Abstract
Epidemic respiratory infections are responsible for extensive morbidity and mortality within both military and civilian populations. We describe a high-throughput method to simultaneously identify and genotype species of bacteria from complex mixtures in respiratory samples. The process uses electrospray ionization mass spectrometry and base composition analysis of PCR amplification products from highly conserved genomic regions to identify and determine the relative quantity of pathogenic bacteria present in the sample. High-resolution genotyping of specific species is achieved by using additional primers targeted to highly variable regions of specific bacterial genomes. This method was used to examine samples taken from military recruits during respiratory disease outbreaks and for follow up surveillance at several military training facilities. Analysis of respiratory samples revealed high concentrations of pathogenic respiratory species, including Haemophilus influenzae, Neisseria meningitidis, and Streptococcus pyogenes. When S. pyogenes was identified in samples from the epidemic site, the identical genotype was found in almost all recruits. This analysis method will provide information fundamental to understanding the polymicrobial nature of explosive epidemics of respiratory disease.
Keywords: genotyping, group A streptococci, infectious disease, Streptococcus pyogenes, pneumonia
Despite the prevalence of epidemic respiratory infections and their important impact on global human health, the molecular underpinnings of these conditions remain poorly understood. Epidemic respiratory infections can be caused by a wide variety of bacteria, including several species of Streptococcus, Haemophilus influenzae, Staphylococcus aureus, Neisseria meningitidis, Mycoplasma pneumoniae, and Chlamydophila pneumoniae, or viruses such as influenza virus, adenovirus, rhinovirus, or coronaviruses (1, 2). Although various culture methods, molecular techniques, and serologic diagnostic tests exist, for many epidemics the causative microorganism(s) are never determined. Furthermore, there has been no practical method for examining the broad bacterial ecology of respiratory infections to dissect the complex polymicrobial interactions that occur during explosive outbreaks of disease.
Group A streptococci (GAS), or Streptococcus pyogenes, is one of the most important organisms associated with respiratory infections because of its prevalence and its ability to cause severe disease with complications such as acute rheumatic fever and acute glomerulonephritis (3). The ability to simultaneously identify GAS and other bacteria and viruses in large numbers of samples would greatly facilitate our understanding of respiratory epidemics. It is also essential to follow the spread of specific virulent strains of GAS in populations and to distinguish virulent strains from avirulent streptococci (3).
Molecular methods have been developed to genotype GAS based on the sequence of the emm gene that encodes the M-protein virulence factor (4–6). More than 150 different emm types have been defined and correlated with phenotypic properties of thousands of GAS isolates by using this molecular classification (www.cdc.gov/ncidod/biotech/strep/strepindex.html) (7). Recently, a strategy known as multilocus sequence typing (MLST) was developed to determine the molecular epidemiology of GAS and other bacterial pathogens. The results from MLST are highly concordant with several other typing methods (8).
Although MLST provides detailed analysis of isolated GAS strains, it provides no information about the other respiratory microbes that may participate in the pathology. We now report a technique that rapidly identifies multiple respiratory microorganisms simultaneously in a quantitative fashion. This allows for broad microbial population analysis and strain tracking of an ongoing, geographically dispersed epidemic on a large scale. We specifically identified the bacterial pathogens present during a respiratory disease outbreak at a military training camp (9), characterized the GAS strain-genotype, and analyzed the spread to other military facilities.
Materials and Methods
Selected isolates used in this research from the Naval Health Research Center were collected in compliance with all applicable federal regulations governing the protection of human subjects in research under approved protocol NHRC.2001.0008.
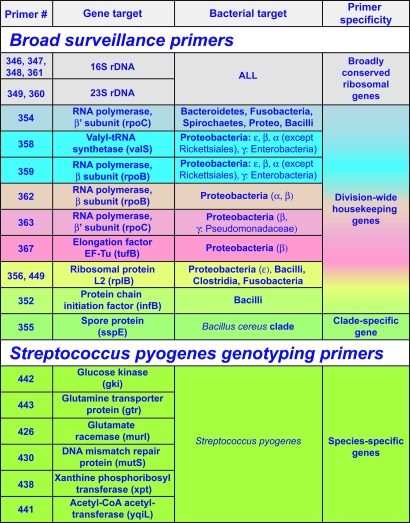
Primer Selection. Broad-range PCR primers for mass spectrometry analysis were designed to target conserved regions of bacterial ribosomal DNA genes (16S and 23S) and genes encoding housekeeping proteins common to all bacteria (Table 1). Primers for genotyping GAS using mass spectrometric analysis were designed to target sequences from each of the seven housekeeping genes used in MLST. The nucleotide sequences for these genes from 212 isolates of GAS (78 distinct emm types) were obtained from www.mlst.net. These correspond to the 100 different allelic profiles referred to by Enright et al. as ST1-ST100 (8). Twenty-four primer pairs were designed and validated against S. pyogenes. A final subset of six primer pairs (sequences are shown in Table 4, which is published as supporting information on the PNAS web site) was chosen based on a theoretical calculation of minimal number of primer pairs that maximized resolution between emm types.
Table 1.
Broad-range and S. pyogenes-specific genotyping primer targets and scope of coverage
Sequences of the primers are provided in Table 4. Primer coverage of bacterial phylogeny is depicted pictorially in Fig. 4. The locations of the primers targeting ribosomal sequences are depicted mapped to the rRNA structures in Fig. 2 (16S) and 3 (23S).
Mass Spectrometry and MLST. After amplification, 15 μl-aliquots of each PCR were desalted and purified by using a weak anion exchange protocol as described (10). Accurate-mass (±1 ppm), high-resolution (M/ΔM > 100,000 full-width half maximal) mass spectra were acquired for each sample by using high-throughput electrospray ionization mass spectrometry (ESI-MS) protocols as described (11, 12). For each sample, 1.5 μl of analyte solution was consumed during the 60-s spectral acquisition. Raw mass spectra were converted to monoisotopic molecular masses. Unambiguous base compositions were derived from the exact mass measurements (13). Quantitative results are obtained by comparing the peak heights with an internal PCR calibration standard present in every PCR well at 500 molecules for the ribosomal DNA-targeted primers and 100 molecules for the protein-encoding gene targets (11). GAS isolates were analyzed by using emm gene-specific PCR as described (4, 14). MLST analysis was performed as described (8).
Results and Discussion
Broad Surveillance, Identification, and Rapid Strain-Genotyping of Bacterial Pathogens. To begin to decipher the polymicrobial dynamics that underlie epidemic outbreaks of respiratory disease, it would be valuable to analyze respiratory samples for a broad range of bacterial pathogens simultaneously and to obtain high-resolution strain-genotyping information on specific species. We have developed a rapid, high-throughput molecular method to achieve these objectives and have tested it on samples obtained from respiratory disease outbreaks associated with S. pyogenes in military training facilities (9). The experimental methodology is based on analysis of multiple PCR amplicons using PCR/ESI-MS to determine base compositions of complex mixtures of amplicons (11, 13). High-resolution genotyping of specific bacterial species, in this case S. pyogenes, was accomplished by analyzing the samples with species-specific primers that interrogate regions of high intraspecies variability to distinguish closely related strains.
To measure the broad landscape of bacteria present in respiratory samples, a set of 16 broad-range surveillance primers was used that allow PCR amplification and quantitative identification of many different bacterial pathogens and respiratory commensal flora. Gene targets of these primers are listed in Table 1, and sequences are shown in Table 4. The surveillance primers were chosen by computational analysis of sequence alignments of the ribosomal DNA operons and 160 broadly conserved protein-encoding housekeeping genes. The ribosomal DNA-targeted primers have the broadest range of bacterial coverage. For example, the four primer pairs targeted to 16S ribosomal DNA match, on average, 98% of the bacterial sequences in the Ribosomal Database Project (http://rdp.cme.msu.edu) allowing for two to three mispairings under permissive PCR cycling conditions. The sites of hybridization and the sequence conservation in these regions are shown on the ribosomal RNA structures in Figs. 2 and 3, which are published as supporting information on the PNAS web site. The primers targeted to protein-encoding housekeeping genes have breadth of coverage at the level of major bacterial subdivisions (e.g., beta proteobacteria, bacilli); their specificity is described in Table 1 and graphically depicted in Fig. 4, which is published as supporting information on the PNAS web site.
Analysis of the amplified regions from major respiratory pathogens showed that the base compositions of these regions unambiguously distinguished all recognized respiratory pathogens from each other and from normal flora, including closely related species of Streptococci and Staphylococci (base compositions are listed in Table 5, which is published as supporting information on the PNAS web site). Although any single primer target region might have an overlap of base compositions with another species, combined information from multiple primer pairs provided unambiguous organism-specific signatures for all major respiratory pathogens. For example, S. pyogenes and Streptococcus pneumoniae have target regions that are amplified by 9 and 10 of the surveillance primers, respectively. The base compositions of these two species are identical in only one target region, and differ in all remaining target regions by up to four base substitutions per region. We confirmed the resolving power of the target regions by determining the base compositions of 120 isolates of respiratory pathogens representing 70 different bacterial species (data not shown). The results showed that the observed variations (usually one or two base substitutions in the amplified region) in base composition amongst multiple isolates of the same species did not prevent correct identification of major pathogenic species.
For high-resolution strain genotyping, we designed a strategy to generate strain-specific signatures that follows the rationale of MLST. We constructed an alignment of concatenated alleles of the seven MLST housekeeping genes from each of 212 previously emm-typed strains (8) and determined the number and location of the primer pairs that would maximize strain discrimination. An initial set of 24 primer pairs was selected that would amplify regions covering >97% of the nucleotide variation in the MLST sequencing targets. We then determined how much strain discrimination could be achieved from a smaller set of primers. Performance calculations for different possible combinations of primer subsets showed that six pairs of primers allowed discrimination at the individual emm-type level of >75% of all of the emm types listed by Enright et al. (8), whereas the remaining 25% clustered into groups of two or more emm types (see Fig. 5, which is published as supporting information on the PNAS web site, for details). This degree of resolution was considered sufficient for applications such as tracking the clonal expansion of a particular strain type during a specific epidemic. However, if complete emm typing is required, 12 primer pairs can be used to completely resolve all emm types.
Identification and Strain Genotyping of GAS Isolates. Four sets of throat samples taken from recruits at different military facilities were examined. The first set was collected at a military training center in 2002 during one of the most severe outbreaks of pneumonia associated with GAS in the U.S. since 1968 (9). Throat swabs were taken from both healthy and hospitalized recruits and plated for selection of putative GAS colonies. A second set of 15 original patient specimens was taken during the height of this disease outbreak. The third set were historical samples from disease outbreaks at this and other military training facilities during previous years. The fourth set of samples was collected from five geographically separated military facilities in the continental U.S. in the winter immediately after the severe 2002 outbreak.
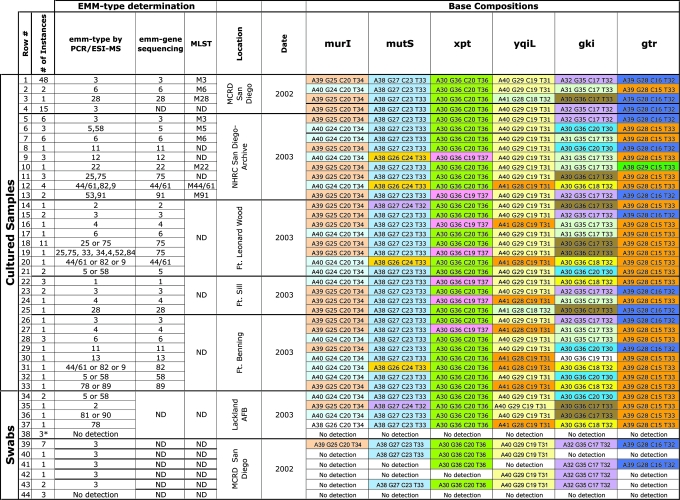
Colonies isolated from GAS selective media from all four collection periods were analyzed with the broad surveillance primers and GAS genotyping primers (Table 1). When the surveillance primers were used, all samples showed base compositions that precisely matched the four completely sequenced strains of S. pyogenes (15–20). The results of the base composition analysis with genotyping primer pairs for samples from all four collection periods are compared to results from 5′-emm gene sequencing and the MLST gene sequencing methods in Table 2. When only these six primer pairs were used, some of the samples could not be resolved to a unique emm type. However, base composition analysis showed identification consistent with (either uniquely or as a member of a small set) 5′-emm gene sequencing or the MLST sequencing method.
Table 2. Base composition analysis of GAS samples.
Base compositions from each primer target site are color-coded so that each unique base composition has its own color. ND, no data.
*Samples were determined to be GAS-negative by independent culture techniques.
Genotyping GAS Isolated from the 2002 Epidemic and Testing of Original Patient Specimens. Of the 51 samples taken during the peak of the November/December 2002 epidemic (Table 2, rows 1–3), all but three had identical base compositions and corresponded to emm3, a GAS genotype previously associated with high respiratory virulence (17, 19). The three outliers (Table 2, rows 2 and 3) were samples from healthy individuals and probably represent nonepidemic strains harbored by asymptomatic carriers. Archived samples (Table 2, rows 5–13) from historical collections showed a much greater heterogeneity of composition signatures and emm types, as would be expected for different epidemics occurring at different times and places.
During the peak of the 2002 outbreak, duplicate throat swabs were taken from military recruits who were not overtly symptomatic but who were living and training in the same community. One of the paired swabs was used to isolate GAS colonies on selective media and the other swab was analyzed directly. Fifteen of the paired swabs that showed at least one colony on GAS-selective media were selected for further study. When the surveillance primers were used, all 15 of these GAS isolates showed base compositions identical to the sequenced GAS genomes, as was observed with all previous GAS isolates in this study. The six GAS-specific genotyping primers indicated that all 15 samples had the same GAS genotype (Table 2, row 4), corresponding to emm3, the identical signature obtained from the symptomatic individuals in this outbreak, consistent with the clonal expansion of a single genotype.
The duplicate swabs were analyzed without culture by using the 16 broad surveillance and six GAS-specific genotyping primers. Analysis using the surveillance primers revealed that these samples had a mixture of microbes, as might be expected from a complex sample (Table 3). Of the 15 samples, six showed evidence of GAS using the broad surveillance primers, and seven showed positive detection using all six genotyping primers (Tables 3 and 2, row 39). Of the remaining eight samples, five were positive with two to four genotyping primers (Table 2, rows 40–43) and three of the samples were negative with all six genotyping primers (Table 2, row 44). These results suggest that GAS was present in these samples at widely varied concentrations.
Table 3. Analysis of 15 throat swabs.
| No. | Organisms identified | Relative ratios | Positive by gentoyping |
|---|---|---|---|
| 1 | H. influenzae | 5 | 6 |
| S. pyogenes | 1 | ||
| 2 | N. meningitidis | 14 | 0 |
| H. influenzae | 10 | ||
| 3 | H. influenzae | 2 | 6 |
| S. pyogenes | 1 | ||
| 4 | H. influenzae | NA | 0 |
| 5 | H. influenzae | 20 | 6 |
| N. meningitidis | 5 | ||
| S. pyogenes | 4 | ||
| 6 | H. influenzae | 5 | 0 |
| C. pseudodiphtheriticum | 1 | ||
| 7 | S. pyogenes | NA | 6 |
| 8 | S. epidermidis | NA | 3 |
| 9 | N. meningitidis | 7 | 6 |
| S. pyogenes | 1 | ||
| 10 | H. influenzae | 2 | 2 |
| S. pneumoniae | 1 | ||
| 11 | N. meningitidis | >20 | 6 |
| S. gordonii | 1 | ||
| 12 | M. catarrhalis | 2 | 0 |
| H. influenzae | 1 | ||
| 13 | N. meningitidis | NA | 0 |
| 14 | S. pyogenes | 5 | 6 |
| S. aureus | 1 | ||
| 15 | N. meningitidis | >20 | 4 |
| S. mitis | 1 |
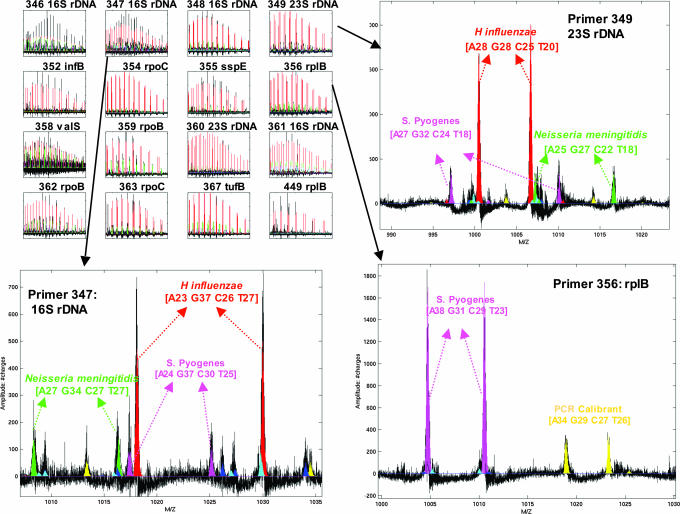
In addition to GAS, other potentially pathogenic organisms were identified. In an exemplary sample (Table 3, sample 5), GAS was identified along with strong signals consistent with N. meningitidis and H. influenzae (Fig. 1). The 16 surveillance primers have a varying degree of breadth in their coverage. The six primers that target 16S and 23S ribosomal DNA were designed to amplify all bacteria from the major divisions (Table 1). Mass spectral analysis of the products from primers that target 16S and 23S ribosomal DNA (Fig. 1 Upper Right and Lower Left) showed that the dominant signals were from H. influenzae, N. meningitidis, and S. pyogenes (GAS) present in a ratio of ≈20:5:4 as determined by comparison of peak heights with that of internal PCR calibration standards for several of the primers. In contrast to the primers that amplify ribosomal DNA genes, the primers that target genes encoding housekeeping proteins were designed to provide coverage of specific divisions of bacteria. For example, primer pair 356 targets the rplB gene (Table 1) and primarily amplifies the bacterial classes Bacilli (which includes S. pyogenes) and Clostridia, but does not amplify Proteobacteria such as N. meningitidis and H. influenzae. Analysis of the spectrum from this primer set shows S. pyogenes as the only major product (Fig. 1 Lower Right). As expected, primers targeted to the proteobacterial species identified N. meningitidis (corroborating evidence for the simultaneous circulation of N. meningitidis during this epidemic was obtained from culture data from a hospitalized patient who subsequently died from pneumonia during this period; K. Russell, unpublished data) and H. influenzae, but not S. pyogenes (data not shown). Although base compositions detected from some of the surveillance primers are consistent with more than one organism, the collective data from the 16 surveillance primers unambiguously identified these three bacterial species as responsible for the bulk of the bacterial load in this throat swab. Thus, using this surveillance panel of primers, it is possible to identify the major bacterial components of a complex sample and to determine their approximate concentrations.
Fig. 1.
Mass spectra from DNA amplified from a throat swab (Table 3, sample 5) using each of the 16 surveillance primers (Table 1). (Upper Left) Thumbnail spectra from 16 primers. Each of the PCR wells was calibrated by using an internal standard identical to the bacterial target sequence except for a 2- to 5-nt internal deletion (calibrant peaks are shown in yellow). (Upper Right) Spectrum from a primer pair that targets 23S rDNA. The paired peaks correspond to the sense and antisense strands of the PCR amplicon that are separated under conditions of ionization. The peaks are labeled with base composition of the amplicons and the organism that matches the composition. (Lower Left) Spectrum from a primer pair that targets 16S rDNA. (Lower Right) Spectrum from a primer pair that targets the Bacilli, but not the Proteobacteria.
To corroborate the results of mass spectrometry analysis of PCR products obtained by using the surveillance primer set, we analyzed two of the samples by broad-range priming followed by cloning and sequencing (21). The results from sequencing ≈700 nucleotides of 16S ribosomal DNA were in good agreement with the mass spectrometry analysis with respect to both identification of species and the relative abundance for the organisms that constituted > 5% of the total microbial load in the sample (details in Table 6, which is published as supporting information on the PNAS web site). However, cloning and sequencing indicated the presence of additional species of bacteria not identified by PCR/ESI-MS. For example, based on sequencing, 5% of the bacterial load in sample 5 (Table 3) was comprised of Corynebacterium fastidiosum. Retrospective analysis of the mass spectra revealed peaks that were consistent with the presence of this organism, but the peak heights across the surveillance primer set were insufficient to make a positive identification. Thus, broad-range primer analysis will be less sensitive on an absolute scale for a low abundance organism than species-specific primers that do not have to compete for PCR resources with multiple microbes (see sensitivity measurements below). The other sample that was analyzed by both PCR/ESI-MS and sequencing (Table 3, sample 14) contained S. pyogenes and Staphylococcus aureus in a ratio of ≈5:1 as determined by PCR/ESI-MS and about 3.5:1 by sequencing.
It is interesting that the 15 throat swabs from military recruits contained a relatively small set of microbes in high abundance (Table 3). The most common were H. influenzae, N. meningitidis, and S. pyogenes; S. epidermidis, M. catarrhalis, C. pseudodiphtheriticum, and S. aureus were present in fewer samples. We also analyzed an equal number of samples from healthy volunteers in the same fashion and did not observe the same pattern of microbes (samples were taken from 23 healthy volunteers from each of three different geographic locations, not from military training settings). Healthy volunteers showed a flora dominated by multiple, commensal non-β-hemolytic Streptococcal species, including viridans group streptococci (Streptococcus parasanguinis, Streptococcus vestibularis, Streptococcus mitis, Streptococcus oralis, and S. pneumoniae; data not shown). Thus, the military recruits in the midst of a respiratory disease outbreak had a dramatically different microbial population than that experienced by the general population in the absence of epidemic disease.
Genotyping GAS Isolated from Geographically Separated Military Facilities in 2003. After the 2002 epidemic associated with a virulent emm3 strain, we surveyed respiratory disease outbreaks at other military facilities. It was possible that the virulent genotype from the epidemic might have spread to these locations later in the winter season. GAS isolated from patients with respiratory disease was examined by base composition analysis and by emm-gene sequencing. The results (Table 2, rows 14–33) showed concordance between base composition analysis and emm gene sequencing. One or two samples from each location had an emm3 genotype. However, the distribution of GAS types at these locations showed a pattern significantly different from the original epidemic, suggesting that the epidemic strain was not dominating the population of GAS at other locations.
Throat swabs from eight individuals showing respiratory symptoms were obtained and analyzed after culture and directly from the swab. Five of the eight patients tested positive for GAS by culture on selective media (Table 2, rows 34–38). Samples that were culture-positive were also GAS-positive by base composition analysis when analyzed directly from the swab, whereas the three culture-negative samples were also negative by PCR/ESI-MS.
Sensitivity, Dynamic Range, and Reproducibility. To evaluate the limit of detection, serial 2-fold dilutions of known amounts of genomic DNA isolated from S. pyogenes were added to water or genomic material isolated from throat swabs from healthy volunteers. In water, both the broad surveillance and genotyping primers reliably detected as few as 15 genome copies of S. pyogenes per PCR well (data not shown). In the presence of normal throat flora, the S. pyogenes genotyping primers, which do not competitively amplify commensal streptococcal species, had the same sensitivity as in water. However, the broad surveillance primers lost sensitivity for S. pyogenes in the presence of floral DNA because of PCR competition with commensal streptococcal organisms. The limit of detection was ≈2,500 genomic copies per well in the presence of the average amount of normal flora taken from a throat swab (pooled from 15 volunteers and divided to one swab-equivalent). On the other hand, when K. pneumoniae and B. anthracis were spiked into normal throat flora, the limit of detection was ≈10–30 genome copies per well for both organisms. The difference in sensitivity for S. pyogenes vs. K. pneumoniae and B. anthracis can be attributed to the predominance of streptococcal organisms in normal flora. Although all of the surveillance primers that amplify S. pyogenes also competitively amplify commensal streptococcal species, several of the surveillance primers (in particular those that target genes encoding housekeeping proteins) amplify K. pneumoniae and B. anthracis and exclude the commensal streptococci. Thus, the lower limit of detection for a particular organism is not absolute, but varies with the level and nature of competing DNA and the coverage of the surveillance primers (see dynamic range experiment below). This is uniquely problematic for S. pyogenes in a throat flora background, which is dominated by commensal streptococcal species. To detect low levels of S. pyogenes in the presence of throat flora, one or more of the S. pyogenes-specific genotyping primers that do not amplify commensal streptococcal species would be necessary.
To determine the dynamic range and linearity of competitive PCR/ESI-MS, we mixed three organisms in varying relative ratios ranging from 10 to 10,000 and analyzed them by using the surveillance primer set. The results (Fig. 6, which is published as supporting information on the PNAS web site) showed a dynamic range of at least 1,000:1, where one organism could be detected in the presence of a 1,000-fold excess of one or two other organisms. The deviation from linearity at 45 cycles of PCR over a 1,000:1 dynamic range was ±60%. The dynamic range would vary somewhat for different mixtures of organisms, depending on their coverage by the surveillance primers. Having a mix of primers with varying breadth in specificities effectively expands the overall dynamic range of the system while assuring that the major bacterial components of a mixture are identified.
To assess swab-to-swab variation, we analyzed duplicate throat swabs from 23 healthy volunteers. The composition of the bacterial flora varied somewhat from individual to individual, but the replicate swabs from the same donor showed virtually identical profiles (Fig. 7, which is published as supporting information on the PNAS web site). The bacteria from these duplicate swabs were all dominated by commensal Streptococcus spp., as expected for normal throat flora from healthy donors (21).
Conclusions
In both developing and developed nations, the leading cause of death by a wide margin is acute respiratory disease (22–24). However, the underlying microbial ecology and the polymicrobial interactions that mediate explosive epidemics remains poorly understood. We have developed a strategy to simultaneously survey respiratory samples for the presence of many different pathogenic agents and to provide high-resolution strain genotyping for selected species of bacteria. Using a set of surveillance primers targeted to broadly conserved regions of bacterial genomes, PCR amplicons were generated and analyzed by ESI-MS, and the identity and relative quantity of microorganisms was determined by using the base compositions of the amplicons. This method allows rapid detection of the abundant microbial flora present in a complex sample. To track a particular bacterial strain that may be the driving force of an epidemic, high-resolution genotyping capability is required. This is accomplished by the use of species-specific primers that target regions of high variation.
In this study, we analyzed four sets of respiratory samples from military settings. Military recruits live in close quarters and are subject to intense physical and vocal stress as a normal part of training. Analysis of respiratory samples from military recruits living in a training community where a high amount of respiratory disease was present showed high concentrations of one or several pathogenic respiratory bacteria, including GAS, H. influenzae, and N. meningitidis. From the epidemic site, the identical GAS genotype was identified in almost all recruits. The respiratory flora present in these recruits was not found in healthy controls.
We have developed a rapid, high-throughput, and cost-effective method for surveying large numbers of samples that provides both a broad view of the bacterial organisms present and a high-resolution genotype of selected species. The PCR/ESI-MS analysis of 96 samples with all surveillance primers takes ≈19 h, and has sufficient speed and throughput to be useful in tracking of an ongoing epidemic. Although this work focused on identification of bacteria, with detailed strain genotyping of GAS, the PCR/ESI-MS method described here can be extended to broad groups of viruses (25), fungi, and pathogenic protozoa. We envision using this methodology to enhance our understanding of the fundamental nature of explosive epidemics of respiratory disease.
Acknowledgments
We acknowledge Dr. Jackie Wyatt for editorial assistance and Defense Advanced Research Planning Agency for financial support.
Author contributions: D.J.E., R.S., and K. Russell designed research; R.S., L.B.B., M.W.E., C.I., J.A.E., B.L., N.F., C.B., J.W., J.C.H., P.M.H., T.A.H., Y.J., R.R., J.A.M., and S.A.H. performed research; V.S., K. Rudnick, A.M.D., E.K.M., D.W.R., C.M., T.A.H., J.J.D., N.W., S.A.H., D.J.K., and D.W.R. contributed new reagents/analytic tools; D.J.E., R.S., L.B.B., M.W.E., C.I., J.A.E., B.L., V.S., K. Russell, N.F., C.B., J.W., A.M.D., E.K.M., D.J.K., J.C.H., P.M.H., C.M., T.A.H., Y.J., R.R., J.J.D., N.W., J.A.M., S.T.C., and S.A.H. analyzed data; and D.J.E. and K. Russell wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: GAS, group A streptococci; MLST, multilocus sequence typing; ESI-MS, electrospray ionization mass spectrometry.
References
- 1.Gwaltney, J. M. (1995) in Principles and Practice of Infectious Diseases, eds. Mandell, G. L., Bennett, J. E. & Dolin, R. (Churchill Livingstone, New York), Vol. 1, pp. 566–572. [Google Scholar]
- 2.Smith, H. & Sweet, C. (2002) in Polymicrobial Diseases, eds. Brogen, K. A. & Guthmiller, J. M. (Am. Soc. Microbiol. Press, Washington, DC), pp. 201–212.
- 3.Bisno, A. L. (1995) in Principles and Practice of Infectious Diseases, eds. Mandell, G. L., Bennett, J. E. & Dolin, R. (Churchill Livingston, New York), Vol. 2, pp. 1786–1799. [Google Scholar]
- 4.Beall, B., Facklam, R. & Thompson, T. (1996) J. Clin. Microbiol. 34 953–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beall, B., Facklam, R., Hoenes, T. & Schwartz, B. (1997) J. Clin. Microbiol. 35 1231–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jamal, F., Pit, S., Facklam, R. & Beall, B. (1999) Emerg. Infect. Dis. 5 182–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Facklam, R., Martin, D. R., Lovgren, M., Johnson, D. R., Efstratiou, A., Thompson, T. A., Gowan, S., Kriz, P., Tyrrell, G. J., Kaplan, E., et al. (2002) Clin. Infect. Dis. 34 28–38. [DOI] [PubMed] [Google Scholar]
- 8.Enright, M. C., Spratt, B. G., Kalia, A., Cross, J. H. & Bessen, D. E. (2001) Infect. Immun. 69 2416–2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crum, N. F., Hale, B. R., Bradshaw, D. A., Malone, J. D., Chun, H. M., Gill, W. M., Norton, D., Lewis, C. T., Truett, A. A., Beadler, C., et al. (2003) Morbid. Mortal. Wkly. Rep. 52 106–109. [Google Scholar]
- 10.Jiang, Y. & Hofstadler, S. A. (2003) Anal. Biochem. 316 50–57. [DOI] [PubMed] [Google Scholar]
- 11.Hofstadler, S. A., Sampath, R., Blyn, L. B., Eshoo, M. W., Hall, T. A., Jiang, Y., Drader, J. J., Hannis, J. C., Sannes-Lowery, K. A., Cummins, L. L., et al. (2005) Int. J. Mass Spectrom. 242 23–41. [Google Scholar]
- 12.Sannes-Lowery, K. A., Drader, J. J., Griffey, R. H. & Hofstadler, S. A. (2000) Trends Anal. Chem. 19 481–491. [Google Scholar]
- 13.Muddiman, D. C., Anderson, G. A., Hofstadler, S. A. & Smith, R. D. (1997) Anal. Chem. 69 1543–1549. [DOI] [PubMed] [Google Scholar]
- 14.Facklam, R., Beall, B., Efstratiou, A., Fischetti, V., Johnson, D., Kaplan, E., Kriz, P., Lovgren, M., Martin, D., Schwartz, B., et al. (1999) Emerg. Infect. Dis. 5 247–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferretti, J. J., McShan, W. M., Ajdic, D., Savic, D. J., Savic, G., Lyon, K., Primeaux, C., Sezate, S., Suvorov, A. N., Kenton, S., et al. (2001) Proc. Natl. Acad. Sci. USA 98 4658–4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tettelin, H., Masignani, V., Cieslewica, M. J., Eisen, J. A., Peterson, S., Wessels, M. R., Paulsen, I. T., Nelson, K. E., Margarit, I., Read, T. D., et al. (2002) Proc. Natl. Acad. Sci. USA 99 12391–12396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tettelin, H., Nelson, K. E., Paulsen, I. T., Eisen, J. A., Read, T. D., Peterson, S., Heidelberg, J., DeBoy, R. T., Haft, D. H., Dodson, R. J., et al. (2001) Science 293 498–506. [DOI] [PubMed] [Google Scholar]
- 18.Smoot, J. C., Barbian, K. D., Van Gompel, J. J., Smoot, L. M., Chaussee, M. S., Sylva, G. L., Sturdevant, D. E., Ricklefs, S. M., Porcella, S. F., Parkins, L. D., et al. (2002) Proc. Natl. Acad. Sci. USA 99 4668–4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beres, S. B., Sylva, G. L., Barbian, K. D., Lei, B., Hoff, J. S., Mammarella, N. D., Liu, M.-Y., Smoot, J. C., Porcella, S. F., Parkins, L. D., et al. (2002) Proc. Natl. Acad. Sci. USA 99 10078–10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ajdic, D., McShan, W. M., McLaughlin, R. E., Savic, G., Chang, J., Carson, M. B., Primeaux, C., Tian, R., Kenton, S., Jia, H., et al. (2002) Proc. Natl. Acad. Sci. USA 99 14434–14439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kroes, I., Lepp, P. W. & Relman, D. A. (1999) Proc. Natl. Acad. Sci. USA 96 14547–14552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murray, C. J. L. & Lopez, A. D. (1996) The Global Burden of Disease (W.H.O., Boston).
- 23.Murray, C. J. L. & Lopez, A. D. (1996) Global Health Statistics (W.H.O., Geneva).
- 24.Berman, S. (1991) Rev. Infect. Dis. 13 Suppl. 6, S454–S462. [DOI] [PubMed] [Google Scholar]
- 25.Sampath, R., Hofstadler, S. A., Blyn, L., Eshoo, M., Hall, T., Massire, C., Levene, H., Hannis, J., Harrell, P. M., Neuman, B., et al. (2005) Emerg. Infect. Dis. 11 373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]