in astrocytes. Neurotoxicology. 2018;64:204–218. doi: 10.1016/j.neuro.2017.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Streifel K.M., Miller J., Mouneimne R., Tjalkens R.B. Manganese inhibits ATP-induced calcium entry through the transient receptor potential channel TRPC3 in astrocytes. Neurotoxicology. 2013;34:160–166. doi: 10.1016/j.neuro.2012.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tjalkens R.B., Zoran M.J., Mohl B., Barhoumi R. Manganese suppresses ATP-dependent intercellular calcium waves in astrocyte networks through alteration of mitochondrial and endoplasmic reticulum calcium dynamics. Brain Res. 2006;1113:210–219. doi: 10.1016/j.brainres.2006.07.053. [DOI] [PubMed] [Google Scholar]
- 8.Henriksson J., Tjälve H. Manganese Taken Up into the CNS via the Olfactory Pathway in Rats Affects Astrocytes. Toxicol. Sci. 2000;55:392–398. doi: 10.1093/toxsci/55.2.392. [DOI] [PubMed] [Google Scholar]
- 9.Horning K.J., Caito S.W., Tipps K.G., Bowman A.B., Aschner M. Manganese Is Essential for Neuronal Health. Annu. Rev. Nutr. 2015;35:71–108. doi: 10.1146/annurev-nutr-071714-034419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonzalez L.E., Juknat A.A., Venosa A.J., Verrengia N., Kotler M.L. Manganese activates the mitochondrial apoptotic pathway in rat astrocytes by modulating the expression of proteins of the Bcl-2 family. Neurochem. Int. 2008;53:408–415. doi: 10.1016/j.neuint.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 11.Aschner M., Gannon M., Kimelberg H.K. Manganese uptake and efflux in cultured rat astrocytes. J. Neurochem. 1992;58:730–735. doi: 10.1111/j.1471-4159.1992.tb09778.x. [DOI] [PubMed] [Google Scholar]
- 12.Wedler F.C., Denman R.B. Glutamine synthetase: the major Mn(II) enzyme in mammalian brain. Curr. Top. Cell. Regul. 1984;24:153–169. doi: 10.1016/b978-0-12-152824-9.50021-6. [DOI] [PubMed] [Google Scholar]
- 13.Streifel K.M., Moreno J.A., Hanneman W.H., Legare M.E., Tjalkens R.B. Gene Deletion of nos2 Protects Against Manganese-Induced Neurological Dysfunction in Juvenile Mice. Toxicol. Sci. 2012;126:183–192. doi: 10.1093/toxsci/kfr335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liddelow S.A., Guttenplan K.A., Clarke L.E., Bennett F.C., Bohlen C.J., Schirmer L., Bennett M.L., Münch A.E., Chung W.-S., Peterson T.C., et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature. 2017;541:481–487. doi: 10.1038/nature21029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liddelow S.A., Barres B.A. Reactive Astrocytes: Production, Function, and Therapeutic Potential. Immunity. 2017;46:957–967. doi: 10.1016/j.immuni.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 16.Tan J., Zhang T., Jiang L., Chi J., Hu D., Pan Q., Wang D., Zhang Z. Regulation of Intracellular Manganese Homeostasis by Kufor-Rakeb Syndrome-associated ATP13A2 Protein. J. Biol. Chem. 2011;286:29654–29662. doi: 10.1074/jbc.M111.233874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Popichak K.A., Afzali M.F., Kirkley K.S., Tjalkens R.B. Glial-neuronal signaling mechanisms underlying the neuroinflammatory effects of manganese. J. Neuroinflammation. 2018;15:324. doi: 10.1186/s12974-018-1349-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen C.-J., Ou Y.-C., Lin S.-Y., Liao S.-L., Chen S.-Y., Chen J.-H. Manganese modulates pro-inflammatory gene expression in activated glia. Neurochem. Int. 2006;49:62–71. doi: 10.1016/j.neuint.2005.12.020. [DOI] [PubMed] [Google Scholar]
- 19.Dobson A.W., Erikson K.M., Aschner M. Manganese neurotoxicity. Ann. N. Y. Acad. Sci. 2004;1012:115–128. doi: 10.1196/annals.1306.009. [DOI] [PubMed] [Google Scholar]
- 20.Aschner M., Erikson K.M., Herrero Hernández E., Tjalkens R. Manganese and its Role in Parkinson’s Disease: From Transport to Neuropathology. NeuroMolecular Med. 2009;11:252–266. doi: 10.1007/s12017-009-8083-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fu Y., Dominissini D., Rechavi G., He C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat. Rev. Genet. 2014;15:293–306. doi: 10.1038/nrg3724. [DOI] [PubMed] [Google Scholar]
- 22.Liu J., Yue Y., Han D., Wang X., Fu Y., Zhang L., Jia G., Yu M., Lu Z., Deng X., et al. A METTL3–METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014;10:93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ping X.-L., Sun B.-F., Wang L., Xiao W., Yang X., Wang W.-J., Adhikari S., Shi Y., Lv Y., Chen Y.-S., et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177–189. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Linder B., Grozhik A.V., Olarerin-George A.O., Meydan C., Mason C.E., Jaffrey S.R. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods. 2015;12:767–772. doi: 10.1038/nmeth.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng G., Dahl J.A., Niu Y., Fedorcsak P., Huang C.-M., Li C.J., Vågbø C.B., Shi Y., Wang W.-L., Song S.-H., et al. ALKBH5 Is a Mammalian RNA Demethylase that Impacts RNA Metabolism and Mouse Fertility. Mol. Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei J., Liu F., Lu Z., Fei Q., Ai Y., He P.C., Shi H., Cui X., Su R., Klungland A., et al. Differential m6A, m6Am, and m1A Demethylation Mediated by FTO in the Cell Nucleus and Cytoplasm. Mol. Cell. 2018;71:973–985.e5. doi: 10.1016/j.molcel.2018.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X., Lu Z., Gomez A., Hon G.C., Yue Y., Han D., Fu Y., Parisien M., Dai Q., Jia G., et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi H., Wang X., Lu Z., Zhao B.S., Ma H., Hsu P.J., Liu C., He C. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 2017;27:315–328. doi: 10.1038/cr.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hou J., Zhang H., Liu J., Zhao Z., Wang J., Lu Z., Hu B., Zhou J., Zhao Z., Feng M., et al. YTHDF2 reduction fuels inflammation and vascular abnormalization in hepatocellular carcinoma. Mol. Cancer. 2019;18:163. doi: 10.1186/s12943-019-1082-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fang C., He M., Li D., Xu Q. YTHDF2 mediates LPS-induced osteoclastogenesis and inflammatory response via the NF-κB and MAPK signaling pathways. Cell. Signal. 2021;85 doi: 10.1016/j.cellsig.2021.110060. [DOI] [PubMed] [Google Scholar]
- 31.Yu R., Li Q., Feng Z., Cai L., Xu Q. m6A Reader YTHDF2 Regulates LPS-Induced Inflammatory Response. Int. J. Mol. Sci. 2019;20:1323. doi: 10.3390/ijms20061323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu R., Ji X., Wu X., Chen J., Li X., Jiang H., Fu H., Wang H., Lin Z., Tang X., et al. Melatonin antagonizes ovarian aging via YTHDF2-MAPK-NF-κB pathway. Genes Dis. 2022;9:494–509. doi: 10.1016/j.gendis.2020.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Exil V., Ping L., Yu Y., Chakraborty S., Caito S.W., Wells K.S., Karki P., Lee E., Aschner M. Activation of MAPK and FoxO by Manganese (Mn) in Rat Neonatal Primary Astrocyte Cultures. PLoS One. 2014;9 doi: 10.1371/journal.pone.0094753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saha P., Guha S., Biswas S.C. P38K and JNK pathways are induced by amyloid-β in astrocyte: Implication of MAPK pathways in astrogliosis in Alzheimer’s disease. Mol. Cell. Neurosci. 2020;108 doi: 10.1016/j.mcn.2020.103551. [DOI] [PubMed] [Google Scholar]
- 35.Zhou J., Wan J., Gao X., Zhang X., Jaffrey S.R., Qian S.-B. Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature. 2015;526:591–594. doi: 10.1038/nature15377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhong L., Liao D., Zhang M., Zeng C., Li X., Zhang R., Ma H., Kang T. YTHDF2 suppresses cell proliferation and growth via destabilizing the EGFR mRNA in hepatocellular carcinoma. Cancer Lett. 2019;442:252–261. doi: 10.1016/j.canlet.2018.11.006. [DOI] [PubMed] [Google Scholar]
- 37.Coimbra-Costa D., Alva N., Duran M., Carbonell T., Rama R. Oxidative stress and apoptosis after acute respiratory hypoxia and reoxygenation in rat brain. Redox Biol. 2017;12:216–225. doi: 10.1016/j.redox.2017.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fei Q., Zou Z., Roundtree I.A., Sun H.-L., He C. YTHDF2 promotes mitotic entry and is regulated by cell cycle mediators. PLoS Biol. 2020;18 doi: 10.1371/journal.pbio.3000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen H., Zhao T., Sun D., Wu M., Zhang Z. Changes of RNA N6-methyladenosine in the hormesis effect induced by arsenite on human keratinocyte cells. Toxicol. Vitro. 2019;56:84–92. doi: 10.1016/j.tiv.2019.01.010. [DOI] [PubMed] [Google Scholar]
- 40.Zhou Y., Zeng P., Li Y.-H., Zhang Z., Cui Q. SRAMP: prediction of mammalian N 6 -methyladenosine (m6 A) sites based on sequence-derived features. Nucleic Acids Res. 2016;44:e91. doi: 10.1093/nar/gkw104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Du H., Zhao Y., He J., Zhang Y., Xi H., Liu M., Ma J., Wu L. YTHDF2 destabilizes m6A-containing RNA through direct recruitment of the CCR4–NOT deadenylase complex. Nat. Commun. 2016;7 doi: 10.1038/ncomms12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park O.H., Ha H., Lee Y., Boo S.H., Kwon D.H., Song H.K., Kim Y.K. Endoribonucleolytic Cleavage of m6A-Containing RNAs by RNase P/MRP Complex. Mol. Cell. 2019;74:494–507.e8. doi: 10.1016/j.molcel.2019.02.034. [DOI] [PubMed] [Google Scholar]
- 43.Li J., Xie H., Ying Y., Chen H., Yan H., He L., Xu M., Xu X., Liang Z., Liu B., et al. YTHDF2 mediates the mRNA degradation of the tumor suppressors to induce AKT phosphorylation in N6-methyladenosine-dependent way in prostate cancer. Mol. Cancer. 2020;19:152. doi: 10.1186/s12943-020-01267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rizor A., Pajarillo E., Nyarko-Danquah I., Digman A., Mooneyham L., Son D.-S., Aschner M., Lee E. Manganese-induced reactive oxygen species activate IκB kinase to upregulate YY1 and impair glutamate transporter EAAT2 function in human astrocytes in vitro. Neurotoxicology. 2021;86:94–103. doi: 10.1016/j.neuro.2021.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He J., Zhou M., Yin J., Wan J., Chu J., Jia J., Sheng J., Wang C., Yin H., He F. METTL3 restrains papillary thyroid cancer progression via m6A/c-Rel/IL-8-mediated neutrophil infiltration. Mol. Ther. 2021;29:1821–1837. doi: 10.1016/j.ymthe.2021.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moerman-Herzog A.M., Barger S.W. A polymorphism in the upstream regulatory region of the interleukin-1α gene confers differential binding by transcription factors of the AP-1 family. Life Sci. 2012;90:975–979. doi: 10.1016/j.lfs.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haeusgen W., Tueffers L., Herdegen T., Waetzig V. Map2k4δ — Identification and functional characterization of a novel Map2k4 splice variant. Biochim. Biophys. Acta. 2014;1843:875–884. doi: 10.1016/j.bbamcr.2014.01.028. [DOI] [PubMed] [Google Scholar]
- 48.Moriguchi T., Toyoshima F., Masuyama N., Hanafusa H., Gotoh Y., Nishida E. A novel SAPK/JNK kinase, MKK7, stimulated by TNFalpha and cellular stresses. EMBO J. 1997;16:7045–7053. doi: 10.1093/emboj/16.23.7045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kyriakis J.M., Avruch J. Mammalian MAPK Signal Transduction Pathways Activated by Stress and Inflammation: A 10-Year Update. Physiol. Rev. 2012;92:689–737. doi: 10.1152/physrev.00028.2011. [DOI] [PubMed] [Google Scholar]
- 50.Watanabe T., Nakagawa K., Ohata S., Kitagawa D., Nishitai G., Seo J., Tanemura S., Shimizu N., Kishimoto H., Wada T., et al. SEK1/MKK4-Mediated SAPK/JNK Signaling Participates in Embryonic Hepatoblast Proliferation via a Pathway Different from NF-κB-Induced Anti-Apoptosis. Dev. Biol. 2002;250:332–347. doi: 10.1006/dbio.2002.0781. [DOI] [PubMed] [Google Scholar]
- 51.Yang D., Tournier C., Wysk M., Lu H.-T., Xu J., Davis R.J., Flavell R.A. Targeted disruption of the MKK4 gene causes embryonic death, inhibition of c-Jun NH 2 -terminal kinase activation, and defects in AP-1 transcriptional activity. Proc. Natl. Acad. Sci. USA. 1997;94:3004–3009. doi: 10.1073/pnas.94.7.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pawate S., Bhat N.R. C-Jun N-Terminal Kinase (JNK) Regulation of iNOS Expression in Glial Cells: Predominant Role of JNK1 Isoform. Antioxidants Redox Signal. 2006;8:903–909. doi: 10.1089/ars.2006.8.903. [DOI] [PubMed] [Google Scholar]
- 53.Kim H.-T., Qiang W., Liu N., Scofield V.L., Wong P.K.Y., Stoica G. Up-regulation of astrocyte cyclooxygenase-2, CCAAT/enhancer-binding protein, glucose-related protein 78, eukaryotic initiation factor 2α, and c-Jun N-terminal kinase by a neurovirulent murine retrovirus. J. Neurovirol. 2005;11:166–179. doi: 10.1080/13550280590922810. [DOI] [PubMed] [Google Scholar]
- 54.Wang X., Li R., He R., Fang F. Effects of repeated manganese treatment on proton magnetic resonance spectra of the globus pallidus in rat brain. NMR Biomed. 2022;35:e4617. doi: 10.1002/nbm.4617. [DOI] [PubMed] [Google Scholar]
- 55.Katsuragi T., Takahashi T., Shibuya K., Nagatomo H., Iwabuchi K. [A patient with parkinsonism presenting hyperintensity in the globus pallidus on T1-weighted MR images: the correlation with manganese poisoning] Rinsho Shinkeigaku. 1996;36:780–782. [PubMed] [Google Scholar]
- 56.Racette B.A., Antenor J.A., McGee-Minnich L., Moerlein S.M., Videen T.O., Kotagal V., Perlmutter J.S. [ 18 F]FDOPA PET and clinical features in parkinsonism due to manganism. Mov. Disord. 2005;20:492–496. doi: 10.1002/mds.20381. [DOI] [PubMed] [Google Scholar]
- 57.Andruska K.M., Racette A.B.A. Neuromythology of Manganism. Curr. Epidemiol. Rep. 2015;2:143–148. doi: 10.1007/s40471-015-0040-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sarkar S., Rokad D., Malovic E., Luo J., Harischandra D.S., Jin H., Anantharam V., Huang X., Lewis M., Kanthasamy A., Kanthasamy A.G. Manganese activates NLRP3 inflammasome signaling and propagates exosomal release of ASC in microglial cells. Sci. Signal. 2019;12 doi: 10.1126/scisignal.aat9900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Harischandra D.S., Rokad D., Neal M.L., Ghaisas S., Manne S., Sarkar S., Panicker N., Zenitsky G., Jin H., Lewis M., et al. Manganese promotes the aggregation and prion-like cell-to-cell exosomal transmission of α-synuclein. Sci. Signal. 2019;12 doi: 10.1126/scisignal.aau4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao F., Cai T., Liu M., Zheng G., Luo W., Chen J. Manganese Induces Dopaminergic Neurodegeneration via Microglial Activation in a Rat Model of Manganism. Toxicol. Sci. 2009;107:156–164. doi: 10.1093/toxsci/kfn213. [DOI] [PubMed] [Google Scholar]
- 61.Krishna S., Dodd C.A., Hekmatyar S.K., Filipov N.M. Brain deposition and neurotoxicity of manganese in adult mice exposed via the drinking water. Arch. Toxicol. 2014;88:47–64. doi: 10.1007/s00204-013-1088-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saunders A., Huang K.W., Sabatini B.L. Globus Pallidus Externus Neurons Expressing parvalbumin Interconnect the Subthalamic Nucleus and Striatal Interneurons. PLoS One. 2016;11 doi: 10.1371/journal.pone.0149798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Escartin C., Galea E., Lakatos A., O’Callaghan J.P., Petzold G.C., Serrano-Pozo A., Steinhäuser C., Volterra A., Carmignoto G., Agarwal A., et al. Reactive astrocyte nomenclature, definitions, and future directions. Nat. Neurosci. 2021;24:312–325. doi: 10.1038/s41593-020-00783-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guttenplan K.A., Weigel M.K., Prakash P., Wijewardhane P.R., Hasel P., Rufen-Blanchette U., Münch A.E., Blum J.A., Fine J., Neal M.C., et al. Neurotoxic reactive astrocytes induce cell death via saturated lipids. Nature. 2021;599:102–107. doi: 10.1038/s41586-021-03960-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Clarke L.E., Liddelow S.A., Chakraborty C., Münch A.E., Heiman M., Barres B.A. Normal aging induces A1-like astrocyte reactivity. Proc. Natl. Acad. Sci. USA. 2018;115:E1896–E1905. doi: 10.1073/pnas.1800165115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Malovic E., Ealy A., Kanthasamy A., Kanthasamy A.G. Emerging Roles of N6-Methyladenosine (m6A) Epitranscriptomics in Toxicology. Toxicol. Sci. 2021;181:13–22. doi: 10.1093/toxsci/kfab021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang X., Zhao B.S., Roundtree I.A., Lu Z., Han D., Ma H., Weng X., Chen K., Shi H., He C. N6-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell. 2015;161:1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Warren E.B., Bryan M.R., Morcillo P., Hardeman K.N., Aschner M., Bowman A.B. Manganese-induced Mitochondrial Dysfunction Is Not Detectable at Exposures Below the Acute Cytotoxic Threshold in Neuronal Cell Types. Toxicol. Sci. 2020;176:446–459. doi: 10.1093/toxsci/kfaa079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rong Y., Ji C., Wang Z., Ge X., Wang J., Ye W., Tang P., Jiang D., Fan J., Yin G., et al. Correction to: Small extracellular vesicles encapsulating CCL2 from activated astrocytes induce microglial activation and neuronal apoptosis after traumatic spinal cord injury. J. Neuroinflammation. 2021;18:285. doi: 10.1186/s12974-021-02336-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nishina H., Fischer K.D., Radvanyi L., Shahinian A., Hakem R., Rubie E.A., Bernstein A., Mak T.W., Woodgett J.R., Penninger J.M. Stress-signalling kinase Sek1 protects thymocytes from apoptosis mediated by CD95 and CD3. Nature. 1997;385:350–353. doi: 10.1038/385350a0. [DOI] [PubMed] [Google Scholar]
- 71.Dong Y., Liu H.D., Zhao R., Yang C.Z., Chen X.Q., Wang X.H., Lau L.T., Chen J., Yu A.C.H. Ischemia activates JNK/c-Jun/AP-1 pathway to up-regulate 14-3-3γ in astrocyte. J. Neurochem. 2009;109:182–188. doi: 10.1111/j.1471-4159.2009.05974.x. [DOI] [PubMed] [Google Scholar]
- 72.Zhang X., Wang J., Qian W., Zhao J., Sun L., Qian Y., Xiao H. Dexmedetomidine Inhibits Tumor Necrosis Factor-Alpha and Interleukin 6 in Lipopolysaccharide-Stimulated Astrocytes by Suppression of c-Jun N-Terminal Kinases. Inflammation. 2014;37:942–949. doi: 10.1007/s10753-014-9814-4. [DOI] [PubMed] [Google Scholar]
- 73.Gao Y.-J., Xu Z.-Z., Liu Y.-C., Wen Y.-R., Decosterd I., Ji R.-R. The c-Jun N-terminal kinase 1 (JNK1) in spinal astrocytes is required for the maintenance of bilateral mechanical allodynia under a persistent inflammatory pain condition. Pain. 2010;148:309–319. doi: 10.1016/j.pain.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wada T., Joza N., Cheng H.y.M., Sasaki T., Kozieradzki I., Bachmaier K., Katada T., Schreiber M., Wagner E.F., Nishina H., Penninger J.M. MKK7 couples stress signalling to G2/M cell-cycle progression and cellular senescence. Nat. Cell Biol. 2004;6:215–226. doi: 10.1038/ncb1098. [DOI] [PubMed] [Google Scholar]
- 75.Nishina H., Vaz C., Billia P., Nghiem M., Sasaki T., De la Pompa J.L., Furlonger K., Paige C., Hui C., Fischer K.D., et al. Defective liver formation and liver cell apoptosis in mice lacking the stress signaling kinase SEK1/MKK4. Development. 1999;126:505–516. doi: 10.1242/dev.126.3.505. [DOI] [PubMed] [Google Scholar]
- 76.Gao Y.-J., Zhang L., Samad O.A., Suter M.R., Yasuhiko K., Xu Z.-Z., Park J.-Y., Lind A.-L., Ma Q., Ji R.-R. JNK-Induced MCP-1 Production in Spinal Cord Astrocytes Contributes to Central Sensitization and Neuropathic Pain. J. Neurosci. 2009;29:4096–4108. doi: 10.1523/JNEUROSCI.3623-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tang J., Zhu C., Li Z.h., Liu X.y., Sun S.k., Zhang T., Luo Z.j., Zhang H., Li W.y. Inhibition of the spinal astrocytic JNK/MCP-1 pathway activation correlates with the analgesic effects of tanshinone IIA sulfonate in neuropathic pain. J. Neuroinflammation. 2015;12:57. doi: 10.1186/s12974-015-0279-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Thompson W.L., Van Eldik L.J. Inflammatory cytokines stimulate the chemokines CCL2/MCP-1 and CCL7/MCP-7 through NFκB and MAPK dependent pathways in rat astrocytes. Brain Res. 2009;1287:47–57. doi: 10.1016/j.brainres.2009.06.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gao K., Wang C.R., Jiang F., Wong A.Y.K., Su N., Jiang J.H., Chai R.C., Vatcher G., Teng J., Chen J., et al. Traumatic scratch injury in astrocytes triggers calcium influx to activate the JNK/c-Jun/AP-1 pathway and switch on GFAP expression: Calcium Upregulates GFAP Via the JNK/c-Jun/AP-1 Pathway. Glia. 2013;61:2063–2077. doi: 10.1002/glia.22577. [DOI] [PubMed] [Google Scholar]
- 80.Lepekhin E.A., Eliasson C., Berthold C.-H., Berezin V., Bock E., Pekny M. Intermediate filaments regulate astrocyte motility: Intermediate filaments regulate astrocyte motility. J. Neurochem. 2001;79:617–625. doi: 10.1046/j.1471-4159.2001.00595.x. [DOI] [PubMed] [Google Scholar]
- 81.Rodier J. Manganese Poisoning in Moroccan Miners. Br. J. Ind. Med. 1955;12:21–35. doi: 10.1136/oem.12.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Blanc P.D. The early history of manganese and the recognition of its neurotoxicity, 1837–1936. Neurotoxicology. 2018;64:5–11. doi: 10.1016/j.neuro.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 83.Pajarillo E., Johnson J., Rizor A., Nyarko-Danquah I., Adinew G., Bornhorst J., Stiboller M., Schwerdtle T., Son D.-S., Aschner M., Lee E. Astrocyte-specific deletion of the transcription factor Yin Yang 1 in murine substantia nigra mitigates manganese-induced dopaminergic neurotoxicity. J. Biol. Chem. 2020;295:15662–15676. doi: 10.1074/jbc.RA120.015552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhao F., Xu Y., Gao S., Qin L., Austria Q., Siedlak S.L., Pajdzik K., Dai Q., He C., Wang W., et al. METTL3-dependent RNA m6A dysregulation contributes to neurodegeneration in Alzheimer’s disease through aberrant cell cycle events. Mol. Neurodegener. 2021;16:70. doi: 10.1186/s13024-021-00484-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ghaisas S., Harischandra D.S., Palanisamy B., Proctor A., Jin H., Dutta S., Sarkar S., Langley M., Zenitsky G., Anantharam V., et al. Chronic Manganese Exposure and the Enteric Nervous System: An in Vitro and Mouse in Vivo Study. Environ. Health Perspect. 2021;129 doi: 10.1289/EHP7877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Perriot S., Canales M., Mathias A., Du Pasquier R. Differentiation of functional astrocytes from human-induced pluripotent stem cells in chemically defined media. STAR Protoc. 2021;2 doi: 10.1016/j.xpro.2021.100902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Salomonis N., Schlieve C.R., Pereira L., Wahlquist C., Colas A., Zambon A.C., Vranizan K., Spindler M.J., Pico A.R., Cline M.S., et al. Alternative splicing regulates mouse embryonic stem cell pluripotency and differentiation. Proc. Natl. Acad. Sci. USA. 2010;107:10514–10519. doi: 10.1073/pnas.0912260107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Emig D., Salomonis N., Baumbach J., Lengauer T., Conklin B.R., Albrecht M. AltAnalyze and DomainGraph: analyzing and visualizing exon expression data. Nucleic Acids Res. 2010;38:W755–W762. doi: 10.1093/nar/gkq405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Olsson A., Venkatasubramanian M., Chaudhri V.K., Aronow B.J., Salomonis N., Singh H., Grimes H.L. Single-cell analysis of mixed-lineage states leading to a binary cell fate choice. Nature. 2016;537:698–702. doi: 10.1038/nature19348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hsu P.J., Zhu Y., Ma H., Guo Y., Shi X., Liu Y., Qi M., Lu Z., Shi H., Wang J., et al. Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017;27:1115–1127. doi: 10.1038/cr.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ramsey F.L., Schafer D.W. 3rd ed. Brooks/Cole; 2013. The Statistical Sleuth: A Course in Methods of Data Analysis. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.