Abstract
The liver has a unique ability to regenerate in response to injury or disease with hepatocytes and biliary epithelial cells (BECs) driving the regenerative response. Liver progenitor cells (LPCs) also play role in regeneration with the ability to differentiate into either hepatocytes or BECs. However, during chronic liver disease, the regenerative capacity of the liver is impaired. The use of LPCs is a promising therapeutic strategy for patients with chronic liver diseases. LPCs can be expanded in vitro as self-renewing organoids, however, most approaches to LPC organoids do not include critical cells from the LPC niche in 3D organoid cultures. In this study, we highlight the role of liver endothelial cells (LiECs), as a part of LPC niche, in supporting the hepatobiliary organoids in long-term culture even in the absence of defined growth supplements, such as Wnt agonists. Furthermore, LiECs alter the gene expression profile of hepatobiliary organoids involved in inflammation, migration, extracellular matrix organization, and receptor signaling pathway through paracrine manner. Our findings expand the role of LiECs for regulating stemness of LPCs and elucidate a role for niche cells in a LPC organoid co-culture model with a reduction in growth supplements.
Keywords: Liver stem/progenitor cell, Hepatobiliary organoid, Liver endothelial cell
1. Introduction
Tissue-resident adult stem cells (AdSCs) play a critical role in maintaining homeostasis of adult tissues through self-renewal and multi-lineage differentiation [1]. However, unlike other organs, the liver exhibits a distinct regeneration strategy with remarkable regenerative capacity. First, during homeostasis and even after partial hepatectomy, regeneration occurs through the proliferation and hypertrophy of resident hepatocytes [2]. Second, adult liver stem/progenitor cells (LPCs), which dedifferentiate from biliary epithelial cells (BECs), undergo self-renewal and differentiation into both hepatocytes and BECs, particularly when hepatocyte-mediated regeneration is impaired [3,4]. Ductular reaction (DR) is the expansion of LPCs after liver injury, and is strongly correlated with the severity of liver diseases [5]. However, the regenerative capacity is eventually compromised in chronic liver diseases due to the eventual dysfunction of LPCs and their niche cells, caused by chronic inflammation and fibrosis. Similar to the use of mesenchymal stem cells or hepatocyte transplantation, promoting liver regeneration utilizing LPCs may be a promising therapeutic strategy for patients with chronic liver diseases.
Organoid culture models, utilizing 3D matrix and specialized growth supplements, enable the massive expansion of AdSCs with genetic stability, emerging as a valuable scientific tool for understanding regenerative biology with potential implications for personalized medicine [6]. Adult liver organoid culture models have been successfully established by isolating Lgr5+ or Epcam+ LPCs or biliary duct fragments [7,8]. However, current approaches for liver organoid models lack interactions with other cellular populations in the liver. Tissue regeneration by AdSCs relies not only on the inherent plasticity of the cells but also on the dynamic interactions with their specialized microenvironment [9,10]. A recent study showed that SCA1+PDGFRα+ mesenchymal cells contribute to the stemness of LPCs through the secretion of paracrine factors or direct cell-to-cell interactions in vitro [11]. However, the specific roles of other niche cells remain largely unknown.
Endothelial cells (ECs), have been demonstrated to be critical in other organ stem cell niches, with specialized tissue-specific angiocrines and contribution to stemness in a tissue-specific manner [12]. Bone marrow ECs are essential for self-renewal and maintenance of hematopoietic stem cells via secretion of stem cell factor [13,14]. Similarly, brain ECs derived vascular endothelial growth factor regulates self-renewal and homeostasis of neural stem cell population [15,16], while lung ECs support the differentiation of lung stem cells into multilineage (alveolar, bronchiolar, bronchioalveolar) through thrombospondin-1 secretion [17]. Our previous study also demonstrated that testicular ECs support the self-renewal of spermatogonial stem cells by producing glial cell-derived neurotrophic factor [18]. However, the role of liver ECs (LiECs) in the LPCs niche remains to be examined.
Growth factor supplements, including FGF10, HGF, Noggin, and Wnt agonists (R-spondin-1 and Wnt3a), mimic the mitogenic environment that promotes the self-renewal of LPCs during liver regeneration, and are essential for the long-term expansion of LPCs in organoid cultures [7,8]. It has been reported that angiocrines, such as BMP2, HGF, and Wnt ligands, of LiECs regulate liver regeneration through proliferation and maturation of hepatocytes [[19], [20], [21], [22]], although the contribution of LiECs in regulating LPCs remains unknown. We propose that LiECs are necessary for the maintenance and expansion of LPCs. However, conventional liver organoid culture medium is supplemented with the defined growth factors or inhibitors for the survival and expansion of LPCs in monoculture [7]. Those factors could hinder to determine the role of LiECs in liver organoid co-culture. Hence, we modified the conventional organoid culture medium by removing FGF10, HGF, Noggin, and Wnt agonists to elucidate the specific role of LiECs in LPC organoid co-cultures. Here, we demonstrate the long-term expansion of hepatobiliary organoids co-cultured with LiECs in a reduced growth medium, revealing that LiECs can support the self-renewal of LPCs by altering their gene expression profile through paracrine interactions.
2. Material and methods
2.1. Mice
12–18 week old wild-type mice (C57BL/6) were used for isolating hepatobiliary ducts. Primary murine liver endothelial cells were isolated from 12 to 18 week old transgenic GFP mice (C57BL/6).
2.2. Primary liver endothelial cell isolation and characterization
Primary liver endothelial cells were isolated from GFP transgenic mice and previously described [18]. In brief, livers were minced and digested in digestion solution containing 2.5 mg/ml collagenase type II (Worthington) and 100 μg/ml DNase I (Roche) in HBSS diluted in 1x (ThermoFisher) for an hour at 37°C. Collagenase activity was quenched with an equal volume of FBS and digested cells were strained through 100 μm then 40 μm strainer. After centrifuging at 930g for 10 min, cells were resuspended and plated on gelatin-coated 150 mm cell culture dish in ECM. After 4 h, media was gently aspirated and replaced with fresh ECM. Once the cells reach confluency, cells are detached with Accutase (Milipore) and washed with MACS buffer containing 2 mM EDTA (Invitrogen) and 0.5 % BSA (Gendepot) in DPBS diluted to 1x (Welgene) followed by centrifuge. Cells are resuspended in MACS buffer and incubated with anti-mouse CD31 antibody conjugated with microbeads (Miltenyi Biotec) and mouse Fc receptor blocker (Miltenyi Biotec) for 15 min at 4°C. After MACS separation, sorted cells were centrifuged at 300g for 5min, then resuspended in DPBS, counted with trypan blue (Gibco) staining, and plated on gelatin-coated 6-well to 100 mm dish according to cell number. Liver endothelial cells were cultured in Advanced DMEM (ThermoFisher) medium supplemented with Glutamax (Gibco), HEPES (ThermoFisher), Penicillin/streptomycin (Welgene), Heparin (Sigma), ECGS (Corning) and 10 % FBS (Gemini), which is referred to as Endothelial cell medium (ECM). Liver endothelial cells were used for experiments between passages 3 and 6.
To confirm the purity of liver endothelial cells, cells were seeded on 10 mm glass coverslips and cultured until reaching confluency, then fixed with neutral buffered formalin (Sigma) for 10min at 4°C. The cells were permeabilized in 0.3 % Triton X-100 (Sigma) in PBS for 10min at RT and blocked in 10 % coat serum (Abcam, ab7481), 3 % BSA and 0.1 % tween 20 (Anatrace) in PBS for 2h at RT, then incubated with primary antibodies (GFP, CD31, VE-cadherin and SMAα) diluted in blocking solution O/N at 4°C. After washing with 0.1 % tween 20 in PBS, cells are incubated with Alexa Fluor-conjugated secondary antibodies diluted in blocking solution for 2h at RT, then mounted in Mounting medium (Abcam). Images were acquired using a Thermo Evos FL Auto 2 microscope and processed using ImageJ software. For tube formation assay, endothelial cells were seeded on Matrigel-coated 96-well plates (15,000 cells/well). Tube formation was observed after 24 h incubation. For acetylated LDL (Ac-LDL) uptake, endothelial cells were incubated with Dil-ac-LDL (10 μg/ml, Invitrogen) for 4 h at 37°C. After washing with DPBS 3 times, images were acquired using a Thermo Evos FL Auto 2 microscope.
2.3. Hepatobiliary organoid culture
Livers isolated from mice were minced into fine pieces and resuspended with washing medium (1 % FBS, 2 mM L-glutamine, 50U/ml penicillin and 50 μg/ml streptomycin in DMEM) and incubated for 5 min on ice. The supernatant was aspirated and liver pieces were resuspended with washing medium and repeated two times. After discarding the supernatant, digestion solution was added and incubated with liver pieces for 1 h at 37°C. An equivalent volume of FBS was added to quench collagenase activity and the supernatant was then strained through a 100 μm strainer. After washing the strainer to flush out remaining single cells and debris, hepatobiliary duct fragments remaining on the strainer were flushed into a conical tube using HBSS and the tube was centrifuged at 300g for 5min. The fragments were resuspended with basal medium (advanced DMEM supplemented with 25 mM HEPES, 100 μg/ml heparin (Sigma), GlutaMAX (Gibco), 5 % Knock-out serum replacement (Gibco), 100 μg/ml ECGS, 10 mM Nicotinamide (Sigma), 100 μg/ml Primocin (Invitrogen), 50U/ml penicillin and 50 μg/ml streptomycin) and mixed with GFR Matrigel (Corning) at a ratio of 1:2 (total 60 μl), then plated on the center of each well of a 24-well cell culture dish. After incubation at 37°C for 50 min, growth medium (basal medium supplemented with 10 μg/ml Y-27632 (Tocris), 50 ng/ml EGF (Peprotech) was added to each well. The medium was changed every 3–4 days. For serial passage, hepatobiliary organoids were harvested and transferred to ice-cold recovery solution (BD) for 10min. The organoids were gently resuspended with using a blunt tip every 5 min until Matrigel was not visible. The organoids were transferred to a 1.5 ml tube with recovery solution, then incubated while gently rotating at 4°C for 30 min. The supernatant was carefully discarded after all the organoids settled to the bottom. TrypLE (Gibco) solution was added and the organoids were mechanically disrupted into small pieces by pipetting, then incubated at 37°C for 10min. An equal volume of basal medium was added and centrifuged at 300g for 5min. Cells were counted with trypan blue staining and 5000 single cells were mixed with Matrigel for serial passages. For co-culture, LiECs (P3∼P6) were harvested prior to serial passages and counted. 100,000 cells were resuspended in basal medium and mixed with hepatobiliary duct fragments (Passage 0) or 5000 single cells (Passage 1∼), then mixed with Matrigel. Hepatobiliary organoid alone (HO) or co-cultured with LiECs (HO:LiEC) were subcultured every 7 days. For co-cultures using Transwell plates, 5000 single cells in Matrigel were seeded on Transwell alone with or without 100,000 LiECs in Matrigel on the bottom beneath the Transwell. All bright-field images were acquired using a Thermo Evos FL Auto 2 microscope. Organoid numbers and sizes were quantified by Celleste Image Analysis software using stitched images, for organoids greater than 100 μm of diameter.
2.4. Whole mount immunofluorescence staining
Hepatobiliary organoids were harvested and gently resuspended in ice-cold recovery solution to removing Matrigel, then incubated for 30 min in ice. Organoids were transferred to ice-cold Neutral buffered formalin (NBF) for 30 min on ice. Organoids were then transferred to ice-cold quenching solution (0.5M glycine, 0.1 % BSA in PBS) for 10 min, transferred to washing solution (0.1 % BSA in PBS), and either stored at 4°C or processed for staining. Organoids were transferred to permeabilization solution (0.2 % tritonX100, 5 % DMSO in PBS) for 15 min, then transferred to PBST solution (0.1 % tween 20 in PBS). After washing three times with PBST, organoids were transferred to blocking solution (10 % coat serum, 3 % BSA, 0.1 % tween 20 in PBS) for 2 h at RT and incubated at 4°C overnight with the following primary antibodies: EpCAM (1:100, eBioscience), CK19 (1:200, Abcam), SOX9 (1:100, Merck Millipore), Lgr5 (1:100, Abcam), Ki67 (1:200, Invitrogen), and F-actin (1:200, Thermo Fisher). Organoids were washed with PBST and incubated with secondary antibodies diluted in blocking buffer at RT for 2 h or overnight at 4°C. Images were obtained using a confocal laser scanning microscope (LSM710, Carl Zeiss).
2.5. RNA isolation, sequencing, and analysis
Total RNA was extracted from HO (p1) or HO:LiEC (p8) cultured in Transwell using Arcturus picopure RNA isolation kit (Thermo fisher) following manufacturer's instructions and processed for mRNA-sequencing. Quant-IT RiboGreen (Invitrogen) was used to determine total RNA concentration. Samples were analyzed on the TapeStation RNA screentape (Agilent), and only samples with RIN greater than 7 were used for RNA library construction. 1ug of total RNA was used to prepare libraries by Illumina TruSeq Stranded mRNA Sample Prep Kit (Illumina, Inc., San Diego, CA, USA). First, the poly‐A containing mRNA were purified using poly‐T‐attached magnetic beads. And then, the mRNA is sheared into small pieces with divalent cations under increasing temperature. The fragmented RNA was copied into first strand cDNA by SuperScript II reverse transcriptase (Invitrogen) and random primers. After that, DNA Polymerase I, RNase H and dUTP were used to synthesize second strand cDNA, which were further proceed into end repair process. The addition of a single ‘A’ base then was ligated with adapters. This was followed by purification and enrichment by PCR to access final cDNA library. The quantification/qualification of the library were measured by KAPA Library Quantification kits (KAPA BIOSYSTEMS) and TapeStation D1000 ScreenTape (Agilent Technologies). Indexed libraries were then uploaded to an Illumina NovaSeq (Illumina, Inc., San Diego, CA, USA), and paired-end (2 × 100 bp) sequencing was conducted by Macrogen Incorporated. The total mRNA sequencing reads were aligned to the mus musculus genome (version GRCm38) utilizing the Spliced Transcripts Alignment to a Reference (STAR) alignment software. The raw read counts and transcripts per kilobase million (TPM) counts were generated through RNA-Seq by Expectation-Maximization (RSEM). RNA-seq analysis in this study was performed using Software R version 4.1.2. Differential expressed genes (DEGs) analysis was executed through R package DESeq2. In the DEGs result, genes for pathway analysis are selected by q-value cut-off (q-value <0.05). For gene ontology (GO) enrichment analysis, the selected genes were subjected to analysis using web based tool (https://genenontology.org). All RNA sequencing data are accessible through GEO:GSE2594 35 in the NCBI Gene Expression Omnibus.
2.6. Measurement of hepatobiliary organoid formation and size
Organoid formation numbers and sizes were quantified by counting the total number of the organoids greater than 100 μm of diameter at 7 days after culture by using Celleste Image Analysis software. Organoid-forming efficiency is normalized by the total number of single cells seeded (5,000).
2.7. Statistical analysis
Data were analyzed as detailed in Figure legends. Significance between two groups was determined using Student's two-tailed unpaired t-test in GraphPad prism.
3. Results
3.1. Liver endothelial cells support formation and growth of adult hepatobiliary organoid
To co-culture hepatobiliary organoids with liver endothelial cells (LiECs), we isolated primary LiECs from an adult GFP transgenic mouse and then characterized them by immunofluorescence staining with the endothelial markers CD31 and VE-cadherin and functionally by tube formation and Dil-Ac-LDL uptake to obtain greater than 98 % LiEC purity (Figs. S1A–C).
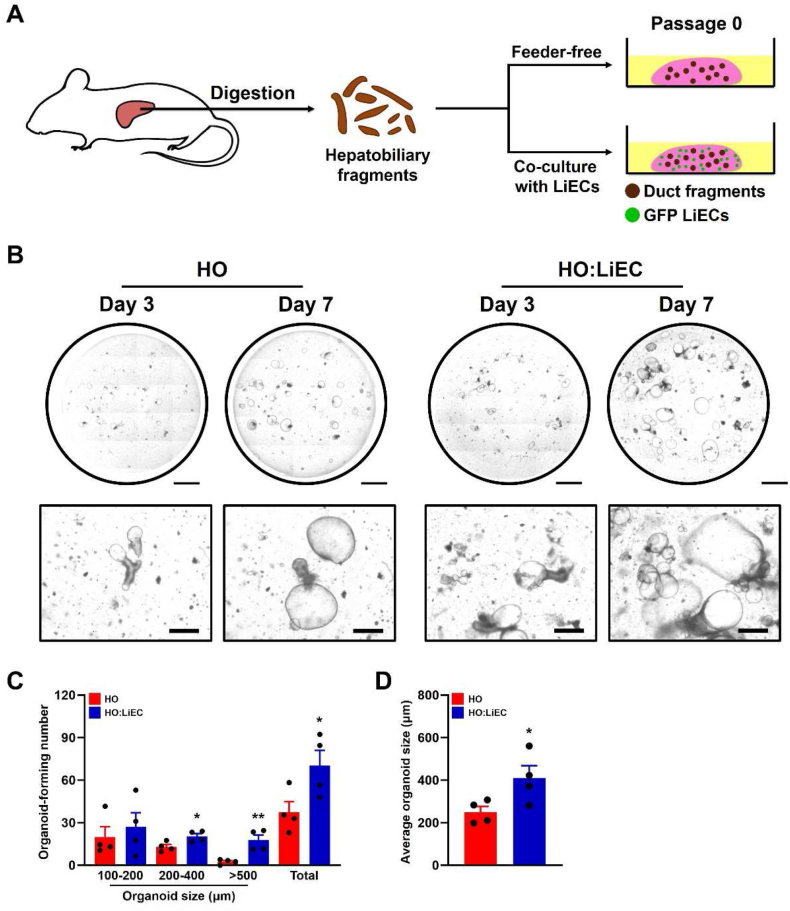
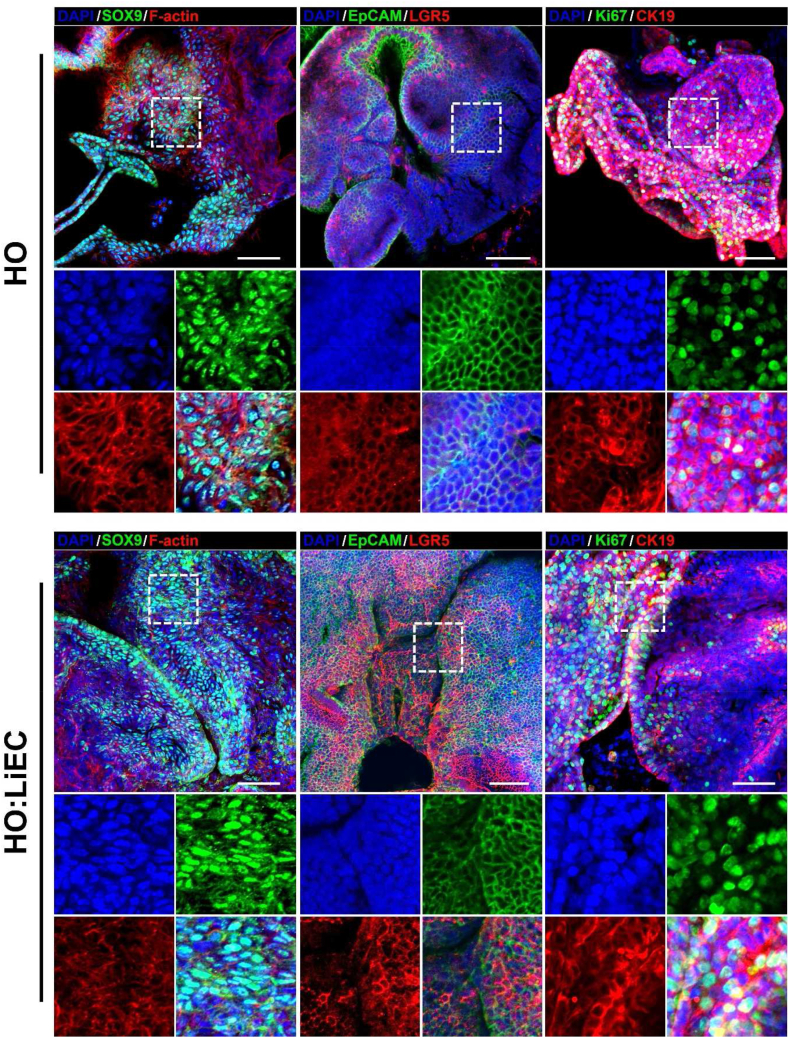
We isolated hepatobiliary duct fragments containing LPCs to establish hepatobiliary organoids, then embedded them alone (HO) or co-cultured with LiECs (HO:LiEC) in Matrigel (Fig. 1A). The organoids were formed at the ends of hepatobiliary duct fragments (Fig. 1B) and we measured the number and size of all organoids at day 7. HO:LiEC demonstrate significantly greater number of organoids and much larger average size (Fig. 1C–D). Characterization of HO and HO:LiEC by immunofluorescence analysis of progenitor cell, epithelial and proliferation markers, demonstrate robust expression of progenitor markers (LGR5, SOX9), biliary epithelial marker (EpCAM, CK19), and proliferation marker (Ki67) in both HOs grown alone or in co-culture with LiECs (Fig. 2). Expression of LGR5 and SOX9 indicate the presence of LPCs in the established organoids. These results also indicate that hepatobiliary organoids can successfully grow in the modified growth medium and that the addition of LiECs enhance both the formation and growth of the organoids.
Fig. 1.
Liver endothelial cells support formation and growth of hepatobiliary organoids. (A) Schematic depicting isolation and organoid culture derived from hepatobiliary ducts. (B) Representative stitched and 40x bright-field images at day 3 and 7 (passage 0) of HO or HO:LiEC. Scale bar, 2 mm (stitch) and 500 μm (40x). (C) Quantitation of the total number of whole organoids formed in HOs alone and HOs + LiECs, categorized by size (100–200 μm, 200–400 μm, >500 μm, and total) and (D) the average organoid size at day 7 (passage 0). Data are means ± SEM obtained from n = 4 independent biological replicates. *p < 0.05; **p < 0.01.
Fig. 2.
Immunofluorescence analysis of HO and HO:LiEC. Z-stack projection of confocal images of intact whole organoids of HO and HO:mLiEC at day 7 (passage 0). Sox9, EpCAM, Ki67 (green), Lgr5, CK19, F-actin (red), and DAPI (blue). White dot squares indicate magnified images. Scale bar, 50 μm.
3.2. Liver endothelial cells maintain hepatobiliary organoids in long-term expansion
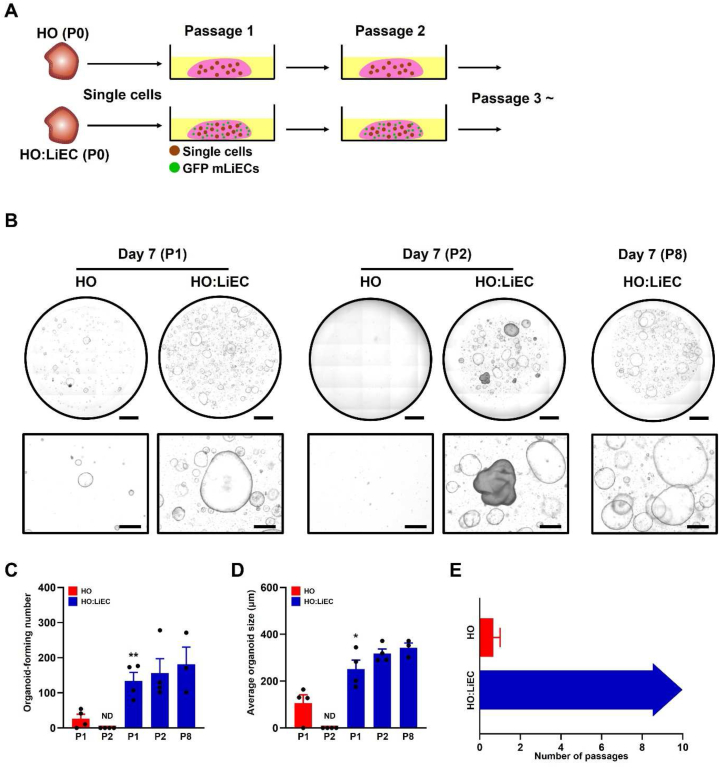
LPCs gradually lose their self-renewal capacity when cultured in a reduced growth medium lacking the defined growth supplements in early passages [7]. To test whether LiECs support the self-renewal and growth of LPCs without the essential growth supplements in long-term expansion, single cells were isolated from HO (P0) or HO:LiEC (P0) and serially passaged (Fig. 3A). Notably, HO showed reduced organoid formation and growth at passage 1, while HO:LiEC exhibited a 5-fold higher efficiency in organoid formation (around 2–3%) and the average size was 2-fold larger compared to HO (Fig. 3B–E). Additionally, we observed that organoid formation of HO cultures alone was significantly reduced at passage 2, whereas HO:LiEC consistently exhibited both organoid formation and expansion in serial passages.
Fig. 3.
LiECs maintain formation and growth of hepatobiliary organoids in long-term expansion. (A) Schematic depicting hepatobiliary organoid passage strategy. 5000 single cells from HO or HO:LiEC (P0) were passaged for long-term expansion. For co-culture, LiECs were harvested between P3 to P6 prior to serial passages. (B) Representative stitched and bright-field images from each group at P1, P2, and P8 (day 7). Scale bar, 2 mm (stitched image) and 500 μm (bright-field image). (C) Quantitation of whole organoid-forming number HO and HO:LiEC and (D) average organoid size at P1, P2, and P8 (day 7). (E) LiECs maintain expansion of hepatobiliary organoids for >8 passages (>2 months). Data are means ± SEM obtained from at least n = 3 independent biological replicates. ND; non-detected. *p < 0.05; **p < 0.01 (compared to HO at P1).
We analyzed expression of progenitor and epithelial cell markers in HO:LiEC after long-term passage and found robust expression of progenitor marker SOX9, biliary epithelial markers EpCAM and CK19 (Fig. S2). Interestingly, there was no LGR5 expression, which is an essential component of the Wnt signaling pathway and a receptor activated by one of the Wnt agonists, R-spondin-1. This suggests that LGR5-independent Wnt signaling pathway or other signaling pathways may be involved in the self-renewal of LPCs in our LiEC co-cultures.
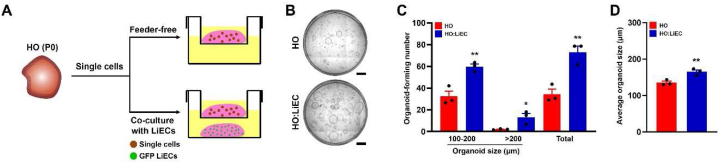
To determine whether LiEC-derived factors was sufficient to support HO maintenance or of hepatocyte-LiEC contact was necessary, we used Transwell culture plates to prevent cell-to-cell contact. We cultured single cells isolated from HO (P0) on top of the Transwell with or without LiEC on the bottom and quantified organoid growth (Fig. 4A). Under these conditions, we consistently observed a significant increase in the number of organoid formation and in the average organoid size of HO:LiEC compared to HO (Fig. 4B–D). Furthermore, we observed significantly increased in the average organoid size of HO:LiEC (mixed co-culture) compared to HO:LiEC using Transwell (Figs. S3A–C), suggesting that the paracrine factors produced by LiECs support self-renewal of LPCs.
Fig. 4.
LiECs support formation and growth of hepatobiliary organoids by paracrine manner. (A) Schematic depicting organoid culture using Transwell from 5000 single cells of HO at P0. (B) Representative stitched image of HO and HO:LiEC at day 7 (passage 1). Scale bar, 1 mm. (C) Quantitation of the total number of whole organoids formed in HO and HO:LiEC, categorized by size (100–200 μm, >200 μm, and total) and (D) the average organoid size at day 7 (passage 1). Data are means ± SEM obtained from n = 3 independent biological replicates. *p < 0.05; **p < 0.01.
3.3. Gene expression analysis of hepatobiliary organoids co-cultured with liver endothelial cells
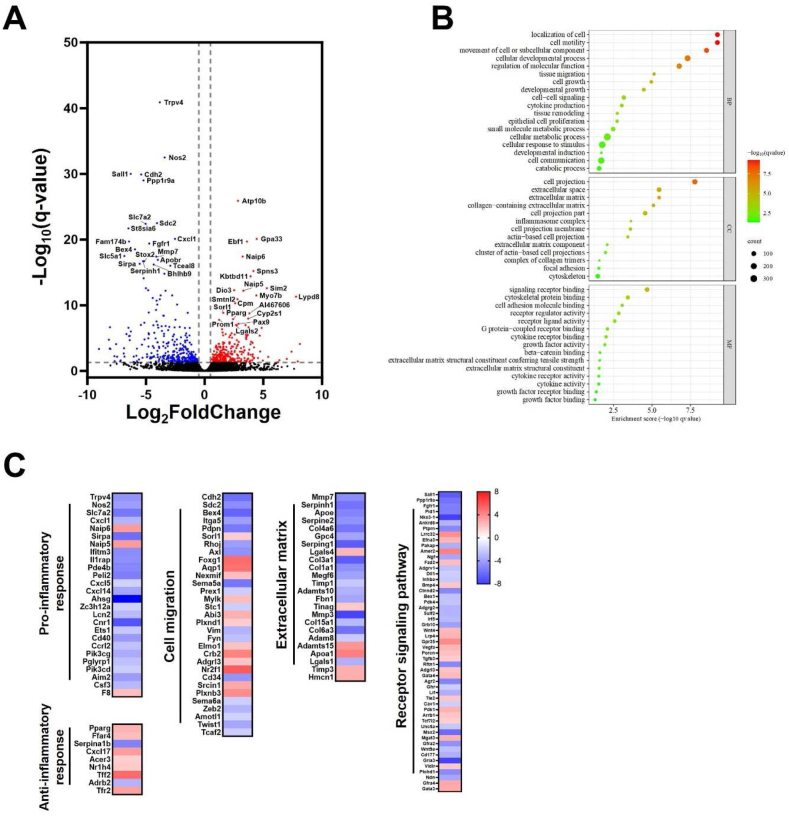
It has been reported that alterations of gene expression profile occurs when stem cells are co-cultured with other types of cells [23,24]. To determine gene expression profile of hepatobiliary organoids co-cultured with LiECs, we isolated total RNA from HO or HO:LiEC in Transwell culture plates. We identified 583 differentially expressed genes (DEGs) using a threshold of log2fold change (FC) > and -log10FDR >1.3. Of these DEGs, 278 genes were up-regulated and 305 genes were down-regulated in HO:LiEC compared to HO (Fig. 5A). Gene ontology (GO) enrichment analysis of DEGs identified the genes strongly associated with cell migration (e.g., cell motility, cell projection, tissue migration), extracellular matrix organization (e.g., tissue remodeling, extracellular space, extracellular matrix), inflammation (e.g., cytokine production, inflammasome complex, cytokine receptor binding), and cellular signaling activity (e.g., signaling receptor binding, receptor regulator activity, receptor ligand activity, growth factor activity) (Fig. 5B). We then analyzed DEGs belonging to selected pathways applying a cutoff of log2(FC) > , in particular we focused on the genes associated with extracellular matrix organization, inflammation, cell migration, receptor signaling pathway (Fig. 5C). Genes associated with inflammation, pro-inflammatory genes were abundantly expressed in HO, such as chemokine-mediated inflammation (Cxcl1, Cxcl5, Cxcl14), while conversely, anti-inflammatory genes (e.g., Pparg, Ffar4, Cxcl17) were enriched in HO:LiEC. Furthermore, HO showed an enrichment of genes involved in cell migration (e.g., Cdh2, Sdc2, Bex4, Itga5) and extracellular matrix organization (e.g., Col4a6, Col3a1, Col1a1). Interestingly, key genes (Cdh2, Axl, Vim, Zeb2, Twist1) associated with epithelial-to-mesenchymal transition (EMT) were down-regulated in HO:LiEC. For receptor signaling pathway, Wnt signaling components (Tcf7l2, Fzd2, Lrp4, Gpr35, Porcn, Tle2) and Pi3K signaling component (PDK1) were up-regulated and genes known as tumor suppressor (Sall1, Nkx3.1, Irf5, Ndn) were down-regulated in HO:LiEC. Moreover, we observed several genes involved in growth factors were up-regulated (Bmp4, Wnt4, Vegfa, Tgfb3) and down-regulated (Ngf, Wnt9a) in HO:LiEC.
Fig. 5.
Differentially expressed genes analysis of HO and HO:LiEC. Transcriptome analysis of HO and HO:LiEC (at least n = 2 independent biological replicates). (A) Volcano plot of DEGs comparing HO and HO:LiEC from RNA-seq analysis. Red (HO:LiEC > HO) and blue (HO:LiEC < HO) dots represent DEGs. Up- and down-regulated top 20 DEGs were indicated. (B) The enrichment bubble plot visualizing selected pathways from GO analysis of DEGs (BP; biological process, CC; cellular component, MF; molecular function). The size of each circle represents the number of genes. Red color indicates lower q-value. (C) Heatmap representing up- and down-regulated DEGs in HO:LiEC for selected signaling pathways of GO analysis involved in pro- and anti-inflammatory response, cell migration, extracellular matrix, and receptor signaling pathway.
4. Discussion
LPCs participate in liver regeneration through self-renewal and differentiation into hepatocytes and biliary epithelial cells when hepatocyte-mediated regeneration is impaired in chronic liver disease [3,4]. However, the regenerative potential of native LPCs is not sufficient to regenerate end-stage liver due to persisted injuries or altered cellular microenvironment, including chronic inflammation and extensive fibrosis. Enhancing stemness or increasing population of LPCs to regenerate liver is alternative treatment strategy for the patients with chronic liver diseases.
LPCs-derived organoid culture models facilitate the massive expansion of LPCs while preserving their genetic stability and the functional characteristics of the original tissue [7,8]. These models are emerging as a promising scientific tool for advancing hepatic biology research and driving therapeutic innovations for chronic liver diseases. Nevertheless, the models face several challenges, encompassing standardized protocols, reproducibility, and the absence of cellular interactions of niche cells.
LPCs are regulated by their surrounding niche, a complex microenvironment composed of diverse cell types, extracellular matrix components, and soluble factors [9,10]. This niche orchestrates dynamic cellular communications that are critical for maintaining the stemness of LPCs and facilitating liver regeneration responses to chronic liver injury. In developing liver organoid culture models, accurately replicating these multifaceted and dynamic niche interactions remains a formidable challenge, yet it is essential for creating physiologically relevant and functional organoids. Here, we first established the long-term expansion of HOs by co-cultured with LiECs and demonstrated that LiECs sustain the formation and proliferation of HOs through a paracrine manner. Further, we analyzed the transcripts, identifying altered gene expression profiles of HOs co-cultured with LiECs compared to HOs alone. Interestingly, LGR5, one of the main components of the Wnt signaling pathway, was no longer expressed in late passages of HOs in our culture model. Furthermore, HOs alone showed low colony formation and proliferation along with up-regulated pro-inflammatory, migration, and deposition of extracellular matrix (ECM) related genes compared to HOs sustained by LiECs.
Following acute liver injury, LGR5-expressing BECs are identified as LPCs and R-spondin-1 and Wnt3a highlighted as crucial Wnt agonists for the establishment and long-term maintenance of liver organoids in vitro [7]. However, the activation of EGFR and VEGFR signaling pathways plays a significant role in LPCs during liver regeneration when hepatocyte ablation [25,26]. Moreover, a recent report indicates that YAP signaling is essential for self-renewal of LPCs after liver injury, while LGR5-mediated Wnt signaling may be dispensable for this process [27]. In our observations, HOs co-cultured with LiECs lack LGR5 expression at late passage, yet exhibit elevated gene expression levels of PDK1 and Wnt signaling components. This suggests that R-spondin-1/LGR5-independent Wnt signaling or alternative signaling pathways could be involved in the self-renewal of LPCs through interactions with LiECs. Interestingly, PDK1 is a major regulator for RTKs/Pi3K-dependent signaling pathways through the activation of AKT and YAP [28,29], implies that our model may more similarly represent the physiological aspects of LPC proliferation during liver regeneration. Future research will focus on elucidating these signaling pathways within HOs co-cultured with LiECs.
Angiogenesis is critical process for tissue regeneration, involving EC proliferation and migration to form new microvasculature [30,31]. ECs play essential roles in nutrients and oxygen delivery and regulating damaged microenvironment to recover via ECM remodeling and interactions with surrounding cells, including AdSCs, through angiocrines [12,32]. Ductular reaction (DR) is a reparative mechanism following liver injury, associated with LPCs proliferation and angiogenesis [[33], [34], [35]]. However, the role of LiECs in regulating of LPCs is poorly understood. Moreover, despite expansion of DR in chronic liver diseases, it paradoxically contributes to disease progression [34,36]. Previous studies have indicated that LPCs during DR exhibit increased gene expression related to inflammation, EMT, and collagen synthesis in chronic liver injury [[37], [38], [39]]. We also observed the downregulation of pro-inflammatory, EMT, and ECM remodeling related genes in HOs co-cultured with LiECs, suggesting that cellular interactions with functional LiECs could mitigate the pathological progression of LPCs and enhance regenerative properties.
In conclusion, our findings expand the role of ECs as a critical niche population for AdSCs and provide new perspective for modifying conventional culture models that elucidate niche cell functions with reducing growth supplements in vitro. Interestingly, liver organoids from non-alcoholic steatohepatitis mice exhibited significantly reduced growth rates and pathological features similar to those observed in vivo according to disease progression stages [40]. Exploring whether HOs from chronic liver diseases could be improved growth or restored pathological characters when co-cultured with LiECs in our culture model presents a promising future research.
Ethics statement
All animal studies were approved by the Institutional Animal Care and Use Committee in Sungkyunkwan University School of Medicine (SUSM) and performed at the Laboratory Animal Research Center (LARC) of SUSM (Permit No. 001004). LARC is a registered research facility with the Association for Assessment and Accreditation of Laboratory Animal Care International and is committed to complying with the guide for the care and use of laboratory animals (National Research Council, USA).
Data availability statement
The data included in this study have not been deposited in any publicly accessible repository. The datasets used and analyzed during this study are available from the corresponding author upon reasonable request.
CRediT authorship contribution statement
Hyun-Soo Roh: Writing – review & editing, Writing – original draft, Visualization, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Da-Eun Kim: Investigation. Gahee Kim: Investigation. Jongsu Kim: Software, Formal analysis. Dengxia Fan: Software, Formal analysis. Hong Sook Kim: Resources, Supervision. Yong-Hee Kim: Resources. Jae-Hee Lee: Resources. Byung Gak Kim: Resources. Min-Ok Ryu: Resources. Hwan Soo Kim: Resources. Kwan-Hyuck Baek: Supervision, Funding acquisition, Resources. Dong Ha Bhang: Supervision, Project administration, Funding acquisition, Resources.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We would like to thank Dr. Sandra Ryeom for helpful advice and editing the manuscript. This research was supported by a grant (20163MFDS120) from Ministry of Food and Drug Safety and also supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2020R1I1A1A0105192213).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e36120.
Contributor Information
Kwan-Hyuck Baek, Email: khbaek@skku.edu.
Dong Ha Bhang, Email: bhang@attislab.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Mohammad R., Jyotsna D., Moustapha K. Concise review: quiescence in adult stem cells: biological significance and relevance to tissue regeneration. Stem Cell. 2015;33:2903–2912. doi: 10.1002/stem.2056. [DOI] [PubMed] [Google Scholar]
- 2.Miyaoka Yuichiro, Ebato Kazuki, Kato Hidenori, Arakawa Satoko, Shimizu Shigeomi, Miyajima Atsushi. Hypertrophy and unconventional cell division of hepatocytes underlie liver regeneration. Curr. Biol. 2012;22:1166–1175. doi: 10.1016/j.cub.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 3.Deng X., Zhang X., Li W., Feng R.X., Li L., Yi G.R., Zhang X.N., Yin C., Yu H.Y., Zhang J.P., Lu B. Chronic liver injury induces conversion of biliary epithelial cells into hepatocytes. Cell Stem Cell. 2018;23:114–122. doi: 10.1016/j.stem.2018.05.022. [DOI] [PubMed] [Google Scholar]
- 4.Raven A., Lu W.-Y., Man T.Y., Ferreira-Gonzalez S., O'Duibhir E., Dwyer B.J., Thomson J.P., Meehan R.R., Bogorad R., Koteliansky V., Kotelevtsev Y., ffrench-Constant C., Boulter L., Forbes S.J. Cholangiocytes act as facultative liver stem cells during impaired hepatocyte regeneration. Nature. 2017;547:350–354. doi: 10.1038/nature23015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banales J.M., Huebert R.C., Karlsen T., Strazzabosco M., LaRusso N.F., Gores G.J. Cholangiocyte pathobiology. Nat. Rev. Gastroenterol. Hepatol. 2019;16:269–281. doi: 10.1038/s41575-019-0125-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clevers H. Modeling development and disease with organoids. Cell. 2016;165:1586–1597. doi: 10.1016/j.cell.2016.05.082. [DOI] [PubMed] [Google Scholar]
- 7.Huch M., Dorrell C., Boj S.F., van Es J.H., Li V.S., van de Wetering M., Sato T., Hamer K., Sasaki N., Finegold M.J., et al. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature. 2013;494:247–250. doi: 10.1038/nature11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huch M., Gehart H., van Boxtel R., Hamer K., Blokzijl F., Verstegen M.M., Ellis E., van Wenum M., Fuchs S.A., de Ligt J., et al. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell. 2015;160:299–312. doi: 10.1016/j.cell.2014.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carpino G., Renzi A., Franchitto A., Cardinale V., Onori P., Reid L., Alvaro D., Gaudio E. Stem/progenitor cell niches involved in hepatic and biliary regeneration. Stem Cell. Int. 2016;2016 doi: 10.1155/2016/3658013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Häussinger D., Kordes C. Space of Disse: a stem cell niche in the liver. Biol. Chem. 2020;401:81–95. doi: 10.1515/hsz-2019-0283. [DOI] [PubMed] [Google Scholar]
- 11.Cordero-Espinoza L., Dowbaj A.M., Kohler T.N., Strauss B., Sarlidou O., Belenguer G., Pacini C., Martins N.P., Dobie R., Wilson-Kanamori J.R., Butler R. Dynamic cell contacts between periportal mesenchyme and ductal epithelium act as a rheostat for liver cell proliferation. Cell Stem Cell. 2021;28:1907–1921. doi: 10.1016/j.stem.2021.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rafii S., Butler J.M., Ding B.S. Angiocrine functions of organ-specific endothelial cells. Nature. 2016;529:316–325. doi: 10.1038/nature17040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Butler J.M., Nolan D.J., Vertes E.L., Varnum-Finney B., Kobayashi H., Hooper A.T., Seandel M., Shido K., White I.A., Kobayashi M., Witte L. Endothelial cells are essential for the self-renewal and repopulation of Notch-dependent hematopoietic stem cells. Cell Stem Cell. 2010;6:251–264. doi: 10.1016/j.stem.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ding L., Saunders T.L., Enikolopov G., Morrison S.J. Endothelial and perivascular cells maintain haematopoietic stem cells. Nature. 2012;481:457–462. doi: 10.1038/nature10783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen Q., Goderie S.K., Jin L., Karanth N., Sun Y., Abramova N., Vincent P., Pumiglia K., Temple S. Endothelial cells stimulate self-renewal and expand neurogenesis of neural stem cells. science. 2004;304:1338–1340. doi: 10.1126/science.1095505. [DOI] [PubMed] [Google Scholar]
- 16.Sato Y., Uchida Y., Hu J., Young-Pearse T.L., Niikura T., Mukouyama Y.S. Soluble APP functions as a vascular niche signal that controls adult neural stem cell number. Development. 2017;144:2730–2736. doi: 10.1242/dev.143370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee J.H., Bhang D.H., Beede A., Huang T.L., Stripp B.R., Bloch K.D., Wagers A.J., Tseng Y.H., Ryeom S., Kim C.F. Lung stem cell differentiation in mice directed by endothelial cells via a BMP4-NFATc1-thrombospondin-1 axis. Cell. 2014;156:440–455. doi: 10.1016/j.cell.2013.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhang D.H., Kim B.J., Kim B.G., Schadler K., Baek K.H., Kim Y.H., Hsiao W., Ding B.S., Rafii S., Weiss M.J., Chou S.T. Testicular endothelial cells are a critical population in the germline stem cell niche. Nat. Commun. 2018;9:1–16. doi: 10.1038/s41467-018-06881-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeLeve L.D. Liver sinusoidal endothelial cells and liver regeneration. J. Clin. Investig. 2013;123(5):1861–1866. doi: 10.1172/JCI66025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding B.-S., Nolan D.J., Butler J.M., James D., Babazadeh A.O., Rosenwaks Z., Mittal V., Kobayashi H., Shido K., Lyden D., Sato T.N., Rabbany S.Y., Rafii S. Inductive angiocrine signals from sinusoidal endothelium are required for liver regeneration. Nature. 2010;468:310–315. doi: 10.1038/nature09493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang J., Li Y., Lu J., Zhao H., Frenkel S., Zhong Z., Le T., Zou J., Yang Y., Christiani D. Angiocrine Wnt signaling controls liver growth and metabolic maturation in mice. Hepatology. 2023;68(2):707–722. doi: 10.1002/hep.29613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koch P.-S., Olsavszky V., Ulbrich F., Sticht C., Demory A., Leibing T., Henzler T., Meyer M., Zierow J., Schneider S., Breitkopf-Heinlein K., Gaitantzi H., Spencer-Dene B., Arnold B., Klapproth K., Schledzewski K., Goerdt S., Géraud C. Angiocrine Bmp2 signaling in murine liver controls normal iron homeostasis. Blood. 2017;129(4):415–419. doi: 10.1182/blood-2016-07-729822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cai Y., Wang W., Liang H., Sun L., Teitelbaum D.H., Yang H. Adipose tissue-derived stem cells in intestinal repair: effects on muscle and nerve regeneration. Nutr. Metabol. 2014;11(1):12. [Google Scholar]
- 24.Gracz A.D., Williamson I.A., Roche K.C., Johnston M.J., Wang F., Wang Y., Attayek P.J., Balowski J., Liu X.F., Laurenza R.J., Gaynor L.T., Sims C.E., Galanko J.A., Li L., Allbritton N.L., Magness S.T. A high-throughput platform for stem cell niche co-cultures and downstream gene expression analysis. Nat. Cell Biol. 2015;17:340–349. doi: 10.1038/ncb3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cai P., Ni R., Lv M., Liu H., Zhao J., He J., Luo L. VEGF signaling governs the initiation of biliary-mediated liver regeneration through the PI3K-mTORC1 axis. Cell Rep. 2023;42 doi: 10.1016/j.celrep.2023.113028. [DOI] [PubMed] [Google Scholar]
- 26.Oderberg I.M., Goessling W. Biliary epithelial cells are facultative liver stem cells during liver regeneration in adult zebrafish. JCI Insight. 2023;8(1) doi: 10.1172/jci.insight.163929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Planas-Paz L., Sun T., Pikiolek M., Cochran N.R., Bergling S., Orsini V., Yang Z., Sigoillot F., Jetzer J., Syed M., Neri M., et al. YAP, but not RSPO-LGR4/5, signaling in biliary epithelial cells promotes a ductular reaction in response to liver injury. Cell Stem Cell. 2019;25(1):39–53. doi: 10.1016/j.stem.2019.04.005. [DOI] [PubMed] [Google Scholar]
- 28.Mora A., Komander D., van Aalten M.F.D., Alessi R.D. PDK1, the master regulator of AGC kinase signal transduction. Semin. Cell Dev. Biol. 2004;15:161–170. doi: 10.1016/j.semcdb.2003.12.022. [DOI] [PubMed] [Google Scholar]
- 29.Xia H., Dai X., Yu H., Zhou S., Fan Z., Wei G., Tang Q., Gong Q., Bi F. EGFR-PI3K-PDK1 pathway regulates YAP signaling in hepatocellular carcinoma: the mechanism and its implications in targeted therapy. Cell Death Dis. 2018;9:269. doi: 10.1038/s41419-018-0302-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tonnesen M.G., Feng X., Clark R.A.F. Angiogenesis in wound healing. J. Invest. Dermatol. Symp. Proc. 2000;5:40–46. doi: 10.1046/j.1087-0024.2000.00014.x. [DOI] [PubMed] [Google Scholar]
- 31.Senger D.R., Davis G.E. Angiogenesis. Cold Spring Harbor Perspect. Biol. 2011;3(8):a005090. doi: 10.1101/cshperspect.a005090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Witjas F.M.R., van den Berg B.M., van den Berg C.W., Engelse M.A., Rabelink T.J. The endothelial cell extracellular matrix regulates tissue homeostasis and repair. Stem Cells Translational Medicine. 2019;8:375–382. doi: 10.1002/sctm.18-0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gouw A.S.H., van den Heuvel M.C., Boot M., Slooff M.J.H., Poppema S., de Jong K.P. Dynamics of the vascular profile of the finer branches of the biliary tree in normal and diseased human livers. J. Hepatol. 2006;45(3):393–400. doi: 10.1016/j.jhep.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 34.Sato K., Marzioni M., Meng F., Francis H., Glaser S., Alpini G. Ductular reaction in liver diseases: pathological mechanisms and translational significances. Hepatology. 2019;69(1):420–430. doi: 10.1002/hep.30150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coll M., Ariño S., Martínez-Sánchez C., Garcia-Pras E., Gallego J., Moles A., Aguilar-Bravo B., Blaya D., Vallverdú J., Rubio-Tomás T., et al. Ductular reaction promotes intrahepatic angiogenesis through Slit2-Roundabout 1 signaling. Hepatology. 2022;75:353–368. doi: 10.1002/hep.32140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams M.J., Clouston A.D., Forbes S.J. Links between hepatic fibrosis, ductular reaction, and progenitor cell expansion. Gastroenterology. 2014;146(2):349–356. doi: 10.1053/j.gastro.2013.11.034. [DOI] [PubMed] [Google Scholar]
- 37.Aguilar-Bravo B., Rodrigo-Torres D., Ariño S., Coll M., Pose E., Blaya D., Graupera I., Perea L., Vallverdú J., Rubio-Tomás T., et al. Ductular reaction cells display an inflammatory profile and recruit neutrophils in alcoholic hepatitis. Hepatology. 2019;69(5):2180–2195. doi: 10.1002/hep.30472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fabris L., Brivio S., Cadamuro M., Strazzabosco M. Revisiting epithelial-to-mesenchymal transition in liver fibrosis: clues for a better understanding of the (reactive) biliary epithelial phenotype. Stem Cell. Int. 2016;2016 doi: 10.1155/2016/2953727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang W.-J., Qiu B.-J., Qi X.-S., Chen C.-Y., Liu W.-M., Zhou S.-A., Ding M., Lu F.-F., Zhao J., Tang D., et al. CD24+LCN2+ liver progenitor cells in ductular reaction contributed to macrophage inflammatory responses in chronic liver injury. Cell Biosci. 2023;13:184. doi: 10.1186/s13578-023-01123-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elbadawy M., Yamanaka M., Goto Y., Hayashi K., Tsunedomi R., Hazama S., Nagano H., Yoshida T., Shibutani M., Ichikawa R., et al. Efficacy of primary liver organoid culture from different stages of non-alcoholic steatohepatitis (NASH) mouse model. Biomaterials. 2020;237 doi: 10.1016/j.biomaterials.2020.119823. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data included in this study have not been deposited in any publicly accessible repository. The datasets used and analyzed during this study are available from the corresponding author upon reasonable request.