Abstract
Although cellular mitochondrial DNA (mtDNA) copy number varies widely among cell lines and tissues, little is known about the mechanism of mtDNA copy number control. Most nascent replication strands from the leading, heavy-strand origin (OH) are prematurely terminated, defining the 3′ boundary of the displacement loop (D-loop). We have depleted mouse LA9 cell mtDNA to ∼20% of normal levels by treating with 2′,3′-dideoxycytidine (ddC) and subsequently allowed recovery to normal levels of mtDNA. A quantitative ligation-mediated PCR assay was used to determine the levels of both terminated and extended nascent OH strands during mtDNA depletion and repopulation. Depleting mtDNA leads to a release of replication termination until mtDNA copy number approaches a normal level. Detectable total nascent strands per mtDNA genome remain below normal. Therefore, it is likely that the level of replication termination plays a significant role in copy number regulation in this system. However, termination of D-loop strand synthesis is persistent, indicating formation of the D-loop structure has a purpose that is required under conditions of rapid recovery of depleted mtDNA.
INTRODUCTION
The mammalian mitochondrial genome is a closed circular molecule of ∼16 kb and exists in multiple copies per cell. Mammalian mitochondrial DNA (mtDNA) is usually present at 103–104 copies per cell, but can range from several hundred in some cells to over 100 000 in oocytes (1–3). In normal tissues, copy number tends to vary according to the oxidative capacity of the cells. This can change depending on the needs of the tissue, as demonstrated by induction through exercise and chronic stimulation of skeletal muscle (4). This loose correlation between mtDNA and oxidative capacity found in normal tissues does not appear to exist in transformed cells (5). The amount of mtDNA also varies in other pathologic states where there are both examples of mtDNA proliferation and depletion. Some mutations within mtDNA can lead to a proliferation of mtDNA and of mitochondria in general (6–8). Patients with mtDNA depletion syndromes apparently carry tissue-specific defects in mtDNA maintenance (9). Recently, mutations in several nuclear genes encoding enzymes involved in nucleotide biosynthesis have been found in patients with mtDNA depletion, indicating that depletion can occur as a result of altered nucleotide pools (10–12). Finally, HIV patients treated with antiviral nucleoside analogs such as AZT and 2′,3′-dideoxycytidine (ddC) also have tissue-specific depletions of mtDNA (13). These drugs are viral replication inhibitors that can also inhibit mtDNA replication (14). However, these pathologic examples have yielded little insight into the regulatory system responsible for maintaining the normally steady tissue-specific levels of mtDNA.
Mitochondrial DNA replication is coupled to transcription and is initiated from the single promoter on the light strand (15). The resulting transcript forms a stable RNA:DNA hybrid which is processed by the enzyme RNase MRP. This enzyme cleaves the RNA into fragments, which are then used as replication primers for the mtDNA polymerase γ (pol γ) at this origin of leading-strand replication (OH). Most of these replication strands are prematurely terminated after being extended by several hundred nucleotides. These terminated strands are known collectively as 7S DNA. A unique structure termed the displacement loop (D-loop) is formed when a 7S DNA remains bound to the parental molecule and displaces one of the duplex parental strands (Fig. 1). In dividing mouse L-cells, ∼75% of the mtDNA molecules contain D-loops. 7S DNA is rapidly turned over, having a half-life of ∼70 min. It has been estimated that 95% of all the synthesized leading replication strands are prematurely terminated in this manner (1). Therefore, much of the DNA synthesis within mitochondria is devoted to regenerating 7S DNA, with consequent formation of the D-loop. Although it is unclear whether or not the 7S strand is subsequently used for further replication, it is in principle possible, since the 3′ end is extendable by a DNA polymerase in vitro (16). The repeated creation of the D-loop indicates that cells have a capacity for replication that is greater than demanded for maintenance of normal levels of mtDNA.
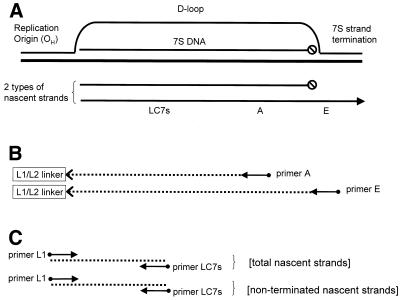
Figure 1.
Strategy for quantification of replication intermediates using ligation-mediated PCR. (A) Top, a newly synthesized 7S strand and the D-loop formed by the displaced H-strand. Bottom, the two fates of nascent replication strands are either to terminate to form 7S DNA or to avoid termination and become a true replication strand. Positions of the primers are indicated below the nascent strands. (B) Two independently primed second-strand runoff reactions were performed to establish the total nascent (primer A) and non-terminated (primer E) strand populations. A linker (L1/L2) was then ligated onto the ends of the double-stranded molecules produced independently from the A and E primed reactions. (C) Each of the two populations was subjected independently to quantitative PCR using the L1 linker primer and the common internal primer, LC7s.
It has been suggested that the termination region is a probable early site for regulation of mtDNA replication (17). This concept has been supported by the identification of a conserved 15-bp region called the termination associated sequence (TAS). TAS elements are potential cis-acting sequences that are frequently present in multiple copies and are located 5′ of the termination sites (18). Phylogenetic comparisons have since extended the region of conservation to include a broader area called the extended or ETAS region (19). More recently, a decrease in termination has been shown to be responsible for the increased replication of mtDNA in proliferating T lymphocytes (20).
As an approach to study the mechanism of mtDNA replication control, we depleted mouse LA9 cells of mtDNA and allowed recovery using the mtDNA replication inhibitor ddC. We have applied a modified ligation-mediated PCR technique used previously (20) to quantify leading-strand replication intermediates during depletion and repopulation of mtDNA. This has provided insight as to how mtDNA copy number is regulated at the leading-strand origin. ddC was chosen to deplete mtDNA because of its relatively specific mode of replication inhibition compared with the commonly used ethidium bromide (21). For ddC to inhibit mtDNA replication, it must be phosphorylated by cytoplasmic kinases to the active triphosphate form before being transported into the mitochondria (22). Inhibition of mtDNA replication is thought to occur by both a competitive inhibition of the mtDNA polymerase as well as by acting as a terminator of nascent strand elongation (23,24). The overall effectiveness of ddC inhibition permitted analysis of mtDNA replication after depletion of the mtDNA population. Examination of the fate of nascent leading-strands suggests that increasing the frequency of D-loop strand elongation is a critical and early response in restoring the normal cellular mtDNA population.
MATERIALS AND METHODS
Cell culture
Mouse LA9 cells (ATCC CCL 1.4) were grown in 100 ml spinner flasks with Spinner minimal essential medium supplemented with 10% newborn calf serum, 10 U penicillin/ml, 10 µg streptomycin/ml, 292 µg glutamine/ml, 1 mM sodium pyruvate, 50 µg uridine/ml, 9.5 mg HEPES/ml and 2.7 mg sodium bicarbonate/ml (Gibco BRL). ddC (Sigma) was prepared as a 4 mM stock solution and used at a final concentration of 20 µM. Cells were centrifuged at passage every 43–48 h and resuspended in fresh medium at a concentration of 1.5–2 × 105 cells/ml. ddC treatment was for 120–125 h to deplete mtDNA. A subsequent complete medium replacement without ddC allowed recovery of mtDNA.
DNA preparation and quantification of mtDNA
Cells were harvested by centrifugation of 1 × 106 cells per sample, and stored at –80°C. DNA was prepared using the High Pure PCR template preparation kit (Roche). Total cellular DNA concentrations were assayed using the double-stranded DNA binding fluorogenic dye Pico-Green (Molecular Probes). Quantification of both mtDNA and each of the nascent replication strands was performed using the quantitative PCR LightCycler apparatus (Roche). Fluorescence data acquisition at each cycle was obtained using a reaction mixture that included the double-stranded DNA binding dye SYBR green I (Roche). Quantification of total mtDNA was done using 2 ng of total cellular DNA per sample. PCR primers were designed to amplify a 116-nt region of the COI gene within mtDNA. Those primer sequences were 5′-gccccagatatagcattccc-3′ and 5′-gttcatcctgttcctgctcc-3′. The Roche SYBR green 1 Master mix was used at 4 mM MgCl2. Temperature cycling was as follows: initial denaturation at 95°C for 30 s, followed by 33 cycles at (95°C for 3 s, 55°C for 6 s, 72°C for 10 s) per cycle. Ramping rates were set at 20°C/s, and data acquisition was at the end of each of the 72°C extensions. Amplification was followed by a melting curve analysis to monitor the homogeneity of amplicons. This consisted of a 3-s denaturation at 95°C followed by a 10-s renaturation step at 70°C and a slow melt approaching 86°C at a ramping rate of 0.2°C/s with continuous data acquisition. Initial samples were also confirmed to be the appropriate size by agarose gel analysis. A standard linear curve was generated using the plasmid p501-1 as the reference, which contains the entire mouse mtDNA genome. One unit of mtDNA is approximately 1 × 106 molecules, based on the calculated molecular weight of the plasmid.
Quantitative ligation-mediated PCR, efficiency normalization
Quantification of nascent replication strands was performed using ligation-mediated PCR essentially as has been described (20), along with the use of the quantitative LightCycler technology. An anchor/adaptor sequence ligated to the free 5′ ends of the nascent replication strands served to ensure the quantification of only nascent strands and not the parental circular mtDNA itself. Two independent second strand extension primers were paired with the ligated linker/adapter to distinguish between terminated and non-terminated replication strands. As shown in Figure 1B, primers A and E were independently annealed to total DNA and extended using Vent polymerase (New England Biolabs). A linker was ligated to the ends of these primer extensions. The linker primer and the internal primer LC7s were then used to quantify the amount of extensions in the reaction using the LightCycler. Primer sequences were as follows: extension primer A, 5′-ggtcataaaataatcatcaac-3′; extension primer E, 5′-gtacataaatttacatagtacaacag-3′; linker sequences L1, 5′-tgtggtgtcatgcatttggt-3′ and L2, 5′-gaatacagatc-3′; and the internal primer LC7s, 5′-atcctccgtgaaaccaacaa-3′. In order to compare the quantification between the two strands, we adjusted the annealing temperatures of the initial extension primers (A and E) so that that they had the same extension efficiency. Cloned mtDNA (p501-1) was linearized with Bsu36 I, which cleaves three nucleotides downstream of the 5′ end of the major replication strand. An equal amount of this linearized template was used to assay for the relative efficiencies of primer A and primer E in annealing and extension. Annealing temperatures of 49°C for primer A and 48°C for primer E were determined to yield equivalent amounts of extension product using the LC7s and linker-primer pair in a subsequent quantitative PCR assay. The amplifications were calculated to range in efficiency from 72 to 80%. Slight efficiency differences between the two primers within an experiment were determined and the data adjusted accordingly. Temperature cycling for the quantitative assay was as follows: initial denaturation and activation of the fast-start polymerase was at 95°C for 10 min, followed by 30 cycles at (95°C for 10 s, 57°C for 6 s, 72°C for 10 s) per cycle. Ramping rates, data acquisition and amplicon quality monitoring were as described above. A representative set of amplification profiles and the resulting standard curve are shown in Figure 2. The standard curve values are generated by extrapolation of the early log–linear phase of each amplification to yield the cycle number of earliest detection.
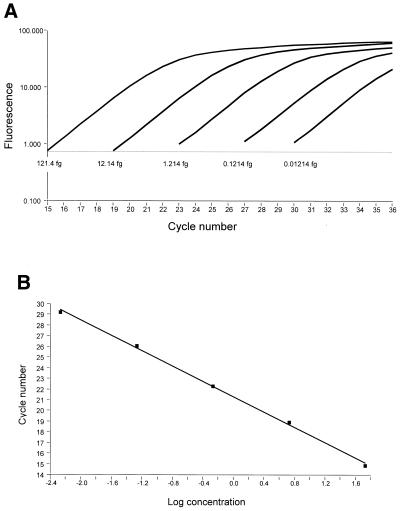
Figure 2.
Quantitative PCR using the LightCycler™ apparatus. (A) A representative standard curve amplification of the replication strands described in Figure 1C. Standard DNA samples with known initial concentrations were amplified and the early linear range of each sample was used to determine the cycle number where the product is first detected. This cycle number correlates with the initial concentration and was used to construct a standard curve (B) that was used to calculate the concentration of unknown samples.
RESULTS
Depletion of mtDNA
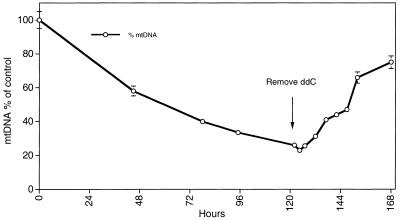
Ethidium bromide treatment results in a complete block of mtDNA replication and is commonly used as a method of depleting cells of mtDNA. At 100% efficiency, every population doubling in ethidium bromide would be expected to yield daughter cells containing half the amount of mtDNA of the immediate parental cell (25). In contrast, ddC was never more than 42% efficient in blocking mtDNA replication, which occurs within the first 2 days of treatment. From day 3 to day 4 this efficiency is further decreased to 17% and from day 4 to day 5 is only 22% further efficient. Figure 3 shows the mtDNA depletion profile and illustrates the shallowing of the depletion rate as the cells approach a state containing 20% of the normal amount of mtDNA. This depletion profile is consistent with two phenomena. The first is that it is likely that ddCTP is rarely incorporated into mtDNA as a chain terminator, since more than half of all progeny mtDNA molecules successfully complete replication. The second is that the declining efficiency of mtDNA depletion as the cells approach 80% depletion indicates that the cells have acquired a ddC resistance or some compensatory response to maintain the remaining level of mtDNA. Treatment of the cells beyond day 5 with repeated ddC-containing media replacement failed to result in further depletion of the mtDNA (T. A. Brown, data not shown).
Figure 3.
Mitochondrial DNA depletion during ddC treatment and removal. Mitochondrial DNA is depleted through day 5 of ddC treatment and subsequently repopulates after removal of the ddC. The arrow shows the point of ddC removal.
Screening for other effects of ddC
Treatment of LA9 cells with ddC could have cellular effects that might complicate the interpretation of the data. An apoptotic response might affect mtDNA replication, considering that the mitochondrion is the center for stress-related apoptotic events. In addition, if the drug treatment caused a growth arrest, we would also have expected at least a lower rate of mtDNA replication. However, Table 1 demonstrates that the concentration of ddC and exposure time used here does not affect the growth rate of this cell line. Population doubling times between treated and untreated cells are comparable. Viability was also unaffected. In addition, we have assayed for gross changes in mitochondrial distribution and cellular amount using an immunofluorescent staining of cytochrome c; we found no differences between untreated and ddC-treated cells (T. A. Brown and A. Santel, data not shown). Therefore, the differences in mtDNA replicative intermediates presented here are not secondary to apoptotic or proliferation changes, nor are they a result of a general global alteration of mitochondrial organelle biogenesis.
Table 1. Population doubling times of untreated and ddC-treated cells.
| Days of ddC treatment | Number of trialsa | PDTb control cells (h) | Number of trials | PDT ddC cells (h) |
|---|---|---|---|---|
| Days 0–2 |
5 |
23.5 (4.9) |
8 |
22.2 (2.1) |
| Days 2–4 |
8 |
22.1 (3.3) |
9 |
25.2 (7.3) |
| Days 4–5 |
6 |
18.5 (2.7) |
8 |
25.4 (5.2) |
| 18 h recovery | 4 | 26.9 (12.0) | 5 | 25.2 (4.5) |
aNumber of trials varies because cell counts were not taken every day for all experiments.
bPopulation doubling time (PDT) was calculated using the formula: hours in culture/log2[final cell density] – log2[initial cell density]. The values are means with SD in parentheses.
Total nascent replication strand assay
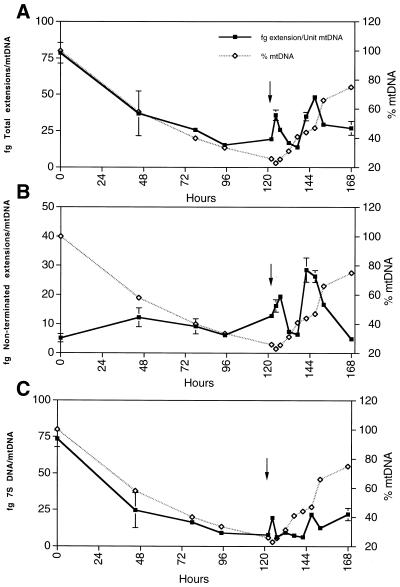
After addition of ddC, total nascent strand levels immediately and progressively decrease (Fig. 4A). Upon removal of ddC at 120 h (Fig. 4A), total nascent strand levels immediately increase. This is followed by a decline in strand levels and then another similar peak and decline. The simplest interpretation is that this cycling is the result of a release of replication inhibition and a synchronization of mtDNA replication. Although normal mtDNA replication is thought to be asynchronous, the removal of replication inhibition may provide the opportunity for synchronization, as occurs after removal of nuclear DNA replication inhibitors. The decline in total nascent strands may be due to the limited availability of replication proteins and appropriate substrates due to their full employment. Figure 4 also displays an overlay of the mtDNA recovery profile. This shows that after ddC removal, mtDNA recovery rates also cycle between slow and fast in a stepwise manner, consistent with the concept of synchronous replication. The mtDNA recovery pattern is in phase with the proposed synchronization of mtDNA replication seen in the assays of total nascent strands.
Figure 4.
Kinetics of replication strand intermediates during mtDNA depletion and repopulation. (A) Total replication strands per mtDNA were quantified, which includes both terminated D-loop and non-terminated replicative strands. (B) Non-terminated replication strands per mtDNA genome, which represent true replicative leading strands. (C) A profile of 7S DNA per mtDNA unit calculated from the data in (A) and (B). Replication strand data are displayed as solid lines and the corresponding levels of mtDNA are overlayed as dashed lines. Arrow indicates removal of ddC.
Non-terminated nascent strands
Unlike the total nascent strand levels, non-terminated strands increase upon depletion of mtDNA (Fig. 4B). This indicates that there has been a release of termination. This occurs by day 2 when the cells are depleted to 55% of normal, and is followed by a return to a near basal level of termination. However, the number of non-terminated strands relative to those that are initiated is still far greater than normal. This is emphasized by the ratio of total nascent to non-terminated strands shown in Table 2 (see below). The release of termination is perhaps more dramatically seen after the removal of ddC. Again the cycling and timing of non-terminated strand levels is similar to that in the total strand plot of Figure 4A. This indicates that the replication cycling is regulated at the level of initiation of mtDNA synthesis. In addition, the termination frequency relative to initiation remains well below normal, and this relaxation of termination is maintained during mtDNA repopulation.
Table 2. Relative levels of nascent strands and their ratios.
| Hours in culture | Total nascent strands | Non-terminated strands | Total/non-terminated |
|---|---|---|---|
|
0a |
78.65 (7.06)b |
5.12 (1.45) |
15.36 |
|
45 |
36.94 (15.32) |
12.26 (3.23) |
3.01 |
|
78 |
25.73 (2.37) |
9.24 (2.62) |
2.78 |
|
95 |
15.39 (2.42) |
6.20 (0.03) |
2.47 |
|
120 |
19.39 (1.92) |
12.83 (1.16) |
1.5 |
| 124.5 |
35.85 (3.58) |
16.29 (2.13) |
2.2 |
| 127 |
25.89 (0.61) |
19.47 (1.02) |
1.33 |
| 132 |
16.85 (0.45) |
7.40 (1.29) |
2.27 |
| 137 |
13.90 (0.55) |
6.45 (0.94) |
2.15 |
| 142 |
34.92 (2.92) |
28.54 (4.15) |
1.22 |
| 147 |
48.11 (1.61) |
26.34 (2.04) |
1.82 |
| 152 |
29.39 (0.59) |
16.72 (1.27) |
1.75 |
| 168 | 26.90 (4.72) | 4.91 (0.49) | 5.47 |
aSamples in the presence of ddC are shown in italics.
bValues are the average of four assays with the SD in parentheses.
Ratio of total/non-terminated strands
The calculated ratio of total nascent strands to non-terminated strands in Table 2 further illustrates the release of replication termination at the end of the D-loop. In untreated control cells there are approximately 15 strands initiated for every one extended beyond the TAS region. This is in close agreement with previous data that estimated approximately 19 strands initiated for every one extended beyond the D-loop (1). In contrast, at day 2 in the presence of ddC, this ratio decreases to three strands initiated for every one extended. Much of this ratio change during depletion is due to an absolute decrease in total nascent strands (numerator), although an increase in non-terminated strand levels (denominator) is contributory. More importantly, when the ddC is removed and total nascent strands become abundant, the now higher level of non-terminated strands is primarily responsible for this lower ratio. The non-terminated strand level per mtDNA molecule remains higher than normal as cells recover to ∼80% of the control level of mtDNA.
7S DNA and D-loop formation
Figure 4C is a plot of calculated 7S DNA levels during depletion and repopulation of mtDNA. The data reinforce the notion that TAS termination is persistent (also seen in Table 2). The replication machinery insists on terminating, even when doing so would presumably retard the ability to recover mtDNA. This suggests that termination and formation of the D-loop structure may be a continually required dynamic process in mtDNA maintenance.
DISCUSSION
Little is known regarding cellular regulation of mtDNA copy number. We have shown here that at least one mechanism is a relaxation of termination at the 3′ end of the D-loop when cells have a decreased amount of mtDNA. This supports the conclusion of Kai et al. (20) who first used a similar technique to demonstrate that proliferating T lymphocytes release mtDNA replication termination in order to keep pace with rapid cell proliferation. Regarding the mechanism of termination, there are several D-loop binding proteins identified with putative roles in this event. Madsen et al. (26) characterized a 48 kDa protein (or protein complex) which binds in vivo to a TAS region in bovine mtDNA. TAS binding regions have also been identified in other species, along with several potentially functional termination proteins (27–31). Cantatore and colleagues (32) have cloned and characterized the mtDBP gene in sea urchin, which is also a regulatory candidate. This protein has sequence homology with the mammalian mitochondrial transcription termination factor, raising the possibility that termination of replication and transcription may have shared elements. This concept and the apparent heterogeneity of the proteins identified to date also indicate that this region of the D-loop may have multiple roles which could include copy number regulation, as well as transcriptional and D-loop structure regulation. Alternatively, this may indicate that the regulation of D-loop formation has rapidly diverged in concert with the control region mtDNA sequence during evolution. Moraes et al. (33) have obtained evidence using ape-human cybrids that a nuclear trans-acting gene and mtDNA sequence play key roles in mtDNA maintenance between species. Termination proteins and cis-acting TAS sequences thus might account for some of the species specificity in the maintenance of mtDNA.
The principal result presented here is that mitochondrial replication termination at the TAS area of the control region is released during cellular recovery of mtDNA. This provides a possible mechanism by which the cell can regulate and perhaps even sense the levels of mtDNA. As cells recover the full ability to replicate mtDNA after ddC removal they do not exhibit the normal level of D-loop strand termination. In the absence of reduced termination the ability of the cells to replenish their mtDNA would be severely limited without an increase in replication initiation. In addition, Figure 3 illustrates that as cells approach 80% depletion of their mtDNA, there is a resistance to further reduction in mtDNA levels. This result could indicate that nearly full abrogation of termination, realized when the cellular mtDNA level is reduced to ∼20%, is sufficient to maintain the mtDNA population at that level. We note that our experiments do not bear on the issue of whether any resection (or rapid loss and resynthesis) of the previously terminated strand is required for continued elongation.
Our data do not address nor exclude other possible modes of mtDNA copy number regulation. In principle, any critical component of the replication machinery could be a limiting factor in determining maximum mtDNA copy number, and the nature of regulation could vary between cell types. Examples of this are the mtDNA depletion syndromes resulting from defects in mitochondrial nucleotide biosynthetic genes (10–12). It has been previously suggested that transcriptional regulation might control mtDNA copy number at the level of replication initiation. The levels of the mitochondrial transcription factor, mtTFA (34,35), and the single-stranded binding protein, mtSSB (36), have been correlated with mtDNA copy number, but a direct demonstration of cause and effect is currently lacking. However, a recent study using isolated mitochondria has revealed a stimulatory effect of mtTFA on the synthesis of D-loop strands (37). In our total nascent strand assay we saw no evidence of a substantial compensatory increase in replication initiation during depletion or recovery, but it is possible that a small increase could have occurred. We also note that the assays employed in this study to examine early replication intermediates engaged in leading-strand DNA synthesis, and our interpretation of the results, are independent of the character of advanced replication intermediates. A measure of the relative amount of early termination has little bearing on advanced replication intermediates containing either extensive single-stranded character or molecules in a similar state of replication that are extensively duplex in character (38). Finally, the upper limit of cellular mtDNA appears to be defined by the total mass of mtDNA, since this is kept constant between isogenic cell lines carrying mitochondrial genomes of different sizes (39). It will be important to determine the relationship between initiation and termination in effecting mtDNA amount.
Our results indicate that ddC is not a profound inhibitor of mtDNA synthesis in these cells. This was perhaps fortuitous, since a more severe effect might have resulted in induction of apoptosis, growth inhibition, or other acute responses. Such effects were not seen in this system, thus limiting the experimental variables. In addition, our data give insight into the mode of mtDNA replication inhibition in this cell line. There are potentially five mechanisms by which ddCTP can inhibit mtDNA replication. First, incorporation of this analog into the 3′ end of a replicating chain would terminate elongation through the inability to form the subsequent 5′-phosphodiester linkage. Secondly, ddCTP can directly inhibit the processivity of the polymerase through competitive occupation of the nucleotide binding site. Thirdly, pol γ has an intrinsic exonuclease domain that could be inhibited by ddCTP or cause the enzyme to repeatedly proofread incorporation of the aberrant chain-terminating nucleotide. Inhibition of the exonuclease function has not been demonstrated for ddC, but does occur with other chain-terminating nucleoside analogs (40,41). Either of these exonuclease effects would also lead to an apparently less processive polymerase. Fourthly, any inhibition of pol γ might also be expected to cripple the role of this enzyme in other DNA repair mechanisms (42). Fifthly, it is possible that the cytoplasmic nucleotide kinases and mitochondrial transporters are saturated or inhibited by the ddC, thereby causing a limitation in mitochondrial nucleotides and a slower replication rate.
We believe that a less processive pol γ is primarily responsible for mtDNA depletion in these cells. For example, even a low rate of irreversible chain termination would lead to a highly inhibitory effect on replication. If a single chain terminator were to be incorporated into each replicating mtDNA molecule, the net efficiency of inhibition would be 100%. Since the genome is 16 295 nt, replication of both strands would require the successful polymerization of 32 590 nt to avoid inhibition of productive replication. Successful chain termination is far rarer than that, since it can be calculated from Figure 3 that the efficiency of inhibition is 42% at most. This means that either ddCTP-mediated chain termination is rare, or that the rate of mtDNA replication has decreased below what is required to keep pace with cell division. Figure 4A shows that the total number of nascent strands per mtDNA progressively decreases. The mtDNA depletion profile and the total nascent replication strand profile are together inconsistent with a high amount of chain termination. The frequency of ddCTP-mediated chain termination would have to be low to rationalize the mtDNA depletion profile but, conversely, would have to be high in order to explain the decline in the levels of total nascent replication strands per mtDNA molecule. A less processive polymerase could account for both the progressive decline in total nascent strands as well as continued depletion of mtDNA.
Although replication termination clearly plays a role in copy number recovery, we believe that the results suggest another function of replication termination, which is the formation of the D-loop structure itself. As seen in Figure 4C and Table 2, there is a persistent level of termination, even when cells are actively recovering a normal level of mtDNA. Persistence of termination was also seen during rapid mtDNA replication in proliferating T cells (20). In its simplest interpretation, a functional termination apparatus should impede efficient mtDNA recovery. However, the available data indicate that the D-loop structure is functionally important and is not simply a reflection of the level of replication termination. Although its full repertoire is unknown, since formation of the D-loop is dependent upon termination, one important function may be related to mtDNA copy number regulation. Previous attempts at correlating this structure to the transcriptional and replicative needs of the cell have only revealed that the relationship is complex (43). More intriguing is the evidence that the D-loop is associated with the membrane and the suggestion that this structure is involved in the formation of the multi-copy mtDNA ‘nucleoids’ (44,45). Such an organization could be the basis of a copy number surveillance system with termination at the center of replication regulation and nucleoid formation.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Kim M. Clark and Jackie N. Doda for ongoing discussions and support of this work, as well as Ansgar Santel for his assistance with immunofluorescence microscopy.
REFERENCES
- 1.Bogenhagen D. and Clayton,D.A. (1978) Mechanism of mitochondrial DNA replication in mouse L-cells: kinetics of synthesis and turnover of the initiation sequence. J. Mol. Biol., 119, 49–68. [DOI] [PubMed] [Google Scholar]
- 2.Pikó L. and Taylor,K.D. (1987) Amounts of mitochondrial DNA and abundance of some mitochondrial gene transcripts in early mouse embryos. Dev. Biol. (NY), 123, 364–374. [DOI] [PubMed] [Google Scholar]
- 3.Robin E.D. and Wong,R. (1988) Mitochondrial DNA molecules and virtual number of mitochondria per cell in mammalian cells. J. Cell Physiol., 136, 507–513. [DOI] [PubMed] [Google Scholar]
- 4.Williams R.S. (1986) Mitochondrial gene expression in mammalian striated muscle. Evidence that variation in gene dosage is the major regulatory event. J. Biol. Chem., 261, 12390–12394. [PubMed] [Google Scholar]
- 5.Van den Bogert C., De Vries,H., Holtrop,M., Muus,P., Dekker,H.L., Van Galen,M.J., Bolhuis,P.A. and Taanman,J.W. (1993) Regulation of the expression of mitochondrial proteins: relationship between mtDNA copy number and cytochrome-c oxidase activity in human cells and tissues. Biochim. Biophys. Acta, 1144, 177–183. [DOI] [PubMed] [Google Scholar]
- 6.Holt I.J., Harding,A.E., Cooper,J.M., Schapira,A.H., Toscano,A., Clark,J.B. and Morgan-Hughes,J.A. (1989) Mitochondrial myopathies: clinical and biochemical features of 30 patients with major deletions of muscle mitochondrial DNA. Ann. Neurol., 26, 699–708. [DOI] [PubMed] [Google Scholar]
- 7.Goto Y., Horai,S., Matsuoka,T., Koga,Y., Nihei,K., Kobayashi,M. and Nonaka,I. (1992) Mitochondrial myopathy, encephalopathy, lactic acidosis and stroke-like episodes (MELAS): a correlative study of the clinical features and mitochondrial DNA mutation. Neurology, 42, 545–550. [DOI] [PubMed] [Google Scholar]
- 8.Shoffner J.M., Lott,M.T., Lezza,A.M., Seibel,P., Ballinger,S.W. and Wallace,D.C. (1990) Myoclonic epilepsy and ragged-red fiber disease (MERRF) is associated with a mitochondrial DNA tRNA(Lys) mutation. Cell, 61, 931–937. [DOI] [PubMed] [Google Scholar]
- 9.Vu T.H., Sciacco,M., Tanji,K., Nichter,C., Bonilla,E., Chatkupt,S., Maertens,P., Shanske,S., Mendell,J., Koenigsberger,M.R. et al. (1998) Clinical manifestations of mitochondrial DNA depletion. Neurology, 50, 1783–1790. [DOI] [PubMed] [Google Scholar]
- 10.Nishino I., Spinazzola,A. and Hirano,M. (1999) Thymidine phosphorylase gene mutations in MNGIE, a human mitochondrial disorder. Science, 283, 689–692. [DOI] [PubMed] [Google Scholar]
- 11.Mandel H., Szargel,R., Labay,V., Elpeleg,O., Saada,A., Shalata,A., Anbinder,Y., Berkowitz,D.G., Hartman,C., Barak,M. et al. (2001) The deoxyguanosine kinase gene is mutated in individuals with depleted hepatocerebral mitochondrial DNA. Nature Genet., 29, 337–340. [DOI] [PubMed] [Google Scholar]
- 12.Saada A., Shaag,A., Mandel,H., Nevo,Y., Eriksson,S. and Elpeleg,O. (2001) Mutant mitochondrial thymidine kinase in mitochondrial DNA depletion myopathy. Nature Genet., 29, 342–344. [DOI] [PubMed] [Google Scholar]
- 13.Yarchoan R., Mitsuya,H. and Broder,S. (1989) Clinical and basic advances in the antiretroviral therapy of human immunodeficiency virus infection. Am. J. Med., 87, 191–200. [DOI] [PubMed] [Google Scholar]
- 14.Martin J.L., Brown,C.E., Matthews-Davis,N. and Reardon,J.E. (1994) Effects of antiviral nucleoside analogs on human DNA polymerases and mitochondrial DNA synthesis. Antimicrob. Agents Chemother., 38, 2743–2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shadel G.S. and Clayton,D.A. (1997) Mitochondrial DNA maintenance in vertebrates. Annu. Rev. Biochem., 66, 409–435. [DOI] [PubMed] [Google Scholar]
- 16.Eichler D.C., Wang,T.S.,Clayton,D.A. and Korn,D. (1977) In vitro replication of mitochondrial DNA. Elongation of the endogenous primer sequence in D loop mitochondrial DNA by human DNA polymerase beta. J. Biol. Chem., 252, 7888–7893. [PubMed] [Google Scholar]
- 17.Clayton D.A. (1982) Replication of animal mitochondrial DNA. Cell, 28, 693–705. [DOI] [PubMed] [Google Scholar]
- 18.Doda J.N., Wright,C.T. and Clayton,D.A. (1981) Elongation of displacement-loop strands in human and mouse mitochondrial DNA is arrested near specific template sequences. Proc. Natl Acad. Sci. USA, 78, 6116–6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sbisà E., Tanzariello,F., Reyes,A., Pesole,G. and Saccone,C. (1997) Mammalian mitochondrial D-loop region structural analysis: identification of new conserved sequences and their functional and evolutionary implications. Gene, 205, 125–140. [DOI] [PubMed] [Google Scholar]
- 20.Kai Y., Miyako,K., Muta,T., Umeda,S., Irie,T., Hamasaki,N., Takeshige,K. and Kang,D. (1999) Mitochondrial DNA replication in human T lymphocytes is regulated primarily at the H-strand termination site. Biochim. Biophys. Acta, 1446, 126–134. [DOI] [PubMed] [Google Scholar]
- 21.King M.P. and Attardi,G. (1988) Injection of mitochondria into human cells leads to a rapid replacement of the endogenous mitochondrial DNA. Cell, 52, 811–819. [DOI] [PubMed] [Google Scholar]
- 22.Chen C.H. and Cheng,Y.C. (1989) Delayed cytotoxicity and selective loss of mitochondrial DNA in cells treated with the anti-human immunodeficiency virus compound 2′,3′-dideoxycytidine. J. Biol. Chem., 264, 11934–11937. [PubMed] [Google Scholar]
- 23.Waqar M.A., Evans,M.J., Manly,K.F., Hughes,R.G. and Huberman,J.A. (1984) Effects of 2′,3′-dideoxynucleosides on mammalian cells and viruses. J. Cell Physiol., 121, 402–408. [DOI] [PubMed] [Google Scholar]
- 24.Starnes M.C. and Cheng,Y.C. (1987) Cellular metabolism of 2′,3′-dideoxycytidine, a compound active against human immunodeficiency virus in vitro. J. Biol. Chem., 262, 988–991. [PubMed] [Google Scholar]
- 25.King M.P. and Attardi,G. (1996) Isolation of human cell lines lacking mitochondrial DNA. Methods Enzymol., 264, 304–313. [DOI] [PubMed] [Google Scholar]
- 26.Madsen C.S., Ghivizzani,S.C. and Hauswirth,W.W. (1993) Protein binding to a single termination-associated sequence in the mitochondrial DNA D-loop region. Mol. Cell. Biol., 13, 2162–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roberti M., Musicco,C., Polosa,P.L., Milella,F., Gadaleta,M.N. and Cantatore,P. (1998) Multiple protein-binding sites in the TAS-region of human and rat mitochondrial DNA. Biochem. Biophys. Res. Commun., 243, 36–40. [DOI] [PubMed] [Google Scholar]
- 28.Qureshi S.A. and Jacobs,H.T. (1993) Characterization of a high-affinity binding site for a DNA-binding protein from sea urchin embryo mitochondria. Nucleic Acids Res., 21, 811–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Polosa P.L., Roberti,M., Mustich,A., Gadaleta,M.N. and Cantatore,P. (1994) Purification and characterization of a mitochondrial DNA-binding protein that binds to double-stranded and single-stranded sequences of Paracentrotus lividus mitochondrial DNA. Curr. Genet., 25, 350–356. [DOI] [PubMed] [Google Scholar]
- 30.Kumar S., Suzuki,H., Onoue,S., Suzuki,S., Hattori,N. and Ozawa,T. (1995) Rat mitochondrial mtDNA-binding proteins to inter-specifically conserved sequences in the displacement loop region of vertebrate mtDNAs. Biochem. Mol. Biol. Int., 36, 973–981. [PubMed] [Google Scholar]
- 31.Suzuki H., Suzuki,S., Sakurai,T., Kumar,S. and Ozawa,T. (1996) A bovine mtDNA-binding protein to a conserved sequence adjacent to the termination associated sequence in the vertebrate mitochondrial displacement loop region. Biochem. Mol. Biol. Int., 38, 275–283. [PubMed] [Google Scholar]
- 32.Loguercio Polosa P., Roberti,M., Musicco,C., Gadaleta,M.N., Quagliariello,E. and Cantatore,P. (1999) Cloning and characterisation of mtDBP, a DNA-binding protein which binds two distinct regions of sea urchin mitochondrial DNA. Nucleic Acids Res., 27, 1890–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moraes C.T., Kenyon,L. and Hao,H. (1999) Mechanisms of human mitochondrial DNA maintenance: the determining role of primary sequence and length over function. Mol. Biol. Cell, 10, 3345–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Larsson N.G., Oldfors,A., Holme,E. and Clayton,D.A. (1994) Low levels of mitochondrial transcription factor A in mitochondrial DNA depletion. Biochem. Biophys. Res. Commun., 200, 1374–1381. [DOI] [PubMed] [Google Scholar]
- 35.Larsson N.G., Wang,J., Wilhelmsson,H., Oldfors,A., Rustin,P., Lewandoski,M., Barsh,G.S. and Clayton,D.A. (1998) Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nature Genet., 18, 231–236. [DOI] [PubMed] [Google Scholar]
- 36.Schultz R.A., Swoap,S.J., McDaniel,L.D., Zhang,B., Koon,E.C., Garry,D.J., Li,K. and Williams,R.S. (1998) Differential expression of mitochondrial DNA replication factors in mammalian tissues. J. Biol. Chem., 273, 3447–3451. [DOI] [PubMed] [Google Scholar]
- 37.Gensler S., Weber,K., Schmitt,W.E., Perez-Martos,A., Enriquez,J.A., Montoya,J. and Wiesner,R.J. (2001) Mechanism of mammalian mitochondrial DNA replication: import of mitochondrial transcription factor A into isolated mitochondria stimulates 7S DNA synthesis. Nucleic Acids Res., 29, 3657–3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holt I.J., Lorimer,H.E. and Jacobs,H.T. (2000) Coupled leading- and lagging-strand synthesis of mammalian mitochondrial DNA. Cell, 100, 515–524. [DOI] [PubMed] [Google Scholar]
- 39.Tang Y., Schon,E.A., Wilichowski,E., Vazquez-Memije,M.E., Davidson,E. and King,M.P. (2000) Rearrangements of human mitochondrial DNA (mtDNA): new insights into the regulation of mtDNA copy number and gene expression. Mol. Biol. Cell, 11, 1471–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gray N.M., Marr,C.L., Penn,C.R., Cameron,J.M. and Bethell,R.C. (1995) The intracellular phosphorylation of (–)-2′-deoxy-3′-thiacytidine (3TC) and the incorporation of 3TC 5′-monophosphate into DNA by HIV-1 reverse transcriptase and human DNA polymerase gamma. Biochem. Pharmacol., 50, 1043–1051. [DOI] [PubMed] [Google Scholar]
- 41.Bridges E.G., Faraj,A. and Sommadossi,J.P. (1993) Inhibition of mammalian DNA polymerase-associated 3′ to 5′ exonuclease activity by 5′-monophosphates of 3′-azido-3′-deoxythymidine and 3′-amino-3′-deoxythymidine. Biochem. Pharmacol., 45, 1571–1576. [DOI] [PubMed] [Google Scholar]
- 42.Pinz K.G. and Bogenhagen,D.F. (2000) Characterization of a catalytically slow AP lyase activity in DNA polymerase gamma and other family A DNA polymerases. J. Biol. Chem., 275, 12509–12514. [DOI] [PubMed] [Google Scholar]
- 43.Annex B.H. and Williams,R.S. (1990) Mitochondrial DNA structure and expression in specialized subtypes of mammalian striated muscle. Mol. Cell. Biol., 10, 5671–5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Albring M., Griffith,J. and Attardi,G. (1977) Association of a protein structure of probable membrane derivation with HeLa cell mitochondrial DNA near its origin of replication. Proc. Natl Acad. Sci. USA, 74, 1348–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nass M.M.K. (1969) Mitochondrial DNA. I. Intramitochondrial distribution and structural relations of single- and double-length circular DNA. J. Mol. Biol., 42, 521–528. [DOI] [PubMed] [Google Scholar]