Abstract
Base insertion mutations in the anticodons of two different Escherichia coli tRNAs have been isolated that allow suppression of a series of +1 frameshift mutations. Insertion of a U between positions 34 and 35 of tRNA![]() or addition of a G between positions 36 and 37 of tRNALys expand the anticodons of both tRNAs similarly to 3′−GUUU−5′ and allow decoding of complementary 5′−CAAA−3′ quadruplets. Analysis of the suppressed mRNA sequences suggests that suppression occurs by pairing of the expanded anticodons to all four bases of the complementary, quadruplet codon. The tRNA
or addition of a G between positions 36 and 37 of tRNALys expand the anticodons of both tRNAs similarly to 3′−GUUU−5′ and allow decoding of complementary 5′−CAAA−3′ quadruplets. Analysis of the suppressed mRNA sequences suggests that suppression occurs by pairing of the expanded anticodons to all four bases of the complementary, quadruplet codon. The tRNA![]() mutants are identical to the sufG class of frameshift suppressors isolated both in Salmonella enterica serovar Typhimurium and E.coli by Kohno and Roth and previously thought to affect tRNALys.
mutants are identical to the sufG class of frameshift suppressors isolated both in Salmonella enterica serovar Typhimurium and E.coli by Kohno and Roth and previously thought to affect tRNALys.
INTRODUCTION
The mechanism of reading frame maintenance has been studied genetically in both bacteria and yeast through the isolation of mutations in components of the protein synthesis apparatus that promote or inhibit switches into alternate reading frames (reviewed in 1). Many of these mutants are altered tRNAs, and have typically been isolated as suppressors of frameshift (suf) mutations. Early analyses of tRNA mutants that promoted quadruplet decoding showed that these tRNAs had single base insertions in their anticodon loops. These results suggested that the translocation step size might be determined simply by the size of the anticodon loop. However, extensive analyses of the decoding properties of many different tRNA mutants indicates that a more complex picture obtains (1–3). In some instances, it is now apparent that it is not the mutant tRNA itself that causes frameshifting, but instead frameshifting occurs when a near-cognate tRNA first binds in-frame to the A site codon and subsequently shifts into the alternate reading frame (4,5). Presumably, the mutations in the cognate tRNA impair its activity and allow the near-cognate tRNA to compete effectively for A site binding. Further unexpected decoding possibilities of tRNAs with enlarged anticodon loops emerged from analysis of tRNA1Valmutants that were isolated as suppressors of a –1 frameshift mutation (3). These tRNAs have single base insertions in the anticodon loop and decode quadruplets, but can also ‘hop’, that is they can become unpaired from the zero frame codon and re-pair on an overlapping cognate codon in an alternate reading frame. The unexpected decoding properties of these altered tRNAs have illuminated unanticipated potentials of the decoding process and have made it of interest to characterize more tRNA mutants and study their decoding properties.
In this study, Escherichia coli mutants that enhance frameshifting into the +1 reading frame at CAA AAA ACC have been isolated and characterized. These frameshift suppressor mutants consist of insertions in the anticodon loops of tRNALys and tRNA1Glnso that the enlarged anticodon of both tRNAs is now 3′-GUUU-5′. These studies also prompted a re-examination of the sufG class of frameshift suppressors isolated by Kohno and Roth in both Salmonella enterica serovar Typhimurium and E.coli (6), that had been thought to cause frameshifting at AAAA and AAAU sequences. Sequencing of two of these sufG mutants showed that they also contained insertions of a single U in the anticodon of tRNA1Glnand characterization of a series of sufG-suppressible frameshift mutations in the his operon of Salmonella showed that each contained a CAAA quadruplet. Together these data suggest that expansion of the anticodon allows the extended anticodon to decode CAAA quadruplets by pairing with all four bases of the complementary 5′-CAAA-3′ sequence in the mRNA.
Historically, most of the mutant tRNAs that can decode quadruplets were isolated genetically during the course of investigations into the mechanism of reading frame maintenance. Recently, novel approaches have been used to identify quadruplet- and quintuplet-decoding tRNAs. These tRNAs all contained expanded anticodons and were constructed with the aim of expanding the genetic code and directing the incorporation of non-natural amino acids (2,7,8). The two quadruplet-decoding tRNAs described here add to these examples of mutant tRNAs with expanded decoding possibilities.
MATERIALS AND METHODS
Bacterial strains and plasmids
The trpE9777-containing strain (9), strain MC141 [F– Δ(lac-Pro) thi-1 trpE9777], was used for the isolation and identification of suppressors of the trpE9777 +1 frameshift mutation. A recA– derivative of this strain, designated MC223, was made by mating MC141 with JC10240 (Hfr srlC300::Tn10 recA56 thr-300 ilv-318 rpsE300). Strains TR4447 (E.coli trpE9777 cysB met nag F’152 nag+ sufG100) and TR4592 (S.enterica serovar Typhimurium hisO1242 hisD6580 nag1 sufG70 F’152 nag+) containing sufG alleles (6) were obtained from Dr John Roth, University of Utah. A strain with suf-3 linked to a kanamycin resistant transposon (10) was constructed to facilitate mapping of the suf-3 suppressor. Hfr mapping was carried out using the Hfr::Tn10 strain kit obtained from the E.coli Genetic Stock Center, Yale University, and the methodology described in Wanner (11). Strains carrying genetic markers in the 10–30 min region of the chromosome were obtained from Dr Barbara Bachmann and Dr Mary Berlyn at the E.coli Genetic Stock Center. P1 transductions, transposon and chemical mutagenesis and other genetic procedures were performed as described (10).
The Salmonella strains TR2706 (hisO1242 C6581 tyr545 sufA2 sufG70), TR3683 (hisO1242 B6480 nag-1), TR2692 (hisO1242 B6575 xyl-540 tyr-545) and TR3696 (hisO1242 F6527 proA622) containing sufG-suppressible his mutations (6) were obtained from Dr John Roth, University of Utah. Because many of these strains contained additional undesired mutations, each his mutation was transferred to a clean genetic background by P22-mediated crosses with TT27 (hisD::Tn10) or TT10286 (hisD9953::mudJ).
Plasmids carrying lacZ frameshift mutations were constructed by ligating complementary oligonucleotides with HindIII and ApaI overhangs into ApaI–HindIII cleaved pLM90.91, a wild-type lacZ plasmid, as described previously (3). Cells to be assayed for β-galactosidase activity were grown to mid-logarithmic phase in minimal E medium (12) containing glucose (0.2%), thiamine, proline, tryptophan and casamino acids (0.2%), tetracycline (12.5 mg/l) and neomycin (50 mg/l), and assayed as described previously (3). Chromosomal and plasmid DNA isolations, construction of genomic libraries and transformations were carried out as described (13). The low copy number, pSC101-derived plasmid, pLG339 (14), was used as a cloning vector for the suf-3 mutant. The kanamycin resistant, pLG339-derived plasmid, p815, contains the wild-type valU operon inserted in the BamHI site of the tetracycline resistance gene of pLG339 (3).
Isolation and identification of frameshift suppressors
Suppressors of trpE9777 were isolated by plating 0.1 ml of an overnight culture of MC141 on minimal (E) medium plates (12) containing thiamine and proline and incubating at 30°C. Intragenic and trpE+ revertants were identified by preparing P1 lysates on each Trp+ isolate, using this phage to transduce strain MC35 [F– Δ(lac-Pro) thi-1 Δ(trpE-C)8] to trpC+ trpD+ (growth on anthranilic acid) and screening for inheritance of tryptophan independence.
RESULTS AND DISCUSSION
Isolation, mapping and sequencing of suppressors of the +1 frameshift mutant trpE9777
The trpE9777 frameshift mutation contains an insertion of a single A residue at codon 4 or 5 of the E.coli trpE gene (Table 1) which abolishes anthranilate synthetase activity. Frameshift suppressor mutations that promoted frameshifting in the trpE9777 mRNA were isolated by selecting for tryptophan-independent derivatives of MC141 (a trpE9777-containing strain). Approximately 50 tryptophan-independent isolates were obtained in this way and five isolates that grew at varying rates on tryptophan-free medium were chosen for further study. Four of these isolates were shown by co-transduction experiments to map close to, or within, the tryptophan operon. These presumptive intragenic suppressors were not studied further. One suppressor, suf-3, was not co-transducible with the trp region of the chromosome and a linked kanamycin resistant Tn10 element, zbe2::Tn10Kan, was isolated to facilitate mapping of this new suppressor. The zbe2::Tn10Kan element was mapped to the 10–30 min region of the chromosome by Hfr crosses. Previous work by Kohno and Roth (6) had suggested that mutants of tRNALys could suppress trpE9777, however, suf-3 showed no linkage by P1 transduction to nadA::Tn10, which is close to the tRNALys-encoded by lysT and lysW genes at 17 min (15). Further transductional mapping with markers in the 10–30 min region showed that zbe2::Tn10Kan was 91% co-transducible with supE44. The UAG suppressor, supE44, is an allele of glnV, encoding a CAG-decoding tRNA2Glnand is located in an operon containing seven tRNA genes, including duplicate genes for tRNA1Gln(glnU and glnW) and tRNA2Gln (glnV and glnX). The close linkage of zbe2::Tn10Kan to supE44 suggested that the suf-3 might be a mutant form of one of these seven tRNAs.
Table 1. Suppression of hisB/C/D/F and trpE frameshifts by mutant tRNA1Gln and tRNALys.
Suppression of his mutants was assessed by growth on minimal, histidine-free media. +, weak suppression; ++, +++ and ++++, increasingly efficient suppression; –, no suppression. hisD6580 was shown to be suppressed by the E.coli sufG100 and Salmonella sufG70 mutants (6) that have the same alteration in tRNA1Gln as the pLG31-encoded mutant analyzed here. CAAN quadruplets are indicated in bold type.
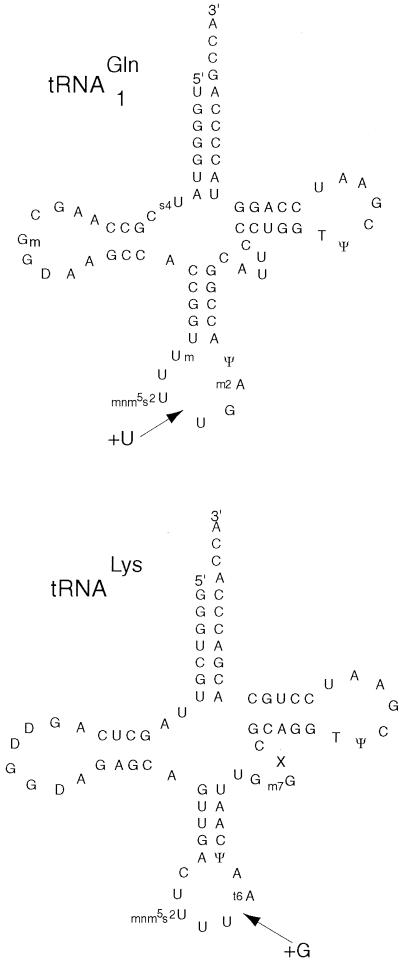
A plasmid carrying the suf-3 suppressor was isolated from a genomic library and the suppressor was localized to a 3.6 kb EcoRI–BamHI DNA fragment. Sequence analysis indicated that the insert was derived from the glnV region of the chromosome. Sequencing of the glnU, V, W and X genes showed that suf-3 contained an insertion of a single thymine residue in the glnU gene between the bases corresponding to U33 and U34 of wild-type tRNA1Gln. This insertion is predicted to enlarge the anticodon loop to eight bases and, according to the classical model for frameshift suppressor action, is predicted to allow the tRNA to decode 5′-CAAA-3′ (Fig. 1).
Figure 1.
Secondary structure of tRNA1Gln and tRNALys showing the base insertions found in the frameshift suppressor derivatives described in the text. The base modifications indicated are those found in the respective wild-type tRNAs, the modification status of the mutant tRNAs has not been determined.
Analysis of sufG frameshift suppressors from Salmonella and E.coli
Kohno and Roth (6) have reported the isolation and partial characterization of frameshift suppressors, designated sufG, that were believed to act at runs of A residues. sufG mutations were isolated both in Salmonella and E.coli and sufG alleles from both organisms suppressed trpE9777. Mapping experiments had shown that sufG was 80% co-transducible (by P22-mediated transduction) with the nag locus and, based on the reported localization of tRNALys-encoding genes to this region of the chromosome, it was proposed that sufG was a mutant form of tRNALys that allowed it to decode AAAA quadruplets. Both the Salmonella and E.coli sufG alleles also suppress the hisD6580 +1 frameshift mutation (Table 1). This hisD mutation has since been sequenced (16) and, based on the presence of an in-frame AAAU sequence, it was inferred that sufG suppressed both AAAA and AAAU quadruplets. However, a comparison of the sequences of both sufG-suppressible frameshift mutations shows that trpE9777 and hisD6580 each contain an in-frame CAAA quadruplet (Table 1), raising the possibility that sufG mutants might be altered forms of a CAA-decoding tRNA1Glnrather than mutants of tRNALys, as had originally been proposed.
Fragments of 200 bp (glnU/W genes) were amplified by PCR from both the E.coli sufG100 and the Salmonella sufG70-containing strains, cloned into the high copy plasmid, pGEM-T, and shown to suppress the trpE9777 frameshift mutation. Nucleotide sequencing of the glnU and W genes showed that sufG100 and sufG70 each had an insertion of a single U between the bases corresponding to U33 and U34 of the mature tRNA1Gln. Thus, the sufG100 and sufG70 mutations are both identical to the suf-3 suppressor isolated in this study, and each suppressor is predicted to have an enlarged 3′-GUUU-5′ anticodon that can pair with the 5′-CAAA-3′ quadruplet found in both trpE9777 and hisD6580 frameshift mutations.
Isolation of CAAA-decoding mutants of tRNALys
tRNALys is involved in several instances of ribosomal frameshifting and its ability to shift frames has been proposed to be due, in part, to an unusual anticodon loop conformation (17,18). The ability of altered forms of this tRNA to promote +1 frameshifting was explored by selecting for tRNALys mutants that could suppress the trpE9777 frameshift mutation. The trpE9777 strain MC141 containing plasmid p815 (3), which carries genes for both valine and lysine tRNAs, was mutagenized with N-methyl-N′-nitro-N-nitrosoguanidine and tryptophan-independent colonies were selected on minimal medium containing kanamycin. Plasmid DNA was extracted from pooled Trp+ colonies, MC141 was transformed with this DNA and Trp+ transformants were again selected on minimal medium. In this way, plasmid-borne suppressors were separated from chromosomal suppressors and trpE revertants. Two plasmid associated suppressors (designated suf-11 and suf-13) were recovered and nucleotide sequencing revealed that each of these plasmids contained an insertion of a single G residue between the bases corresponding to U36 and t6A37 in the anticodon loop of the tRNALys (Fig. 1). The simplest interpretation of this result is that, as is seen with the tRNA1Gln-derived sufG mutants described above, the insertion in the anticodon of tRNALys generates a four base 3′-GUUU-5′ anticodon that can decode 5′-CAAA-3′ quadruplets.
Decoding properties of the mutant tRNAs
Analysis of the suppression spectrum of sufG70 by Kohno and Roth (6) identified several mutations in the his operon of Salmonella that were suppressed by the E.coli sufG100 and Salmonella sufG70 alleles. Since the tRNA1Gln mutant isolated in this study is identical to sufG100 and sufG70, several of these his mutations were sequenced and tested for their ability to be suppressed by the tRNA1Gln-and tRNALys-derived tRNAs. The sequences of hisB6480 and hisB6575 mutations were identical; insertion of a C residue generated a CAAA quadruplet that was suppressed by both mutant tRNAs. Curiously, it had been reported previously (6,19) that while hisB6575 was suppressed by sufB, hisB6480 was not. When the hisB6480 and hisB6575 mutations were transferred to a clean genetic background by P22-mediated transductions with TT27 (strains MS44 and MS50, respectively) or TT10286 (strains MS52 and MS53, respectively), all the resultant strains were detectably leaky on histidine-free media, particularly at 30°C. However, of the original hisB6575/hisB6480-containing strains, only TR2692 (hisO1242 B6575 tyr545 xyl–) was leaky. Conceivably, additional mutations are present in the original hisB6480-containing strain TR3683 (which had been subjected to proflavin mutagenesis) that restrict both its leakiness and its ability to be suppressed by certain mutant tRNAs. Sequencing of the hisC6581 mutant indicated that it also contained a CAAA quadruplet in its frameshift window (Table 1). This mutant was strongly suppressed by the altered tRNA1Gln but only poorly suppressed by tRNALys. It had been reported that hisF6527 was suppressed by sufG70 (6). However, neither the tRNA1Gln mutant analyzed here (which is identical in primary sequence to the Salmonella sufG70 allele) or the tRNALys mutant suppressed hisF6527. Suppression was not observed either in the original TR3696 strain or in MS51, a reconstructed version of this hisF mutant. The reason for this discrepancy is unclear.
Several other frameshift mutations of known sequence and containing CAAN sequences in their frameshifting windows were also tested for suppression by the mutant tRNAs. The hisD3068 mutation contains a CAAU quadruplet 10 codons downstream of the +G insertion but is not suppressed. The hisD85 mutation contains a CAAG quadruplet 2 codons downstream of the +C insertion, however, no suppression by either tRNA mutant was observed. A CAAA quadruplet 33 codons downstream of the frameshift is also found in hisD85 but even if suppression occurs at this site, it is unlikely to yield an active protein given the number of miscoded amino acids that would be incorporated between the potentially suppressible CAAA quadruplet and the site of the insertion mutation. Thus, suppression by mutant sufG/tRNA1Gln and mutant tRNALys appears to be limited to CAAA quadruplets located close to the sites of base insertion in +1 frameshift mutations.
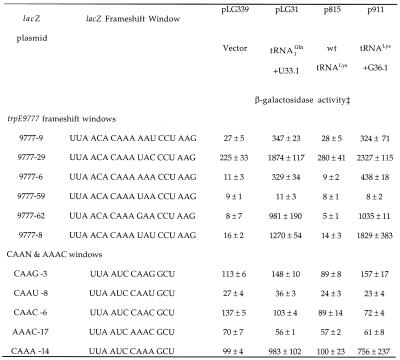
Quadruplet decoding of CAAA quadruplets and related sequences was analyzed further by constructing lacZ frameshift mutants containing the desired target sequences within a frameshifting window and assaying the effects of mutant tRNAs on β-galactosidase activity. In the first series of synthetic lacZ mutants (trpE9777 windows), the CAAA target sequence was embedded in the sequence context found in the trpE9777 frameshift and the 3′ codon was varied. The data presented in Table 2 show that almost all CAAA-containing sequences were well suppressed by both mutant tRNAs. The sole exception occurred when a CAAA quadruplet was followed by a UAA stop codon. The efficiency of suppression varied considerably, depending on the identity of the sense codon 3′ to the CAAA quadruplet. A further series of constructs (CAAN and AAAC windows) indicated that AAAC, CAAC or CAAU quadruplets were not suppressed, suggesting that suppression required codon–anticodon complementarity at the first and fourth positions. The single species of wild-type tRNALys in E.coli decodes AAG as well as AAA and wild-type tRNA1Glnmay decode CAG as well as CAA (another isoacceptor, tRNA2Gln, exists that decodes CAG). Surprisingly, however, the CAAG quadruplet was only poorly suppressed by either mutant tRNA, consistent with the failure to observe suppression of the CAAG-containing hisD85 frameshift described above. In both wild-type tRNALys and tRNA1Gln, position U34 is similarly modified with a mnm5s2 modification (20). This modification has been proposed to limit wobble pairing, however, recent experimental results with unmodified tRNAs are inconsistent with this proposal (21,22).
Table 2. Effects of mutant tRNAs on +1 frameshifting.
‡Numbers represent Miller units of β-galactosidase activity and each is the mean ± SE of at least three independent determinations.
Recent analyses of random libraries of tRNASer mutants that can suppress four base codons in the β-lactamase mRNA indicated that the highest suppression efficiency was obtained with four base codon–anticodon interactions involving Watson–Crick pairings at all positions. In this system, G–U wobble pairings at the fourth codon position led to 10–15-fold decreases in four base reading. It may be that in contrast to triplet decoding, efficient four base reading requires that the extended codon–anticodon helix be stabilized by canonical, Watson–Crick pairings. In conclusion, only 5′-CAAA-3′ quadruplets are decoded efficiently by mutant tRNAs that have complementary 3′-GUUU-5′ anticodons.
Decoding by the mutant tRNAs was further characterized by peptide sequencing of β-galactosidase isolated from strains expressing a lacZ frameshift reporter gene construct and a mutant tRNA. N-terminal sequencing of β-galactosidase isolated from MC141 carrying the p9777–29 lacZ frameshift construct (Table 2) and plasmid pLG31 encoding the mutant tRNA1Gln showed that the sequence ACA CAAA UAC was decoded as threonine, glutamine, tyrosine. Similarly, sequencing of β-galactosidase isolated from MC141 carrying the p9777–8 lacZ frameshift construct (Table 2) and plasmid p911 encoding the mutant tRNALys showed that the sequence ACA CAAA UAU was decoded as threonine, lysine, tyrosine. These data indicate that single amino acids, glutamine and lysine, are inserted by the mutant tRNA1Gln and tRNALys, respectively, at the CAAA quadruplet. Insertion of lysine at CAAA by the mutant tRNALys provides an explanation for the weak suppression of hisC6581 by this tRNA; presumably substitution of lysine for glutamine at this position in the polypeptide adversely affects the folding and/or enzymic activity of the HisC polypeptide.
Mechanism of frameshifting: dual error versus quadruplet decoding models
Early studies on altered tRNAs that suppressed +1 frameshift mutants suggested that four base translocations were effected by tRNAs with correspondingly larger anticodons (1), leading to the proposal of a simple relationship between anticodon loop size and translocation step size. However, the available evidence indicates that the situation is considerably more complex. Rare wild-type tRNAs exist with eight-membered anticodon loops that presumably do not cause rampant frameshifting (23). Some of the characterized tRNA mutants that effect doublet or quadruplet decoding have normal-sized anticodon loops and yeast tRNA mutants with nine-membered loops have been isolated that can decode quadruplets (1,2,24). Moreover, at least some of the tRNA suppressor mutations (including the classical sufB class derived from tRNA2Pro)cause frameshifting indirectly, by allowing a near-cognate tRNA to decode an in-frame triplet in the A site. However, this partially mismatched codon–anticodon helix is unstable and the tRNA is prone to slippage into alternate reading frames. The availability of a cognate codon in an alternate reading frame stabilizes the shifted tRNA, thereby re-phasing translation. According to this mechanism, two sequential events bring about shifts in reading frame. This ‘dual error’ model has since been expanded to explain the action of all frameshift suppressor tRNAs; in the case of the sufA and sufJ type suppressors, base insertions in the anticodon are proposed to promote slippage of the tRNA or isomerization of the codon–anticodon complex, again leading to re-phasing of the reading frame.
Several considerations argue against the universal applicability of such slippage models. In many instances, either a suitable near-cognate tRNA does not exist, the possibility of a mutant tRNA or a near-cognate tRNA binding stably in an alternate reading frame cannot be easily envisioned, and/or there is a strict requirement for Watson–Crick base pairing at each of the four codon–anticodon pairs (2,25). The work described here indicates that the 5′-CAAA-3′ quadruplet can be decoded by either of two different tRNAs with complementary 3′-GUUU-5′ anticodons, and quadruplets that are not perfectly complementary are not decoded by the mutant tRNAs. Moreover, peptide sequencing shows that glutamine and lysine, respectively, are inserted by these mutant tRNAs at CAAA quadruplets, indicating that the mutant tRNAs themselves (and not some near-cognate species) are responsible for suppression. Suitable near-cognate tRNAs do not exist that can cause suppression of CAAA by a dual error mechanism and the possibility of either mutant tRNA binding, and subsequently re-pairing in the +1 reading frame is extremely unlikely, given the number of codon–anticodon mismatches required.
The example of four base decoding described here is similar to quadruplet decoding of 5′-CCGU-3′ by either mutant tRNA2Arg(sufT) or tRNA1Prowith enlarged 3′-GGCI-5′ and 3′-GGCA-5′ anticodons, respectively (25). In both cases, the decoded quadruplet is precisely complementary to the enlarged anticodon and, where tested, there is the necessity for Watson–Crick pairing at the first and fourth codon–anticodon pairs. Both sets of suppressors are dominant and the possibility of pairing and subsequent slippage by the mutant (or a near-cognate) tRNA does not exist. Together, these data argue in favor of four base decoding by direct pairing of the quadruplet codon with the enlarged anticodon.
Acknowledgments
ACKNOWLEDGEMENTS
I am indebted to Dr Albert Dahlberg for providing the support to carry out these experiments. Thanks are due to Drs John Roth and John Atkins for supplying numerous bacterial strains and to John Atkins and Bob Scheckman for carrying out protein sequencing analyses. Hugo Strait is acknowledged for his antiquarian insights. This work was supported by grant GMS19756 to Albert E. Dahlberg from the National Institutes of Health and the initial experiments were aided by funds from the Rhode Island Foundation (to M.O’C.).
REFERENCES
- 1.Atkins J.F., Weiss,R.B., Thompson,S. and Gesteland,R.F. (1991) Towards a genetic dissection of the basis of triplet decoding and its natural subversion: programmed reading frame shifts and hops. Annu. Rev. Genet., 25, 201–228. [DOI] [PubMed] [Google Scholar]
- 2.Magliery T.J., Anderson,J.C. and Schultz,P.G. (2001) Expanding the genetic code: selection of efficient suppressors of four-base codons and identification of ‘shifty’ four-base codons with a library approach in Escherichia coli. J. Mol. Biol., 307, 755–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Connor M., Gesteland,R.F. and Atkins,J.F. (1989) tRNA hopping: enhancement by an expanded anticodon. EMBO J., 8, 4315–4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qian Q., Li,J.N., Zhao,H., Hagervall,T.G., Farabaugh,P.J. and Bjork,G.R. (1998) A new model for phenotypic suppression of frameshift mutations by mutant tRNAs. Mol. Cell., 1, 471–482. [DOI] [PubMed] [Google Scholar]
- 5.Farabaugh P.J. and Bjork,G.R. (1999) How translational accuracy influences reading frame maintenance. EMBO J., 18, 1427–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kohno T. and Roth,J.R. (1978) A Salmonella frameshift suppressor that acts at runs of A residues in the messenger RNA. J. Mol. Biol., 126, 37–52. [DOI] [PubMed] [Google Scholar]
- 7.Hohsaka T., Ashizuka,Y., Taira,H., Murakami,H. and Sisido,M. (2001) Incorporation of nonnatural amino acids into proteins by using various four-base codons in an Escherichia coli in vitro translation system. Biochemistry, 40, 11060–11064. [DOI] [PubMed] [Google Scholar]
- 8.Hohsaka T., Ashizuka,Y., Murakami,H. and Sisido,M. (2001) Five-base codons for incorporation of nonnatural amino acids into proteins. Nucleic Acids Res., 29, 3646–3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bronson M.J. and Yanofsky,C. (1974) Characterization of mutations in the tryptophan operon of Escherichia coli by RNA nucleotide sequencing. J. Mol. Biol., 88, 913–915. [DOI] [PubMed] [Google Scholar]
- 10.Miller J.H. (1991) A Short Course in Bacterial Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 11.Wanner B.L. (1986) Novel regulatory mutants of the phosphate regulon in Escherichia coli K-12. J. Mol. Biol., 191, 399–358. [DOI] [PubMed] [Google Scholar]
- 12.Vogel H.J. and Bonner,D.M. (1956) Acetylornithinase of Escherichia coli: partial purification and some properties. J. Biol. Chem., 218, 97–106. [PubMed] [Google Scholar]
- 13.Silhavy T.J., Berman,M.L. and Enquist,L.W. (1984) Experiments with Gene Fusions. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 14.Stoker N.G., Fairweather,N.F. and Spratt,B.G. (1982) Versatile low-copy-number plasmid vectors for cloning in Escherichia coli. Gene, 18, 335–341. [DOI] [PubMed] [Google Scholar]
- 15.Berlyn M.K., Low,K.B. and Rudd,K. (1996) Linkage map of Escherichia coli K-12, edition 9. In Neidhardt,F.C., Cuttiss,R., Ingraham,J.L., Lin,E.C., Low,K.B., Magasanik,B., Reznikoff,W.S., Riley,M., Schaechter,M. and Umbarger,H.E. (eds), Escherichia coli and Salmonella. Cellular and Molecular Biology, 2nd Edn. ASM Press, Washington, DC, pp. 1715–1902.
- 16.Bossi L. and Roth,J.R. (1981) Four-base codons ACCA, ACCU and ACCC are recognized by frameshift suppressor sufJ. Cell, 25, 489–496. [DOI] [PubMed] [Google Scholar]
- 17.Agris P.F., Guenther,R., Ingram,P.C., Basti,M.M., Stuart,J.W., Sochacka,E. and Malkiewicz,A. (1997) Unconventional structure of tRNA(Lys)SUU anticodon explains tRNA’s role in bacterial and mammalian ribosomal frameshifting and primer selection by HIV-1. RNA, 3, 420–428. [PMC free article] [PubMed] [Google Scholar]
- 18.Watanabe K., Hayashi,N., Oyama,A., Nishikawa,K., Ueda,T. and Miura,K. (1994) Unusual anticodon loop structure found in E.coli lysine tRNA. Nucleic Acids Res., 22, 79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sroga G.E., Nemoto,F., Kuchino,Y. and Bjork,G.R. (1992) Insertion (sufB) in the anticodon loop or base substitution (sufC) in the anticodon stem of tRNA(Pro)2 from Salmonella typhimurium induces suppression of frameshift mutations. Nucleic Acids Res., 20, 3463–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steinberg S., Misch,A. and Sprinzl,M. (1993) Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res., 21, 3011–3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kruger M.K., Pedersen,S., Hagervall,T.G. and Sorensen,M.A. (1998) The modification of the wobble base of tRNAGlu modulates the translation rate of glutamic acid codons in vivo. J. Mol. Biol., 284, 621–631. [DOI] [PubMed] [Google Scholar]
- 22.Hagervall T.G., Pomerantz,S. and McCloskey,J.A. (1998) Reduced misreading of asparagine codons by Escherichia coli tRNALys with hypomodified derivatives of 5-methylaminomethyl-2-thiouridine in the wobble position. J. Mol. Biol., 284, 33–42. [DOI] [PubMed] [Google Scholar]
- 23.Li M. and Tzagoloff,A. (1979) Assembly of the mitochondrial membrane system: sequences of yeast valine and an unusual threonine tRNA gene. Cell, 18, 47–53. [DOI] [PubMed] [Google Scholar]
- 24.Winey M., Mendenhall,M.D., Cummins,C.M., Culbertson,M.R. and Knapp,G. (1986) Splicing of a yeast proline tRNA containing a novel suppressor mutation in the anticodon stem. J. Mol. Biol., 192, 49–63. [DOI] [PubMed] [Google Scholar]
- 25.Tuohy T.M.F., Thompson,S., Gesteland,R.F. and Atkins,J.F. (1992) Seven, eight and nine-membered anticodon loop mutants of tRNA Arg which cause +1 frameshifting. Tolerance of DHU arm and other secondary mutations. J. Mol. Biol., 228, 1042–1054. [DOI] [PubMed] [Google Scholar]