Abstract
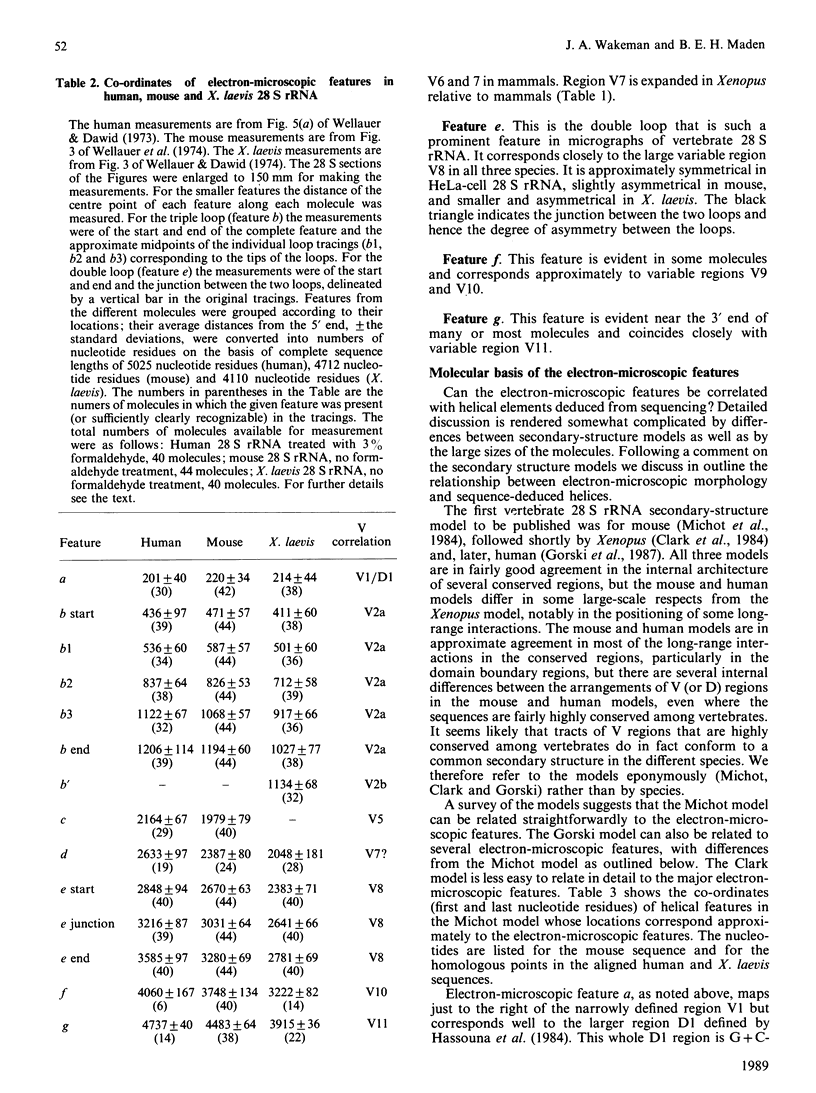
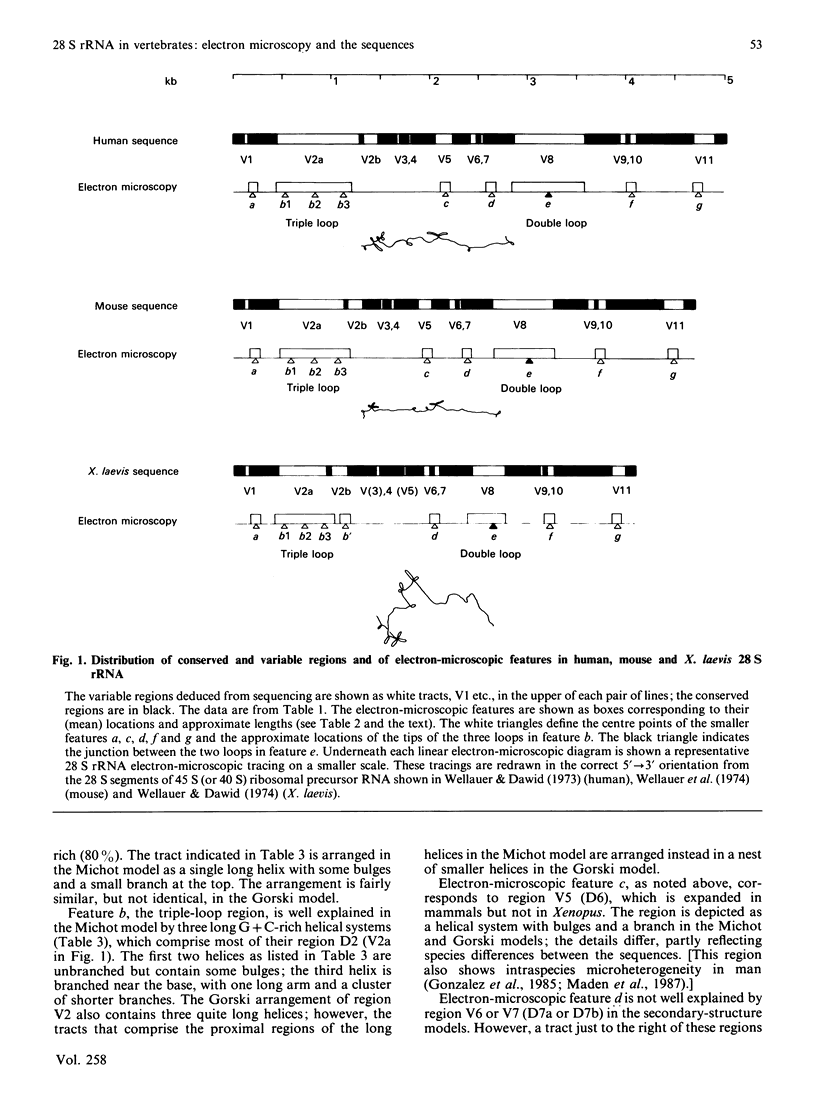
The 28 S rRNA from several vertebrate species, when examined by electron microscopy, is seen to contain regions of extensive secondary structure, as first reported for HeLa-cell 28 S rRNA by Wellauer & Dawid [(1973) Proc. Natl. Acad. Sci. U.S.A. 70, 2827-2831]. Here we correlate the locations of these regions, determined from the electron-microscopic data, with the primary structure of 28 S rRNA from human, mouse and Xenopus laevis determined by sequence analysis of rDNA. The secondary-structure features observed by electron microscopy correspond closely to phylogenetically variable G + C-rich regions that largely comprise the eukaryotic expansion segments in these three species. In most if not all cases the features can be identified with long G + C-rich helices deduced from sequence data. Correlations are given between the locations of the secondary-structure features and several 'landmark' restriction sites in 28 S rDNA. By correlating the locations of the rRNA methyl groups reported elsewhere [Maden (1988) J. Mol. Biol. 201, 289-314] with the present findings it is concluded that the rRNA secondary-structure features revealed by electron microscopy are largely or wholly unmethylated.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Clark C. G., Tague B. W., Ware V. C., Gerbi S. A. Xenopus laevis 28S ribosomal RNA: a secondary structure model and its evolutionary and functional implications. Nucleic Acids Res. 1984 Aug 10;12(15):6197–6220. doi: 10.1093/nar/12.15.6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawid I. B., Wellauer P. K. A reinvestigation of 5' leads to 3' polarity in 40S ribosomal RNA precursor of Xenopus laevis. Cell. 1976 Jul;8(3):443–448. doi: 10.1016/0092-8674(76)90157-4. [DOI] [PubMed] [Google Scholar]
- Furlong J. C., Forbes J., Robertson M., Maden B. E. The external transcribed spacer and preceding region of Xenopus borealis rDNA: comparison with the corresponding region of Xenopus laevis rDNA. Nucleic Acids Res. 1983 Dec 10;11(23):8183–8196. doi: 10.1093/nar/11.23.8183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlong J. C., Maden B. E. Patterns of major divergence between the internal transcribed spacers of ribosomal DNA in Xenopus borealis and Xenopus laevis, and of minimal divergence within ribosomal coding regions. EMBO J. 1983;2(3):443–448. doi: 10.1002/j.1460-2075.1983.tb01442.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez I. L., Gorski J. L., Campen T. J., Dorney D. J., Erickson J. M., Sylvester J. E., Schmickel R. D. Variation among human 28S ribosomal RNA genes. Proc Natl Acad Sci U S A. 1985 Nov;82(22):7666–7670. doi: 10.1073/pnas.82.22.7666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski J. L., Gonzalez I. L., Schmickel R. D. The secondary structure of human 28S rRNA: the structure and evolution of a mosaic rRNA gene. J Mol Evol. 1987;24(3):236–251. doi: 10.1007/BF02111237. [DOI] [PubMed] [Google Scholar]
- Hadjiolov A. A., Georgiev O. I., Nosikov V. V., Yavachev L. P. Primary and secondary structure of rat 28 S ribosomal RNA. Nucleic Acids Res. 1984 Apr 25;12(8):3677–3693. doi: 10.1093/nar/12.8.3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassouna N., Michot B., Bachellerie J. P. The complete nucleotide sequence of mouse 28S rRNA gene. Implications for the process of size increase of the large subunit rRNA in higher eukaryotes. Nucleic Acids Res. 1984 Apr 25;12(8):3563–3583. doi: 10.1093/nar/12.8.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maden B. E., Dent C. L., Farrell T. E., Garde J., McCallum F. S., Wakeman J. A. Clones of human ribosomal DNA containing the complete 18 S-rRNA and 28 S-rRNA genes. Characterization, a detailed map of the human ribosomal transcription unit and diversity among clones. Biochem J. 1987 Sep 1;246(2):519–527. doi: 10.1042/bj2460519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maden B. E. Identification of the locations of the methyl groups in 18 S ribosomal RNA from Xenopus laevis and man. J Mol Biol. 1986 Jun 20;189(4):681–699. doi: 10.1016/0022-2836(86)90498-5. [DOI] [PubMed] [Google Scholar]
- Maden B. E. Locations of methyl groups in 28 S rRNA of Xenopus laevis and man. Clustering in the conserved core of molecule. J Mol Biol. 1988 May 20;201(2):289–314. doi: 10.1016/0022-2836(88)90139-8. [DOI] [PubMed] [Google Scholar]
- Maden B. E., Salim M., Summers D. F. Maturation pathway for ribosomal RNA in the Hela cell nucleolus. Nat New Biol. 1972 May 3;237(70):5–9. doi: 10.1038/newbio237005a0. [DOI] [PubMed] [Google Scholar]
- Maden B. E., Salim M. The methylated nucleotide sequences in HELA cell ribosomal RNA and its precursors. J Mol Biol. 1974 Sep 5;88(1):133–152. doi: 10.1016/0022-2836(74)90299-x. [DOI] [PubMed] [Google Scholar]
- Michot B., Hassouna N., Bachellerie J. P. Secondary structure of mouse 28S rRNA and general model for the folding of the large rRNA in eukaryotes. Nucleic Acids Res. 1984 May 25;12(10):4259–4279. doi: 10.1093/nar/12.10.4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salim M., Maden B. E. Nucleotide sequence of Xenopus laevis 18S ribosomal RNA inferred from gene sequence. Nature. 1981 May 21;291(5812):205–208. doi: 10.1038/291205a0. [DOI] [PubMed] [Google Scholar]
- Schibler U., Wyler T., Hagenbüchle O. Changes in size and secondary structure of the ribosomal transcription unit during vertebrate evolution. J Mol Biol. 1975 May 25;94(3):503–517. doi: 10.1016/0022-2836(75)90217-x. [DOI] [PubMed] [Google Scholar]
- Smith G. P. Evolution of repeated DNA sequences by unequal crossover. Science. 1976 Feb 13;191(4227):528–535. doi: 10.1126/science.1251186. [DOI] [PubMed] [Google Scholar]
- Stewart M. A., Hall L. M., Maden B. E. Multiple heterogeneities in the transcribed spacers of ribosomal DNA from Xenopus laevis. Nucleic Acids Res. 1983 Feb 11;11(3):629–646. doi: 10.1093/nar/11.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware V. C., Tague B. W., Clark C. G., Gourse R. L., Brand R. C., Gerbi S. A. Sequence analysis of 28S ribosomal DNA from the amphibian Xenopus laevis. Nucleic Acids Res. 1983 Nov 25;11(22):7795–7817. doi: 10.1093/nar/11.22.7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellauer P. K., Dawid I. B., Kelley D. E., Perry R. P. Secondary structure maps of ribosomal RNA. II. Processing of mouse L-cell ribosomal RNA and variations in the processing pathway. J Mol Biol. 1974 Oct 25;89(2):397–407. doi: 10.1016/0022-2836(74)90527-0. [DOI] [PubMed] [Google Scholar]
- Wellauer P. K., Dawid I. B. Secondary structure maps of RNA: processing of HeLa ribosomal RNA. Proc Natl Acad Sci U S A. 1973 Oct;70(10):2827–2831. doi: 10.1073/pnas.70.10.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellauer P. K., Dawid I. B. Secondary structure maps of ribosomal RNA and DNA. I. Processing of Xenopus laevis ribosomal RNA and structure of single-stranded ribosomal DNA. J Mol Biol. 1974 Oct 25;89(2):379–395. doi: 10.1016/0022-2836(74)90526-9. [DOI] [PubMed] [Google Scholar]


