ABSTRACT
We detected highly pathogenic avian influenza A(H5N1) virus in wild rats collected from a rural area in Giza, Egypt, near poultry farms, markets, and backyard flocks. Sequence and phylogenetic analyses indicated that the virus from the rats belonged to clade 2.3.4.4b, which has been the predominant virus genotype circulating in Egypt and worldwide since 2021-2022. Active surveillance of avian influenza viruses in wild and domestic mammals is recommended to prevent further spread to mammals and humans.
KEYWORDS: Influenza, HPAI, rat, egypt, surveillance
Avian influenza viruses (AIVs) pose continuous challenges to human and animal health worldwide. Wild birds are considered the natural reservoir for AIVs and play a major role in spreading influenza viruses over long distances [1]. Highly pathogenic avian influenza (HPAI) H5N1 activity has increased globally causing mass mortality in wild birds and poultry and incidental infections in mammals. H5 clade 2.3.4.4 of the H5N1 subtype emerged in China in 2014 due to reassortment and then diversified into several clades [2]. Clade 2.3.4.4b A(H5N1) viruses were detected in birds across the five continents [3,4]. The spread of clade 2.3.4.4b of A(H5N1) viruses caused high mortality among domestic and wild birds. H5N1 clade 2.3.4.4b viruses have spilled over to several non-avian species, they were detected and isolated from domestic dogs and cats [5] and from different species of marine mammals and minks [6]. Epidemiological investigations based on serological testing showed that H5-specific antibodies were detected in foxes, polecats, and stone martens. These data showed that undetected and clinically mild HPAIV infections have occurred in wild carnivores in the Netherlands [7].
Some influenza viruses can propagate in rodents without adaptation [8]. Previous studies showed the detection of (HPAI) H5N8 virus in a mouse that was found dead in a depopulated poultry house [9] and antibodies against HPAI H5N1 virus were detected in rat sera during the initial outbreak of HPAI H5N1 virus in Hong Kong in 1997 [10]. Rodents can be abundant around poultry houses and share their habitat and may be contributing to the transmission of AIVs across poultry production sectors and across species [11]. The recent outbreak of clade 2.3.4.4b of A(H5N1) viruses in dairy cattle across several states in the United States has raised significant concern nationally and globally [12,13]. Human cases of H5N1 clade 2.3.4.4b virus infection from dairy farm workers were reported [14]. Transmission of the virus from dairy cattle to other mammals including domestic cats was reported [15].
In 2021, the first HPAI H5N1 clade 2.3.4.4b viruses were detected in wild birds and domestic ducks from live bird markets in Egypt [16]. Based on the genetic analysis of HPAI viruses isolated in Egypt in winter 2021–2022, most H5N1 HPAI viruses were genetically close to H5 HPAI circulating in Europe, Africa, and the Middle East. Here, we report the first detection of HPAI H5N1 clade 2.3.4.4b viruses in wild rats in Egypt.
In July 2023, we collected oropharyngeal and paw swab samples from wild rats from a rural area in Giza near poultry farms, markets, and backyard flocks and performed influenza A virus detection via reverse transcription PCR (universal M-gene) [17]. This work was approved by the Medical Research Ethics Committee of the National Research Centre, Egypt (protocol number 1-4-6). A total of 20 rodents, eight Rattus norvegicus, nine Rattus rattus, and three Acomys cahirinus, were trapped alive, euthanized, and then sampled. Four oropharyngeal samples from Rattus norvegicus and four oropharyngeal samples from Rattus rattus were positive for influenza A virus but none of the paw swabs (Table S1). The eight positive samples were inoculated into the allantoic cavity of 10-day-old embryonated chicken eggs (ECEs) and incubated for 48 h post-injection at 37°C, and then chilled at 4°C overnight, and analysed by hemagglutination assay (HA) using 0.5% chicken red blood cells (RBCs). From the four positive oropharyngeal samples from Rattus norvegicus, two were HA positive and confirmed by reverse transcription PCR of M gene (Table S1). Whole genome sequencing was attempted on RNA from the two isolates using Illumina's Nextera XT DNA Sample Preparation kit as previously described [16].
The two positive samples were confirmed to be positive for H5N1 using the whole genome sequencing and their full genomes (A/Rat/Egypt/STK001/2023 and A/Rat/Egypt/STK003/2023) were generated and submitted to GISAID under accession numbers EPI3276046-EPI3276053 and EPI3276054-EPI3276061. We compared the complete sequence of the HPAI H5N1 virus found in the rats (A/Rat/Egypt/STK001/2023 and A/Rat/Egypt/STK003/2023) with other highly pathogenic avian influenza (HPAI) H5N1 virus sequences available in GISAID selected based on a BLAST search. Sequence analysis to determine nucleotide identity between the two isolates of HPAI A(H5N1) viruses revealed that the eight segments had 99.9% to 100% sequence similarity. Also, the two viruses had a high nucleotide identity (99–100%) with the HPAI A(H5N1) viruses of clade 2.3.4.4b from Egypt circulating since 2021 (Table S2).
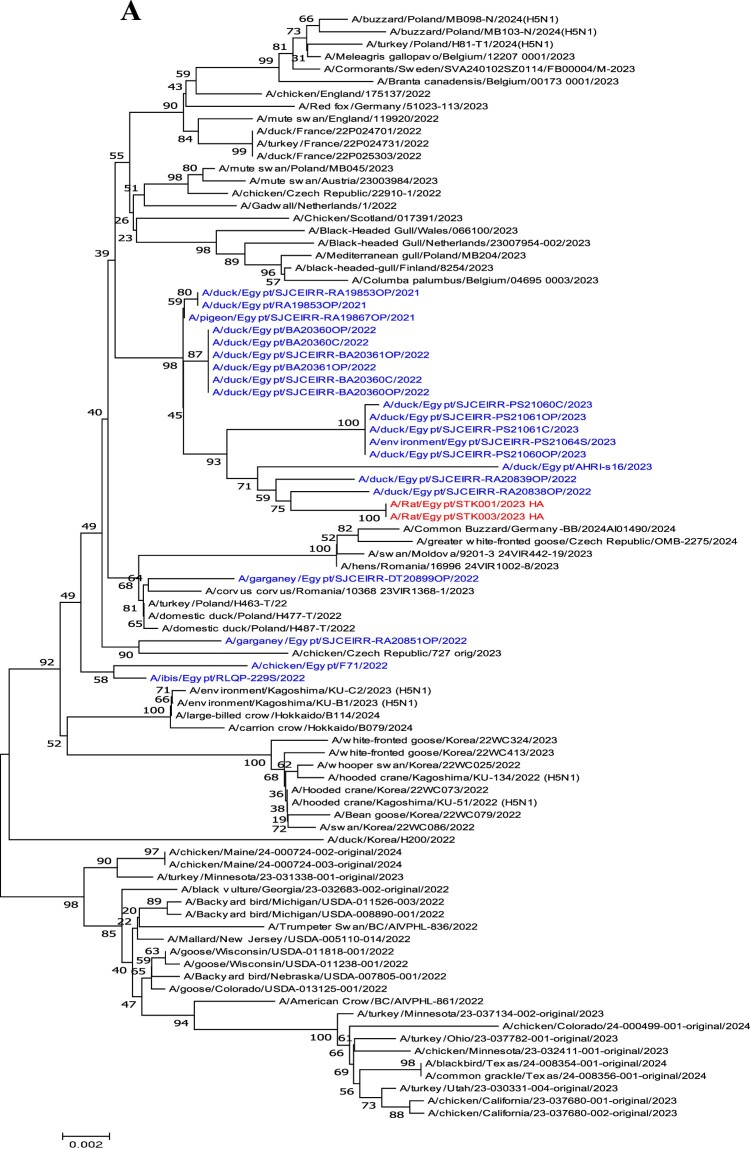
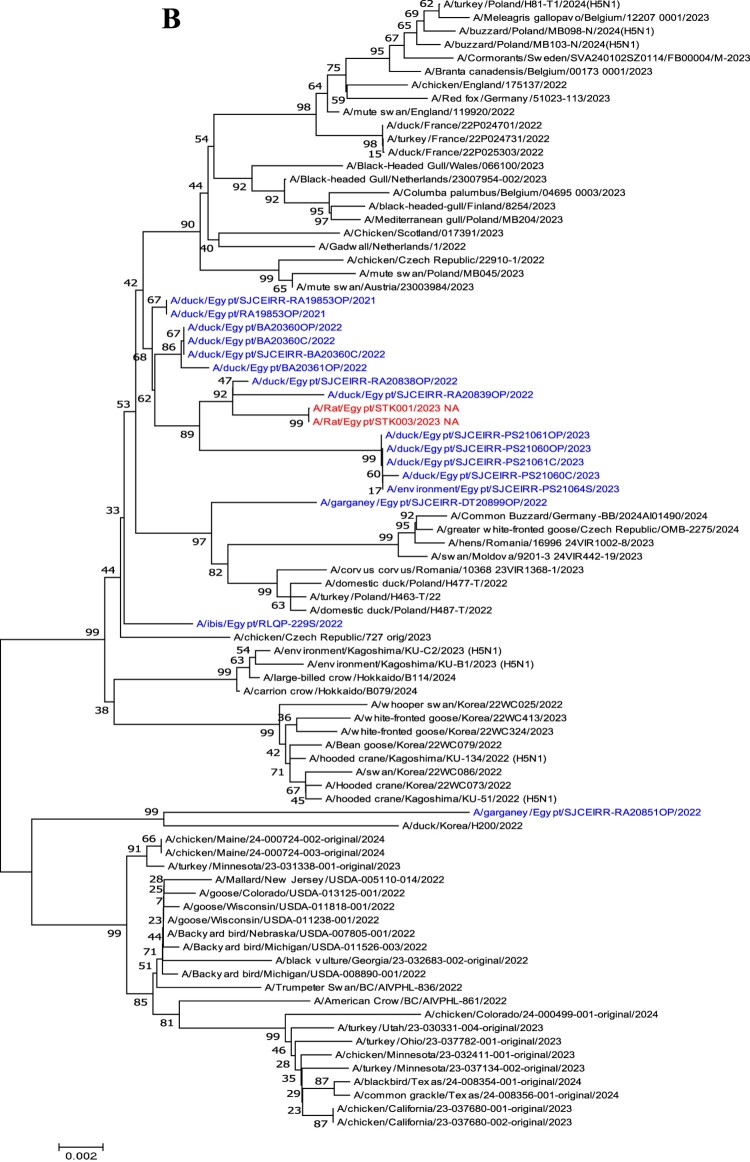
Phylogenetic analyses of the HA and NA genes (Figure 1(A,B)) show that the two HPAI H5N1 viruses cluster together and with HPAI H5N1 viruses collected from ducks (A/Muscovy duck/Egypt/RA20838OP/2022 and A/Muscovy duck/Egypt/RA20839OP/2022). Further examination of the HA gene from the two isolates revealed the presence of a polybasic amino acid sequence at the cleavage site (PLREKRRKR/GLF). The receptor binding sites in the viral HA gene of the two viruses possess the conservative amino acid residues (including 226Q, and 228G; H3 numbering), which indicates that these viruses would preferentially bind to α−2,3-sialic acid linkage, the avian-like receptors (Table S3). The analysis of the NA gene of rat viruses revealed that none of these viruses displayed NA inhibitor – associated resistance markers (Table S3).
Figure 1.
Phylogenetic tree of the (A) HA segment and (B) NA segment obtained from this study. Phylogenetic analysis was performed using the neighbour-joining algorithm with the Kimura two-parameter model. The reliability of phylogenetic inference at each branch node was estimated by the bootstrap method with 1000 replications. Evolutionary analyses were conducted using MEGA 7. Rat viruses isolated in this study are shown in red and Egyptian H5N1viruses clade 2.3.4.4b are shown in blue.
Phylogenetic analyses of the six internal genes (PB2, PB1, PA, NP, M, and NS) of the H5N1 isolates showed that it belonged to the Egyptian lineage (Figure S1). Amino acid mutations at positions K526R, E627K, and D701N in the PB2 protein have been associated with enhanced pathogenicity and transmission of influenza virus in mammalian hosts [18]. The rat viruses maintained the avian virus signature at positions 526, 627, and 701 of PB2 (R, K, and N, respectively). However, the amino acid L89V, G309D, and T339K substitutions were observed in the PB2 gene, and N66S mutation in PB1-F2 gene, which play a role in the replication process of AIVs in mammals [19]. P42S and V149A substitutions in the NS1 protein were detected in two viruses (Table S3).
Serum samples from the rats were analysed for the detection of antibodies against A/pintail/Egypt/RA19853OP/2021 clade 2.3.4.4b H5N1 virus using microneutralization (MN) assay. All sera were negative.
Low biosecurity measures, especially in backyards and markets, provide more chances for close contact between backyard birds and different animal species, creating an opportunity for interspecies transmission of influenza viruses. Dead birds infected with H5N1 are frequently discarded in open surfaces and consumed by stray animals. Such practices typically increase the chances of virus endemicity and increase viral loads on farms and surrounding environments.
Here, we characterized HPAI viruses from rodents. This has important implications on the management and control of the disease as rodents may be a potential route of virus transmission. To reduce the dissemination of AIV on poultry farms or backyard flocks, control measures including facilities’ maintenance, habitat management, removal of food sources, and pest control must be applied. This applies to Egypt and other countries with similar issues especially that the species sampled are wide spread almost globally. Limitations of this study include the small sample size, variation of the types of wild mammals sampled, and the data’s inadequacy to distinguish infection from contamination. HPAI H5N1 cases in mammals continue to be reported globally. Enhanced active surveillance in wild animals and understanding the role of wild mammals in the maintenance and spread of influenza viruses are needed.
Supplementary Material
Acknowledgements
The authors would like to acknowledge Human Link DMCC for providing administrative and technical support to this project. This publication is funded by Human Link DMCC.
Funding Statement
This study was financially supported by the STDF for the research fund under the Center of Scientific Excellence upgrading Fund (46654) and by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, US Department of Health and Human Services (under contract 5N93021C00016).
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Webster RG, Bean WJ, Gorman OT, et al. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56(1):152–179. doi: 10.1128/mr.56.1.152-179.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Role for migratory wild birds in the global spread of avian influenza H5N8. Science. 2016;354(6309):213–217. doi: 10.1126/science.aaf8852 [DOI] [PMC free article] [PubMed]
- 3.Tian J, Bai X, Li M, et al. Highly pathogenic avian influenza virus (H5N1) clade 2.3.4.4b introduced by wild birds, People’s Republic of China, 2021. Emerg Infect Dis. 2023;29(7):1367–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xie R, Edwards KM, Wille M, et al. The episodic resurgence of highly pathogenic avian influenza H5 virus. Nature. 2023;622(7984):810–817. doi: 10.1038/s41586-023-06631-2 [DOI] [PubMed] [Google Scholar]
- 5.Domańska-Blicharz K, Świętoń E, Świątalska A, et al. Outbreak of highly pathogenic avian influenza A(H5N1) clade 2.3.4.4b virus in cats, Poland, June to July 2023. Eurosurveillance. 2023;28(31):2300366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leguia M, Garcia-Glaessner A, Muñoz-Saavedra B, et al. Highly pathogenic avian influenza A (H5N1) in marine mammals and seabirds in Peru. Nat Commun. 2023;14(1):5489. doi: 10.1038/s41467-023-41182-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chestakova IV, van der Linden A, Bellido Martin B, et al. High number of HPAI H5 virus infections and antibodies in wild carnivores in the Netherlands, 2020–2022. Emerg Microbes Infect. 2023;12(2):2270068. doi: 10.1080/22221751.2023.2270068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thangavel RR, Bouvier NM.. Animal models for influenza virus pathogenesis, transmission, and immunology. J Immunol Methods. 2014;410:60–79. doi: 10.1016/j.jim.2014.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Velkers F, Elbers ARW, Bouwstra RJ, et al. H5N8 in Nederland in 2014: Een nadere blik op de uitbraken. 2015.
- 10.Shortridge KF, Gao P, Guan Y, et al. Interspecies transmission of influenza viruses: H5N1 virus and a Hong Kong SAR perspective. Vet Microbiol. 2000;74(1-2):141–147. doi: 10.1016/S0378-1135(00)00174-7 [DOI] [PubMed] [Google Scholar]
- 11.Velkers FC, Blokhuis SJ, Veldhuis Kroeze EJB, et al. The role of rodents in avian influenza outbreaks in poultry farms: a review. Vet Q. 2017;37(1):182–194. doi: 10.1080/01652176.2017.1325537 [DOI] [PubMed] [Google Scholar]
- 12.Abdelwhab EM, Beer M.. Panzootic HPAIV H5 and risks to novel mammalian hosts. npj Viruses. 2024;2(1):22. doi: 10.1038/s44298-024-00039-z [DOI] [Google Scholar]
- 13.Sah R, Srivastava S, Kumar S, et al. Concerns on H5N1 avian influenza given the outbreak in U.S. dairy cattle. Lancet Reg Health Am. 2024;35:100785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.WHO . Influenza at the human-animal interface summary and assessment, 7 June 2024. 2024; Available from: https://www.who.int/publications/m/item/influenza-at-the-human-animal-interface-summary-and-assessment–7-june-2024
- 15.Burrough ER, Magstadt DR, Petersen B, et al. Highly pathogenic avian influenza A(H5N1) clade 2.3.4.4b virus infection in domestic dairy cattle and cats, United States, 2024. Emerg Infect Dis. 2024;30(7):1335–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.El-Shesheny R, Moatasim Y, Mahmoud SH, et al. Highly pathogenic avian influenza A(H5N1) virus clade 2.3.4.4b in wild birds and live bird markets, Egypt. Pathogens. 2022;12(1):36. doi: 10.3390/pathogens12010036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Centers for Disease Control and Prevention . CDC realtime RT-PCR (rRTPCR) protocol for detection and characterization of influenza (version 2007), in CDC REF# I-007-05. 2007.
- 18.Steel J, Lowen AC, Mubareka S, et al. Transmission of influenza virus in a mammalian host is increased by PB2 amino acids 627K or 627E/701N. PLoS Pathog 2009;5:e1000252. doi: 10.1371/journal.ppat.1000252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmolke M, Manicassamy B, Pena L, et al. Differential contribution of PB1-F2 to the virulence of highly pathogenic H5N1 influenza A virus in mammalian and avian species. PLoS Pathog. 2011;7(8):e1002186. doi: 10.1371/journal.ppat.1002186 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.