Abstract
Objective
Clinical validity of genome sequencing (GS) (>30×) has been preliminarily verified in the post-natal setting. This study is to investigate the potential utility of trio-GS as a prenatal test for diagnosis of central nervous system (CNS) anomalies.
Methods
We performed trio-based GS on a prospective cohort of 17 foetuses with CNS abnormalities. Single nucleotide variation (SNV), small insertion and deletion (Indel), copy number variation (CNV), structural variant (SV), and regions with absence of heterozygosity (AOH) were analyzed and classified according to ACMG guidelines.
Results
Trio-GS identified diagnostic findings in 29.4% (5/17) of foetuses, with pathogenic variants found in SON, L1CAM, KMT2D, and ASPM. Corpus callosum (CC) and cavum septum pellucidum (CSP) abnormalities were the most frequent CNS abnormalities (47.1%, 8/17) with a diagnostic yield of 50%. A total of 29.4% (5/17) foetuses had variants of uncertain significance (VUS). Particularly, maternal uniparental disomy 16 and a de novo mosaic 4p12p11 duplication were simultaneously detected in one foetus with abnormal sulcus development. In addition, parentally inherited chromosomal inversions were identified in two foetuses.
Conclusion
GS demonstrates its feasibility in providing genetic diagnosis for foetal CNS abnormalities and shows the potential to expand the application to foetuses with other ultrasound anomalies in prenatal diagnosis.
Keywords: Prenatal diagnosis, genome sequencing, central nervous system, genomic variant
Introduction
Foetal structural malformations of the brain and other parts of the central nervous system (CNS) are among the most common congenital abnormalities, which have a prevalence at birth of 1 to 2:1000 [1,2]. Most of them can be detected by imaging strategies especially ultrasonography and magnetic resonance imaging (MRI) [3]. In addition to the severity of the imaging findings, making pregnancy decisions requires considering a combination of causes, the prognosis for neurodevelopment and function, neonatal care requirements, and recurrence risk [4,5]. However, most anomalies involving brain growth and cortical differentiation might occur during the second or third trimesters of pregnancy [5]. In order to have enough time for genetic counselling and pregnancy management, it is crucial to have genetic diagnosis as early as possible.
Conventional genetic tests include karyotyping analysis, fluorescence in situ hybridization (FISH), and chromosomal microarray analysis (CMA)/low-coverage genome sequencing (CNV-seq) are commonly used to detect numerical disorder, copy number variant (CNV), and structural variant (SV). The estimated proportion of foetuses with CNS who have chromosomal abnormalities and pathogenic CNVs was 6.5 ∼ 10.9% [6–8]. In recent years, exome sequencing (ES) was also used for the prenatal diagnosis of monogenic diseases with CNS abnormalities. Combined data from various studies, the incremental diagnostic yield of ES over karyotype and/or CMA in foetuses with CNS anomalies was 32% (95% Cl 27–36; I2 = 72%) [9].
Although through current methods, 30 ∼ 40% pathogenic genomic variants including aneuploidy, single nucleotide variant (SNV)/small insertion and deletion (Indel), CNV, and a part of SV could be detected in foetuses with CNS anomalies [6–9], there still exists a significant proportion of foetuses with CNS abnormalities beyond routine detection methods. Therefore, performing a comprehensive genetic testing like high-depth genome sequencing (GS) (>30×) [10,11] for prenatal diagnosis, might be a feasible approach to unravel the genetic aetiology of foetal CNS abnormalities. The clinical effectiveness of GS has been preliminarily verified in the postpartum environment, showing the ability to detect the above variants, as well as SV, regions with absence of heterozygosity (ROH) and other genomic variants. However, its use in the prenatal diagnosis has not yet been fully developed and there is still insufficient data to support it due to complexity of interpretation and high cost. Based on this, this purpose of this study is to perform GS in foetuses with CNS abnormalities to investigate the diagnostic efficacy and feasibility of GS in foetuses with CNS abnormalities.
Methods
Study design and participants
The study enrolled pregnant women with foetal central nervous system anomalies detected by ultrasonography or MRI from February to September 2022.
Inclusion criteria were as following: (1) Foetal CNS abnormalities (including absence of corpus callosum, except for single choroid cyst, mild ventriculomegaly and other ultrasound soft markers). (2) Samples were processed for rapid aneuploidy detection (RAD) and maternal contamination by QF-PCR (quantitative fluorescence polymerase chain reaction). Only samples with normal QF-PCR results for chromosomes 13, 18, 21, and sex chromosomes were included. (3) Genomic DNA samples of the foetuses and parents were available. Exclusion criteria were pregnancies with known history of intrauterine infection, exposure to teratogenic factors during this pregnancy or family history of known genetic disorders.
Refer to the classification criteria of Blayney et al. [9], foetal central nervous system abnormalities were divided into (1) abnormal brain development; (2) posterior fossa anomalies; (3) abnormal ventricular; (4) abnormal midline; (5) malformations of cortical development. CNS anomaly subgroups were divided into single CNS abnormality, complex CNS abnormalities (two or more types of abnormalities) and non-isolated CNS abnormalities (combined with other system abnormalities, such as skeletal system, cardiovascular system, etc.).
All the enrolled cases have received clinical genetic tests including CNV-Seq and ES, independent of GS. CNV-Seq and ES were performed by our in-house methods previously published [12,13]. Parents underwent pre-test counselling including the detection scope and limitations of GS. All the parents provided written informed consent. This study was approved by the ethical committee of the First Affiliated Hospital of Zhengzhou University (Ethics No. 2021-KY-0290-002) and conformed to the ethical standards for medical research involving human subjects, as laid out in the 1964 Declaration of Helsinki and its later amendments.
DNA preparation
Foetal DNA was obtained from one chorionic villi sampling (CVS), thirteen amniotic fluid (AF) and three umbilical cord blood. In case 9, CVS was performed at 11+6 weeks of gestation by CNV-seq due to increased nuchal translucency. The case was later enrolled in the study at 29 weeks of gestation because of foetal CNS abnormalities. Parental peripheral blood samples were collected for DNA extraction for trio‐based analyses. The genomic DNA of foetuses and their parents were extracted using the QIAamp DNA extraction kit (Qiagen, Germany), and then quantified by using Qubit 4.0 (Thermo Fisher Scientific Inc., USA). In all prenatal samples, maternal contamination was excluded by QF-PCR.
Genome sequencing
DNA libraries were constructed by using DNA PCR-Free library Prep (Illumina, San Diego, CA, USA), followed by sequencing on the NovaSeq6000 platform (Illumina, San Diego, CA, USA) with 150 bps paired-end reads. Quality control (QC) information included: (1) average output data amount per case was 100 Gb; (2) average sequencing depth for nuclear exome regions was over 30×, more than 90% of the regions reached 20× sequencing coverage.
The paired-end reads were assessed by using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and were subsequently aligned to the human reference genome (hg19) by Burrows–Wheeler Aligner (BWA) [14] and reformatted with SAMtools [15]. SNV and Indel detection were performed with HaplotypeCaller v3.4 from the Genome Analysis Toolkit (GATK, Broad Institute) and annotation by ANNOVAR and InterVA with public and our internal database [16–20]. CNV (read‐depth‐based and chimeric‐read‐based detection method), structural rearrangement (SV) analyses (such as translocations, inversions, and insertions) and regions with absence of homozygosity/uniparental disomy (AOH/UPD) were performed using our in-house bioinformatics analysis pipelines previously published [21–24].
Variant interpretation
Pathogenicity of SNVs, Indels, and CNVs were evaluated in accordance with guidelines of the American College of Medical Genetics and Genomics (ACMG) and the Association for Molecular Pathology (AMP). Variants were classified into I to V classes according to standards and guidelines for interpretation of sequence variants of ACMG (class V: pathogenic; class IV: likely pathogenic; class III: VUS; class II: likely benign; class I: benign). For the foetuses, we only reported pathogenic (P, class V) and likely pathogenic variants (LP, class IV) associated with foetal phenotypes.
SVs were interpreted according to gene disruption or potential gene dysregulation by the disruption of regulatory elements or topological associated domains (TAD, 3D Genome browser, http://3dgenome.fsm.northwestern.edu/index.html) [25].
For VUS findings, ACMG recommends to report VUS in genes related to the foetal phenotype, especially for autosomal recessive conditions if a VUS is found in ‘trans’ with a pathogenic/likely pathogenic variant. We reported these VUS results after reviewing and discussing by multidisciplinary team (MDT), which comprised of clinical geneticists, genetic laboratory specialist, paediatricians and obstetricians. For incidental findings in the foetus, we followed the ACMG recommendation to report pathogenic/expected pathogenic variants in genes known to cause moderate or severe childhood-onset disorders [26]. Additionally, carrier status was analyzed for the enrolled couples, P/LP heterozygous variants in autosomal or X-linked recessive disorders as well as clinically actionable disorders were reported as parental incidental findings, not limited to the ACMG SF3.0 [27]. PCR amplification and Sanger sequencing were used to confirm candidate SNV/Indel and SV (e.g. spurious alignment signals) results.
Statistical analysis
Statistical analysis was performed using SPSS 26.0 software. McNemar test was used to compare the VUS findings of GS and clinical genetic tests. p < 0.05 was considered to indicate statistical significance.
Results
Demographics of enrolled cases
Trio-GS was performed on 17 families with different types of foetal CNS abnormalities. The clinical information of all enrolled foetuses, including the gestational age, foetal imaging phenotype, and family history was shown in Table 1 and Supplementary Table 1. The maternal age ranged from 19 to 35 years old, with median maternal age of the cohort was 27 years old and average age was 28.4 years old. The paternal age ranged from 19 to 35 years old, with median paternal age of 28 years old and average age of 28.6 years old. The gestational age for enrolled cases ranged from 19+4 weeks to 32+1 weeks, with the median gestational age at 25+3 weeks.
Table 1.
Diagnostic findings identified by trio-GS in the cohort of foetuses with central nervous system abnormalities.
| No | CNS phenotype | Other phenotypes | GA (wk) | Family history | Gene (OMIM) | Variant | Inheritance | Variant classification (ACMG criteria) | Related disease and inheritance mode | Outcome |
|---|---|---|---|---|---|---|---|---|---|---|
| 3 | ACC, CSP anomalies | Tetralogy of fallot, prenasal skin thickening | 22 + 3 | – | SON (617140) |
NM_032195.3 c.6127C > T (p. Arg2043Ter) |
De novo | P(PVS1 + PS2 + PM2_ Supporting) | ZTTK syndrome, AD | TOP |
| 4 | Hydrocephalus, ACC | – | 32 + 1 | – | L1CAM (308840) |
NM_000425.5 c.3531-12G > A |
Maternal | LP(PS3+ PM2_ Supporting + PP4) | Hydrocephalus, XL | NA |
| 7 | Hydrocephalus | – | 24 | Previous two pregnancies of hydrocephalus | L1CAM (308840) |
NM_000425.5 c.1258del (p. Tyr420ThrfsTer10) |
Maternal | LP(PVS1 + PM2_ Supporting) | Hydrocephalus, XL | TOP |
| 9 | CSP anomalies | Increased NT, increased NF, increased left renal parenchyma echogenicity, hydronephrosis, pelvic ectopic kidney with pyelectasis, and small gallbladder | 29 | – | KMT2D (602113) |
NM_003482.4 c.8401C > T (p. Arg2801Ter) |
De novo | P(PVS1 + PS2 + PP5 + PM2_ Supporting) | Kabuki syndrome 1, AD | TOP |
| 12 | Microcephaly, CSP anomalies | Increased NF, Increased nasofrontal angle, prefrontal skin thickening, TR | 25 + 5 | – | ASPM (605481) |
NM_018136.5 c.1592_1595del (p. Val531GlufsTer17) /c.9744del (p. Lys3248AsnfsTer14) |
Maternal/Paternal | LP(PVS1 + PM2_ Supporting)/ LP(PVS1 + PM2_ Supporting) | Microcephaly 5 primary autosomal recessive, AR | NA |
ACC, aplasia of the corpus callosum; CSP, cavum septum pellucidum; NF, nuchal fold, TR, tricuspid regurgitation; TOP, termination of pregnancy; NA, not available.
The foetal imaging phenotypes included CNS anomalies with abnormalities out of CNS in seven cases (41.2%, 7/17), single CNS abnormalities in four cases (23.5%, 4/17) and complex CNS abnormalities in six cases (35.3%, 6/17). The most frequent type of CNS anomalies was abnormal midline, especially was the corpus callosum (CC) and cavum septum pellucidum (CSP) (47.1%, 8/17) (Table 1 and Supplementary Tables 1 and 2). Besides various CNS anomalies, cardiovascular anomalies were the most commonly observed extra‐CNS findings (3/17). Average GS sequencing read‐depth reached at least 30× for each sample.
Diagnostic yield
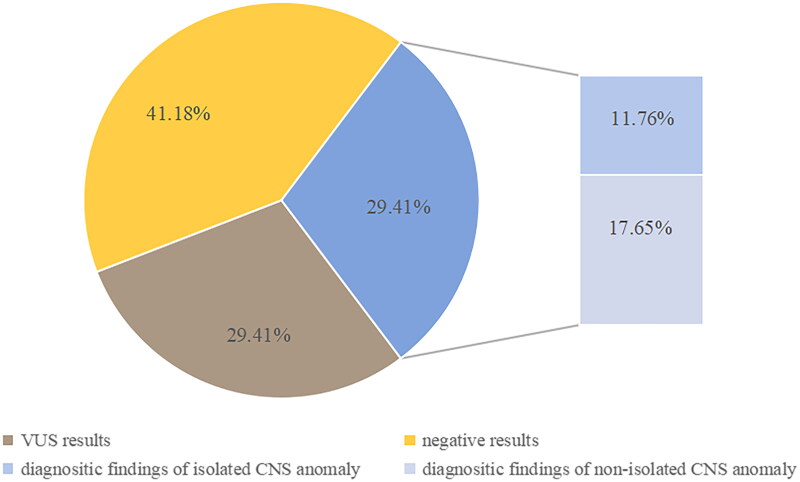
Overall, our study detected diagnostic variants in 5 of the 17 trios (29.4%), including six pathogenic variants detected in SON, L1CAM, KMT2D and ASPM. All these variants were associated with genetic syndromes (Table 1 and Figure 1). Two of these diagnoses were from autosomal dominant (AD) genes, including SON and KMT2D and one was from autosomal recessive (AR) gene, ASPM. Variants from an X-linked gene, L1CAM, were identified in two male foetuses. All the AD genetic variants were de novo variants, all the XLR genetic variants were inherited from the mothers, and the complex heterozygous variants of the foetus with AR genetic diseases were inherited from their parents. Two of the six pathogenic variants were nonsense variants in SON and KMT2D genes, three were frameshift variants in ASPM and KMT2D genes, and one splicing variant in L1CAM gene (Table 1). The diagnostic rate of foetuses with non-isolated CNS anomalies was 3/7(42.9%). The diagnosis rate of isolated CNS anomalies (single CNS anomalies and complex CNS anomalies) was 2/10 (20%).
Figure 1.
Diagnostic rate of trio-based GS in 17 foetuses with Central nervous system abnormalities. VUS, variant of uncertain significance; CNS, central nervous system abnormalities
Among the diagnosed foetuses, case 4 and case 7 had isolated CNS anomalies, and both of them showed ventriculomegaly or hydrocephalus. Case 3, 9 and 12 had ultrasound anomalies beside CNS, therefore, they were likely affected by genetic syndromes involving multiple systems. In addition, four cases (80%) including case 3, 4, 9 and 12 all had CSP or CC abnormalities. There were totally 8 cases of CSP or CC abnormalities in this study, and the diagnostic rate of isolated ACC was 25% (1/4), that with non-isolated was 75% (3/4).
SNVs/Indels of unknown significance
Four cases carried uncertain significant heterozygous variants, including three cases with one pathogenic/likely pathogenic variant and one VUS in trans detected (Supplementary Table 2 and Figure 1). For example, case2-F presented with cerebral haemorrhage and clubfoot. One maternally inherited likely pathogenic variant and one paternally inherited VUS variant were detected by GS. However, LAMC3 defects lead to occipital epidermal malformation [OMIM:614115], and case2-F have no particularly typical imaging findings such as malformations of cortical development characterized by polymicrogyria and pachygyria of the occipital lobes till 31 weeks of gestation. Therefore, there is insufficient evidence to support the pathogenicity of the variants.
Chromosome structural variants
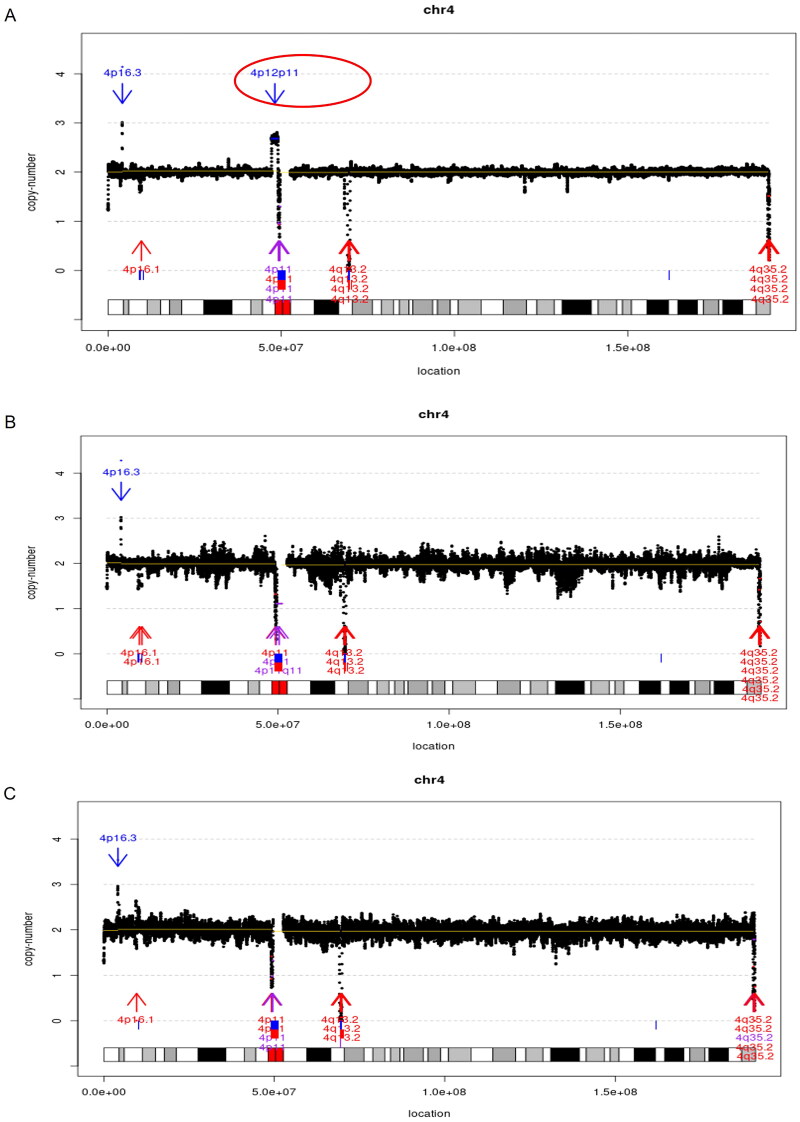
In particular, a de novo 4p12p11 duplication at a mosaic level of 68% ([GRCh37] dup(4)(p12p11)dn chr4:g.47235624_49094057dup [0.7]) and maternal uniparental disomy 16 were simultaneously detected in case16 (Figure 2 and Supplementary Table 2). In addition, a 1.78 Mb inv (4) in case3 and a 3.22 Mb inv (1) in case10 were incidentally identified in the foetus and found to be parentally inherited.
Figure 2.
Chromosomal structural variants identified in case16. A, mosaic 4p12p11 duplication detected in the foetus (red circle, mosaic level: 68%). B&C, parental confirmation results indicated de novo occurrence of the duplication.
Incidental findings
Trio-based GS also identified the carrier status of the 17.6% (3/17) couples, including a likely pathogenic SNV in EVC2 in case7‐P (father), a pathogenic SNV in CFTR in case7-M (mother), a pathogenic SNV in NF1 in case14-M (mother), a likely pathogenic SNV in BLM in case16‐P (Supplementary Table 3).
Comparison of results between GS and CNV-Seq plus ES
Among the 17 families with diagnosed findings, 29.4% (5/17) of the foetuses were detected by GS to have SNVs/Indels. In the families with negative results, 5 cases acquired VUS results (29.4%, 5/17), and incidental findings of carrier status were found in 3 couples. Compared with CNV-Seq plus ES strategy, GS showed consistent positive results (29.4%, 5/17) and incidental findings. However, GS found more VUS findings, but there was no significant difference (29.4% vs 11.8%, p = 0.25).
The additional VUS findings of GS included variants in noncoding regions in three cases. In case 2, case 11 and case 14, trio-GS could detect compound heterozygous variants with pathogenic/likely pathogenic variants and VUS. All the VUS variants were detected in non-coding region (Supplementary Table 2), which could not be found by CNV-Seq or ES.
In case 16, a 4p12p11 duplication with 68% mosaicism and an extra maternal UPD (16) of undetermined significance were detected by GS at the same time. By contrast, CNV-Seq only detected [GRCh37]dup(4)(p12p11) chr4:g.47307108_49053030dup), the UPD(16) were not identified by both CNV-Seq and ES.
In case 3 and 10, trio-GS demonstrated the extra ability to identify chromosomal inversions compared with CNV-Seq and ES. However, the two inversions were inherited from the parents and considered to be clinically unsignificant.
Follow-up results
Among the 17 cases, 76.5% (13/17) pregnancies were termination of pregnancy (TOP), 11.8% (2/17) cases lost to follow-up, 11.8% (2/17) had a live birth including one case with neonatal death. 5 cases with positive results all chose TOP. And the both foetuses of live births had negative results, one of which with cerebellar haemorrhage and foot varus live birthed. The other case born prematurely due to intrauterine distress and the newborn boy did not survive due to neonatal respiratory distress syndrome.
Discussion
In our cohort, GS was performed in foetuses with CNS abnormalities, and a total of 29.4% (5/17) foetuses were diagnosed. The diagnostic rate of non-isolated CNS anomalies was higher (42.9%, 3/7) than isolated CNS anomalies (20.0%, 2/10), which is in line with other studies [28–30]. There were four cases with single CNS anomaly with a diagnostic rate of 25% (1/4) (Table 1 and Supplementary Table 1).
In this study, 40.0% foetuses had de novo AD disorders. The advanced parental age may lead to gonadal mosaicism, which could influence the identification of de novo variants [31]. However, in the two samples with AD disorders, the ages of both parents were lower than 30 years old. Therefore, the possibility of parental mosaicism occurring in cases with de novo variants is low though the risk cannot be ruled out.
Agenesis of the corpus callosum (ACC) was the most frequent cerebral malformation in this cohort. Similar to other studies, the diagnostic rate of isolated ACC in our study is lower than when it is non-isolated, which probably have good prognosis related to learning and social deficits, such as neurodevelopmental delay [32–35]. ACC is a component of one of more than 200 different genetic syndromes, such as ZTTK syndrome, X-linked recessive; corpus callosum partial agenesis of, X-linked recessive Hydrocephalus; Kabuki syndrome; primary Microcephaly 5 found in this study. We suggest that the risk of genetic syndromes should be ruled out when syndromic ACC is suspected.
In foetuses with confirmed genetic diagnosis, two foetuses with ventriculomegaly or hydrocephalus were both diagnosed with LICAM variants. L1CAM (L1 cell adhesion molecule) gene is involved in regulating the embryonic brain development, which is the most common causative gene of hydrocephalus (1:30000), and the pathogenic variant of male patients is mostly inherited from the mother. In case7, the pregnant woman had two children with recurrent hydrocephalus in the past, and the previous children did not perform genetic testing. However, according to the inheritance mode of the L1CAM, it was more likely that the previous abnormal gestation history is caused by the variant on L1CAM.
In this study, the detection rates of trio-GS and CNV-Seq plus trio-ES were consistent, and the detection results were all SNVs/Indels, which indicated that the detection of SNV/Indel by GS was accurate. The current conventional sequencing depth of high-depth GS technology is above 30×, which allows for effectively detecting SNV/Indel, SV and other variants. A few of studies shown that the sequencing depth of GS for SNV/Indel is lower than that of ES, which may affect the detection of mosaic sequence variants [36]. Therefore, it needs to take caution when candidate SNVs/Indels with low variant allele frequency are found, these variants should be further verified by sanger sequencing or other verification techniques.
Except for the positive diagnostic findings, GS also detected SNVs of unknown significance, including three in noncoding regions in case 2, 11 and 14. It is worth noting that in case 11, the pregnant woman had the history of two pregnancies with foetuses having hydrocephalus, indicating the foetus is at risk for genetic disorders. GS detected variants in non-coding region of CRPPA and the foetal phenotypes overlapped with some patients with the gene-related diseases [37]. Many studies have reported that variants in non-coding region and deep intronic region could cause genetic disorders, and the ACMG guidelines have been applied to the exploration of non-coding sequence variants [38]. However, the evidence for the pathogenicity of non-coding region variants is still limited. Finally, due to the limitation of prenatal phenotypes, the variants were not reported as prenatal diagnostic results. It can be seen that the interpretation of GS results might be more complicated than CNV-Seq and ES due to such variants of unknown significance. These VUS results whether should be reported is a major concern, especially those in non-coding regions, which making genetic counselling more challenging. Therefore, carrying out long-term postpartum follow-up and functional study might provide valuable information for clinical diagnosis, and is essential for interpretation of genetic variants [39].
Besides SNV/Indels, GS also shows the ability to detect structural variants including chromosomal inversion and small CNV, which echoes the findings from the other studies [20,40,41]. In case 3 and case 10, inversions were detected by utilizing paired-end reads from GS. Interestingly, maternal uniparental disomy 16 and a de novo 4p12p11 duplication with mosaic level around 68% were simultaneously found in case16. Therefore, we demonstrated the feasibility of comprehensively detecting genome variants by using GS as one-stop test in the prenatal cohort.
Notably, clinical presentations in foetuses with maternal UPD (16) are known to be variable, ranging from normal to malformations, but CNS abnormalities is not involved in the symptoms of reported cases [42]. There was also no proof that the phenotypes of case16 were caused by unbalanced expression of imprinted genes encoded on chromosome 16. In addition, no pathogenic compound heterozygous variants or homozygous variants of autosomal recessive (AR) disease genes were found in these regions. Currently, there is insufficient evidence supporting the relationship between UPD (16) and inversions and foetal phenotypes. This suggests that the genomic structural variants detected by GS could bring challenges to variant interpretation and genetic counselling. Large cohort studies are need to further explore the clinical significance of GS in detecting structural variants.
Previous studies have demonstrated that GS has a larger detection scope and higher detection sensitivity than combination of ES and CNV analysis. For CNVs, compared with CMA, these approaches may be affected by the platform adopted and the probe-distribution strategy which may lead to missed detection. GS has higher sequencing depth and resolution than CNV-Seq, which can stably detect small CNV (< 50 kb), and pinpoint the boundaries of each CNV, as well as to detect mosaicism with higher sensitivity. GS could also detect intragenic CNVs while ES may miss the detection of CNVs due to the capture bias. For chromosomal balance rearrangements (BCRs), although karyotyping combined with CMA/CNV-seq could confirm BCRs and determine whether there are any cryptic copy number variants, GS could further delineate the breakpoint junctions. Though we did not find disruption of disease-causing genes in this study because of the small sample size, several studies have shown the ability of GS to detect balanced chromosomal translocations. Both Cao et al. and Zhou et al. [29,43] found parents for BCRs carriers by GS, and clarified the detailed information and causes of foetal CNVs. Therefore, we believe that GS can be used as a one-stop solution to detect most types of genetic abnormalities.
In addition, GS is suitable for implementation in prenatal diagnosis because less amount of foetal DNA is required when compared with routine genetic tests, such as ES and low coverage GS. Especially, PCR-Free assay was used in this study, which could reduce the input DNA to 50-100ng in the GS experiment. According to a recent meta-analysis, the pooled median of TAT for GS was shown to be 18 days [44]. A 13.5-h diagnostic TAT of GS led to a clinical diagnosis of thiamine metabolism dysfunction syndrome 2 (THMD2) in a true clinical case was previously reported [45], indicating that GS yields great potential in shortening the TAT for clinical management and decision making.
There are also some limitations in our study. We included only 17 samples, the case number in each phenotype category was limited. Among these cases, VUS results were found in 5 samples, these results brought great challenge for interpretation and genetic counselling during the prenatal period. Other genetic tests such as transcriptome analysis and functional study may be solutions to demonstrate the potential pathogenic mechanism. Additionally, TOP was chosen in most cases, making it limited to further discuss about the clinical utility of GS for prenatal diagnosis.
Conclusion
In summary, trio-GS provided a diagnostic yield in 29.4% foetuses with negative QF-PCR results. This study was the first to compare trio-GS with CNV-Seq plus ES to obtain the diagnostic effect. This study demonstrates its feasibility in providing genetic diagnosis for foetuses with CNS abnormalities and it shows the potential to expand the application to foetuses with other ultrasound anomalies in prenatal diagnosis. Trio-GS could simultaneously identify various types of genomic variants in the families with foetal CNS abnormalities, providing comprehensive genetic information for these families. Studies with larger sample sizes of trio-GS could provide statistical power and robust basis for the utility of GS in prenatal diagnosis.
Supplementary Material
Acknowledgements
The authors would like to thank all the patients who contributed their samples and information for this study.
Funding Statement
This research is supported by grants from Key Scientific Research Project of Henan Province Colleges and Universities (22B320011), Guangxi Key Laboratory of birth defects open project (GXWCH-ZDKF-2022-05), Henan Provincial Science and Technology Research Project (242102310113) and Henan Province Medical Science and Technology Research Joint Project (LHGJ20210340). The sequencing kits were supplied by Illumina through a research grant program.
Ethics statement
The project passed the review of the ethics committee by the Ethics Committee for Scientific Research and Clinical Trials of the First Affiliated Hospital of Zhengzhou University (Ethics No.: 2021-KY-0290-002) and conformed to the ethical standards for medical research involving human subjects, as laid out in the 1964 Declaration of Helsinki and its later amendments. All patients and their family members signed informed consent.
Author contributions
All authors contributed to the article and approved the submitted version. Yanfei Wang, Zhi Gao, Chunxiao Hua, Meimei Liu and Jinna Jiang collected the samples. Zirui Dong, Ye Cao, Yuting Zheng, Kwong Wai Choy, Zhi Gao and Yanfei Wang performed the analysis and data interpretation. Yanfei Wang, Zhi Gao and Xiaofan Zhu wrote the manuscript. Xiangdong Kong and Xiaofan Zhu supervised the project and provided critical revisions.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data is not available to the public because of privacy. The corresponding author can be contacted if necessary.
References
- 1.Di Mascio D, Sileo FG, Khalil A, et al. Role of magnetic resonance imaging in fetuses with mild or moderate ventriculomegaly in the era of fetal neurosonography: systematic review and meta-analysis. Ultrasound Obstet Gynecol. 2019;54(2):164–171. doi: 10.1002/uog.20197. [DOI] [PubMed] [Google Scholar]
- 2.Chitty LS, Pilu G.. The challenge of imaging the fetal central nervous system: an aid to prenatal diagnosis, management and prognosis. Prenat Diagn. 2009;29(4):301–302. doi: 10.1002/pd.2242. [DOI] [PubMed] [Google Scholar]
- 3.Tanacan A, Ozgen B, Fadiloglu E, et al. Prenatal diagnosis of central nervous system abnormalities: neurosonography versus fetal magnetic resonance imaging. Eur J Obstet Gynecol Reprod Biol. 2020;250:195–202. doi: 10.1016/j.ejogrb.2020.05.013. [DOI] [PubMed] [Google Scholar]
- 4.Grosse SD, Berry RJ, Mick Tilford J, et al. Retrospective assessment of cost savings from prevention: folic acid fortification and spina bifida in the U.S. Am J Prev Med. 2016;50(5 Suppl 1):S74–s80. doi: 10.1016/j.amepre.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Onkar D, Onkar P, Mitra K.. Evaluation of fetal central nervous system anomalies by ultrasound and its anatomical co-relation. J Clin Diagn Res. 2014;8(6):AC05–AC07. doi: 10.7860/JCDR/2014/8052.4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shaffer LG, Rosenfeld JA, Dabell MP, et al. Detection rates of clinically significant genomic alterations by microarray analysis for specific anomalies detected by ultrasound. Prenat Diagn. 2012;32(10):986–995. doi: 10.1002/pd.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhi Y, Liu L, Cui S, et al. Pathogenic/likely pathogenic copy number variations and regions of homozygosity in fetal central nervous system malformations. Arch Gynecol Obstet. 2023;308(6):1723–1735. doi: 10.1007/s00404-022-06866-w. [DOI] [PubMed] [Google Scholar]
- 8.Sun L, Wu Q, Jiang SW, et al. Prenatal diagnosis of central nervous system anomalies by high-resolution chromosomal microarray analysis. Biomed Res Int. 2015;2015:426379–426379. doi: 10.1155/2015/426379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blayney GV, Laffan E, Jacob PA, et al. Monogenic conditions and central nervous system anomalies: a prospective study, systematic review and meta‐analysis. Prenat Diagn. 2023;44(4):422–431. doi: 10.1002/pd.6466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stavropoulos DJ, Merico D, Jobling R, et al. Whole genome sequencing expands diagnostic utility and improves clinical management in pediatric medicine. NPJ Genom Med. 2016;1:15012–15012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marshall CR, Chowdhury S, Taft RJ, et al. Best practices for the analytical validation of clinical whole-genome sequencing intended for the diagnosis of germline disease. NPJ Genom Med. 2020;5(1):47. doi: 10.1038/s41525-020-00154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu X, Gao Z, Wang Y, et al. Utility of trio‐based prenatal exome sequencing incorporating splice‐site and mitochondrial genome assessment in pregnancies with fetal ultrasound anomalies: prospective cohort study. Ultrasound Obstet Gynecol. 2022;60(6):780–792. doi: 10.1002/uog.24974. [DOI] [PubMed] [Google Scholar]
- 13.Shi P, Liang H, Hou Y, et al. The uncertainty of copy number variants: pregnancy decisions and clinical follow-up. Am J Obstet Gynecol. 2023;229(2):170.e1-170–e8. doi: 10.1016/j.ajog.2023.01.022. [DOI] [PubMed] [Google Scholar]
- 14.Li H, Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Handsaker B, Wysoker A, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhen Q, Yang Z, Wang W, et al. Genetic study on small insertions and deletions in psoriasis reveals a role in complex human diseases. J Invest Dermatol. 2019;139(11):2302.e14–2312.e14. doi: 10.1016/j.jid.2019.03.1157. [DOI] [PubMed] [Google Scholar]
- 17.McKenna A, Hanna M, Banks E, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang K, Li M, Hakonarson H.. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164–e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Q, Wang K.. InterVar: clinical interpretation of genetic variants by the 2015 ACMG-AMP guidelines. Am J Hum Genet. 2017;100(2):267–280. doi: 10.1016/j.ajhg.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choy KW, Wang H, Shi M, et al. Prenatal diagnosis of fetuses with increased nuchal translucency by genome sequencing analysis. Front Genet. 2019;10:761. doi: 10.3389/fgene.2019.00761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong Z, Jiang L, Yang C, et al. A robust approach for blind detection of balanced chromosomal rearrangements with whole-genome low-coverage sequencing. Hum Mutat. 2014;35(5):625–636. doi: 10.1002/humu.22541. [DOI] [PubMed] [Google Scholar]
- 22.Dong Z, Wang H, Chen H, et al. Identification of balanced chromosomal rearrangements previously unknown among participants in the 1000 Genomes Project: implications for interpretation of structural variation in genomes and the future of clinical cytogenetics. Genet Med. 2018;20(7):697–707. doi: 10.1038/gim.2017.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi M, Leng X, Li Y, et al. Genome sequencing reveals the role of rare genomic variants in Chinese patients with symptomatic intracranial atherosclerotic disease. Stroke Vasc Neurol. 2022;7(3):182–189. doi: 10.1136/svn-2021-001157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dong Z, Chau MHK, Zhang Y, et al. Low-pass genome sequencing-based detection of absence of heterozygosity: validation in clinical cytogenetics. Genet Med. 2021;23(7):1225–1233. doi: 10.1038/s41436-021-01128-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dixon JR, Selvaraj S, Yue F, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485(7398):376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Monaghan KG, Leach NT, Pekarek D, et al. The use of fetal exome sequencing in prenatal diagnosis: a points to consider document of the American College of Medical Genetics and Genomics (ACMG). Genet Med. 2020;22(4):675–680. doi: 10.1038/s41436-019-0731-7. [DOI] [PubMed] [Google Scholar]
- 27.Miller DT, Lee K, Chung WK, et al. ACMG SF v3.0 list for reporting of secondary findings in clinical exome and genome sequencing: a policy statement of the American College of Medical Genetics and Genomics (ACMG). Genet Med. 2021;23(8):1381–1390. doi: 10.1038/s41436-021-01172-3. [DOI] [PubMed] [Google Scholar]
- 28.Yang Y, Zhao S, Sun G, et al. Genomic architecture of fetal central nervous system anomalies using whole-genome sequencing. NPJ Genom Med. 2022;7(1):31. doi: 10.1038/s41525-022-00301-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou J, Yang Z, Sun J, et al. Whole genome sequencing in the evaluation of fetal structural anomalies: a parallel test with chromosomal microarray plus whole exome sequencing. Genes (Basel). 2021;12(3):376. doi: 10.3390/genes12030376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhi Y, Liu L, Wang H, et al. Prenatal exome sequencing analysis in fetuses with central nervous system anomalies. Ultrasound Obstet Gynecol. 2023;62(5):721–726. doi: 10.1002/uog.26254. [DOI] [PubMed] [Google Scholar]
- 31.Jónsson H, Sulem P, Kehr B, et al. Parental influence on human germline de novo mutations in 1,548 trios from Iceland. Nature. 2017;549(7673):519–522. doi: 10.1038/nature24018. [DOI] [PubMed] [Google Scholar]
- 32.Heide S, Spentchian M, Valence S, et al. Prenatal exome sequencing in 65 fetuses with abnormality of the corpus callosum: contribution to further diagnostic delineation. Genet Med. 2020;22(11):1887–1891. doi: 10.1038/s41436-020-0872-8. [DOI] [PubMed] [Google Scholar]
- 33.D’Antonio F, Pagani G, Familiari A, et al. Outcomes associated with isolated agenesis of the corpus callosum: a meta-analysis. Pediatrics. 2016;138(3):e20160445. doi: 10.1542/peds.2016-0445. [DOI] [PubMed] [Google Scholar]
- 34.Mangione R, Fries N, Godard P, et al. Neurodevelopmental outcome following prenatal diagnosis of an isolated anomaly of the corpus callosum. Ultrasound Obstet Gynecol. 2011;37(3):290–295. doi: 10.1002/uog.8882. [DOI] [PubMed] [Google Scholar]
- 35.Van den Veyver IB. Prenatally diagnosed developmental abnormalities of the central nervous system and genetic syndromes: a practical review. Prenat Diagn. 2019;39(9):666–678. doi: 10.1002/pd.5520. [DOI] [PubMed] [Google Scholar]
- 36.Miceikaite I, Fagerberg C, Brasch-Andersen C, et al. Comprehensive prenatal diagnostics: exome versus genome sequencing. Prenat Diagn. 2023;43(9):1132–1141. doi: 10.1002/pd.6402. [DOI] [PubMed] [Google Scholar]
- 37.Bayram N, Bayram AK, Per H, et al. Analysis of genotype-phenotype correlation in Walker-Warburg syndrome with a novel CRPPA mutation in different clinical manifestations. Eur J Ophthalmol. 2022;32(5):Np71–np76. doi: 10.1177/11206721211016306. [DOI] [PubMed] [Google Scholar]
- 38.Ellingford JM, Ahn JW, Bagnall RD, et al. Recommendations for clinical interpretation of variants found in non-coding regions of the genome. Genome Med. 2022;14(1):73. doi: 10.1186/s13073-022-01073-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu P, Vossaert L.. Emerging technologies for prenatal diagnosis: the application of whole genome and RNA sequencing. Prenat Diagn. 2022;42(6):686–696. doi: 10.1002/pd.6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qi Q, Jiang Y, Zhou X, et al. Whole genome sequencing analysis in fetal structural anomalies: novel phenotype-genotype discoveries. Ultrasound Obstet Gynecol. 2023;63(5):664–671. doi: 10.1002/uog.27517. [DOI] [PubMed] [Google Scholar]
- 41.Hu P, Zhang Q, Cheng Q, et al. Whole genome sequencing vs chromosomal microarray analysis in prenatal diagnosis. Am J Obstet Gynecol. 2023;229(3):302 e1–302 e18. doi: 10.1016/j.ajog.2023.03.005. [DOI] [PubMed] [Google Scholar]
- 42.Scheuvens R, Begemann M, Soellner L, et al. Maternal uniparental disomy of chromosome 16 [upd(16)mat]: clinical features are rather caused by (hidden) trisomy 16 mosaicism than by upd(16)mat itself. Clin Genet. 2017;92(1):45–51. doi: 10.1111/cge.12958. [DOI] [PubMed] [Google Scholar]
- 43.Cao Y, Chau MHK, Zheng Y, et al. Exploring the diagnostic utility of genome sequencing for fetal congenital heart defects. Prenat Diagn. 2022;42(7):862–872. doi: 10.1002/pd.6151. [DOI] [PubMed] [Google Scholar]
- 44.Shreeve N, Sproule C, Choy KW, et al. Incremental yield of whole genome sequencing over chromosome microarray and exome sequencing for congenital anomalies in prenatal period and infancy: systematic review and meta-analysis. Ultrasound Obstet Gynecol. 2023;63(1):15–23. doi: 10.1002/uog.27491. [DOI] [PubMed] [Google Scholar]
- 45.Owen MJ, Niemi AK, Dimmock DP, et al. Rapid sequencing-based diagnosis of thiamine metabolism dysfunction syndrome. N Engl J Med. 2021;384(22):2159–2161. doi: 10.1056/NEJMc2100365. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data is not available to the public because of privacy. The corresponding author can be contacted if necessary.