Abstract
We have recently described three group I introns inserted into a single gene, orf142, of the staphylococcal bacteriophage Twort and suggested the presence of at least two additional self-splicing introns in this phage genome. Here we report that two previously uncharacterized introns, 429 and 1087 nt in length, interrupt the Twort gene coding for the large subunit of ribonucleotide reductase (nrdE). Reverse transcription–polymerase chain reaction (RT–PCR) of RNA isolated from Staphylococcus aureus after phage infection indicates that the introns are removed from the primary transcript in vivo. Both nrdE introns show sequence similarity to the Twort orf142 introns I2 and I3, suggesting either a common origin of these introns or shuffling of intron structural elements. Intron 2 encodes a DNA endonuclease, I-TwoI, with similarity to homing endonucleases of the HNH family. Like I-HmuI and I-HmuII, intron-encoded HNH endonucleases in Bacillus subtilis phages SPO1 and SP82, I-TwoI nicks only one strand of its DNA recognition sequence. However, whereas I-HmuI and I-HmuII cleave the template strand in exon 2, I-TwoI cleaves the coding strand in exon 1. In each case, the 3′ OH created on the cut strand is positioned to prime DNA synthesis towards the intron, suggesting that this reaction contributes to the mechanism of intron homing. Both nrdE introns are inserted in highly conserved regions of the ribonucleotide reductase gene, next to codons for functionally important residues.
INTRODUCTION
Group I introns and inteins are intervening sequences (IVSs) that autocatalytically splice from a precursor RNA or polypeptide, respectively (1,2). A large number of these IVSs encode site-specific DNA endonucleases (3). Rather than being involved in the splicing process, these endonucleases promote the transfer of the IVS from an IVS-plus gene to a IVS-minus gene, in a process known as homing. In general, a double-strand break generated by the endonuclease initiates a unidirectional and non-reciprocal transfer of the IVS (4,5). Therefore, the intron/intein-encoded endonucleases are frequently referred to as homing endonucleases (6).
Although group I introns and inteins are rarely encountered in microbial genomes, there is a striking bias for the genes in which they are inserted. Whereas the introns in eubacterial genomes generally interrupt tRNA genes, the phage introns are inserted in protein coding genes, most of which are involved in some aspect of DNA metabolism (7). A preferential insertion into genes involved in DNA synthesis is also seen for inteins in bacteria and archaea (8). These observations suggested either a regulatory role for IVSs in nucleic acid metabolism or, alternatively, a particular susceptibility of these loci for IVS insertion (7,9).
Ribonucleotide reductase (RR) provides the building blocks for DNA replication and repair and plays an important role in the regulation of DNA synthesis. Three distinct classes of RRs have been defined on the basis of their primary structure, subunit composition, the metallo-cofactor and the mechanism used for radical generation. Aerobic class I enzymes generate a tyrosyl radical with an iron–oxygen center, class II utilize adenosylcobalamin, and the anaerobic class III generate a glycyl radical from S-adenosylmethionine (10). Despite these differences in the catalytic mechanism, similarities in the tertiary structure and allosteric control suggest these enzymes might have evolved from a common, most likely anaerobic, ancestor (11,12). RRs of all three classes have been found in eubacteria and bacteriophages, with some organisms encoding enzymes of more than one class. Archaea code for either class II or III enzymes only (10).
Interestingly, several RR genes of eubacteria (and their phages) and archaea are interrupted by IVSs. For one of the enzymes of Deinococcus radiodurans, the large subunit is interrupted by an intein (13). In Aquifex aeolicus, an intein is inserted in the small subunit gene (14). In Escherichia coli bacteriophage T4, the genes for the small subunit of the aerobic RR (nrdB) and for the anaerobic enzyme (nrdD) both contain a self-splicing group I intron (15–17). In Spβ, and related prophages integrated into the Bacillus subtilis genome, the genes for the large subunit (bnrdE) and the small subunit (bnrdF) harbor group I introns (18). In addition, an intein has been found in the structural gene for the large subunit. In the archaeon genus Pyrococcus, several species have one or more inteins inserted in the RR genes (11).
In this paper we describe two group I introns in the large subunit RR gene of Staphylcoccus aureus bacteriophage Twort, making a total of at least five introns in this genome. One intron encodes a nicking, homing-type DNA endonuclease of the HNH family that cleaves the intronless cognate gene and is the first example of an intron-encoded endonuclease in this phage genome. We further provide a detailed comparison of the insertion sites of the Twort introns and previously reported IVSs, with respect to functionally important regions of the large subunit of the RR.
MATERIALS AND METHODS
Bacterial and bacteriophage strains and growth conditions
Phage Twort (HER48) and its host S.aureus Twort (HER1048) were obtained from the Felix d’Herelle Reference Center maintained by H. -W. Ackermann. Staphylococcus aureus and its phage were maintained as described (19). Escherichia coli XL-1 Blue (Stratagene) was used as a recipient strain for high-frequency plasmid electroporation. Escherichia coli ER2566 (New England Biolabs) was used as host for protein expression.
Plasmids
Genomic plasmid minilibraries were generated as follows. Twort DNA was digested with DraI and DNA fragments of sizes 0.4–0.7 kb and 0.5–1.2 kb were gel purified from a low melting point agarose gel and ligated to a XbaI linker. After digestion with XbaI, the DNA fragments were ligated into the XbaI site of pBSM13+ (Stratagene). Minilibraries were transformed into E.coli XL-1 Blue. pTDZ10 (accession no. AF485080; Supplementary Material, Fig. S1, residues 3065–3699), derived from a 0.4–0.7 kb library, was isolated by colony hybridization (20) using radiolabeled degenerate oligonucleotide S1403, complementary to the S sequence of phage introns. pTDX120 (residues 1319–2025), derived from a 0.5–1.2 kb library, was isolated by colony hybridization using RNA extracted from cells after 10 min of infection by phage Twort, and labeled in vitro with [α-32P]GTP (19).
I-TwoI expression vector pET19bCl5 was generated by polymerase chain reaction (PCR) of Twort DNA using primers S2561 and S2562, followed by digestion with BamHI. Ligation was into pET19b (Novagen), which had been digested with NcoI, filled in with Klenow DNA polymerase, and digested with BamHI.
Sequencing
All sequences were determined on both DNA strands. Intron 2 sequence was obtained by primer walking, on two independently amplified products of Twort DNA. PCR primers S2398 and S2399 were inferred from the previously determined sequences of pTDX120 and pTDZ10, respectively.
Intron 1 sequence was obtained by PCR amplification of Twort DNA using the degenerate upstream primer S2471, and sequence-specific downstream primer S2470. Two independent PCR products were sequenced. The remaining parts of the nrdE gene were obtained by cycle sequencing of Twort DNA extracted from CsCl-purified phage particles (20).
Reverse transcription (RT) and PCR
RNA (4 µg) isolated from S.aureus 5 min after infection with Twort was reverse transcribed and PCR amplified as described (19), and products were sequenced to determine splice junctions.
Protein expression
Plasmid pET19bCl5, for overexpression of I-TwoI, was introduced into ER2566. Cells were grown in LB supplemented with ampicillin (50 µg/ml) at 37°C to A600 = 0.65 and expression was induced by addition of IPTG to a final concentration of 1 mM. Incubation was continued at 37°C for 3 h.
Preparation of protein extract
Cells were harvested by centrifugation at 6000 g for 20 min. The harvested cells were resuspended in ice-cold 50 mM Tris–HCl (pH 7.2), 1 mM EDTA, 1 mM PMSF, 2 mg/ml leupeptin and 200 mM KCl at a concentration of 6 ml/g cells. The resuspension was sonicated to complete lysis and centrifuged at 12 000 g for 1 h. The protein was present in the pellet fraction. The pellet was washed with chilled deionized H2O, resuspended at 1 ml/g of cells in 6 M guanidine hydrochloride, and renatured by dialyzing twice against 100 vol of 50 mM potassium phosphate (pH 7.2), 100 mM NaCl and 1 mM DTT, and stored with 10% glycerol at –80°C.
Endonuclease assay
PCR-generated DNA fragments, radiolabeled at their 5′ termini (with T4 polynucleotide kinase) or internally labeled, were used as substrates. Labeled PCR products were purified with QIAquick spin PCR purification. Reactions were performed with 5 × 103–3 × 104 c.p.m. of labeled target DNA in a 5 µl reaction, in 50 mM Tris–HCl (pH 7.9), 10 mM MgCl2, 100 mM NaCl, 1mM DTT, 20 µg/ml poly(dI–dC), with 2 µl I-TwoI protein extract. Reactions were allowed to proceed for 10 min at 30°C and terminated by the addition of 2 µl 95% formamide, 20 mM EDTA, 0.05% bromphenol blue and 0.05% xylene cyanol. The reaction products were separated on a 4 or 5% polyacrylamide gel under denaturing conditions and visualized by autoradiography.
Mapping of I-TwoI cleavage sites
The RT–PCR product was reamplified with individually [32P] 5′ end-labeled primers S2458 and S2399. Labeled PCR product was incubated with 2 µl of a 1:10 dilution of I-TwoI protein extract, phenol extracted and separated on 5% polyacrylamide/6 M urea gel.
Oligonucleotides
S1403, 5′-CCAT(G/A)NAGNNCAGACTATATC (complement: 3331–3352); S2398, 5′-TACGAGCTATAGGTCTTGGAGC (1945–1966); S2399, 5′-TATGTATGTGTCCATGTTACCA (complement: 3477–3498); S2470, 5′-AATTCATGAATATCTGGGTG (complement: 1329–1348); S2471, 5′-CA(A/G) CCN(T/G)CNACNCCNAC (582–597); S2458, 5′-AAGTCACATGAACCTAG (2172–2188); S2561, 5′-GAGGAGTTATGGAAAGAAATACCTG (2508–2532); S2562, 5′-CTTGTAAGGATCCTTAATGTGTTTGTT (complement: 3223–3249); S2575, 5′-AGACTCCGTACGACAAGC (695–712). The underlined sequence indicates an introduced BamHI restriction site.
RESULTS
Two group I introns in the nrdE gene of bacteriophage Twort
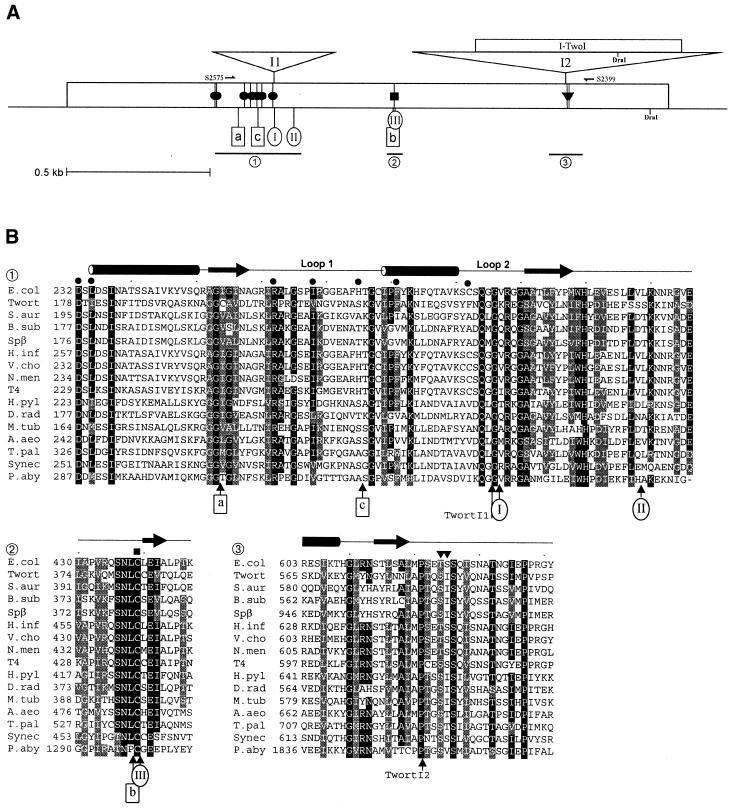
We have previously shown that the late transcribed gene, orf142, of the S.aureus phage Twort is interrupted by three group I introns, and GTP-labeling of phage RNA indicated the presence of at least two additional self-splicing introns in this phage genome (19). In spite of differences in sequence and RNA secondary structure, the introns in orf142 and the previously described group I introns in other bacteriophages are highly similar in the S sequence, comprising the junction between pairing elements P8 and P7 and the 3′ part of P7 (19,21). We used an end-labeled degenerate oligonucleotide (S1403), complementary to the consensus phage S sequence and the 5′ part of P9, to screen a Twort genomic plasmid library for additional introns. In this screen we isolated a 641-bp DraI DNA fragment (pTDZ10). The distal end of the fragment could be translated into a stretch of 103 amino acids which, in a BLASTP search (22), showed a high similarity to a portion of the large subunit RR (NrdE) of S.aureus. Since the coding region of the isolated DNA fragment could not be extended further, we examined the sequence for the presence of an IVS. The sequence upstream of the coding region could be folded into an RNA secondary structure that is typical for the 3′ ends of group I introns, suggesting that the Twort gene for the large subunit of RR is interrupted by an intron. When the complete DNA sequence of this gene was obtained, translation of the nucleotide sequence revealed that the coding sequence is interrupted by a second group I intron, which is inserted further upstream. In summary, the Twort nrdE gene is interrupted by two group I introns, nrdE-I1 and nrdE-I2, 429 and 1087 bp in length, inserted after codons 241 and 583, respectively (Fig. 1).
Figure 1.
Gene map and partial amino acid sequence alignment of RRs. (A) Map of large subunit RR gene and respective IVS insertion sites. Twort nrdE intron insertion sites are shown on top. Intron insertion sites in Spβ and related phages are shown as ovals and named I to III (18). Intein insertion sites are indicated by boxes and named a to c (38). Three conserved regions are underlined, with sequences shown in (B). Residues in the E.coli RR structure that are involved in binding of the allosteric effector dTTP, in generation of the transient protein radical, and in substrate binding are indicated by filled circles, squares and inverted triangles, respectively. Relevant DraI restiction sites are shown. Positions of oligonucleotide primers S2399 and S2575 are shown. (B) Alignment was generated with ClustalW1.8 (40) using RR amino acid sequences with GenBank accession nos: S.aur (S.aureus, CAB38642), B.sub (B.subtilis, P50620), Spβ (Phage Spβ, AAC13134), H.inf (Haemophilus influenzae, AAC23305), E.col (E.coli, AAG57363), V.cho (Vibrio cholerae, AAF94415), N.men (Neisseria meningitidis, AAF41667), T4 (Phage T4, AAA32527), H.pyl (Helicobacter pylori, AAD06201), D.rad (D.radiodurans, AAF12547), M.tub (Mycobacterium tuberculosis, AAA64444), A.aeo (A.aeolicus, AAC06460), T.pal (Treponema pallidum, AAC65956), Synec (Synechocystis PCC 6803, P74240), P.aby (P.abyssi, NP_127282). Three regions of the alignment, intron and intein insertion sites, and symbols indicting functionally important residues are as indicated in (A). Numbers preceding the aligned sequence indicate position of the first amino acid. Protein secondary structure assignments are shown at the top.
Splicing in vivo
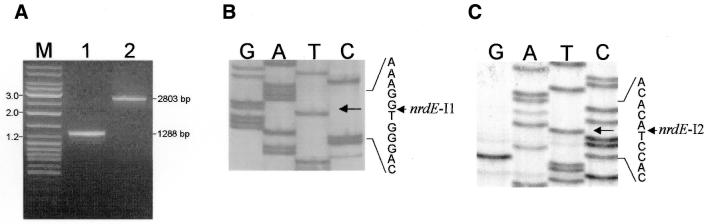
Splicing of the Twort nrdE introns in vivo was tested by RT of RNA followed by amplification by PCR. RNA was isolated from S.aureus 5 min after infection with phage Twort and reverse transcribed using oligonucleotide S2399. The cDNA was amplified by PCR, using oligonucleotides S2399 and S2575. The electrophoretic mobility of the reaction product is consistent with 1288 bp, the size expected for the cDNA made from the spliced mRNA (Fig. 2A). The nucleotide sequence of the cDNA confirms that the introns are removed in vivo (Fig. 2B and C), precisely as predicted from the secondary structures (Fig. 3).
Figure 2.
In vivo splicing. (A) Electrophoretic analysis of (1) RT–PCR product obtained by using primers S2399 and S2575 (see Fig. 1) on 5 min Twort RNA and (2) PCR product obtained by using the same primers on Twort DNA. Predicted sizes of the PCR products are indicated to the right. The Gene-Ruler Mix (MBI Fermentas) was used as marker (M), with sizes given on the left. Sequencing ladders of the regions around insertion sites of nrdE-I1 (B) and nrdE-I2 (C) are shown. Lanes are labeled with the dideoxynucleotides. The sequence of the ligated exons is given to the right of the ladders, with arrows indicating the intron insertion sites.
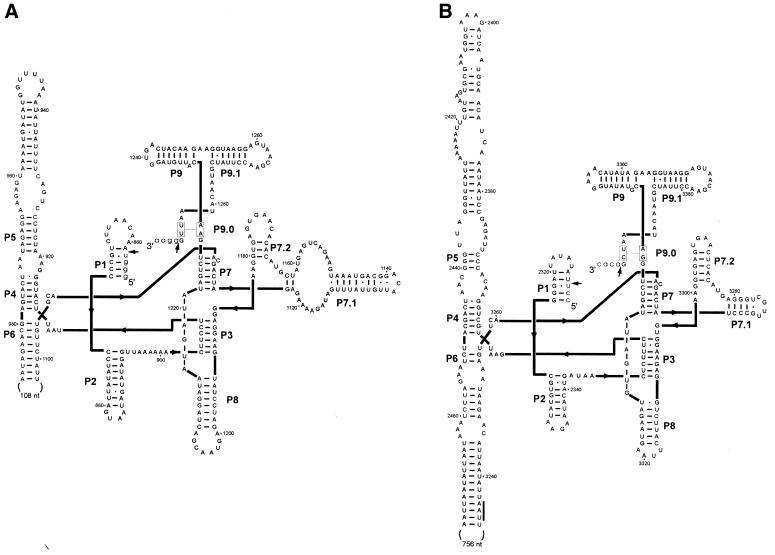
Figure 3.
Secondary structures of introns nrdE-I1 (A) and nrdE-I2 (B). Exon sequences are in lower case and intron sequences in upper case letters. Arrows indicate splice sites. Conserved structural elements P1 to P9 are shown. The line next to UAA in P6 of nrdE-I2 indicates the stop codon of the I-TwoI coding sequence. Numbering corresponds to accession no. AF485080 (Supplementary Material, Fig. S1).
Evidence for genetic exchange between Twort introns
Figure 3 shows the putative secondary structures of the Twort nrdE introns. Both group I introns follow the consensus secondary structure with base pairings P1 to P9. Based on the presence of extra nucleotides between P3 and P7, which fold into two stable stem–loop structures (P7.1 and P7.2), and conserved nucleotides in the S sequence, the Twort nrdE introns belong to subgroup IA2 (21), comprising the previously described bacteriophage introns. Like most other phage introns, the group I introns in the Twort nrdE gene have additional nucleotides inserted into terminal loops of paired regions of the secondary structure. In introns 1 and 2, the loop of P6 contains 106 and 756 extra nt, respectively, suggesting the presence of intron-encoded ORFs.
A comparison of the nucleotide sequences of nrdE-I1 and I2 shows 77% sequence identity in the 3′ part of the introns over 117 aligned nt, whereas the overall identity is 62% (125 out of 200 aligned nt). BLASTN searches of the database (22) indicate that the 3′ parts of both nrdE introns are highly similar to the corresponding region of the closely related Twort orf142 introns I2 and I3, which are identical in the 71 nt at their 3′ ends. In particular, intron nrdE-I1 shares 89% identity with orf142 intron 3 over 188 aligned nt (including the phylogenetically non-conserved pairings P7.1, P7.2 and P9.1), suggesting a common origin or a genetic exchange between these introns.
nrdE-I2 encodes a nicking DNA endonuclease
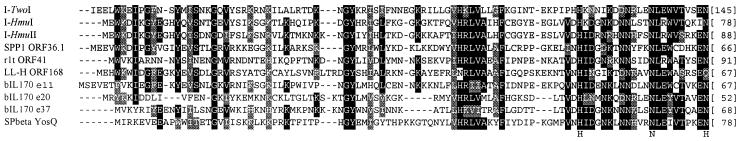
Group I introns frequently encode maturases and/or DNA endonucleases (23). Both nrdE introns have additional nucleotides inserted in the loop of P6 as indicated in Figure 3. In nrdE-I1, 106 nt of the loop and sequences upstream thereof code for a 52 amino acid peptide (accession no. AF485080; Supplementary Material, Fig. S1, residues 958–1113), which revealed no similarities to proteins in the database using BLASTP (22) with default settings. The additional nucleotides inserted in nrdE-I2 code for 257 amino acids (residues 2463–3233). BLASTP searches with this ORF, which lacks a start codon in the context of a recognizable ribosomal binding site (RBS), indicated similarities to phage proteins with the conserved HNH homing endonuclease motif (Fig. 4), including the group I intron encoded endonucleases I-HmuI and I-HmuII (24). Based on the sequence alignment, we defined an AUA isoleucine codon (residues 2505–2507), which is preceded by a putative RBS, as the start of this 243 amino acid ORF.
Figure 4.
Amino acid sequence alignment I-TwoI and highly similar ORFs. Alignment is shown to DNA endonucleases I-HmuI and I-HmuII, and other ORFs with the HNH motif from phages that infect Gram-positive bacteria. GenBank accession nos: I-HmuI (AAA64536), I-HmuII (AAA56884), SPP1 ORF36.1 (CAA48056), r1t ORF41 (AAB18716), LL-H ORF168 (AAC41637), bIL170 e11 (AAC27227), e20 (AAC27218), e37 (AAC27202) and SPβ YosQ (AAC13136). Alignment was generated with ClustalW1.8. Conserved residues are shaded. Brackets indicate number of residues following the aligned sequence. Letters below the alignment indicate positions of conserved residues in the original definition of the HNH motif.
To address the question of whether the ORF in nrdE-I2 encodes a functional endonuclease, we expressed the protein in E.coli by changing the presumed isoleucine start codon to AUG, in plasmid pET19bC15. The expressed protein was partially purified for DNA endonuclease assays. Intron-encoded DNA endonucleases generally recognize and cleave the intronless version of their cognate genes. However, since I-HmuI and I-HmuII are exceptional in this respect, also cleaving the intron-containing gene (24), we used the intronless allele and the exon–intron boundaries of nrdE as DNA substrates. Figure 5 shows that protein from cells expressing the intronic ORF had endonucleolytic activity, cleaving only the intronless Twort nrdE gene. No activity was detected using protein derived from cells harboring the expression plasmid without the insert. Based on this activity the intron-encoded ORF was designated I-TwoI. The similarity to I-HmuI and I-HmuII further suggested that I-TwoI might be a nicking DNA endonuclease. Differentially end-labeled substrates were used to determine the strand specificity (Fig. 6). Indeed, I-TwoI cleaves only the coding strand. No cleavage of the template strand was detectable under conditions in which the coding strand is completely turned into product. Precise mapping of the cleavage site places it on the coding strand, 3 nt upstream of the intron insertion site (Fig. 7).
Figure 5.
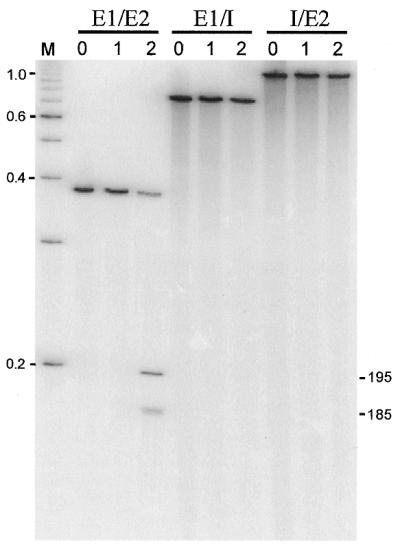
Endonuclease assay. Internally radiolabeled intronless (E1/E2), exon1/intron boundary (E1/I) and intron/exon2 boundary (I/E2) substrate DNAs were generated by PCR amplification. Substrates were incubated with: (0) no protein; (1) protein from cells harboring pET19b; and (2) protein from cells harboring pET19bCl5 expressing the intronic ORF. Products of the cleavage reactions were separated on a 4% denaturing polyacrylamide gel. Sizes of 100-bp DNA ladder markers (M) are indicated in kb. Estimated sizes of cleavage products are indicated in bp.
Figure 6.
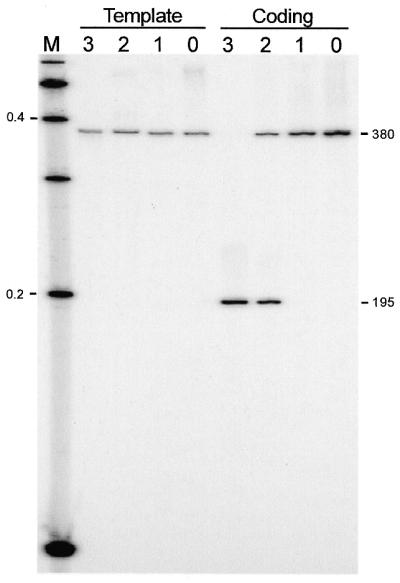
Strand-specificity assays. Substrate DNAs (380 bp) were generated by PCR amplification with primers that were individually 5′ end-labeled, to differentially label the template and coding strands. Substrates were incubated with: (0) no protein; (1) protein from cells harboring pET19b; (3) protein from cells harboring pET19bCl5 expressing the intronic ORF; and (2) a 1/10 dilution thereof. Products of the cleavage reactions were separated on a 4% denaturing polyacrylamide gel. Sizes of 100-bp DNA ladder markers (M) are indicated in kb. Predicted sizes of the substrate and cleavage product (based on data from Fig. 7) are shown in bp.
Figure 7.
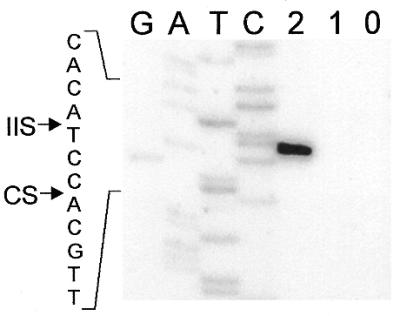
Cleavage site mapping. Intronless DNA, which was 5′ end-labeled on the coding strand, was generated by PCR and incubated with protein from E.coli ER2566 harboring pET19bCl5 (2), from cells harboring pET19b (1), and with no protein (0). Cleavage products were separated on a denaturing polyacrylamide gel together with a sequencing ladder generated with the end-labeled primers used in the PCR. Sequence around the cleavage site is indicated to the left. Arrows point to the positions of the intron insertion site (IIS) and cleavage site (CS).
DISCUSSION
Five introns in the genome of phage Twort
In this work we describe two self-splicing group I introns in the large subunit RR gene (nrdE) of the S.aureus phage Twort, corresponding in size to the two GTP-labeled RNA species present maximally at 3 min after infection (19). Including the previously reported introns in the gene orf142, which are transcribed during the late period of infection (15–20 min after infection), the Twort genome harbors five self-splicing group I introns, the largest number of introns in a single phage genome. In addition, a major GTP-labeled RNA species of about 1400 nt, which reaches peak levels at about 10 min of infection, suggests the presence of at least one additional ORF-containing intron, belonging to a third transcriptional class.
Interestingly, the nrdE introns are highly similar to the orf142 introns I2 and I3. The existence of a homing endonuclease in one of the Twort introns raises the possibility that they may have spread by an endonuclease-mediated transposition. However, the similarity of these introns is most pronounced in their 3′ portions, which suggests that genetic exchange might have also contributed to their similarity. Bryk and Belfort (25) isolated domain-switch variants (from the nrdD intron) as genetic suppressors of a splicing-defective P7.1 deletion in the phage T4 td intron. This novel example of suppression through inter-intron sequence substitution indicated that the introns are in a state of genetic flux and confirmed the functional equivalence of homologous regions.
A homing endonuclease encoded by nrdE-I2
In contrast to the orf142 introns, whose structures consist entirely of conserved elements of secondary structure, the nrdE introns contain insertions in structural element P6. The short ORF in nrdE-I1 shows no similarity to known protein sequences. However, it remains possible that it is the remnant of a partially deleted DNA endonuclease, as has been shown for the truncated coding sequences in the Bacillus phage β22 thy intron (26) and the phage T4 nrdB intron (27).
nrdE-I2 encodes an ORF highly similar to phage proteins with the HNH homing endonuclease motif. The similarity among these proteins is restricted to the N-terminal portion of the proteins comprising the HNH motif, which has been suggested to form the active site of these endonucleases (28,29). The presence of C-termini, variable in length and sequence, suggests a two-domain structure analogous to the one in the T4 td intronic endonuclease I-TevI, with N-terminal catalytic and C-terminal DNA-binding domains (30). Of the proteins most closely related to the nrdE-I2 ORF, only two, I-HmuI and I-HmuII (encoded by the DNA polymerase introns in Bacillus phages SPO1 and SP82, respectively), have been shown to be DNA endonucleases (24). These enzymes have properties that are atypical of the group I intron homing enzymes. A hallmark of homing endonucleases is specific double-strand cleavage of their intronless cognate gene, close to the site of intron insertion (23). This cleavage initiates the mobility of the intron into the intron-minus DNA, as a consequence of double-strand break repair (4). I-HmuI and I-HmuII differ by being able to cleave the intron-containing and intronless DNA. In addition, I-HmuI and I-HmuII are the only group I intron endonucleases that nick the template strand in exon 2, instead of generating a double-strand break (24).
The endonuclease encoded by nrdE-I2 (I-TwoI) behaves like a typical homing endonuclease, with specificity exclusively for the intronless nrdE gene. However, like its relatives, I-TwoI cuts only one strand of the DNA substrate. Rather than cutting the template strand it nicks the coding strand in exon 1, just 3 nt from the intron insertion site. The nicking activity of these phage intronic endonucleases is puzzling with respect to the initiating events of group I intron homing (4). Either a nick is sufficient to initiate the mobility of these introns in vivo, or second strand cleavage is achieved by some intron endonuclease-independent component. Group II intron homing is initiated by a ribonucleoprotein complex that includes the excised intron. An endonuclease in the protein component nicks the template strand, whereas the coding strand is cleaved by reverse splicing of intron RNA into the DNA (31). The similarity of group I and group II intron endonucleases, sharing the HNH nuclease motif (32,33), raises the possibility that second strand cleavage by group I intron RNA is also involved in the mobility of these phage introns. However, nicking of the coding strand by I-TwoI argues strongly against involvement of intron RNA in the homing mechanism (Fig. 8).
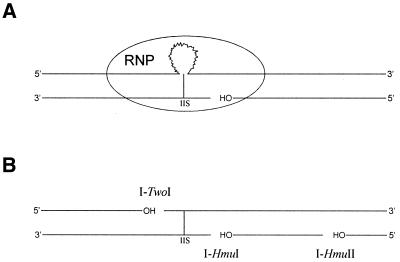
Figure 8.
Comparison of DNA cleavage reactions of group II intron ribonucleoprotein particles (RNPs) and I-TwoI and related HNH endonucleases. (A) A RNP carries out template strand nicking by protein endonuclease and reverse splicing by the intron RNA (jagged line) into the DNA coding strand. (B) Cleavage sites of three HNH endonucleases from phage group I introns are shown, with respect to their intron insertion sites (IIS). Either the coding strand or the template strand can be nicked.
Interestingly, independent of which stand is cleaved, the incision that each of these endonucleases generates is 5′ of the intron insertion site (Fig. 8). Furthermore, since I-HmuII cleavage products can be ligated by T4 DNA ligase (our unpublished results), the 3′ OH groups generated by this family of endonucleases are positioned to prime DNA synthesis towards the intron insertion site. It certainly will be interesting to determine the steps by which these phage HNH endonuclease nicks result in intron mobility.
The insertion sites of nrdE introns are highly conserved and functionally important
The initial discovery of group I introns in early transcribed genes involved in DNA metabolism of E.coli phage T4 and B.subtilis phage SPO1 prompted the idea that splicing might be involved in a global regulatory mechanism involving DNA replication. However, subsequent findings of introns in various late transcribed phage genes not involved in DNA metabolism (19,34–36), and the paucity of group I introns in close relatives of T4 phages (7), weakened that hypothesis. Edgell et al. (7) suggested that phage-encoded homing endonucleases use highly conserved nucleotide sequences, found in genes that function in replication and transcription, as recognition sequences to maximize their spread to related phages. Their suggestion that recognition sequences of homing endonucleases correspond to important protein functional domains is supported by the insertion of the phage T4 td intron (and its endonuclease) close to the coding sequence corresponding to the thymidylate synthase active site.
The Twort introns described in this work provide additional examples of phage group I introns inserted into RRs, highly conserved genes involved in DNA metabolism. We compared the Twort intron insertion sites with those of previously reported IVSs in RR genes, and with the location of potential functionally important residues as determined from the crystal structure of the E.coli class I RR (37). The Twort nrdE-I1 is inserted one codon upstream of the insertion site of an unrelated group I intron in B.subtilis prophage SPβ, within a region coding for highly conserved amino acids (Fig. 1, site I). In the E.coli structure these residues make up loop 2, which plays a critical role in the allosteric regulation of the enzyme. The loop interacts via Cys292 with the effector nucleotide bound to the allosteric specificity site (37). Another important mediator in this allosteric control is loop 1 in the E.coli RR structure, with three residues interacting with the effector nucleotide. Intriguingly, the corresponding region in the RR of Pyrococcus abyssi (class II) is interrupted by an intein (Fig. 1, site c). Similarly, nrdE-I2 is inserted in the vicinity of functionally important amino acids, in a region of high sequence conservation. Immediately upstream of the insertion site are codons for two residues in the E.coli RR, Thr624 and Ser625 (Fig. 1B), which are involved in binding of the ribonucleotide substrate (37).
Several RR genes of Bacillus prophages have a group I intron inserted after the codon of an active site residue, a universally conserved cysteine (Fig. 1, site III), suggested to be involved in generation of a transient substrate radical in the E.coli enzyme (10). Interestingly, in some cases where no intron was found, an intein was inserted (Fig. 1, site b) before this cysteine codon (18). RR genes in various archaea (class II), the bacterium D.radiodurans (class II) and the Chilo iridescent virus (class I) have inteins inserted in the homologous position (38) indicating the preference of IVSs to target this region of RR genes. Surprisingly, the intein in the eukaryotic C.iridescent virus is most closely related to the intein in RR of the B.subtilis prophage, providing a dramatic example of lateral transfer of these mobile elements (8). In the Bacillus phage RRs, which may contain either an intron or an intein, these IVSs are presumably competing for the same insertion site, since presence of one IVS at this location would interrupt the recognition site of the homing endonuclease of the other (18). A similar scenario could be envisioned for unrelated introns, like Twort nrdE-I1 and the B.subtilis phage introns inserted at site I (Fig. 1B), which interrupt sites separated by a single codon.
Since RR genes are highly conserved and ubiquitously present in every organism (and also some bacterial and eukaryotic viruses) they present susceptible targets for mobile IVSs such as introns and inteins. Despite their different modes of splicing, group I introns and inteins share an analogous mobility (homing) mechanism. These IVSs move from an intron-containing into an intronless allele by an endonuclease initiated recombination–repair process. Dalgaard et al. (39) pointed out that inteins are often situated in close proximity to critical active site residues. Though restricted by their splicing mechanism to a certain sequence context, mobile inteins might follow the same strategy as mobile group I introns to maximize their spread by targeting highly conserved sequences.
Ongoing sequencing efforts will certainly present more examples of introns and inteins, which will provide further insights in the genes these IVS interrupt, the sites of insertion within these genes, the host genomes they invade, and the dynamics and stability of these mobile elements within populations.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank David Edgell and Richard Bonocora for critical reading of the manuscript. This work was supported by grant GM37746 from the National Institutes of Health.
REFERENCES
- 1.Cech T.R. (1990) Self-splicing of group I introns. Annu. Rev. Biochem., 59, 543–568. [DOI] [PubMed] [Google Scholar]
- 2.Paulus H. (2000) Protein splicing and related forms of protein autoprocessing. Annu. Rev. Biochem., 69, 447–496. [DOI] [PubMed] [Google Scholar]
- 3.Chevalier B.S. and Stoddard,B.L. (2001) Homing endonucleases: structural and functional insight into the catalysts of intron/intein mobility. Nucleic Acids Res., 29, 3757–3774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belfort M. and Perlman,P.S. (1995) Mechanisms of intron mobility. J. Biol. Chem., 270, 30237–30240. [DOI] [PubMed] [Google Scholar]
- 5.Liu X.Q. (2000) Protein-splicing intein: genetic mobility, origin and evolution. Annu. Rev. Genet., 34, 61–76 [DOI] [PubMed] [Google Scholar]
- 6.Belfort M. and Roberts,R.J. (1997) Homing endonucleases: keeping the house in order. Nucleic Acids Res., 25, 3379–3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edgell D.R., Belfort,M. and Shub,D.A. (2000) Barriers to intron promiscuity in bacteria. J. Bacteriol., 182, 5281–5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pietrokovski S. (2001) Intein spread and extinction in evolution. Trends Genet., 17, 465–472. [DOI] [PubMed] [Google Scholar]
- 9.Derbyshire V. and Belfort,M. (1998) Lightning strikes twice: intron–intein coincidence. Proc. Natl Acad. Sci. USA, 95, 1356–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jordan A. and Reichard,P. (1998) Ribonucleotide reductases. Annu. Rev. Biochem., 67, 71–98. [DOI] [PubMed] [Google Scholar]
- 11.Riera J., Robb,F.T., Weiss,R. and Fontecave,M. (1997) Ribonucleotide reductase in the archaeon Pyrococcus furiosus: a critical enzyme in the evolution of DNA genomes? Proc. Natl Acad. Sci. USA, 94, 475–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stubbe J. (2000) Ribonucleotide reductases: the link between an RNA and a DNA world? Curr. Opin. Struct. Biol., 10, 731–736. [DOI] [PubMed] [Google Scholar]
- 13.White O., Eisen,J.A., Heidelberg,J.F., Hickey,E.K., Peterson,J.D., Dodson,R.J., Haft,D.H., Gwin,M.L., Nelson,W.C., Richardson,D.L. et al. (1999) Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science, 286, 1571–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deckert G., Warren,P.V., Gaasterland,T., Young,W.G., Lenox,A.L., Graham,D.E., Overbeek,R., Snead,M.A., Keller,M., Aujay,M. et al. (1998) The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature, 392, 353–358. [DOI] [PubMed] [Google Scholar]
- 15.Gott J.M., Shub,D.A. and Belfort,M. (1986) Multiple self-splicing introns in bacteriophage T4: evidence from autocatalytic GTP labeling of RNA in vitro. Cell, 47, 81–87. [DOI] [PubMed] [Google Scholar]
- 16.Sjöberg B.M., Hahne,S., Mathews,C.Z., Mathews,C.K., Rand,K.N. and Gait,M.J. (1986) The bacteriophage T4 gene for the small subunit of ribonucleotide reductase contains an intron. EMBO J., 5, 2031–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Young P., Ohman,M., Xu,M.-Q., Shub,D.A. and Sjöberg,B.M. (1994) Intron-containing T4 bacteriophage gene sunY encodes an anaerobic ribonucleotide reductase. J. Biol. Chem., 269, 20229–20232. [PubMed] [Google Scholar]
- 18.Lazarevic V. (2001) Ribonucleotide reductase genes of Bacillus prophages: a refuge to introns and intein coding sequences. Nucleic Acids Res., 29, 3212–3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Landthaler M. and Shub,D.A. (1999) Unexpected abundance of self-splicing introns in the genome of bacteriophage Twort: introns in multiple genes, a single gene with three introns and exon skipping by group I ribozymes. Proc. Natl Acad. Sci. USA, 96, 7005–7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York.
- 21.Michel F. and Westhof,E. (1990) Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J. Mol. Biol., 216, 585–610. [DOI] [PubMed] [Google Scholar]
- 22.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jurica M.S. and Stoddard,B.L. (1999) Homing endonucleases: structure, function and evolution. Cell. Mol. Life Sci., 55, 1304–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goodrich-Blair H. and Shub,D.A. (1996) Beyond homing: competition between intron endonucleases confers a selective advantage on flanking genetic markers. Cell, 84, 211–221. [DOI] [PubMed] [Google Scholar]
- 25.Bryk M. and Belfort,M. (1990) Spontaneous shuffling of domains between introns of phage T4. Nature, 346, 394–396. [DOI] [PubMed] [Google Scholar]
- 26.Bechhofer D.H., Hue,K.K. and Shub,D.A. (1994) An intron in the thymidylate synthase gene of Bacillus bacteriophage β22: evidence for independent evolution of a gene, its group I intron and the intron open reading frame. Proc. Natl Acad. Sci. USA, 91, 11669–11673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eddy S.R. and Gold,L. (1991) The phage T4 nrdB intron: a deletion mutant of a version found in the wild. Genes Dev., 5, 1032–1041. [DOI] [PubMed] [Google Scholar]
- 28.Kleanthous C., Kuhlmann,U.C., Pommer,A.J., Ferguson,N., Radford,S.E., Moore,G.R., James,R. and Hemmings,A.M. (1999) Structural and mechanistic basis of immunity toward endonuclease colicins. Nature Struct. Biol., 6, 243–252. [DOI] [PubMed] [Google Scholar]
- 29.Kuhlmann U.C., Moore,G.R., James,R., Kleanthous,C. and Hemmings,A.M. (1999) Structural parsimony in endonuclease active sites: should the number of homing endonuclease families be redefined? FEBS Lett., 463, 1–2. [DOI] [PubMed] [Google Scholar]
- 30.Derbyshire V., Kowalski,J.C., Dansereau,J.T., Hauer,C.R. and Belfort,M. (1997) Two-domain structure of the td intron-encoded endonuclease I-TevI correlates with the two-domain configuration of the homing site. J. Mol. Biol., 265, 494–506 [DOI] [PubMed] [Google Scholar]
- 31.Yang J., Zimmerly,S., Perlman,P.S. and Lambowitz,A.M. (1996) Efficient integration of an intron RNA into double-stranded DNA by reverse splicing. Nature, 381, 332–335. [DOI] [PubMed] [Google Scholar]
- 32.Shub D.A., Goodrich-Blair,H. and Eddy,S.R. (1994) Amino acid sequence motif of group I intron endonucleases is conserved in open reading frames of group II introns. Trends Biochem. Sci., 19, 402–404. [DOI] [PubMed] [Google Scholar]
- 33.Gorbalenya A.E. (1994) Self-splicing group I and group II introns encode homologous (putative) DNA endonucleases of a new family. Protein Sci., 3, 1117–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mikkonen M. and Alatossava,T. (1995) A group I intron in the terminase gene of Lactobacillus delbrueckii subsp. lactis phage LL-H. Microbiology, 141, 2183–2190. [DOI] [PubMed] [Google Scholar]
- 35.van Sinderen D., Karsens,H., Kok,J., Terpstra,P., Ruiters,M.H., Venema,G. and Nauta,A. (1996) Sequence analysis and molecular characterization of the temperate lactococcal bacteriophage r1t. Mol. Microbiol., 19, 1343–1355. [DOI] [PubMed] [Google Scholar]
- 36.Foley S., Bruttin,A. and Brussow,H. (2000) Widespread distribution of a group I intron and its three deletion derivatives in the lysin gene of Streptococcus thermophilus bacteriophages. J. Virol., 74, 611–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eriksson M., Uhlin,U., Ramaswamy,S., Ekberg,M., Regnström,K., Sjöberg,B.M. and Eklund,H. (1997) Binding of allosteric effectors to ribonucleotide reductase protein R1: reduction of active-site cysteines promotes substrate binding. Structure, 5, 1077–1092. [DOI] [PubMed] [Google Scholar]
- 38.Perler F.B. (2002) InBase: the Intein Database. Nucleic Acids Res., 30, 383–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dalgaard J.Z., Moser,M.J., Hughey,R. and Mian,I.S. (1997) Statistical modeling, phylogenetic analysis and structure prediction of a protein splicing domain common to inteins and hedgehog proteins. J. Comput. Biol., 4, 193–214. [DOI] [PubMed] [Google Scholar]
- 40.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.