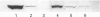Abstract
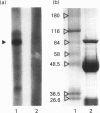
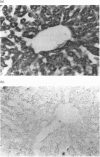
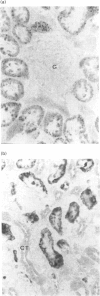
Expression of mitochondrial dimethylglycine dehydrogenase (Me2GlyDH) was analysed in various tissues, liver cell types and developmental stages of the rat. Total RNA extracted from liver, spleen, brain, kidney, lung and heart was reverse-transcribed into cDNA and amplified with Me2GlyDH cDNA-specific oligonucleotides by PCR. Expression of the enzyme was observed mainly in liver and kidney. In addition, Me2GlyDH mRNA could be demonstrated in total RNA samples of lung, heart and brain but was barely detectable in spleen total RNA. In RNA prepared from 14-day rat embryos, Me2GlyDH-specific mRNA was clearly present. Among various liver cell types, besides hepatocytes, endothelial cells showed a high level of Me2GlyDH mRNA expression. There was no amplification product detectable in liver macrophages (Kupffer cells) and only a very faint one in fat-storing cells (Ito cells). Western blots confirmed at the protein level the predominant expression of the enzyme in liver and kidney, but Me2GlyDH protein was also present in the protein extract of lung, heart, spleen and brain. Immunohistochemical staining of liver slices with Me2GlyDH-specific antiserum revealed that expression of this enzyme is evenly distributed throughout the liver tissue. In the kidney, expression of the enzyme was located in the proximal tubule cells. Our results demonstrate that, contrary to the previously assumed liver-restricted expression, this enzyme is specifically expressed predominantly in the liver and kidney, but, in addition, it is detectable in many other tissues of the rat.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Cook R. J., Wagner C. Dimethylglycine dehydrogenase and sarcosine dehydrogenase: mitochondrial folate-binding proteins from rat liver. Methods Enzymol. 1986;122:255–260. doi: 10.1016/0076-6879(86)22179-5. [DOI] [PubMed] [Google Scholar]
- Eyhorn S., Schlayer H. J., Henninger H. P., Dieter P., Hermann R., Woort-Menker M., Becker H., Schaefer H. E., Decker K. Rat hepatic sinusoidal endothelial cells in monolayer culture. Biochemical and ultrastructural characteristics. J Hepatol. 1988 Feb;6(1):23–35. doi: 10.1016/s0168-8278(88)80459-8. [DOI] [PubMed] [Google Scholar]
- Garcia-Perez A., Burg M. B. Renal medullary organic osmolytes. Physiol Rev. 1991 Oct;71(4):1081–1115. doi: 10.1152/physrev.1991.71.4.1081. [DOI] [PubMed] [Google Scholar]
- Haubrich D. R., Gerber N. H. Choline dehydrogenase. Assay, properties and inhibitors. Biochem Pharmacol. 1981 Nov 1;30(21):2993–3000. doi: 10.1016/0006-2952(81)90265-3. [DOI] [PubMed] [Google Scholar]
- Kawada N., Klein H., Decker K. Eicosanoid-mediated contractility of hepatic stellate cells. Biochem J. 1992 Jul 15;285(Pt 2):367–371. doi: 10.1042/bj2850367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai E., Darnell J. E., Jr Transcriptional control in hepatocytes: a window on development. Trends Biochem Sci. 1991 Nov;16(11):427–430. doi: 10.1016/0968-0004(91)90169-v. [DOI] [PubMed] [Google Scholar]
- Lang H., Polster M., Brandsch R. Rat liver dimethylglycine dehydrogenase. Flavinylation of the enzyme in hepatocytes in primary culture and characterization of a cDNA clone. Eur J Biochem. 1991 Jun 15;198(3):793–799. doi: 10.1111/j.1432-1033.1991.tb16083.x. [DOI] [PubMed] [Google Scholar]
- McKeever M. P., Weir D. G., Molloy A., Scott J. M. Betaine-homocysteine methyltransferase: organ distribution in man, pig and rat and subcellular distribution in the rat. Clin Sci (Lond) 1991 Oct;81(4):551–556. doi: 10.1042/cs0810551. [DOI] [PubMed] [Google Scholar]
- Porter R. K., Scott J. M., Brand M. D. Characterization of betaine efflux from rat liver mitochondria. Biochim Biophys Acta. 1993 Mar 1;1141(2-3):269–274. doi: 10.1016/0005-2728(93)90052-h. [DOI] [PubMed] [Google Scholar]
- Porter R. K., Scott J. M., Brand M. D. Choline transport into rat liver mitochondria. Characterization and kinetics of a specific transporter. J Biol Chem. 1992 Jul 25;267(21):14637–14646. [PubMed] [Google Scholar]
- Tran-Thi T. A., Phillips J., Falk H., Decker K. Toxicity of D-galactosamine for rat hepatocytes in monolayer culture. Exp Mol Pathol. 1985 Feb;42(1):89–116. doi: 10.1016/0014-4800(85)90021-8. [DOI] [PubMed] [Google Scholar]
- Wilken D. R., McMacken M. L., Rodriquez A. Choline and betaine aldehyde oxidation by rat liver mitochondria. Biochim Biophys Acta. 1970 Sep 1;216(2):305–317. doi: 10.1016/0005-2728(70)90222-7. [DOI] [PubMed] [Google Scholar]
- Wittwer A. J., Wagner C. Identification of folate binding protein of mitochondria as dimethylglycine dehydrogenase. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4484–4488. doi: 10.1073/pnas.77.8.4484. [DOI] [PMC free article] [PubMed] [Google Scholar]