Abstract
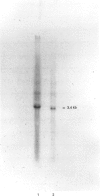
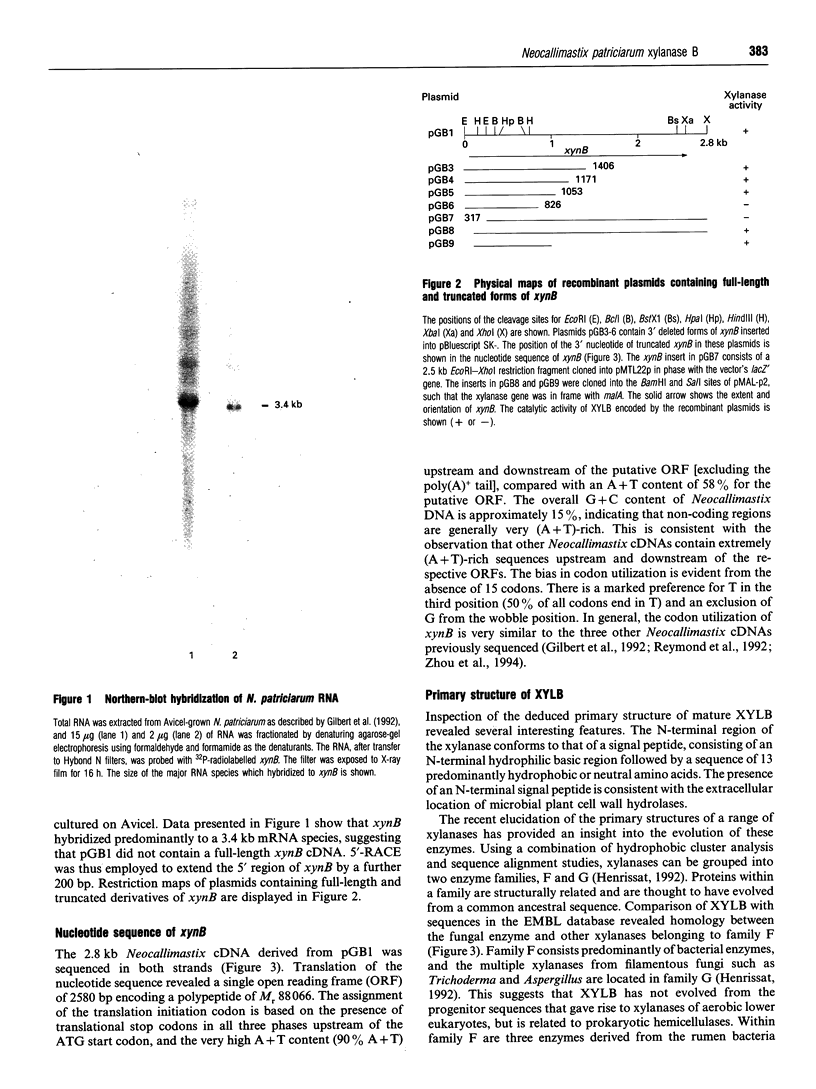
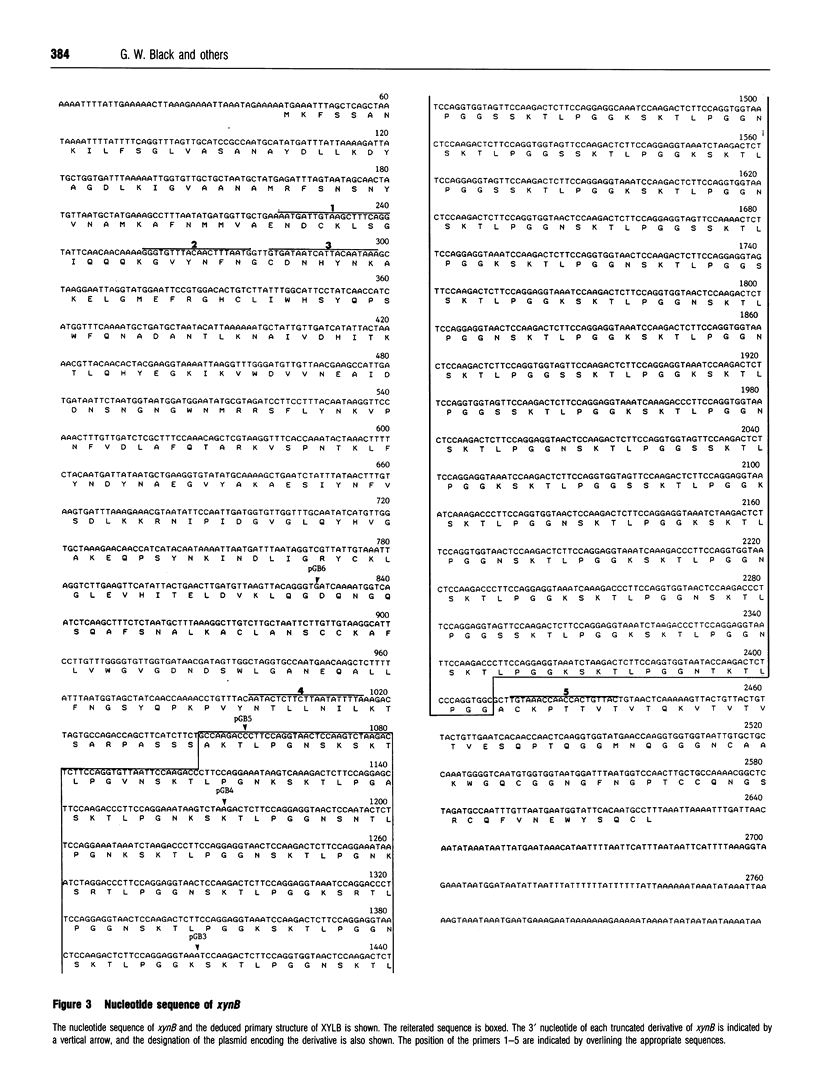
A Neocallimastix patriciarum cDNA library was screened for xylanase-expressing clones, which were distinct from the previously characterized N. patriciarum xynA cDNA encoding xylanase A. A single cDNA, designated xynB, which did not exhibit homology with xynA, was isolated. Northern-blot analysis of mRNA from Avicel-grown N. patriciarum showed that xynB hybridized to a 3.4 kb mRNA species. The nucleotide sequence of xynB revealed a single open reading frame of 2580 bp coding for a protein designated xylanase B (XYLB), of M(r) 88,066. The primary structure of XYLB was comprised of a 21-residue N-terminal signal peptide, followed by a 304-amino acid sequence that exhibited substantial homology with the catalytic domains of family F xylanases. The N-terminal domain was linked to a C-terminal 70-residue sequence by a putative linker region, comprising 12 tandem repeats of a sequence containing TLPG as the core sequence, followed by an octapeptide XSKTLPGG where X can be S, K or N, which was repeated in tandem 45 times. Truncated derivatives of xynB encoding the N-terminal 338 residues directed the synthesis of a functional xylanase, confirming that the region of XYLB, which exhibited homology with family F xylanases, constitutes the catalytic domain. To investigate the catalytic properties of XYLB, the catalytic domain was fused to the Escherichia coli maltose-binding protein, and the fusion protein purified by amylose affinity chromatography. The purified enzyme hydrolysed oat, rye and wheat arabinoxylan releasing primarily xylobiose, xylotriose and some xylose. The XYLB fusion did not cleave any cellulosic substrates. The data presented in this report suggest that the multiple xylanases of N. patriciarum arose, not through the duplication of a single gene, but by the transfer of distinct xylanase-encoding DNA sequences into the anaerobic fungus. The possible origin of the xynB gene is discussed.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chambers S. P., Prior S. E., Barstow D. A., Minton N. P. The pMTL nic- cloning vectors. I. Improved pUC polylinker regions to facilitate the use of sonicated DNA for nucleotide sequencing. Gene. 1988 Aug 15;68(1):139–149. doi: 10.1016/0378-1119(88)90606-3. [DOI] [PubMed] [Google Scholar]
- Clarke J. H., Laurie J. I., Gilbert H. J., Hazlewood G. P. Multiple xylanases of Cellulomonas fimi are encoded by distinct genes. FEMS Microbiol Lett. 1991 Oct 15;67(3):305–309. doi: 10.1016/0378-1097(91)90493-t. [DOI] [PubMed] [Google Scholar]
- Ferreira L. M., Durrant A. J., Hall J., Hazlewood G. P., Gilbert H. J. Spatial separation of protein domains is not necessary for catalytic activity or substrate binding in a xylanase. Biochem J. 1990 Jul 1;269(1):261–264. doi: 10.1042/bj2690261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint H. J., McPherson C. A., Bisset J. Molecular cloning of genes from Ruminococcus flavefaciens encoding xylanase and beta(1-3,1-4)glucanase activities. Appl Environ Microbiol. 1989 May;55(5):1230–1233. doi: 10.1128/aem.55.5.1230-1233.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert H. J., Hazlewood G. P., Laurie J. I., Orpin C. G., Xue G. P. Homologous catalytic domains in a rumen fungal xylanase: evidence for gene duplication and prokaryotic origin. Mol Microbiol. 1992 Aug;6(15):2065–2072. doi: 10.1111/j.1365-2958.1992.tb01379.x. [DOI] [PubMed] [Google Scholar]
- Gilbert H. J., Jenkins G., Sullivan D. A., Hall J. Evidence for multiple carboxymethylcellulase genes in Pseudomonas fluorescens subsp. cellulosa. Mol Gen Genet. 1987 Dec;210(3):551–556. doi: 10.1007/BF00327211. [DOI] [PubMed] [Google Scholar]
- Gilbert H. J., Sullivan D. A., Jenkins G., Kellett L. E., Minton N. P., Hall J. Molecular cloning of multiple xylanase genes from Pseudomonas fluorescens subsp. cellulosa. J Gen Microbiol. 1988 Dec;134(12):3239–3247. doi: 10.1099/00221287-134-12-3239. [DOI] [PubMed] [Google Scholar]
- Gilkes N. R., Henrissat B., Kilburn D. G., Miller R. C., Jr, Warren R. A. Domains in microbial beta-1, 4-glycanases: sequence conservation, function, and enzyme families. Microbiol Rev. 1991 Jun;55(2):303–315. doi: 10.1128/mr.55.2.303-315.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J., Hazlewood G. P., Barker P. J., Gilbert H. J. Conserved reiterated domains in Clostridium thermocellum endoglucanases are not essential for catalytic activity. Gene. 1988 Sep 15;69(1):29–38. doi: 10.1016/0378-1119(88)90375-7. [DOI] [PubMed] [Google Scholar]
- Hazlewood G. P., Davidson K., Laurie J. I., Romaniec M. P., Gilbert H. J. Cloning and sequencing of the celA gene encoding endoglucanase A of Butyrivibrio fibrisolvens strain A46. J Gen Microbiol. 1990 Oct;136(10):2089–2097. doi: 10.1099/00221287-136-10-2089. [DOI] [PubMed] [Google Scholar]
- Kellett L. E., Poole D. M., Ferreira L. M., Durrant A. J., Hazlewood G. P., Gilbert H. J. Xylanase B and an arabinofuranosidase from Pseudomonas fluorescens subsp. cellulosa contain identical cellulose-binding domains and are encoded by adjacent genes. Biochem J. 1990 Dec 1;272(2):369–376. doi: 10.1042/bj2720369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp P., Lander D. J., Orpin C. G. The lipids of the rumen fungus Piromonas communis. J Gen Microbiol. 1984 Jan;130(1):27–37. doi: 10.1099/00221287-130-1-27. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lüthi E., Love D. R., McAnulty J., Wallace C., Caughey P. A., Saul D., Bergquist P. L. Cloning, sequence analysis, and expression of genes encoding xylan-degrading enzymes from the thermophile "Caldocellum saccharolyticum". Appl Environ Microbiol. 1990 Apr;56(4):1017–1024. doi: 10.1128/aem.56.4.1017-1024.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maglione G., Matsushita O., Russell J. B., Wilson D. B. Properties of a genetically reconstructed Prevotella ruminicola endoglucanase. Appl Environ Microbiol. 1992 Nov;58(11):3593–3597. doi: 10.1128/aem.58.11.3593-3597.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matte A., Forsberg C. W. Purification, characterization, and mode of action of endoxylanases 1 and 2 from Fibrobacter succinogenes S85. Appl Environ Microbiol. 1992 Jan;58(1):157–168. doi: 10.1128/aem.58.1.157-168.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy G., Kavanagh T. Speeding-up the sequencing of double-stranded DNA. Nucleic Acids Res. 1988 Jun 10;16(11):5198–5198. doi: 10.1093/nar/16.11.5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond P., Geourjon C., Roux B., Durand R., Fevre M. Sequence of the phosphoenolpyruvate carboxykinase-encoding cDNA from the rumen anaerobic fungus Neocallimastix frontalis: comparison of the amino acid sequence with animals and yeast. Gene. 1992 Jan 2;110(1):57–63. doi: 10.1016/0378-1119(92)90444-t. [DOI] [PubMed] [Google Scholar]
- Sakka K., Kojima Y., Kondo T., Karita S., Ohmiya K., Shimada K. Nucleotide sequence of the Clostridium stercorarium xynA gene encoding xylanase A: identification of catalytic and cellulose binding domains. Biosci Biotechnol Biochem. 1993 Feb;57(2):273–277. doi: 10.1271/bbb.57.273. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Saul D. J., Williams L. C., Grayling R. A., Chamley L. W., Love D. R., Bergquist P. L. celB, a gene coding for a bifunctional cellulase from the extreme thermophile "Caldocellum saccharolyticum". Appl Environ Microbiol. 1990 Oct;56(10):3117–3124. doi: 10.1128/aem.56.10.3117-3124.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedmak J. J., Grossberg S. E. A rapid, sensitive, and versatile assay for protein using Coomassie brilliant blue G250. Anal Biochem. 1977 May 1;79(1-2):544–552. doi: 10.1016/0003-2697(77)90428-6. [DOI] [PubMed] [Google Scholar]
- Staden R. A new computer method for the storage and manipulation of DNA gel reading data. Nucleic Acids Res. 1980 Aug 25;8(16):3673–3694. doi: 10.1093/nar/8.16.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teunissen M. J., de Kort G. V., Op den Camp H. J., Huis in 't Veld J. H. Production of cellulolytic and xylanolytic enzymes during growth of the anaerobic fungus Piromyces sp. on different substrates. J Gen Microbiol. 1992 Aug;138(Pt 8):1657–1664. doi: 10.1099/00221287-138-8-1657. [DOI] [PubMed] [Google Scholar]
- Tokatlidis K., Salamitou S., Béguin P., Dhurjati P., Aubert J. P. Interaction of the duplicated segment carried by Clostridium thermocellum cellulases with cellulosome components. FEBS Lett. 1991 Oct 21;291(2):185–188. doi: 10.1016/0014-5793(91)81279-h. [DOI] [PubMed] [Google Scholar]
- Ujiie M., Roy C., Yaguchi M. Low-molecular-weight xylanase from Trichoderma viride. Appl Environ Microbiol. 1991 Jun;57(6):1860–1862. doi: 10.1128/aem.57.6.1860-1862.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. X., Flint H. J. A bifunctional xylanase encoded by the xynA gene of the rumen cellulolytic bacterium Ruminococcus flavefaciens 17 comprises two dissimilar domains linked by an asparagine/glutamine-rich sequence. Mol Microbiol. 1992 Apr;6(8):1013–1023. doi: 10.1111/j.1365-2958.1992.tb02167.x. [DOI] [PubMed] [Google Scholar]
- Zhou L., Xue G. P., Orpin C. G., Black G. W., Gilbert H. J., Hazlewood G. P. Intronless celB from the anaerobic fungus Neocallimastix patriciarum encodes a modular family A endoglucanase. Biochem J. 1994 Jan 15;297(Pt 2):359–364. doi: 10.1042/bj2970359. [DOI] [PMC free article] [PubMed] [Google Scholar]