Abstract
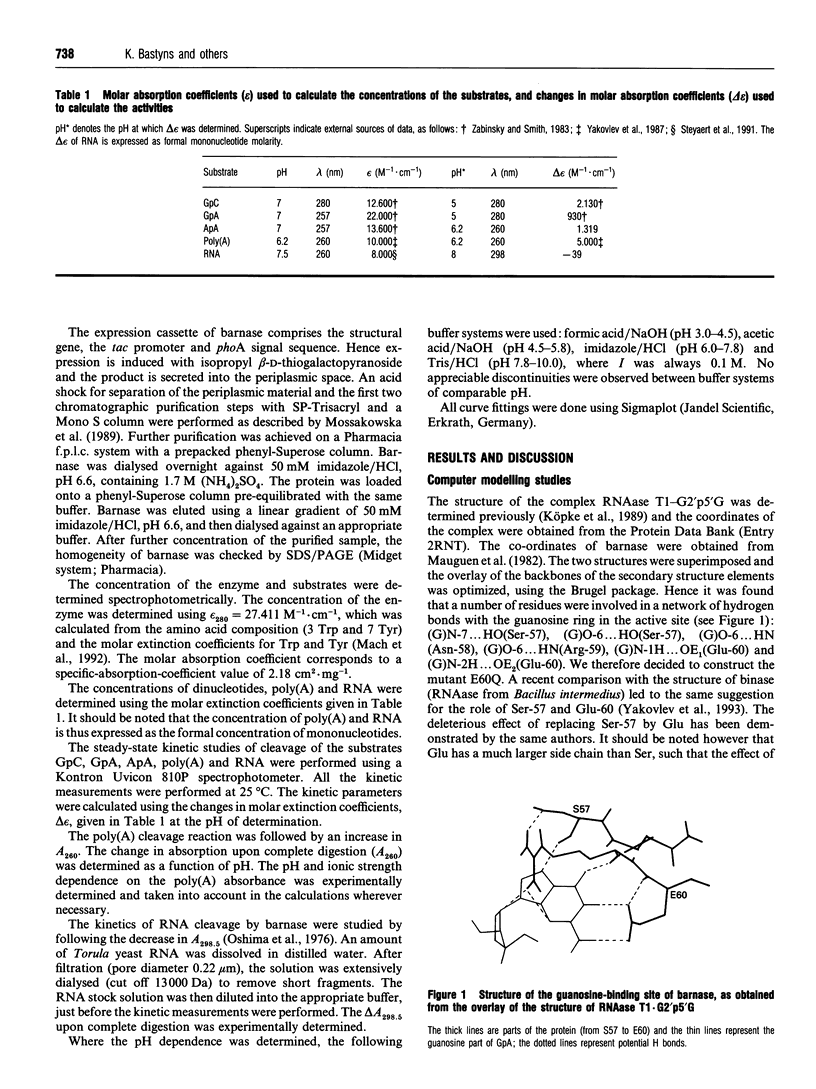
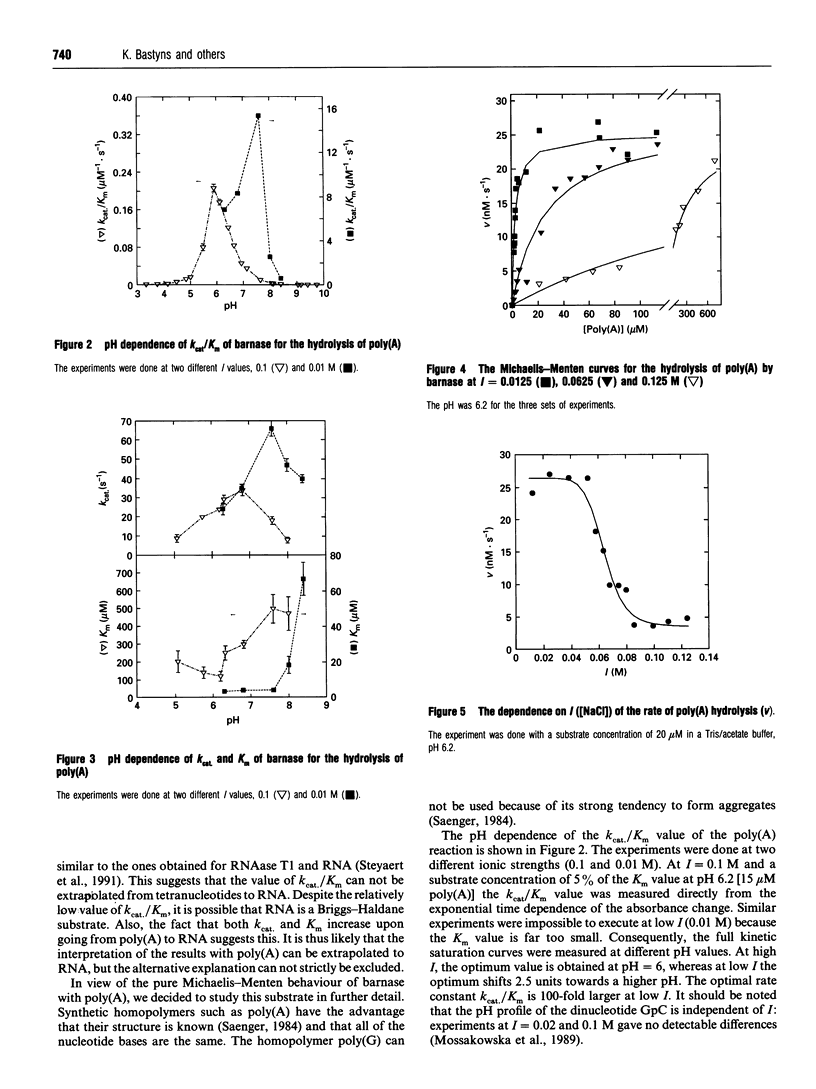
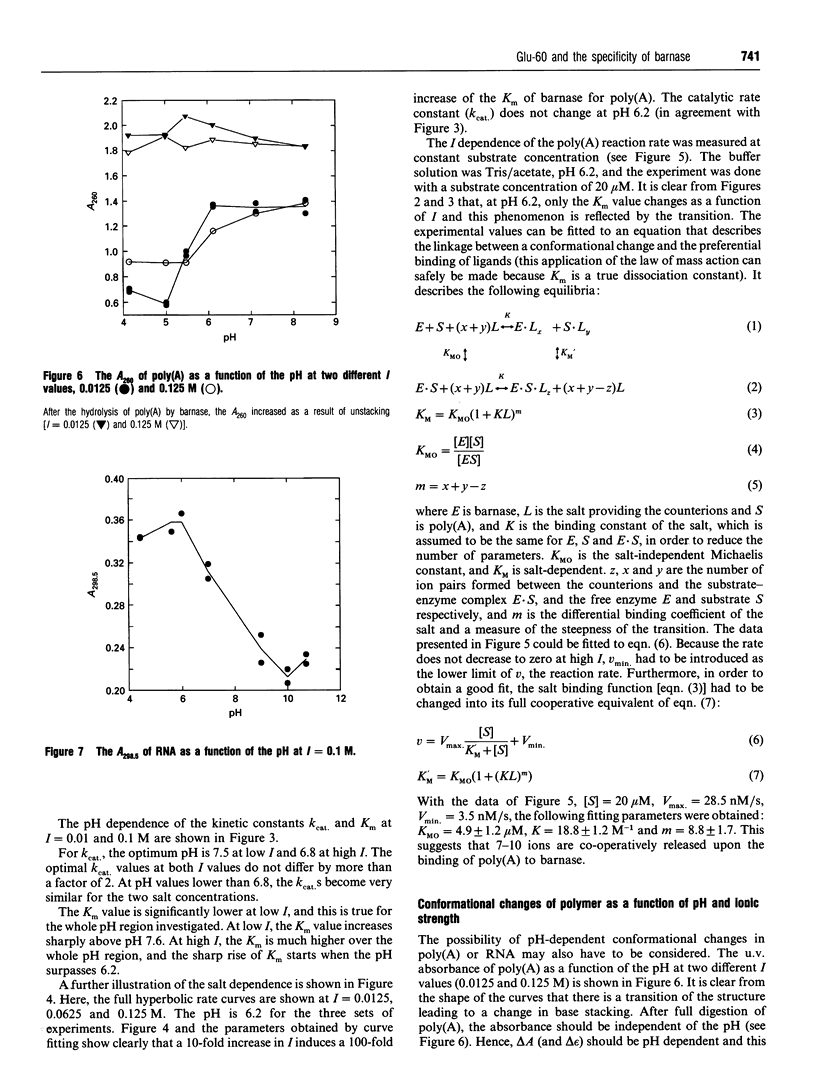
A computer model of the complex between G2'p5'G and barnase, the recombinant ribonuclease of Bacillus amyloliquefaciens, was constructed, based on the known structure of the complex RNAase T1.G2'p5'G. This model suggests that the conserved residue Glu-60 plays an important role in the specificity of barnase for guanosine. A barnase mutant was therefore made in which Glu-60 was replaced by Gln. This mutation increases the Km for the dinucleotides GpC and GpA, by a factor of 10, but does not change the kcat. For ApA, the kcat/Km decreases by a similar factor, but the individual parameters could not be determined. The mutation, however, has no influence on the kcat and the Km of barnase action towards RNA and poly(A). This demonstrates that the interactions between the substrate and the residue at position 60 must be different in the case of ApA and poly(A). For RNA, this conclusion is also likely, but not absolutely certain, because barnase/RNA might be a Briggs-Haldane type enzyme/substrate pair. Therefore, if the effect of the mutation were limited to an increase of the dissociation rate constant of the substrate (k-1), this would not be evident in Km or kcat/Km. In view of the clear cut situation with poly(A), the pH profile for and the effect of salt concentration on the kinetic parameters of the mutant barnase were studied for this substrate. The influence of salt on the Km can be interpreted via the linked function concept and shows a cooperative dissociation of 7-10 counterions upon poly(A) binding. The binding of the substrate is strongly reduced at high pH, and the pKa involved decreases strongly at high salt concentrations. Poly(A) and RNA show a pH dependency of their absorbance spectrum, indicating a pH-dependent change of base stacking, which may influence the catalytic parameters.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Buckle A. M., Fersht A. R. Subsite binding in an RNase: structure of a barnase-tetranucleotide complex at 1.76-A resolution. Biochemistry. 1994 Feb 22;33(7):1644–1653. doi: 10.1021/bi00173a005. [DOI] [PubMed] [Google Scholar]
- Day A. G., Parsonage D., Ebel S., Brown T., Fersht A. R. Barnase has subsites that give rise to large rate enhancements. Biochemistry. 1992 Jul 21;31(28):6390–6395. doi: 10.1021/bi00143a005. [DOI] [PubMed] [Google Scholar]
- Hartley R. W., Rogerson D. L., Jr Production and purification of the extracellular ribonuclease of Bacillus amyloliquefaciens (barnase) and its intracellular inhibitor (barstar). I. Barnase. Prep Biochem. 1972;2(3):229–242. doi: 10.1080/00327487208061473. [DOI] [PubMed] [Google Scholar]
- Koepke J., Maslowska M., Heinemann U., Saenger W. Three-dimensional structure of ribonuclease T1 complexed with guanylyl-2',5'-guanosine at 1.8 A resolution. J Mol Biol. 1989 Apr 5;206(3):475–488. doi: 10.1016/0022-2836(89)90495-6. [DOI] [PubMed] [Google Scholar]
- LENNOX E. S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955 Jul;1(2):190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- Mach H., Middaugh C. R., Lewis R. V. Statistical determination of the average values of the extinction coefficients of tryptophan and tyrosine in native proteins. Anal Biochem. 1992 Jan;200(1):74–80. doi: 10.1016/0003-2697(92)90279-g. [DOI] [PubMed] [Google Scholar]
- Mauguen Y., Hartley R. W., Dodson E. J., Dodson G. G., Bricogne G., Chothia C., Jack A. Molecular structure of a new family of ribonucleases. Nature. 1982 May 13;297(5862):162–164. doi: 10.1038/297162a0. [DOI] [PubMed] [Google Scholar]
- Mossakowska D. E., Nyberg K., Fersht A. R. Kinetic characterization of the recombinant ribonuclease from Bacillus amyloliquefaciens (barnase) and investigation of key residues in catalysis by site-directed mutagenesis. Biochemistry. 1989 May 2;28(9):3843–3850. doi: 10.1021/bi00435a033. [DOI] [PubMed] [Google Scholar]
- Oshima T., Uenishi N., Imahori K. Simple assay methods for ribonuclease T1, T2, and nuclease P1. Anal Biochem. 1976 Apr;71(2):632–634. doi: 10.1016/s0003-2697(76)80039-5. [DOI] [PubMed] [Google Scholar]
- Paddon C. J., Hartley R. W. Expression of Bacillus amyloliquefaciens extracellular ribonuclease (barnase) in Escherichia coli following an inactivating mutation. Gene. 1987;53(1):11–19. doi: 10.1016/0378-1119(87)90088-6. [DOI] [PubMed] [Google Scholar]
- Stanssens P., Opsomer C., McKeown Y. M., Kramer W., Zabeau M., Fritz H. J. Efficient oligonucleotide-directed construction of mutations in expression vectors by the gapped duplex DNA method using alternating selectable markers. Nucleic Acids Res. 1989 Jun 26;17(12):4441–4454. doi: 10.1093/nar/17.12.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steyaert J., Opsomer C., Wyns L., Stanssens P. Quantitative analysis of the contribution of Glu46 and Asn98 to the guanosine specificity of ribonuclease T1. Biochemistry. 1991 Jan 15;30(2):494–499. doi: 10.1021/bi00216a027. [DOI] [PubMed] [Google Scholar]
- Yakovlev G. I., Moiseyev G. P., Struminskaya N. K., Romakhina E. R., Leshchinskaya I. B., Hartley R. W. Increase of specificity of RNase from Bacillus amyloliquefaciens (barnase) by substitution of Glu for Ser57 using site-directed mutagenesis. Eur J Biochem. 1993 Jul 1;215(1):167–170. doi: 10.1111/j.1432-1033.1993.tb18019.x. [DOI] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis of DNA fragments cloned into M13 vectors. Methods Enzymol. 1983;100:468–500. doi: 10.1016/0076-6879(83)00074-9. [DOI] [PubMed] [Google Scholar]


