Abstract
tmRNA is found in the usual one-piece form in most cyanobacteria, but a small clade has an unusual two-piece form, resulting from processing of a circularly permuted precursor RNA. Here, the secondary structure of the cyanobacterial one-piece tmRNA is established by phylogenetic sequence comparison; it deviates from most tmRNAs in having an extra (fifth) pseudoknot and a branched structure at the end of the tag reading frame. Patterns of sequence conservation between the two cyanobacterial tmRNA types suggest particular events in the evolution of the two-piece form. A simple gene permutation model of tandem duplication followed by loss of the outermost segments appears inadequate. An additional rearrangement is proposed, in which a structure impaired by deletion was regenerated at the expense of a neighboring structure.
INTRODUCTION
Bacterial tmRNA contains both a tRNA-like domain, with a CCA tail accepting alanine, and an mRNA-like region (1,2). It frees ribosomes that have stalled during translation; the ribosome switches from translating the stalled mRNA and resumes translation on the reading frame in tmRNA. The tmRNA encoded peptide serves as a tag that promotes degradation of the tagged protein.
tmRNA is found in two forms in cyanobacteria, a one-piece form (3) similar to that of most bacteria and a two-piece form with its alanine-accepting tail on one RNA piece and the tag-coding sequence on the other piece (4). The gene for the two-piece tmRNA is circularly permuted, i.e. the segment normally at the 3′ end of tmRNA genes is instead found upstream of the segment normally at the 5′ end (5). The mature two-piece tmRNA results from removal of an intervening segment in the permuted precursor RNA, presumably by the same enzymes used for tRNA processing. A secondary structure model was recently developed for the two-piece tmRNA based on structural probing and phylogenetic sequence comparison (4). Here, a secondary structure model is developed for the one-piece form based on phylogenetic sequence comparison.
Permutation has been observed for several protein coding genes (6,7) but only a single case has been reported for the gene of an RNA other than tmRNA (8). It is therefore remarkable that gene permutation has occurred independently for tmRNA genes in two different bacterial lineages; in addition to the cyanobacterial example, it has also been observed in the α-proteobacteria, where it produces a roughly similar two-piece mature tmRNA (5). This suggests that tmRNA function derives some benefit from the two-piece composition, for which there have been specific proposals. A detailed reconstruction of the events of permutation may not be possible for the α-proteobacteria, where the gene has only been found in the permuted form, leaving no strong candidate for an example of the parental unpermuted sequence. The cyanobacteria instead offer an opportunity to deduce specific events that occurred during gene permutation, because segments of the one-piece and two-piece forms bear an extremely close relationship at the level of nucleotide sequence.
A very simple general model for gene permutation, i.e. tandem duplication of the original gene followed by loss of the outer segments of each copy, can explain much but not all of this particular case. Apparently, one of the pieces of the two-piece tmRNA suffered an internal deletion relative to the one-piece form and a pairing lost due to this deletion was restored at the expense of a neighboring pseudoknot, which somewhat complicates the permutation model.
MATERIALS AND METHODS
Bacterial growth
Oscillatoria PCC6304 stock was purchased from the Pasteur Culture Collection and cultured in BG-11 medium under green light and CO2 bubbling, with the kind help of David Kehoe (Indiana University).
Sequences
tmRNA sequences from Synechocystis PCC6803 and Synechococcus PCC6301 and WH8102 were previously identified (9,3,4). Sequences from Nostoc punctiforme and Anabaena PCC7120 were identified in genomic data from the DOE Joint Genome Institute (http://www.jgi.doe.gov/JGI_microbial/html) and Kazusa DNA Research Institute (http://www.kazusa.or.jp/cyano/anabaena), respectively. Genomic DNAs were kind gifts of Séan Turner (Chroococcidiopsis PCC6712), David Kehoe (Fremyella diplosiphon), Martina Celerin (Plectonema boryanum UTEX485) and Yuichi Fujita (P.boryanum IAM-101), all of Indiana University, or prepared by French pressure cell lysis from Oscillatoria PCC6304 and used for PCR with 5′-GGGGCTGTTTAGGTTTCGAC and 5′-TGGAGCTGATGGGAGTCGAAC. The products were sequenced directly after removing excess primers. Sequences were aligned manually. New sequences have been deposited in GenBank (accession nos AY082650–AY082653) and alignments are available at the tmRNA website (http://www.indiana.edu/~tmrna).
Probability calculation for faint homology
The hypothesis of homology for a 43-nt block in the P2–P4 region of the WH8102 and PCC6301 tmRNAs, with base matches at only 20 positions, was evaluated through the null hypothesis that jumbled versions of the 43-nt WH8102 segment and the comparable 69-nt segment of PCC6301 (nucleotides 24–92, see Fig. 3) would reach this level of matching in at least one of the 27 (1 + 69 – 43) possible registers. Using the base composition of the starting segments, the probability of a match at a single position was calculated (PM = 0.2511). This was used to calculate the probability of 20 or more matches for random 43-nt blocks with the compositions of the two starting segments, as the tail of the binomial distribution (P20,43 = 0.001902). Finally, the probability that at least one of the 27 registers would produce 20 or more matches was calculated as P = 1 – (1 – P20,43)27 = 0.0501.
Figure 3.
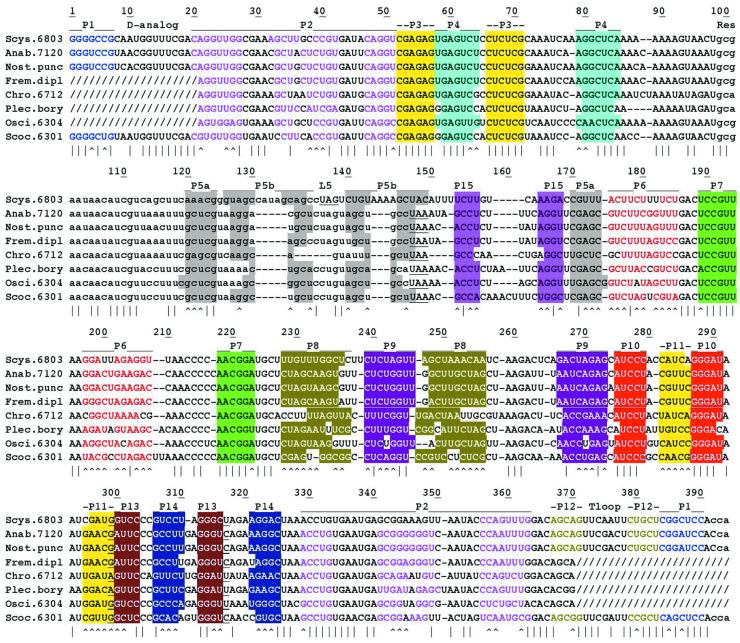
Alignment of cyanobacterial one-piece tmRNA sequences. Tag reading frames and the CCA tails expected to be added post-transcriptionally are in lower case. Slashes show segments of unverified sequence where PCR priming occurred. Vertical lines mark positions conserved among all sequences and carats mark positions where proposed pairings (color coded) exhibit Watson–Crick base pair co-variation. Underlines show the stop codon of the tag reading frame and base co-variation that suggests either extension of P13 at the expense of P11 or a base triple at the pseudoknot junction. Full species names in order: Synechocystis PCC6803; Anabaena PCC7120; Nostoc punctiforme; Fremyella diplosiphon; Chroococcidiopsis PCC6712; Plectonema boryanum; Oscillatoria PCC6304; Synechococcus PCC6301. Numbering refers to the PCC6301 sequence.
RESULTS AND DISCUSSION
Phylogenetic analysis of cyanobacterial one-piece tmRNA
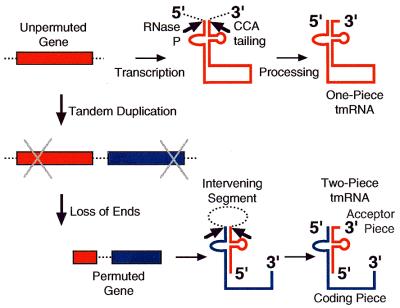
Figure 1 shows how a two-piece tmRNA is thought to be generated from a permuted gene, with a simple model for how permutation might occur. Evaluating models for this instance of gene permutation requires an understanding of tmRNA structure, not only for the two-piece form, as recently established (4) but, equally importantly, for its likely progenitor. The secondary structure of cyanobacterial one-piece tmRNA was examined by phylogenetic sequence comparison.
Figure 1.
One- and two-piece tmRNAs from unpermuted and permuted genes and an oversimplified gene permutation model.
Most tmRNA sequences can be threaded through the standard secondary structure that has been established for Escherichia coli tmRNA by phylogenetic analysis and structural probing (10–12). In this standard structure, the ends form a tRNA-like domain, from which emerges a long stem (P2), capped by a large loop; around this loop occur a pseudoknot (PK1), the tag reading frame with a stem–loop (P5) at its end, and a string of three more pseudoknots (PK2–PK4). Most cyanobacterial species have a tmRNA gene that largely follows this standard form (Fig. 2). Phylogenetic analysis of eight such sequences supports most of the pairings of the standard structure by Watson–Crick base pair co-variation (Fig. 3). PK1 is not supported in that P3 is invariant, but the proposed P3 does resemble those in other tmRNAs; it would appear to abut P2, so that P2 and PK1 could derive mutual stabilization by coaxial stacking.
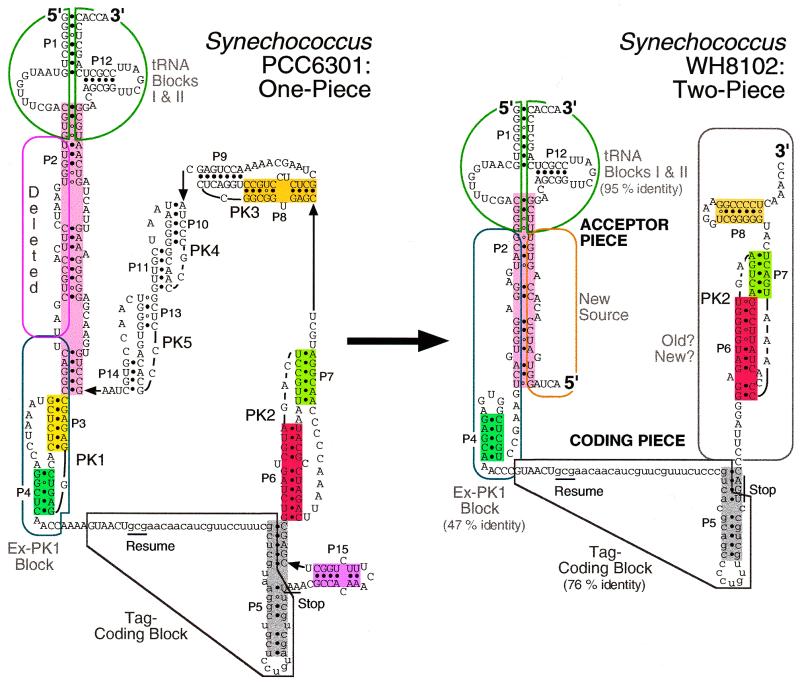
Figure 2.
Related one- and two-piece tmRNAs. Secondary structures were determined here (Fig. 3) and in Gaudin et al. (4).
Two main differences from the standard tmRNA structure occur in these cyanobacteria. In place of the last pseudoknot there are two short abutting pseudoknots (P10 and P11 and P13 and P14), bringing the total pseudoknot count to five. This had been noted previously (9), but now the Watson–Crick co-variation pattern firmly establishes each of the stems, except for P10, which has little sequence variation; only one position in P10 shows any base pair changes; these simply exchange U for C in pairing with G. An interesting pattern of base co-variation occurs at the junction where these two pseudoknots abut, suggesting either alternation with a form in which P13 is extended by 1 bp at the expense of P11 or that a base triple (or more than one) forms at the pseudoknot junction. Both the P11 base pair as drawn and the P13 extension are supported by Watson–Crick co-variation. These two short pseudoknots may well coaxially stack and together fill space like the single pseudoknot that they replace.
The second main difference between these tmRNAs and the standard form is at P5, which is normally a simple stem–loop but in these cyanobacteria takes a branched form, interrupted by a short stem termed P15. P15 is a 4-bp stem with a loop of 4–9 nt; the largest example of this loop, from Synechococcus 6301, may be stabilized by additional internal base pairs (Fig. 2). One end of P5 abuts PK2, which raises the possibility of mutual stabilization by coaxial stacking and is seen frequently, but not consistently, with other tmRNAs.
Among the cyanobacterial one-piece tmRNAs, the central portion of P5 exhibits two different relationships to the stop codon of the tag reading frame. The stop codon is either in the loop of P5 (L5), as in Synechocystis PCC6803 and as in standard tmRNA, or the stop codon is at the end of the central portion of P5 with L5 in the reading frame. Structural homology between these two cyanobacterial forms of P5 is suggested by the block of sequence identity (CUAGU) shared between L5 of PCC6803 and L5 of some other cyanobacteria. It will be necessary to collect sequences from additional species with the stop codon in L5 to confirm this point.
These observations suggest early events in the evolution of cyanobacterial tmRNA structure. Two occurred very early, doubling of the last pseudoknot and formation of a branched P5; we have no cyanobacterial examples of the standard forms of these structures. Presumably the L5 originally contained the stop codon, as in other bacterial phyla and as exemplified by PCC6803. Then P5 shifted in an ancestor of all the other sequences. This could be explained by a 5-nt deletion in P5b in the tag reading frame that threw the original UAG stop codon out of frame and led to the use of a UAA codon in another frame. Subsequent deletion of a codon on the other side of P5 may have trimmed the new P5 and improved the proteolysis-inducing function of the tag.
Clear homologies with the cyanobacterial two-piece tmRNA
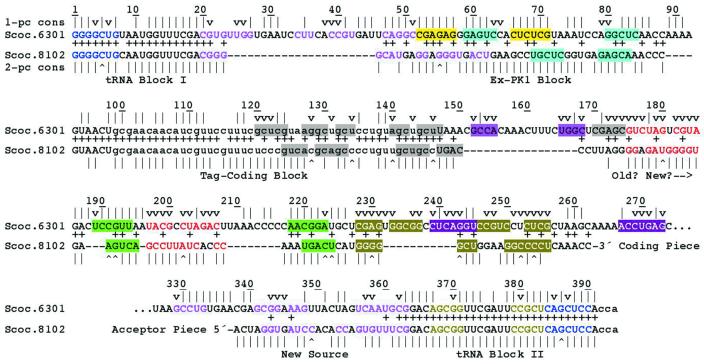
Surprising levels of sequence similarity occur between three sequence blocks in one- and two-piece tmRNAs. Among the known one-piece tmRNAs, the closest relative to the two-piece tmRNAs is that from Synechococcus PCC6301 (5), whose phylogenetic proximity to species with two-piece tmRNAs is confirmed by rRNA sequence analysis (13). tmRNA sequences have been determined for six cyanobacteria with permuted genes and they are very similar to each other; only one gap was necessary to align them and they showed base identity at 75% of the nucleotide positions (4). Clearly they are all descendents of the same original gene permutation event, but among them the sequence from Synechococcus WH8102 can be taken as the representative that is closest to the unpermuted sequence of PCC6301 (Fig. 4). The WH8102 and PCC6301 sequences differ at only three positions in the tRNA-like domain (95% identity), which is composed of two separate sequence blocks in each gene. Throughout a third block (PCC6301 nt 94–147) that contains the tag reading frame they differ at only eight positions (76% identity).
Figure 4.
Alignment of the closest available cyanobacterial one-piece and two-piece tmRNA sequences. Plus signs mark base identities between one-piece Synechococcus PCC6301 and two-piece Synechococcus WH8102 sequences. Conservation summaries for one-piece (Fig. 3) and two-piece (4) tmRNAs are shown. Indications are as in Figure 3. The alignment of the ‘Ex-PK1 Block’ maximizes base identity matches but shifts comparable structural elements. Alignment is instead forced for pairings P6–P8 and may not represent true sequence homology.
Some of the identity in the tRNA-like domain can be discounted, since approximately 28 of its 55 encoded positions are conserved among almost all tmRNAs. Likewise, in the tag reading frame some of the conservation can be attributed to selection for a functional amino acid sequence of the tag. However, the levels of nucleotide sequence conservation in these regions exceeds what can be ascribed to such functional constraints. This is demonstrated by evaluating the matches of these same regions from PCC6301 to those from other one-piece cyanobacterial tmRNAs (Fig. 2); none exceeds the match to WH8102, except for the reading frame segment from Oscillatoria 6304. Thus the PCC6301 gene appears to represent well the ancestral gene that underwent permutation.
Replacement of deleted stem RNA with former pseudoknot RNA
The presence of two blocks that are highly conserved between the one-piece tmRNA and the coding piece of the two-piece tmRNA, i.e. the tag-coding block and one of the tRNA blocks, suggests that there could be a homologous relationship for the segments in between these blocks. These segments have different sizes, 43 nt in WH8102 and 69 nt in PCC6301. One ungapped alignment of these two segments produces 20 base matches (47% identity) (Fig. 4), whose significance must be assessed carefully since random sequences of balanced base composition should have 25% identity. The hypothesis that this alignment represents a homologous block was evaluated against the null hypothesis that unrelated sequences with the lengths and base compositions of these segments would reach this level of matching in at least one of the possible registers; such would occur with probability P = 0.05 (Materials and Methods). Although faint, this appears a significant homology.
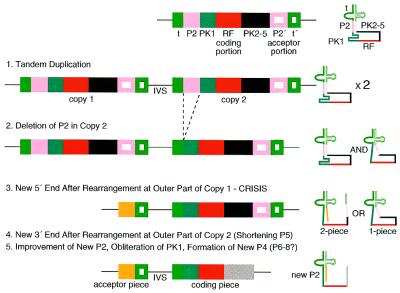
This alignment throws the comparable structural features P2 and P4 out of alignment (Fig. 4). Accepting it as homology requires proposing a deletion of most of the P2 portion in the downstream copy of the presumed tandemly duplicated parent gene (corresponding to 22 nt of PCC6301) (Fig. 5, step 2). The P2 equivalents in the two tmRNA forms come from different portions of the parental sequence, with the two-piece P2 formed from sequence that was originally part of PK1. The concomitant obliteration of PK1 allowed the apparently de novo formation of P4 from the remainder of PK1. Three selective forces can be postulated to explain the many base changes between these segments in one- and two-piece tmRNAs: (i) improvement of a P2 pairing between originally unmatched partners; (ii) elimination of competing PK1 structure; (iii) formation of a new P4 that may compensate functionally for the loss of PK1. PK1 has been shown to be essential for E.coli tmRNA function in vitro (14). A subsequent small deletion at the other end of this sequence block may have improved utilization of the resume codon.
Figure 5.
Proposed steps in gene permutation.
Other rearrangements of gene permutation
The simple model of permutation, with tandem gene duplication followed by loss of outer portions, is complicated by the deletion of a P2 segment because P2 is important, among other things, for holding together the two pieces of tmRNA. Still, the idea of change at the outermost segments of a tandem duplicate seems to hold. The segment of the acceptor piece forming P2 in WH8102 has no obvious sequence relationship to the comparable segment of PCC6301; a reasonable explanation is that it arrived from an exogenous source through a chromosomal rearrangement (Fig. 5, step 3).
Change is also clear at the other outermost segment of the presumed original tandem, downstream from the tag-coding homology block. Interestingly, structural similarity is nonetheless preserved to the extent that a pseudoknot is positioned downstream of the tag-coding region. However, sequence alignments between PK2 of the one- and two-piece tmRNAs is unconvincing and requires several gaps (Fig. 4), and the same is true when two-piece PK2 is forced to align with one-piece PK3 (not shown). It may be that the remaining pseudoknot is homologous to one from the parent, with several deletions and loss of the sequence traces. Alternatively, the whole region may have arrived from an exogenous source through a chromosomal rearrangement, as proposed above for the other end of the tandem. The current data are insufficient to favor either alternative. The latter has interesting corollaries; either that rearrangement brought a preformed pseudoknot into very nearly the same position as the original PK2 or that a new PK2 developed de novo in the new sequence; either corollary implies that a pseudoknot at this position is functionally important in two-piece tmRNA, in contrast to the idea (4,5) that the topology of tmRNA translation makes pseudoknots important for the one-piece form but unimportant in the two-piece form.
One clear result of the changes in this portion of the permuted gene is that a downstream portion of P5 including P15 is lost, such that the long branched form in the parent has become a simple stem–loop. The remaining stem–loop retains sequence similarity and structural homology to the central portion of P5 from the one-piece parent.
A family of gene permutation models
According to the model for permutation of the cyanobacterial tmRNA gene as presented in Figure 5, the first step is a tandem gene duplication, which leaves an intervening segment that (judging from its size in modern permuted tmRNA genes) is too short to provide either promoter or terminator signals for transcription. Thus a dimeric precursor molecule is transcribed and processed to form two identical mature one-piece tmRNA molecules. The next step is deletion of part of P2 from the downstream copy, which inactivates one of the products of the precursor, but the other product is still a functional tmRNA. Next is the change at the upstream end of the tandem, which engenders a crisis phase in evolution. Now the tag reading frame and one of the blocks of the tRNA-like domain occur only once in the precursor RNA, but two copies of the partner block that completes the tRNA-like domain are present, allowing two competing outcomes of processing, one leaving a two-piece tmRNA and the other a one-piece form. The crisis arises because neither of these forms has optimal activity, since there has been no prior selection for regenerating the P2 pairing. If the incipient P2 of the two-piece form were better than that of the one-piece form, this could drive evolutionary resolution of the ambivalent precursor to favor and improve the two-piece form.
Many variations of the Figure 5 model would be consistent with the observations. For example, the original tandem duplication need have been only a partial duplication, which might eliminate the need for a later end rearrangement step. Also, many different (but not all) orders for the post-duplication steps are possible; the ambivalent precursor does not occur with some of these orders. A crisis phase would not be essential for tmRNA gene permutation by first principles (Fig. 1), but in modeling this particular instance it is obligate due to the deletion event that damages P2 and eventually forces a new P2 pairing to form from segments that had not previously paired.
A strain in such a crisis phase would not survive if maximally active tmRNA were essential for growth. The detection of a tmRNA gene in all tested bacteria implies its importance, but the point has been tested only twice in cyanobacteria. An attempt to disrupt the Synechococcus PCC6301 gene failed, implying but not proving that tmRNA is essential in this species (3). In contrast, the Synechococcus PCC6803 has been disrupted with no noticeable effect on growth rate (15). In other bacteria, tmRNA has been found to be essential for growth (Neisseria and Caulobacter), non-essential but required for optimal growth (Escherichia and Bacillus) or non-essential for growth in culture medium yet essential for invasion of eukaryotic hosts (Bradyrhizobium and Salmonella) (5,16–20). These varied phenotypes suggest that selective pressure for a fully active tmRNA gene may not be absolute, which would make the proposed crisis phase plausible.
NOTE ADDED IN PROOF
Genomic sequence data from Synechococcus WH8102 (http://www.jgi.doe.gov) reveal a 12 279-bp genetic element (termed here Syn12X) integrated into the 3" end of the permuted tmRNA gene. Among other features characteristic of elements integrated into bacterial chromosomes, Syn12X encodes an integrase (from the AS subfamily of tyrosine recombinases, not previously known to specify RNA genes) and it has replaced the fragment of its target gene that it displaced upon integration; a 33-nt block of tmRNA sequence is repeated at the other end of Syn12X, with only two base changes that constitute a compensatory base-pair change in the P8 pairing. No such element is found at the permuted tmRNA gene in either of the two complete Prochlorococcus genomes. The question arises whether an ancestor of Syn12X participated in the gene permutation process, perhaps creating the new 3" end of the gene. The position of the attachment site identity block (far from the tRNA-like domain DNA) suggests that the integrase promotes strand crossover in the DNA corresponding to the downstream portion of the single pseudoknot, which is an unprecedented region for crossover in tmRNA genes; moreover, this region may not have been present in the ancestral tmRNA gene. An alternative hypothesis is that Syn12X evolved its site-specificity subsequent to gene permutation, at the new 3" end presented in the permuted gene, which itself has interesting implications for the general question of how integrases so frequently evolve specificity for RNA genes.
Acknowledgments
ACKNOWLEDGEMENTS
I thank David Kehoe, Séan Turner, Martina Celerin and Yuichi Fujita for gifts of genomic DNAs, D. Kehoe for help growing cyanobacteria, S. Turner for useful discussions of cyanobacterial phylogeny and Brice Felden for critical reading of the manuscript and for the suggestion of a base triple at the pseudoknot junction. This work was supported by National Institutes of Health grant GM59881.
DDBJ/EMBL/GenBank accession nos AY082650–AY082653
REFERENCES
- 1.Karzai A.W., Roche,E.D. and Sauer,R.T. (2000) The SsrA-SmpB system for protein tagging, directed degradation and ribosome rescue. Nature Struct. Biol., 7, 449–455. [DOI] [PubMed] [Google Scholar]
- 2.Gillet R. and Felden,B. (2001) Emerging views on tmRNA-mediated protein tagging and ribosome rescue. Mol. Microbiol., 42, 879–885. [DOI] [PubMed] [Google Scholar]
- 3.Watanabe T., Sugita,M. and Sugiura,M. (1998) Identification of 10Sa RNA (tmRNA) homologues from the cyanobacterium Synechococcus sp. strain PCC6301 and related organisms. Biochim. Biophys. Acta, 1396, 97–104. [DOI] [PubMed] [Google Scholar]
- 4.Gaudin C., Zhou,X., Williams,K.P. and Felden,B. (2002) Two-piece tmRNA in cyanobacteria and its structural analysis. Nucleic Acids Res. 30, 2018–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keiler K.C., Shapiro,L. and Williams,K.P. (2000) tmRNAs that encode proteolysis-inducing tags are found in all known bacterial genomes: a two-piece tmRNA functions in Caulobacter. Proc. Natl Acad. Sci. USA, 97, 7778–7783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lindqvist Y. and Schneider,G. (1997) Circular permutations of natural protein sequences: structural evidence. Curr. Opin. Struct. Biol., 7, 422–427. [DOI] [PubMed] [Google Scholar]
- 7.Jung J. and Lee,B. (2001) Circularly permuted proteins in the protein structure database. Protein Sci., 10, 1881–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heinonen T.Y., Schnare,M.N., Young,P.G. and Gray,M.W. (1987) Rearranged coding segments, separated by a transfer RNA gene, specify the two parts of a discontinuous large subunit ribosomal RNA in Tetrahymena pyriformis mitochondria. J. Biol. Chem., 262, 2879–2887. [PubMed] [Google Scholar]
- 9.Williams K.P. and Bartel,D.P. (1998) The tmRNA website. Nucleic Acids Res., 26, 163–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williams K.P. and Bartel,D.P. (1996) Phylogenetic analysis of tmRNA secondary structure. RNA, 2, 1306–1310. [PMC free article] [PubMed] [Google Scholar]
- 11.Felden B., Himeno,H., Muto,A., McCutcheon,J.P., Atkins,J.F. and Gesteland,R.F. (1997) Probing the structure of the Escherichia coli 10Sa RNA (tmRNA). RNA, 3, 89–103. [PMC free article] [PubMed] [Google Scholar]
- 12.Kelley S.T., Harris,J.K. and Pace,N.R. (2001) Evaluation and refinement of tmRNA structure using gene sequences from natural microbial communities. RNA, 7, 1310–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turner S., Pryer,K.M., Miao,V.P. and Palmer,J.D. (1999) Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis. J. Eukaryot. Microbiol., 46, 327–338. [DOI] [PubMed] [Google Scholar]
- 14.Nameki N., Felden,B., Atkins,J.F., Gesteland,R.F., Himeno,H. and Muto,A. (1999) Functional and structural analysis of a pseudoknot upstream of the tag-encoded sequence in E. coli tmRNA. J. Mol. Biol., 286, 733–744. [DOI] [PubMed] [Google Scholar]
- 15.de la Cruz J. and Vioque,A. (2001) Increased sensitivity to protein synthesis inhibitors in cells lacking tmRNA. RNA, 7, 1715–1720. [PMC free article] [PubMed] [Google Scholar]
- 16.Huang C., Wolfgang,M.C., Withey,J., Koomey,M. and Friedman,D.I. (2000) Charged tmRNA but not tmRNA-mediated proteolysis is essential for Neisseria gonorrhoeae viability. EMBO J., 19, 1098–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oh B.K. and Apirion,D. (1991) 10Sa RNA, a small stable RNA of Escherichia coli, is functional. Mol. Gen. Genet., 229, 52–56. [DOI] [PubMed] [Google Scholar]
- 18.Muto A., Fujihara,A., Ito,K.I., Matsuno,J., Ushida,C. and Himeno,H. (2000) Requirement of transfer-messenger RNA for the growth of Bacillus subtilis under stresses. Genes Cells, 5, 627–635. [DOI] [PubMed] [Google Scholar]
- 19.Ebeling S., Kundig,C. and Hennecke,H. (1991) Discovery of a rhizobial RNA that is essential for symbiotic root nodule development. J. Bacteriol., 173, 6373–6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Julio S.M., Heithoff,D.M. and Mahan,M.J. (2000) ssrA (tmRNA) plays a role in Salmonella enterica serovar Typhimurium pathogenesis. J. Bacteriol., 182, 1558–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]