Abstract
The most current in vitro genetic methods, including gene preservation, gene editing and developmental modelling, require a significant number of healthy cells. In poultry species, primordial germ cells (PGCs) are great candidates for all the above-mentioned purposes, given their easy culturing and well-established freezing method for chicken. However, the constant monitoring of cultures can be financially challenging and consumes large amounts of solutions and accessories. This study aimed to introduce the Fluorescent Ubiquitination-based Cell Cycle Indicator (FUCCI) complex into the chicken PGCs. FUCCI is a powerful transgenic tool based on the periodic protein expression changes during the cell cycle. It includes chromatin licensing and DNA replication factor 1 attached monomeric Kusabira-Orange and Geminin-attached monomeric Azami-Green fluorescent proteins, that cause the cells to express a red signal in the G1 phase and a green signal in S and G2 phases. Modification of the chicken PGCs was done via electroporation and deemed to be successful according to confocal microscopy, DNA sequencing and timelapse video analysis. Stable clone cell lines were established, cryopreserved, and injected into recipient embryos to prove the integrational competency. The cell health monitoring was tested with medium change experiments, that proved the intended reactions of the FUCCI transgene. These results established the future for FUCCI experiments in chicken, including heat treatment and toxin treatment.
Key words: chicken, PGC, FUCCI, transgenesis, cell cycle monitoring
INTRODUCTION
Gene preservation for avian species has its difficulties due to the unique nature of the birds’ reproductive system. While sperm freezing is a well-established technique in mammalian species (Ugur et al., 2019; Yánez-Ortiz et al., 2022), it is not as effective in avian species due to the male birds’ homogametic nature. Since the male birds have the ZZ sexual chromosome pair, sperm freezing only preserves the genetic information coded on the Z-chromosome, while the genes on the W-chromosome are not conserved. The preservation of the egg or embryo is even more problematic due to the relatively large size, water and protein content of the egg. The freezing of embryonic gonadal tissues (Liptoi et al., 2013; Tiambo et al., 2021; Hu et al., 2022) is lethal to the donor animal, which makes the method unsuitable for endangered species.
A promising tool for avian gene preservation (Naito et al., 1994; Tajima et al., 1998; Setioko et al., 2007; Liu et al., 2012, 2013; Silversides et al., 2013; Nakamura, 2016; Nandi et al., 2016; Lázár et al., 2021; Chaipipat et al., 2022) is the freezing of primordial germ cells (PGCs). PGCs are the precursors of germ cells (Kim and Han, 2018) and they have the unipotent ability to develop into eggs and sperms. They can differentiate both in vivo and in vitro (Matsui et al., 1992; Petitte et al., 2004; Xiong et al., 2015) which makes them a valuable model for developmental biology. They are located in the ventral side of the epiblast layer when the primitive streak forms (Eyal-Giladi et al., 1981). After 18 hours of incubation, they migrate to the germinal crescent (Kochav, et al., 1980; Nakamura et al., 2007), where they start the proliferation, and by the 40th hour, they enter the circulation (circulating PGCs, cPGC) and migrate towards the developing gonads (Hamburger–Hamilton stage 12–17), which they colonize afterwards (gonadal PGCs, gPGC)(Ginsburg and Eyal-Giladi, 1986). cPGCs are the easiest to isolate and the majority of donor embryos survive the procedure.
Primordial germ cells have a wide spectrum of potential usage in various fields of research. In addition to gene preservation, another highly important aspect of the PGCs is their part in avian genome editing and chimaera production (Kino et al., 1997; Ono et al., 1998; Woodcock et al., 2017, 2019; Divya et al., 2021). This advantage is based on their integration capability into recipient animals and their ability to be cultured in vitro. They have been also used in embryonic developmental research (Trefil et al., 2017; Bednarczyk et al., 2021). However, there are only a few species with an already established culturing medium and developing media for new species proved to be challenging.
Most of the techniques, for which PGCs are used, require a substantial quantity of viable cells. To produce and monitor these cultures, there are important methods, such as cell line characterization (Macdonald et al., 2010; Whyte et al., 2015; Lázár et al., 2018; Kong et al., 2018; Ezaki et al., 2020), development of cell isolation and in vitro culture (Zhao and Kuwana, 2003), and cell health control (Lázár et al., 2022). Usually, markers such as annexin, ethidium homodimer or propidium iodide are used to evaluate the cell health, however, these methods cause the death of the cells and can only be used in vitro.
Fluorescent Ubiquitination-based Cell Cycle Indicator (FUCCI) is a set of fluorescent probes. It is based on the observation that during the cell cycle, different proteins are expressed in different stages (Blow and Dutta, 2005). This periodic change is mostly regulated by phosphorylation and ubiquitylation (Nakayama and Nakayama, 2006). FUCCI uses the chromatin licensing and DNA replication factor 1 (Cdt1) (Nishitani et al., 2000) and Geminin (Nishitani et al., 2004) replication licensing factors since they are only present at specific stages of the cell cycle. Cdt1 is active during G1, while Geminin is present in the S, G2, and M phases. To visualize the phase change, a Cdt1 protein degron is attached to the monomeric Kusabira-Orange 2 (mKO2) fluorescent protein (Karasawa et al., 2004), whilst a Geminin protein degron is combined with the monomeric Azami-Green 1 (mAG1) fluorescent protein (Karasawa et al., 2003). This way the cell's color changes between red and green fluorescence, according to their present cell cycle phase (Zielke and Edgar, 2015).
The FUCCI transgene complex has already been used for several experiments. It can be used to track cells inside living animals (Pfeiffer et al., 2018; Tomura et al., 2021), to visualize the cell proliferation (Abdulhasan et al., 2022; Koh et al., 2017; Sakaue-Sawano et al., 2008), or even to investigate the nature of hazardous cell malfunctions, such as cancer (Prasedya et al., 2016; Yano and Hoffman, 2018). An interesting question would also be the effect of medium change on the cells, because this is one of the most basic method in cell culturing. A delayed medium change can cause problems in the culture, such as apoptosis or even necrosis, so the research of this aspect is important.
However, FUCCI has only been available to a few species yet, including zebrafish (Sugiyama et al., 2009), Ciona intestinalis (Ogura et al., 2011), mouse (Abe et al., 2013), human (Bao et al., 2013), Drosophila (Zielke et al., 2014), and axolotl (Duerr et al., 2022). This study aimed to introduce this powerful tool to another important vertebrate model species, the chicken.
MATERIALS AND METHODS
Ethical Statement
Animals were kept according to general animal welfare prescriptions of the Hungarian Animal Protection Law (1998; XXVIII). All experimental techniques described here were approved by the Institutional Ethics Review Board of the Institute for Farm Animal Gene Conservation (no. 7/2011).
Maintenance of Domestic Fowl Experimental Stocks
The White Hungarian chicken (Gallus gallus domesticus) breed was used in this experiment. The birds were kept at the National Centre for Biodiversity and Gene Conservation in Gödöllő, Hungary, in barns with large outdoor areas. The stocking density was 5–6 animals/m2, and the sex ratio was 7 hens with 1 cockerel. There were nest boxes (5 hens/nest) for the collection of eggs. Breeding flocks were fed with laying mash in addition to limestone grit. The eggs were collected twice a day and stored in a cold room. The eggs were transported to the Institute of Genetics and Biotechnology of the Hungarian University of Agriculture and Life Sciences, where the experiments took place.
Isolation, Establishment, and Maintenance of PGC Lines
The eggs were incubated in a MIDI F500S hatchery machine (PL Machine Ltd. Tárnok, Hungary) with two 45° rotations per hour, with 37.8°C temperature and 70% relative humidity. At the age of 2.5 d (Hamburger&Hamilton stage 13–17) (Hamburger and Hamilton, 1951) the surface of the eggs was sterilized with 70% alcohol and opened into a petri dish. The blood was harvested from the embryo's dorsal aorta using a microcapillary connected to a mouth pipette. Approximately 1 µL blood was taken from every embryo. Small tissue samples were collected from the embryos for sex determination.
The blood samples were transferred into the culturing medium on a 48-well plate. The composition of the medium was the following: calcium-free DMEM (Gibco, Billings, MT, 21068-028), tissue culture-grade water (Gibco, Billings, MT, A12873-01), pyruvate (Gibco, Billings, MT, 11360039), MEM vitamin solution (Gibco, Billings, MT, 11120052), MEM amino acids (Sigma, St. Louis, MO, M5550), a B27 supplement (Gibco, Billings, MT, 17504044), Glutamax (Gibco, Billings, MT, 35050038), non-essential amino acids (Gibco, Billings, MT, 11140035), nucleosides (EmbryoMax, Munich, Germany, ES-008-D), B-mercaptoethanol (Gibco, Billings, MT, 31350010), CaCl2 (Sigma, St. Louis, MO, C4901-100G), ovalbumin (Sigma, St. Louis, MO, A5503), Na heparin (Sigma, St. Louis, MO, H3149-25KU), a penicillin–streptomycin mixture (Gibco, Billings, MT, 15070-063), chicken serum (Sigma, St. Louis, MO, C5405), human activin (Invitrogen, Waltham, MA, PHC9564), bFGF2 (Gibco, Billings, MT, 13256-029), and ovotransferrin (Sigma, St. Louis, MO, C7786)(Whyte et al., 2015). The medium was changed on every second day. The cultures were kept in a Sanyo MCO-19AIC (UV) CO2 Incubator (Sanyo, Osaka, Japan, 10040162) at 38°C temperature with 5% CO2 concentration.
PGC Line Characterization
The High Pure PCR Template Preparation Kit (Roche Diagnostics, Indianapolis, IN, 11796828001) was used for the DNA isolation, following the manufacturer's instruction. The CHD1 primer set (10 mM) was used to determine the sex of each PGC line (Griffiths et al., 1996; Lee et al., 2010). The isolated DNA was diluted to 25 ng/µL concentration and was added to the MyTaq Red Mix (Bioline Reagents Ltd. London, UK, BIO-25044). The PCR products were then separated in agarose gel stained with ethidium bromide. Afterwards, the bands were illuminated by UV light and photographed. Based on the results, the 1111 ZW (female) and the 1116 ZZ (male) lines were chosen for further experiments.
Cell pellets were used for the RNA isolation. 125 µL ethanol was added to the samples and after that, the isolation was carried out based on the RNAquarious-Micro Total RNA Isolation Kit (Thermo Fischer Scientific, Waltham, MA, AM1931) protocol. The concentration of the isolated RNA was measured with a NanoDrop One Spectrophotometer (Thermo Scientific). The samples were then diluted to a 25 ng/ µL concentration. 375 ng RNA was used to reverse transcription, prepared according to the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystem, Waltham, MA, 4368814).
The q-RT-PCR mixture for one sample consisted of 5.75 µL nuclease-free water, 7.5 µL Power SYBRGreen PCR Master Mix (Applied Biosystems, Waltham, MA, 4368575), 0.75 µL from both forward and reverse primer, and 0.5 µL cDNAsolution. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Integrated DNA Technologies, Coralville, IA, NCBI: NM_204305.1, FW: GACGTGCAGCAGGAACACTA, RV: CTTGGACTTTGCCAGAGAGG) primer was used as a housekeeping reference gene, while the chicken vasa homolog (CVH; Integrated DNA Technologies, Coralville, IA, NCBI: NM_204708.1, FW: GAACCTACCATCCACCAGCA, RV: ATGCTACCGAAGTTGCCACA) and Deleted in Azoospermia Like gene primer (DAZL; Integrated DNA Technologies, Coralville, IA, NCBI: NM_204218.1, FW: TTGTCTTGAAGGCCTCGTTT, RV: ATCCTTGGCAGGTTGTTGAC) ensured the germ cell specificity (Tsunekawa et al., 2000; Smorag et al., 2014; Lejong et al., 2020). A Mastercycler Realplex Real-Time PCR System (Eppendorf, Hamburg, Germany) was used for the experiments. The q-RT-PCR cycle contained the following steps: 10 min 95 °C; 40x (15 s 95 °C, 40 s 60 °C, and 20 s 68 °C); 15 s 95 °C, 15 s 48 °C, and 15 s 95 °C with 10 min heating before the last state. The reference dye was ROX.
Freezing and Thawing of PGC Lines
The PGC lines were homogenous and stable after four weeks of culturing. The optimal cell number (80,000 cell/300 µL medium) and purity were checked by the NanoEntek Arthur Novel Fluorescence Cell Counter (NanoEntek, Seoul, Korea) based on the shape and size of the cells. The freezing medium contained FBS (Gibco, Billings, MT, 10108-165) with 10% DMSO (Sigma, St. Louis, MO, D2650) (Kong et al., 2018).
The cells were collected, centrifuged and resuspended in 250 µL 2:1 DMEM-water mixture. 250 µL 2x freezing medium was added to the cell suspension. Then the samples were put into the Mr. Frosty Freezing Container (Thermo Fischer) to provide the controlled freezing rate. The container was put into the -80°C freezer immediately. In case of long-term storage, the next day the samples were moved to a -150°C freezer. The cell lines used in this experiment were part of the institute's gene bank.
The samples were thawed on 38°C for 90 s and instantly diluted with 900 µL culture medium to prevent the negative effect of the DMSO on room temperature. The lines were centrifuged on 100 G to eliminate the DMSO and resuspended in 300 µL culture medium before adding them to the culturing plates.
FUCCI Transgenesis in Chicken PGCs
The FUCCI transgene, the hyperactive PiggyBac transposon and the hyPBase transposase plasmids were kindly provided by András Nagy from the Lunenfeld-Tanenbaum Research Institute, Toronto, Canada. The plasmids were transformed into E. coli originated competent cells, which were cultured to gain independent colonies. The colonies were treated with ampicillin to separate the ampicillin resistant transfected colonies. From the colonies, liquid bacterium cultures were established and cultured to reach the required cell mass. These cell suspensions were used for the plasmid isolation done with the Qiagen EndoFree Plasmid Maxi Kit (Qiagen, Hilden, Germany, 169026122). The isolated plasmids were diluted with OptiMEM (Gibco, Billings, MT, 11058-021) to 1200 ng/µL.
The electroporation was carried out with the Neon Transfection System (Invitrogen, Waltham, MA, MPK5000) with a 1:1 FUCCI - hyPBase ratio. 100 µL cell suspension containing approximately 1 million cells was electroporated from each cell line. The electroporation was carried out on 1,300 V, for 10 ms, with 4 repetitions, as described by Altgilbers (Altgilbers et al., 2021). After the electroporation, the suspension was placed on a 12-well culture plate with 1 mL of culture medium. Absolute controls, OptiMEM controls, and electroporated controls were used to compare the effects of OptiMEM treatment and electroporation on the PGCs. On the next day, the green/red fluorescence was verified by the Leica DFC 7000T Stereo Microscope (Leica, Wetzlar, Germany) analysis.
Establishing Transgenic PGC Lines
The transgenic cells were cultured for 2 wk. Samples from the cultures were diluted to 1:18 ratio and individual cells were isolated from these diluted cultures. The isolation was done with microcapillary (approximately 40 µm diameter), under a stereo microscope to keep track of the fluorescent cells. The isolated cells were transferred into a 50 µL medium drop and were checked under stereo microscope to ensure its individuality and fluorescence, before being transferred onto a 96-welled plate in 200 µL culture medium. After 3 wk, all the remaining six lines were cryopreserved for further use. For the experiments, the FCF5 and FCM5 clone lines were chosen due to their high proliferation rate and marker expression.
Immunostaining and Confocal Analysis
The cells were placed on glass slides in 10 µL PBS (Gibco, Billings, MT, 14190-144) containing 0.1% BSA (Sigma, St. Louis, MO, A3311) and were dried onto the surface. 4% PFA (Fluka, Buchs, Switzerland, 30525-89-4) was used to fix the samples and then blocking solution (PBS with 0.1% BSA, 0.1% Triton-X-100 (Fluka, Buchs, Switzerland, 93426), and 2.5% donkey serum) was added to make the cell membrane permeable for nucleus and cytoplasm staining and to prevent the unspecific binding. Stem cell-specific anti-SSEA-1 (Millipore, Munich, Germany, MC480) and primordial germ cell-specific anti-CVH (kindly provided by Bertrand Pain from the Stem Cell and Brain Research Institute (SBRI), Lyon, France) primary antibodies were used for the immunostaining. The anti-SSEA-1 was paired with Anti-Mouse-IgM-rD549 (Jackson ImmunoResearch, West Grove, PA, 715-505-140) secondary antibody, while green Alexa Fluor 488 Anti-Rabbit-IgG (H+L) (Life Technologies/Molecular Probes, Carlsbad, CA, A-21207) was attached to the anti-CVH. Far-red TO-PRO™-3 iodide (642-661) (Invitrogen, Waltham, MA, T3605) was used as a nuclear stain. The samples were then covered with ProLong Diamond Antifade Mountant with DAPI (Invitrogen, Waltham, MA, P36962) and a cover glass.
Samples from the clone cultures were also placed in ProLong Diamond Antifade Mountant with DAPI under a cover glass for immediate analysis. They were examined by the Leica TCS SP8 Confocal Microscope (Leica, Wetzlar, Germany).
When examining the FUCCI cultures, only TO-PRO™-3 nucleus stain was used. Since the maximal emission is at 505 nm for the mAG1 and 565 nm for the mKO2, the TO-PRO™-3 (642-661) could be detected in parallel. However, the TO-PRO™-3 expression was labelled blue on the pictures to help the visual distinction from the mKO2’s signal.
Injection of FUCCI Transgenic PG Cells
The cultures were collected and transferred to 2:1 DMEM - H2O mixture. Their purity and the cell number were measured by the NanoEntek Arthur Novel Fluorescence Cell Counter and the dilution was done accordingly. The concentration for the injection was 5000 cells/µL. White Hungarian chicken eggs were incubated for 60 h before the experiment. The incubation was carried out as previously described. A small hole was made on the eggs’ air chamber and then a 1 cm diameter window was created above the embryo. One µL cell suspension was injected directly into the embryo's heart using a microcapillary and mouth pipette. The eggs were closed with sterilized parafilm and were put back into the incubator.
On the 6th day of the incubation, the eggs were opened and the gonads were dissected from the embryos. The gonads were examined under the Leica DFC 7000T Stereo Microscope to verify the presence of the FUCCI cells based on their fluorescence. Tissue samples from the embryos were collected for sex determination.
Chromosomal Analysis
Methanol-acetic acid fixation and air-drying technique (Alfi et al., 1973) were used to prepare the metaphase chromosomes. One drop of KaryoMAX™ colcemid solution (10 µg/mL, Gibco, Billings, MT, 15212012) was added to every culture with high cell number. The cells were incubated for 2 h and the cell suspension was collected into centrifuge tubes. After a 7-minute centrifugation (300 g) the supernatant was removed and the cells were resuspended in cooled 0.56% KCl solution as a hypotonic treatment. After 10 minutes at room temperature (RT) the cells were fixed in three steps with a 3:1 methanol - acetic acid mixture. The cell suspension was dropped to moist slides, dried at RT and stained with a 5% Giemsa – phosphate buffer mixture (Anand et al., 2018).
DNA Sequencing and Alignment
Genomic DNA was isolated from cell pellets with the Roche High Pure PCR Template Preparation Kit and sequenced on an Illumina NovaSeq (Illumina, San Diego, CA) instrument, paired end, with a read length of 150 nt.
Sequencing quality was controlled using FastQC, before alignment (Andrews, 2010). Reads were then aligned to the whole genome (bGalGal1.mat.broiler.GRCg7b, Ensembl release 110)(Martin et al., 2023), complemented with the fasta sequence of the FUCCI transgene system. The PB_CAG_FUCCI sequence was appended to the top level genomic fasta as an extra contig. The complemented genome was then indexed and the reads were aligned using bwa-mem2 (Vasimuddin et al., 2019). The alignment parameters were set to default.
First, aligned reads, that were at least partially aligned to the FUCCI sequence, were selected, after which the whole set of aligned reads was filtered based on the selected read IDs using Samtools 1.16 (Danecek et al., 2021). Reads that were chimerically aligned to the FUCCI sequence and a genomic scaffold or read pairs of which one read was aligned to the FUCCI sequence and the other to a genomic scaffold were used to determine the exact location of the transgene integrations, using coverage and clipping information. In the cases we judged as true positive hits, multiple reads were overlapping the integration site. In these cases, coverage data and read alignment positions were extracted from the alignment files to identify the exact coordinates.
Affected genes were determined by intersecting the integration coordinates with the relevant annotation file (Ensembl release 110). Position of the integration sites and relevant reads were visually controlled using IGV as well (Robinson et al., 2011).
Environmental Effect Test
To check the FUCCI cells’ reaction to environmental effects, a medium withdrawal test was carried out (the cells did not get fresh medium for 72h) on a 96 well plate, where each occupied well contained around 2500 cells. The cell count was identified via Arthur Fluorescent Cell Counter and was adjusted with dilution. A ImageXpress Micro XLS Imaging System with a built-in incubator (Molecular Devices, San José, CA) was used for serial cell number monitoring (kindly provided by László Homolya from the Molecular Cell Biology Research Group, Institute of Enzymology, Research Centre of Natural Sciences). For each sample, the cell number was checked in 6 parallel wells, with 16 fields of view/well. The measurement lasted 72h with a cell count in every 4h.
The green/red ratio during the 3-day-long lack of medium change was monitored with the Leica TCS SP8 Confocal Microscope. The cells were incubated and after 24h and 72h droplets were put onto microscope slides, covered with cover glass and examined under confocal microscope. Each time, 5 images from random positions of the sample slides were taken with a 10x objective. The cell counts of the different expressions were calculated using these images.
Creating Timelapse Videos of the Cell Cycle
For the imaging of the cell cycle, the ImageXpress Pico Cell Imaging System (Molecular Devices, San José, CA) was used (kindly provided by Ferenc Uher from the National Institute of Hematology and Infectology). The cells were placed on a 96-well plate with 2500 cells/well concentration and pictures were taken of them for 29 hours in every 10 minutes. The timelapse video was created with the Fiji software (Schindelin et al., 2012).
Statistical Analysis
The ratio of the target gene expression to the internal control gene expression was calculated for each sample. GenEx 7.0 program (Multiday, SE) using the formula 2−∆∆Ct, where ∆Ct = Ct target gene—Ct internal control and ∆∆Ct = ∆Ct test sample—∆Ct control sample. Statistical differences between groups were analyzed via t-test using GenEx 7.0. The data are presented as mean ± SD and p-values of less than 0.05 were considered significant. Levels of significance were applied as follows: p < 0.05 *, p < 0.01 **, p < 0.001 ***.
RESULTS
Characteristics of the Original Cell Lines
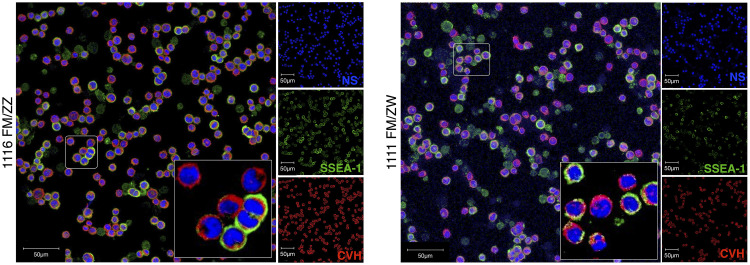
The 1111 ZW and 1116 ZZ cell lines were isolated from White Hungarian chicken embryos and cultured until homogenous, stable cell cultures were produced. The sex was determined based on the CHD1 primers (Lee et al., 2010). RNA was isolated from the cells and qPCR was carried out using CVH and DAZL as germ-cell specific markers. Both cell lines expressed the previous markers, proving that the cells were indeed PGCs. The immunostaining gave further proof of this, since both the SSEA1 stem cell-specific marker, and the CVH germ cell-specific marker expressed in the 1111 and 1116 cells (Figure 1.).
Figure 1.
Immunohistochemical staining on the original cell lines. Both the 1116 male and 1111 female lines were tested with the anti-SSEA-1 stem cell specific, and the anti-CVH germ cell specific antibodies. The SSEA-1 was labeled red, while the CVH was marked with green fluorescence. The far-red TO-PRO-3 nuclear stain was recolored to blue.
Cell Cycle Changes During Cell Culture in the Transgenic Lines
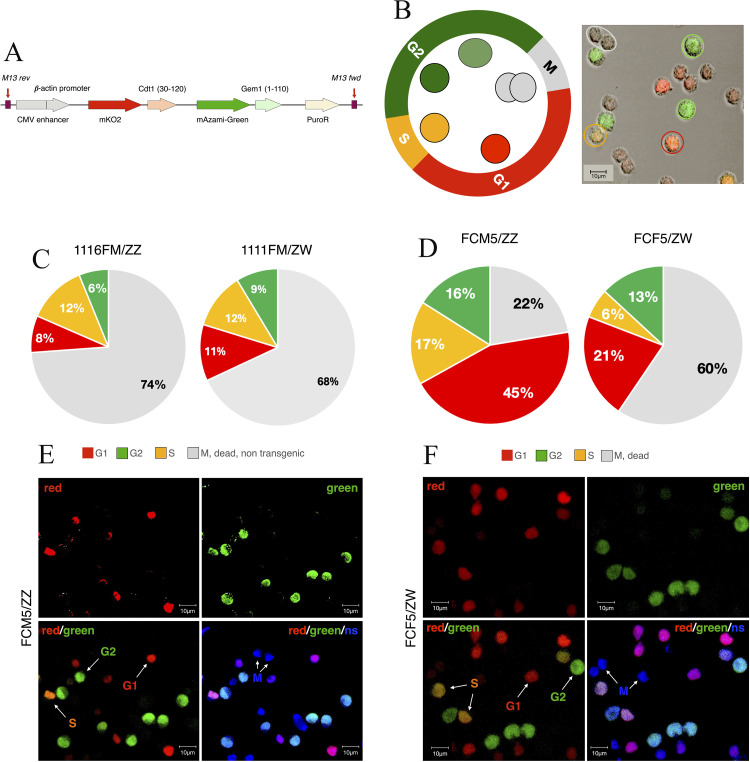
After the electroporation, the lines expressed both mKO2 and mAG fluorescent markers (Figure 2A and 2B.). The expression was analyzed with the NanoEntek Arthur Novel Fluorescence Cell Counter and confocal imaging (Figure 2C.).
Figure 2.
The FUCCI system. (A) The transgene contains the Cdt1-linked mKO2 and the Geminin-linked mAG markers. (B) This way the cells express red fluorescence in the G1 phase, changes to orange in S and to green in G2. At the beginning of mitosis (M) the fluorescence fades and the cell divides. (C) After the electroporation, the fluorescent cell ratio was 26% in the male, and 32% in the female line, but this was not equal to the transgenic ratio. Since during the M phase, the cells lose their fluorescence, part of the non-fluorescent cell mass is a group of transgenic cells under mitosis. (D) In the clone cell lines, the ratio changed compared to the original transgenic lines. The male line contained twice as many fluorescent cells compared to the whole cell number, than the female line. (E,F) With the TO-PRO-3 nuclear staining, the non-fluorescent cells were also visualized to show the presence of cells in different cell cycle stages simultaneously in the culture.
When the transgenic lines reached the optimal stability and cell number, 40 clone lines were established from both cultures, which were then maintained. After three weeks, six lines remained stable: the FCF3, FCF4 and FCF5 female clones (originated from the 1111 ZW line), and the FCM5, FCM7 and FCM8 male clones (originated from the 1116 ZZ line). The fluorescent analysis showed that only the FCF5 female and the three male lines expressed the green/red reporter proteins. The strongest signal was detected in the FCF5 and FCM5 lines (Figures 2D-F.). A far higher ratio of fluorescent cells was observed compared to the original 1116 and 1111 transgenic lines (78% to 26% in the male lines, and 40% to 32% in the female lines).
Characteristics of the Selected Single-cell Cultures
The cells were evaluated with qPCR for CVH and DAZL expression for the second time to prove their PGC status. In both cases, the expression was detected (Figure 3A.).
Figure 3.
Characterization of the clone cell lines. (A) Germ- and stem cell specific gene expression in the original transgenic and clone cell lines. Both the original transgenic 1116 and 1111 lines expressed the CVH, DAZL and POUV markers. (B) The chromosome staining showed no degradation of the sexual chromosomes due to the long term culture in the clone cell lines. (C,D) Their integration into recipient gonads was successful, therefore transgenic gonadal chimaera embryos could be produced using the clone lines. (E) Two copies of the FUCCI construct were found in the FCM5 male line, both of them in an intron region. The insertion on chromosome 12 was a full insertion, while the one on chromosome 2 cannot be proven as a whole integration. (F) In case of FCF5, only one integration was found in an exon region.
Since at this point the cultures were already maintained for a long term, chromosomal analysis was used to ensure the unchanged characteristics of the karyotypes. The karyotype indicated that the number and characteristics of chromosomes have not changed. (Figure 3B.).
To check the cells’ integrational capacity, transgenic gonadal chimera embryos were created, and the gonads were dissected and examined (Figures 3C and 3D). From five surviving embryos injected with the FCM5 male cells two carried the green/red fluorescence, while this rate was six FUCCI-positive gonads out of twenty surviving embryos by the FCF5 female line. In some cases, the cells were visible under the microscope, but they were not integrated into the gonads. On these occasions, the cells were located in the blood vessels surrounding the gonads.
Proving the Presence and Color Change of FUCCI in Chicken PGCs
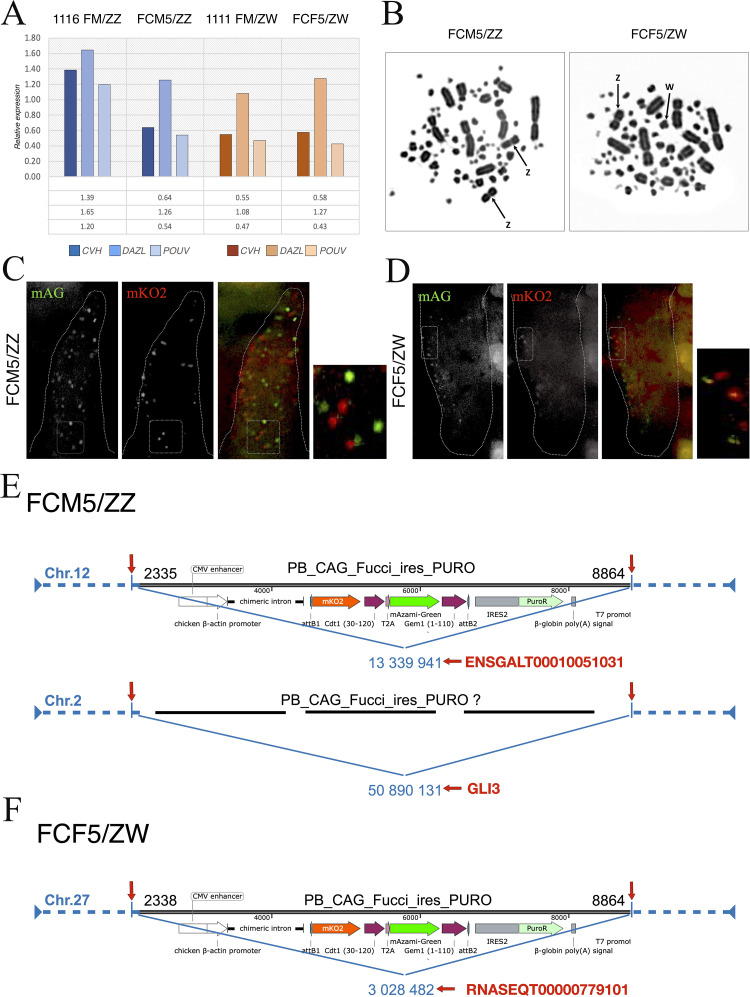
We identified plasmid integration in 2 genomic loci of the FCM5 PGC line, at the genomic coordinates of NC052533.1: 50 890 131 (chr2) and NC_052543.1: 13 339 941 (chr12), which are located in the genes GLI3 and CADPS, respectively (Figures 3E–3F). Both integrations occurred in an intronic sequence. In the FCF5 PGC line, we detected one integration site at the coordinates (chr27) NC052558.1:3028482. It is located in an intergenic region, the closest genes being (Ensembl ID, no name) LOC121107680 and NGFR (annotation version: Ensembl release 110). In all three cases, the insertion is positioned after a TTAA sequence.
In the case of the FCM5 PGC line, at the integration sites on chromosome 12 and in the FCF5 PGC line, the whole FUCCI transgene was integrated (vector coordinates: PB_CAG_FUCCI:2338-8864). It is likely the case in the other integration in the FCM5 PGC line as well, although it cannot be stated with certainty, due to a few overlapping reads at the integration site. Overall, aligned reads are within the vector coordinates PB_CAG_FUCCI:2335-8864. Interestingly, we observed consistent positive alignment across the region at the (chr14) NC_052545.1:4183720-4185523 coordinates in all – transgenic and non-transgenic - samples. This site is within the ACTB gene, and shows a similar number of reads aligned in all samples.
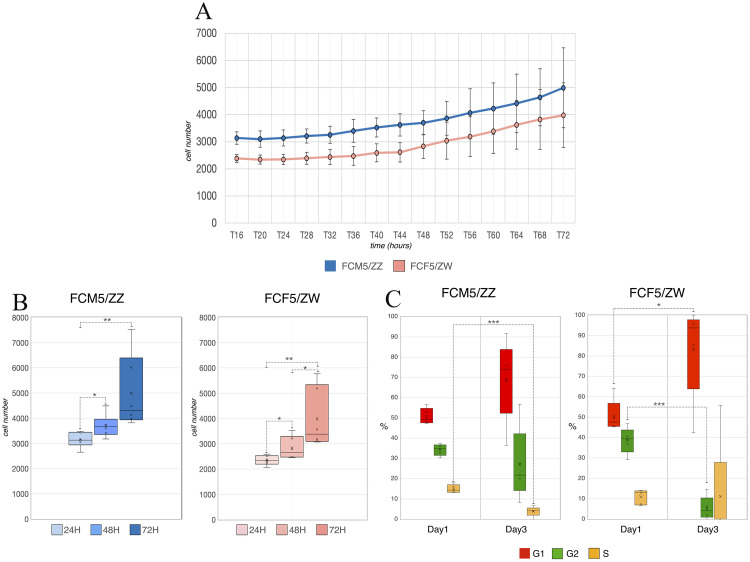
To check the cell cultures’ reaction to the environmental effects, a 3 days long culturing took place without medium change. The cell proliferation level remained for 72h (Figure 4A), with significant elevation between 24h and 48h, and 24h and 72h for both sexes, and between 48h and 72h for FCF5 (Figure 4B). However, there were changes in the percentage for cells in different cell cycle phases during the experiment. The analysis revealed that the delay of medium change elevated the ratio of mKO2 expressing cells and decreased the mAG positive cells in the culture in both cases. This difference was significant in the female culture. The yellow cells also showed a small decrease, but that was significant only in case of the male cell line (Figure 4C.).
Figure 4.
The effects of delayed medium change on the cell number and the cell cycle phase ratio. (A) While the cell number was elevated during the 3 d, (B) and the difference between 24h - 48h and 24h - 72h was significant in each sample, (C) the fluorescent analysis showed that the cell cycle phase ratios changed. The ratio of red fluorescent cells increased and the percentage of green fluorescent cells was decreased in both lines and were significant in the female line. The ratio of yellow cells was also decreased in both sexes, but that was significant only in case of the male line. The standard deviation was also elevated during the culturing in all cases. (D,E) The FUCCI color change was visible in the circuled area in case of both lines. The cells divide, the new cells turn red at the beginning of G1 phase, and as the cell cycle proceeds to S phase, the cells start to emit green fluorescent signal.
Both cell lines were inspected with the ImageXpress Pico Cell Imaging System (Molecular Devices). Timelapse videos were made from the pictures taken by the machine (Figures 4D and 4E.), which show proof of the green G2 phase cells dividing and turning red, while they enter G1 phase.
DISCUSSION
This study aimed to establish stable chicken PGC lines induced with the FUCCI transgene complex. One male and one female PGC line were selected for this purpose. These lines expressed the germ cell and stem cell specific markers, and supported our previous research on RNA expression changes after cell freezing. Compared to the original line (1116), immediately after thawing the expression levels of the male cells (FCM5) mimicked the levels of both female samples (1111 and FCF5) (Ecker et al., 2023).
After reaching stable lines, the FCF5 female and the FCM5 male lines were tested to prove the FUCCI insertion into the genome and its intended function. The DNA sequencing showed that the cells contain the FUCCI sequence in both the FCF5 line (on chr27) and the FCM5 line (on chr2 and chr12). There were consistent positive alignments in both transgenic and wild type samples. This could be explained with the fact, that the FUCCI construct contained the chicken beta actin promoter, which can be found in the chicken genome too. All in all, the DNA sequencing showed that the cells contain the FUCCI sequence in both the FCF5 line (on chr27) and the FCM5 line (on chr2 and chr12).
With PGC injection, gonadal chimaeras were created to prove the integrational capability of the lines. The medium withdrawal test also showed that the cells react to the environment, and this reaction can be monitored with FUCCI via the color change. These findings indicate the negative effect of lacking medium change on cell proliferation and proves that the FUCCI complex works in the cells as intended. We observed a lower effect on the female line, which could have happened due to the fact, that female PGC cultures have a higher probability of creating aggregations and slower proliferation.
FUCCI is a powerful tool, which can help in visualizing cell cycle stages and facilitate monitoring the quality of the cultures. Since its invention in 2008, it was introduced to human cells in 2008 (Sakaue-Sawano et al., 2008), zebrafish in 2009, mouse in 2012, fly in 2014 (Zielke and Edgar, 2015) and axolotl in 2022 (Duerr et al., 2022). Our results made the FUCCI complex available for one more important model animal, the chicken. Thus, the FUCCI technique is now available for a species with a high economical value.
FUCCI positive cell cultures have already been used to indicate toxic effects on the cell cycle, but these experiments usually focused on diseases like Alzheimer (Ippati S et al., 2021) and cancer (Yano et al., 2014; Zhang et al., 2015; Miwa et al., 2015). The findings presented in this article can be used for various applications, like medium tests, toxin- and heat stress studies.
Our future goal is to use the newly established FUCCI chicken PGC lines to investigate the effects of mycotoxins with the largest economical damage, such as T2, on the chicken embryonic development, and to create FUCCI transgenic gonadal chimaera animals to examine toxin effects on adult fertility. Another important topic for the FUCCI PGCs could be the effect of heat treatment, since embryonic heat exposure can cause huge changes in avians. Furthermore, we would like to investigate the length of the cell cycle period in chicken PGCs and create a protocol to synchronize the cells.
DISCLOSURES
The authors declare no conflicts of interest.
ACKNOWLEDGMENTS
This research was funded by the Hungarian National Research, Development and Innovation Office, grant number 2019-2.1.11-TÉT-2019-00036 (E.G.), VEKOP-2.3.2-16-2016-00012 (E.V., E.G.); the Hungarian National Laboratory Project, grant number RRF-2.3.1-21-2022-00007 (E.G.): TKP2021-NKTA-34 funding scheme (E.B). Special thanks to András Nagy for the transgene, to Zoltán Hegyi for his help in calibrating the ImageXpress Pico Cell Imaging System, to László Homolya for providing the ImageXpress Micro XLS Imaging System, to Bertrand Pain for providing the CVH antibody, to Mariann Molnár for her help in the chromosomal staining, and to Nándor Nagy for his help in proofreading.
Author Contributions: Conceptualization: A.E., E.V., E.G.; Methodology: A.E., B.L., R.I.T.; Software: E.G., ZS.F., E.B.; Formal analysis: A.E., E.G., ZS.F., E.B.; Investigation: A.E., R.I.T., M.U., ZS.M.; Writing - original draft: A.E., B.L.; Writing - review & editing: A.E., O.I.H., ZS.F., E.V., E.G.; Supervision: O.I.H., E.B., F.U., E.V., E.G.; Project administration: E.V., E.G.; Funding acquisition: E.B., E.V., E.G.
Data Availability: All relevant data can be found within the article and its supplementary information. Any other information can be provided by the authors.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.psj.2024.104144.
Appendix. Supplementary materials
References
- Abdulhasan M., Ruden X., Marben T., Harris S., Ruden D.M., Awonuga A.O., Puscheck E.E., Rappolee D.A. Using live imaging and fluorescence ubiquitinated cell cycle indicator embryonic stem cells to distinguish g1 cell cycle delays for general stressors like perfluoro-octanoic acid and hyperosmotic sorbitol or G2 cell cycle delay for mutagenic stressors like Benzo(a)pyrene. Stem Cells Dev. 2022;31:296–310. doi: 10.1089/scd.2021.0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe T., Sakaue-Sawano A., Kiyonari H., Shioi G., Inoue K.I., Horiuchi T., Nakao K., Miyawaki A., Aizawa S., Fujimori T. Visualization of cell cycle in mouse embryos with Fucci2 reporter directed by Rosa26 promoter. Development (Cambridge) 2013;140:237–246. doi: 10.1242/dev.084111. [DOI] [PubMed] [Google Scholar]
- Alfi O.S., Donnell G.N., Derencsenyi A. C-banding of human chromosomes produced by D.N.ase. The Lancet. 1973;302:505. doi: 10.1016/s0140-6736(73)92106-5. [DOI] [PubMed] [Google Scholar]
- Altgilbers S., Klein S., Dierks C., Weigend S., Kues W.A. Cultivation and characterization of primordial germ cells from blue layer hybrids (Araucana crossbreeds) and generation of germline chimeric chickens. Sci Rep. 2021;11 doi: 10.1038/s41598-021-91490-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand M., Lázár B., Tóth R., Páll E., Patakiné Várkonyi E., Liptói K., Homolya L., Hegyi Z., Hidas A., Gócza E. Enhancement of chicken primordial germ cell in vitro maintenance using an automated cell image analyser. Acta Vet. Hung. 2018;66:518–529. doi: 10.1556/004.2018.046. [DOI] [PubMed] [Google Scholar]
- Andrews S. FastQC: A quality control tool for high throughput sequence data. Babraham Bioinformatics. 2010 https://www.scirp.org/reference/referencespapers?referenceid=2781642 Accessed May 2024. [Google Scholar]
- Bao Y., Mukai K., Hishiki T., Kubo A., Ohmura M., Sugiura Y., Matsuura T., Nagahata Y., Hayakawa N., Yamamoto T., Fukuda R., Saya H., Suematsu M., Minamishima Y.A. Energy management by enhanced glycolysis in G1-phase in human colon cancer cells in vitro and in vivo. Mol. Cancer Res. 2013;11:973–985. doi: 10.1158/1541-7786.MCR-12-0669-T. [DOI] [PubMed] [Google Scholar]
- Bednarczyk M., Dunislawska A., Stadnicka K., Grochowska E. Chicken embryo as a model in epigenetic research. Poult. Sci. 2021;100 doi: 10.1016/j.psj.2021.101164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow J.J., Dutta A. Preventing re-replication of chromosomal DNA. Nat. Rev. Mol. Cell Biol. 2005;6:476–486. doi: 10.1038/nrm1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaipipat S., Sritabtim K., Piyasanti Y., Prukudom S., Jurutha J., Phetpila V., Sinsiri R., Kammongkun J., Molee A., Thiangtum K., Siripattarapravat K. Initiative on avian primordial germ cell Cryobanking in Thailand. Biopreserv Biobank. 2022;21:458–466. doi: 10.1089/bio.2022.0043. [DOI] [PubMed] [Google Scholar]
- Danecek P., Bonfield J.K., Liddle J., Marshall J., Ohan V., Pollard M.O., Whitwham A., Keane T., McCarthy S.A., Davies R.M. Twelve years of SAMtools and BCFtools. Gigascience. 2021;10 doi: 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divya D., Shukla R., Chatterjee R., Sagar G., Rajendra Prasad A., Bhattacharya T. Production of transgenic chimeric chicken from cryopreserved primordial germ cells and its validation by developing shRNA transgenic chicken chimera. Res Sq. 2021 https://www.jcpr.in/index.php/journal/article/download/41/26 Accessed May 2024. [Google Scholar]
- Duerr T.J., Jeon E.K., Wells K.M., Villanueva A., Seifert A.W., McCusker C.D., Monaghan J.R. A constitutively expressed fluorescent ubiquitination-based cell-cycle indicator (FUCCI) in axolotls for studying tissue regeneration. Development (Cambridge) 2022;149 doi: 10.1242/dev.199637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker A., Lázár B., Tóth R.I., Urbán M., Tokodyné Szabadi N., Salinas Aponte M.T., Adnan M., Várkonyi E., Gócza E. The effects of freezing media on the characteristics of male and female chicken primordial germ cell lines. Life. 2023;13 doi: 10.3390/life13040867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyal-Giladi H., Ginsburg M., Farbarov A. Avian primordial germ cells are of epiblastic origin. J. Embryol. Exp. Morph. 1981;65:9. [PubMed] [Google Scholar]
- Ezaki R., Hirose F., Furusawa S., Horiuchi H. An improved protocol for stable and efficient culturing of chicken primordial germ cells using small-molecule inhibitors. Cytotechnology. 2020;72:397–405. doi: 10.1007/s10616-020-00385-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsburg M., Eyal-Giladi H. Temporal and spatial aspects of the gradual migration of primordial germ cells from the epiblast into the germinal crescent in the avian embryo. J. Embryol. Exp. Morphol. 1986;95:53–71. [PubMed] [Google Scholar]
- Griffiths R., Daan S., Dijkstra C. Sex identification in birds using two CHD genes. Proc. Royal Soc B: Biol. Sci. 1996;263:1251–1256. doi: 10.1098/rspb.1996.0184. [DOI] [PubMed] [Google Scholar]
- Hamburger V., Hamilton H.L. A series of normal stages in the development of the chick embryo. J Morphol. 1951;88:49–92. [PubMed] [Google Scholar]
- Hu T., Taylor L., Sherman A., Tiambo C.K., Kemp S.J., Whitelaw B., Hawken R.J., Djikeng A., McGrew M.J. A low-tech, cost-effective and efficient method for safeguarding genetic diversity by direct cryopreservation of poultry embryonic reproductive cells. Elife. 2022;11 doi: 10.7554/eLife.74036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ippati S., Deng Y., van der Hoven J., Heu C., van Hummel A., Chua S W., Paric E., Chan G., Feiten A., Fath T., Ke Y D., Haass N K., Ittner L M. Rapid initiation of cell cycle reentry processes protects neurons from amyloid-β toxicity. PNAS. 2021;118 doi: 10.1073/pnas.2011876118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasawa S., Araki T., Nagai T., Mizuno H. Cyan-emitting and orange-emitting fluorescent proteins as a donor/acceptor pair for fluorescence resonance energy transfer. Biochem J. 2004;381:307–312. doi: 10.1042/BJ20040321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasawa S., Araki T., Yamamoto-Hino M., Miyawaki A. A green-emitting fluorescent protein from galaxeidae coral and its monomeric version for use in fluorescent labeling. J. Biol. Chem. 2003;278:34167–34171. doi: 10.1074/jbc.M304063200. [DOI] [PubMed] [Google Scholar]
- Kim Y.M., Han J.Y. The early development of germ cells in chicken. Int. J. Develop. Biol. 2018;62:141–148. doi: 10.1387/ijdb.170283jh. [DOI] [PubMed] [Google Scholar]
- Kino K., Pain B., Leibo S.P., Cochran M., Clark M.E., Etches R.J. Production of chicken chimeras from injection of frozen-thawed blastodermal cells. Poult Sci. 1997 doi: 10.1093/ps/76.5.753. [DOI] [PubMed] [Google Scholar]
- Kochav S., Ginsburg M., Eyal-Giladi H. From Cleavage to primitive streak formation: a complementary normal table and a new look at the first stages of the development of the chick II. Microscopic anatomy and cell population dynamics. Dev Biol. 1980;79:296–308. doi: 10.1016/0012-1606(80)90117-7. [DOI] [PubMed] [Google Scholar]
- Koh S.B., Mascalchi P., Rodriguez E., Lin Y., Jodrell D.I., Richards F.M., Lyons S.K. A quantitative FastFUCCI assay defines cell cycle dynamics at a single-cell level. J Cell Sci. 2017;130:512–520. doi: 10.1242/jcs.195164. [DOI] [PubMed] [Google Scholar]
- Kong L., Qiu L., Guo Q., Chen Y., Zhang X., Chen B., Zhang Y., Chang G. Long-term in vitro culture and preliminary establishment of chicken primordial germ cell lines. PLoS One. 2018;13 doi: 10.1371/journal.pone.0196459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lázár B., Anand M., Tóth R., Várkonyi E.P., Liptói K., Gócza E. Comparison of the MicroRNA expression profiles of male and female avian primordial germ cell lines. Stem Cells Int. 2018;2018 doi: 10.1155/2018/1780679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lázár B., Molnár M., Sztán N., Végi B., Drobnyák Á., Tóth R., Tokodyné Szabadi N., McGrew M.J., Gócza E., Patakiné Várkonyi E. Successful cryopreservation and regeneration of a partridge colored Hungarian native chicken breed using primordial germ cells. Poult Sci. 2021;100 doi: 10.1016/j.psj.2021.101207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lázár B., Szabadi N.T., Anand M., Tóth R., Ecker A., Urbán M., Aponte M.T.S., Stepanova G., Hegyi Z., Homolya L., Várkonyi E.P., Pain B., Gócza E. Effect of miR-302b MicroRNA inhibition on chicken primordial germ cell proliferation and apoptosis rate. Genes (Basel) 2022;13 doi: 10.3390/genes13010082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.C.I., Tsai L.C., Hwa P.Y., Chan C.L., Huang A., Chin S.C., Wang L.C., Lin J.T., Linacre A., Hsieh H.M. A novel strategy for avian species and gender identification using the CHD gene. Mol. Cell Probes. 2010;24:27–31. doi: 10.1016/j.mcp.2009.08.003. [DOI] [PubMed] [Google Scholar]
- Lejong M., Choa-Duterre M., Vanmuylder N., Louryan S. Is Vasa such a highly specific marker for primordial germ cells? A comparison of VASA and HSP90 proteins expression in young chicken embryos. Morphologie. 2020;104:20–26. doi: 10.1016/j.morpho.2020.01.003. [DOI] [PubMed] [Google Scholar]
- Liptoi K., Horvath G., Gal J., Varadi E., Barna J. Preliminary results of the application of gonadal tissue transfer in various chicken breeds in the poultry gene conservation. Anim. Reprod. Sci. 2013;141:86–89. doi: 10.1016/j.anireprosci.2013.06.016. [DOI] [PubMed] [Google Scholar]
- Liu J., Cheng K.M., Silversides F.G. Novel needle-in-straw vitrification can effectively preserve the follicle morphology, viability, and vascularization of ovarian tissue in Japanese quail (Coturnix japonica) Anim. Reprod. Sci. 2012;134:197–202. doi: 10.1016/j.anireprosci.2012.08.002. [DOI] [PubMed] [Google Scholar]
- Liu J., Cheng K.M., Silversides F.G. Fundamental principles of cryobiology and application to ex situ conservation of avian species. Avian Biol Res. 2013;6:187–197. [Google Scholar]
- Macdonald J., Glover J.D., Taylor L., Sang H.M., McGrew M.J. Characterisation and germline transmission of cultured avian primordial germ cells. PLoS One. 2010;5 doi: 10.1371/journal.pone.0015518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin F.J., Amode M.R., Aneja A., Austine-Orimoloye O., Azov A.G., Barnes I., Becker A., Bennett R., Berry A., Bhai J., Bhurji S.K., Bignell A., Boddu S., Branco Lins P.R., Brooks L., Ramaraju S.B., Charkhchi M., Cockburn A., Da Rin Fiorretto L., Davidson C., Dodiya K., Donaldson S., El Houdaigui B., El Naboulsi T., Fatima R., Giron C.G., Genez T., Ghattaoraya G.S., Martinez J.G., Guijarro C., Hardy M., Hollis Z., Hourlier T., Hunt T., Kay M., Kaykala V., Le T., Lemos D., Marques-Coelho D., Marugán J.C., Merino G.A., Mirabueno L.P., Mushtaq A., Hossain S.N., Ogeh D.N., Sakthivel M.P., Parker A., Perry M., Piliota I., Prosovetskaia I., Perez-Silva J.G., Salam A.I.A., Saraiva-Agostinho N., Schuilenburg H., Sheppard D., Sinha S., Sipos B., Stark W., Steed E., Sukumaran R., Sumathipala D., Suner M.M., Surapaneni L., Sutinen K., Szpak M., Tricomi F.F., Urbina-Gómez D., Veidenberg A., Walsh T.A., Walts B., Wass E., Willhoft N., Allen J., Alvarez-Jarreta J., Chakiachvili M., Flint B., Giorgetti S., Haggerty L., Ilsley G.R., Loveland J.E., Moore B., Mudge J.M., Tate J., Thybert D., Trevanion S.J., Winterbottom A., Frankish A., Hunt S.E., Ruffier M., Cunningham F., Dyer S., Finn R.D., Howe K.L., Harrison P.W., Yates A.D., Flicek P. Ensembl 2023. Nucleic Acids Res. 2023;51:D933–D941. doi: 10.1093/nar/gkac958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui Y., Zsebo K., Hogan B.L.M. Derivation of pluripotential embryonic stem cells from murine primordial germ cells in culture. Cell. 1992;70:841–847. doi: 10.1016/0092-8674(92)90317-6. [DOI] [PubMed] [Google Scholar]
- Miwa S., Yano S., Kimura H., Yamamoto M., Toneri M., Matsumoto Y., Uehara F., Hiroshima Y., Murakami T., Hayashi K., Yamamoto N., Bouvet M., Fujiwara T., Tsuchiya H., Hoffman R.M. Cell-cycle fate-monitoring distinguishes individual chemosensitive and chemoresistant cancer cells in drug-treated heterogeneous populations demonstrated by real-time fucci imaging. Cell Cycle. 2015;14:621–629. doi: 10.4161/15384101.2014.991604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito M., Tajima A., Tagami T., Yasuda Y., Kuwana T. Preservation of chick primordial germ cells in liquid nitrogen and subsequent production of viable offspring. J. Reprod. Fertil. 1994;102:321–325. doi: 10.1530/jrf.0.1020321. [DOI] [PubMed] [Google Scholar]
- Nakamura Y. Poultry genetic resource conservation using primordial germ cells. J Reprod Develop. 2016;62:431–437. doi: 10.1262/jrd.2016-052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y., Yamamoto Y., Usui F., Mushika T., Ono T., Setioko A.R., Takeda K., Nirasawa K., Kagami H., Tagami T. Migration and proliferation of primordial germ cells in the early chicken embryo. Poult. Sci. 2007;86:2182–2193. doi: 10.1093/ps/86.10.2182. [DOI] [PubMed] [Google Scholar]
- Nakayama K.I., Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nat Rev Cancer. 2006;6:369–381. doi: 10.1038/nrc1881. [DOI] [PubMed] [Google Scholar]
- Nandi S., Whyte J., Taylor L., Sherman A., Nair V., Kaiser P., Mcgrew M.J. Cryopreservation of specialized chicken lines using cultured primordial germ cells. Poult Sci. 2016;95:1905–1911. doi: 10.3382/ps/pew133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani H., Lygerou Z., Nishimoto T. Proteolysis of DNA replication licensing factor Cdt1 in S-phase is performed independently of Geminin through its N-terminal region. J. Biol. Chem. 2004;279:30807–30816. doi: 10.1074/jbc.M312644200. [DOI] [PubMed] [Google Scholar]
- Nishitani H., Lygerou Z., Nishimoto T., Nurse P. The Cdt1 protein is required to license DNA for replication in fission yeast. Nature. 2000;404:625–628. doi: 10.1038/35007110. [DOI] [PubMed] [Google Scholar]
- Ogura Y., Sakaue-Sawano A., Nakagawa M., Satoh N., Miyawaki A., Sasakura Y. Coordination of mitosis and morphogenesis: role of a prolonged G2 phase during chordate neurulation. Development. 2011;138:577–587. doi: 10.1242/dev.053132. [DOI] [PubMed] [Google Scholar]
- Ono T., Matsumoto T., Arisawa Y. Production of donor-derived offspring by transfer of primordial germ cells in Japanese quail. Exp Anim. 1998;47:215–219. doi: 10.1538/expanim.47.215. [DOI] [PubMed] [Google Scholar]
- Petitte J.N., Liu G., Yang Z. Avian pluripotent stem cells. Mech. Dev. 2004;121:1159–1168. doi: 10.1016/j.mod.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Pfeiffer J., Tarbashevich K., Bandemer J., Palm T., Raz E. Rapid progression through the cell cycle ensures efficient migration of primordial germ cells – the role of Hsp90. Dev. Biol. 2018;436:84–93. doi: 10.1016/j.ydbio.2018.02.014. [DOI] [PubMed] [Google Scholar]
- Prasedya E.S., Miyake M., Kobayashi D., Hazama A. Carrageenan delays cell cycle progression in human cancer cells in vitro demonstrated by FUCCI imaging. BMC Complement Altern Med. 2016;16 doi: 10.1186/s12906-016-1199-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J.T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaue-Sawano A., Kurokawa H., Morimura T., Hanyu A., Hama H., Osawa H., Kashiwagi S., Fukami K., Miyata T., Miyoshi H., Imamura T., Ogawa M., Masai H., Miyawaki A. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 2008;132:487–498. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.Y., White D.J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setioko A.R., Tagami T., Tase H., Nakamura Y., Takeda K., Nirasawa K. Cryopreservation of premordial germ cells (PGCs) from White Leghorn embryos using commercial cryoprotectants. J. Poult. Sci. 2007;44:73–77. [Google Scholar]
- Silversides F.G., Robertson M.C., Liu J. Cryoconservation of avian gonads in Canada. Poult Sci. 2013;92:2613–2617. doi: 10.3382/ps.2013-03185. [DOI] [PubMed] [Google Scholar]
- Smorag L., Xu X., Engel W., Pantakani D.V.K. The roles of DAZL in RNA biology and development. Wiley Interdiscip Rev RNA. 2014;5:527–535. doi: 10.1002/wrna.1228. [DOI] [PubMed] [Google Scholar]
- Sugiyama M., Sakaue-Sawano A., Iimura T., Fukami K., Kitaguchi T., Kawakami K., Okamoto H., Higashijima S.-I., Miyawaki A. Illuminating cell-cycle progression in the developing zebrafish embryo. PNAS. 2009;106:20812–20817. doi: 10.1073/pnas.0906464106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima A., Naito M., Yasuda Y., Kuwana T. Production of germ-line chimeras by transfer of cryopreserved gonadal primordial germ cells (gPGCs) in chicken. J Exp Zool. 1998;280:265–267. [PubMed] [Google Scholar]
- Tiambo C.K., Kibui P.W., Kamidi C., Muteti C., Hu T., Kemp S., McGrew M. ILRI Manual 53. ILRI; Nairobi, Kenya: 2021. Laboratory training manual on biobanking and recovery of indigenous poultry genetic resources by cryopreservation of primordial germ cells (PGCs) [Google Scholar]
- Tomura M., Ikebuchi R., Moriya T., Kusumoto Y. Tracking the fate and migration of cells in live animals with cell-cycle indicators and photoconvertible proteins. J Neurosci Methods. 2021;355 doi: 10.1016/j.jneumeth.2021.109127. [DOI] [PubMed] [Google Scholar]
- Trefil P., Aumann D., Koslová A., Mucksová J., Benešová B., Kalina J., Wurmser C., Fries R., Elleder D., Schusser B., Hejnar J. Male fertility restored by transplanting primordial germ cells into testes: a new way towards efficient transgenesis in chicken. Sci Rep. 2017;7 doi: 10.1038/s41598-017-14475-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunekawa N., Naito M., Sakai Y., Nishida T., Noce T. Isolation of chicken vasa homolog gene and tracing the origin of primordial germ cells. Development. 2000;127:2741–2750. doi: 10.1242/dev.127.12.2741. [DOI] [PubMed] [Google Scholar]
- Ugur M.R., Saber Abdelrahman A., Evans H.C., Gilmore A.A., Hitit M., Arifiantini R.I., Purwantara B., Kaya A., Memili E. Advances in cryopreservation of bull sperm. Front. Vet. Sci. 2019;6 doi: 10.3389/fvets.2019.00268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasimuddin M.D., Misra S., Li H., Aluru S. Pages 314–324 in Proceedings - 2019 IEEE 33rd International Parallel and Distributed Processing Symposium, IPDPS 2019. Institute of Electrical and Electronics Engineers Inc; 2019. Efficient architecture-aware acceleration of BWA-MEM for multicore systems. [Google Scholar]
- Whyte J., Glover J.D., Woodcock M., Brzeszczynska J., Taylor L., Sherman A., Kaiser P., McGrew M.J. FGF, Insulin, and SMAD signaling cooperate for avian primordial germ cell self-renewal. Stem Cell Reports. 2015;5:1171–1182. doi: 10.1016/j.stemcr.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock M.E., Gheyas A.A., Mason A.S., Nandi S., Taylor L., Sherman A., Smith J., Burt D.W., Hawken R., McGrew M.J. Reviving rare chicken breeds using genetically engineered sterility in surrogate host birds. Proc Natl Acad Sci U S A. 2019;116:20930–20937. doi: 10.1073/pnas.1906316116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock M.E., Idoko-Akoh A., McGrew M.J. Gene editing in birds takes flight. Mammalian Genome. 2017;28:315–323. doi: 10.1007/s00335-017-9701-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong H., Pu Y., Hu Q., Shan Z., Hu P., Guan W., Ma Y. Embryoid bodies formation from chicken primordial germ cells. Anim Cells Syst (Seoul) 2015;19:168–174. [Google Scholar]
- Yánez-Ortiz I., Catalán J., Rodríguez-Gil J.E., Miró J., Yeste M. Advances in sperm cryopreservation in farm animals: Cattle, horse, pig and sheep. Anim Reprod Sci. 2022:246. doi: 10.1016/j.anireprosci.2021.106904. [DOI] [PubMed] [Google Scholar]
- Yano S., Hoffman R.M. Real-time determination of the cell-cycle position of individual cells within live tumors using FUCCI cell-cycle imaging. Cells. 2018;7 doi: 10.3390/cells7100168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano S., Zhang Y., Miwa S., Tome Y., Hiroshima Y., Uehara F., Yamamoto M., Suetsugu A., Kishimoto H., Tazawa H., Zhao M., Bouvet M., Fujiwara T., Hoffman R.M. Spatial-temporal FUCCI imaging of each cell in a tumor demonstrates locational dependence of cell cycle dynamics and chemoresponsiveness. Cell Cycle. 2014;13:2110–2119. doi: 10.4161/cc.29156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Wu C., Bouvet M., Yano S., Hoffman R.M. Traditional Chinese medicine herbal mixture LQ arrests FUCCI-expressing HeLa cells in G 0 /G 1 phase in 2D plastic, 2.5D Matrigel ®, and 3D Gelfoam ® culture visualized with FUCCI imaging. Oncotarget. 2015;6:5292–5298. doi: 10.18632/oncotarget.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D.F., Kuwana T. Purification of avian circulating primordial germ cells by Nycodenz density gradient centrifugation. Br. Poult. Sci. 2003;44:30–35. doi: 10.1080/0007166031000085382. [DOI] [PubMed] [Google Scholar]
- Zielke N., Edgar B.A. FUCCI sensors: Powerful new tools for analysis of cell proliferation. Wiley Interdiscip Rev. Dev. Biol. 2015;4:469–487. doi: 10.1002/wdev.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielke N., Korzelius J., vanStraaten M., Bender K., Schuhknecht G.F.P., Dutta D., Xiang J., Edgar B.A. Fly-FUCCI: a versatile tool for studying cell proliferation in complex tissues. Cell Rep. 2014;7:588–598. doi: 10.1016/j.celrep.2014.03.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.