Abstract
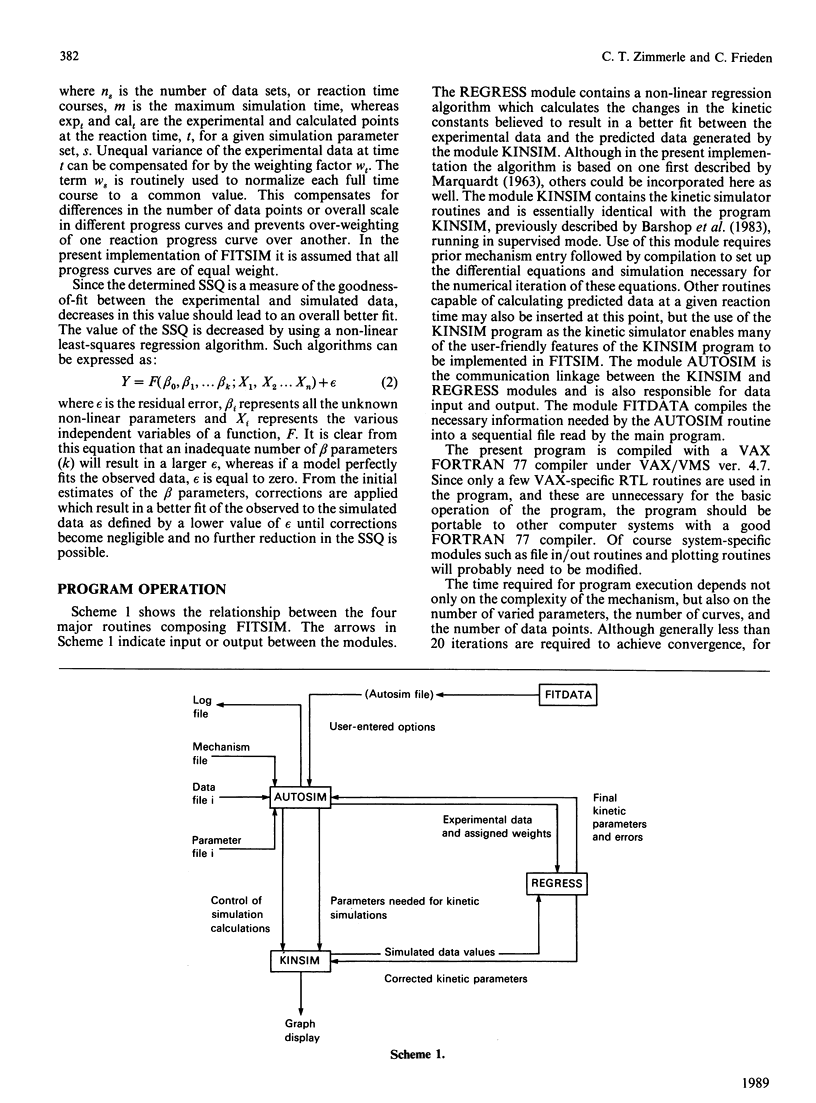
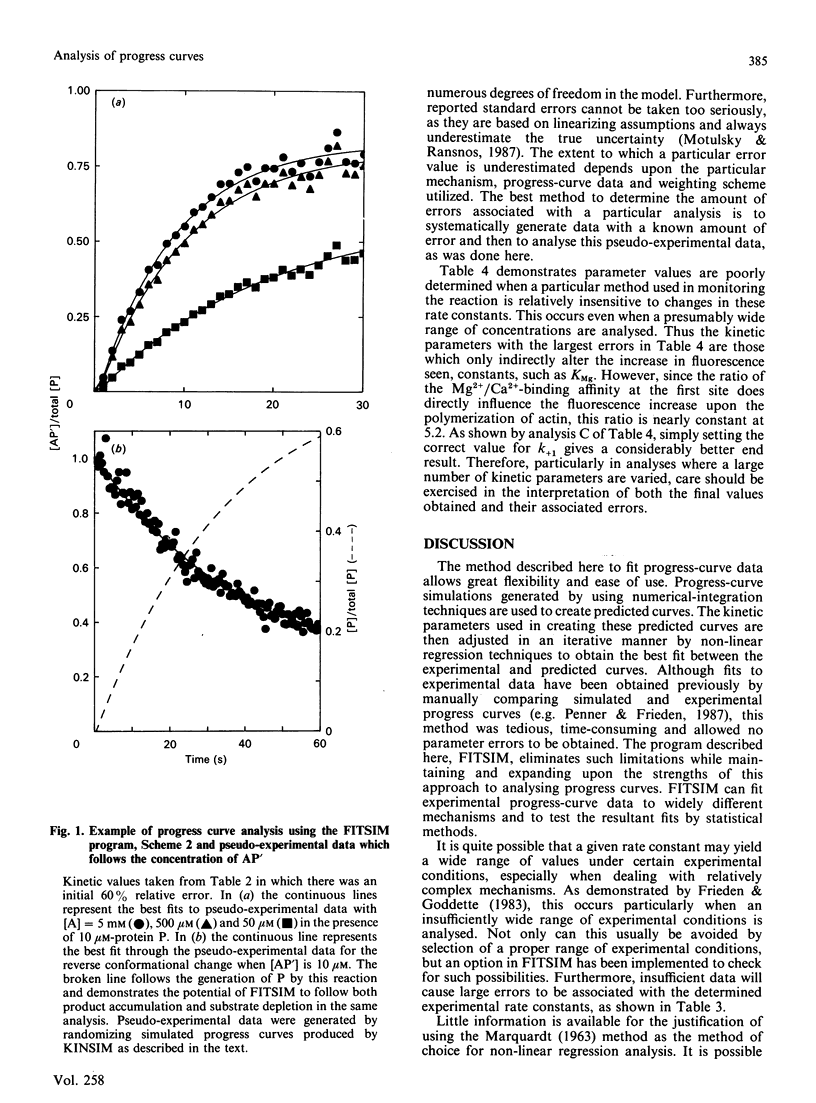
A highly flexible computer program written in FORTRAN is presented which fits computer-generated simulations to experimental progress-curve data by an iterative non-linear weighted least-squares procedure. This fitting procedure allows kinetic rate constants to be determined from the experimental progress curves. Although the numerical integration of the rate equations by a previously described method [Barshop, Wrenn & Frieden (1983) Anal. Biochem. 130, 134-145] is used here to generate predicted curves, any routine capable of the integration of a set of differential equations can be used. The fitting program described is designed to be widely applicable, easy to learn and convenient to use. The use, behaviour and power of the program is explored by using simulated test data.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barshop B. A., Wrenn R. F., Frieden C. Analysis of numerical methods for computer simulation of kinetic processes: development of KINSIM--a flexible, portable system. Anal Biochem. 1983 Apr 1;130(1):134–145. doi: 10.1016/0003-2697(83)90660-7. [DOI] [PubMed] [Google Scholar]
- Canela E. I., Franco R. Enzyme kinetic studies from progress curves. Biochem J. 1986 Jan 15;233(2):599–605. doi: 10.1042/bj2330599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox T. T., Boeker E. A. Analysis of enzyme kinetics by using integrated rate equations. Arginine decarboxylase. Biochem J. 1987 Jul 1;245(1):59–65. doi: 10.1042/bj2450059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco R., Gavaldà M. T., Canela E. I. A computer program for enzyme kinetics that combines model discrimination, parameter refinement and sequential experimental design. Biochem J. 1986 Sep 15;238(3):855–862. doi: 10.1042/bj2380855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieden C., Goddette D. W. Polymerization of actin and actin-like systems: evaluation of the time course of polymerization in relation to the mechanism. Biochemistry. 1983 Dec 6;22(25):5836–5843. doi: 10.1021/bi00294a023. [DOI] [PubMed] [Google Scholar]
- Gordon E. L., Pearson J. D., Slakey L. L. The hydrolysis of extracellular adenine nucleotides by cultured endothelial cells from pig aorta. Feed-forward inhibition of adenosine production at the cell surface. J Biol Chem. 1986 Nov 25;261(33):15496–15507. [PubMed] [Google Scholar]
- Kellershohn N., Laurent M. Analysis of progress curves for a highly concentrated Michaelian enzyme in the presence or absence of product inhibition. Biochem J. 1985 Oct 1;231(1):65–74. doi: 10.1042/bj2310065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koerber S. C., Fink A. L. The analysis of enzyme progress curves by numerical differentiation, including competitive product inhibition and enzyme reactivation. Anal Biochem. 1987 Aug 15;165(1):75–87. doi: 10.1016/0003-2697(87)90203-x. [DOI] [PubMed] [Google Scholar]
- Kurz L. C., Weitkamp E., Frieden C. Adenosine deaminase: viscosity studies and the mechanism of binding of substrate and of ground- and transition-state analogue inhibitors. Biochemistry. 1987 Jun 2;26(11):3027–3032. doi: 10.1021/bi00385a012. [DOI] [PubMed] [Google Scholar]
- Lee K. M., Dahlhauser K. F., Plapp B. V. Reactivity of horse liver alcohol dehydrogenase with 3-methylcyclohexanols. Biochemistry. 1988 May 3;27(9):3528–3532. doi: 10.1021/bi00409a060. [DOI] [PubMed] [Google Scholar]
- Lutz R. A., Bull C., Rodbard D. Computer analysis of enzyme-substrate-inhibitor kinetic data with automatic model selection using IBM-PC compatible microcomputers. Enzyme. 1986;36(3):197–206. doi: 10.1159/000469292. [DOI] [PubMed] [Google Scholar]
- Mannervik B. Regression analysis, experimental error, and statistical criteria in the design and analysis of experiments for discrimination between rival kinetic models. Methods Enzymol. 1982;87:370–390. doi: 10.1016/s0076-6879(82)87023-7. [DOI] [PubMed] [Google Scholar]
- Matyska L., Kovár J. Comparison of several non-linear-regression methods for fitting the Michaelis-Menten equation. Biochem J. 1985 Oct 1;231(1):171–177. doi: 10.1042/bj2310171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motulsky H. J., Ransnas L. A. Fitting curves to data using nonlinear regression: a practical and nonmathematical review. FASEB J. 1987 Nov;1(5):365–374. [PubMed] [Google Scholar]
- Penner M. H., Frieden C. Kinetic analysis of the mechanism of Escherichia coli dihydrofolate reductase. J Biol Chem. 1987 Nov 25;262(33):15908–15914. [PubMed] [Google Scholar]
- Sekhar V. C., Plapp B. V. Mechanism of binding of horse liver alcohol dehydrogenase and nicotinamide adenine dinucleotide. Biochemistry. 1988 Jul 12;27(14):5082–5088. doi: 10.1021/bi00414a020. [DOI] [PubMed] [Google Scholar]
- Zimmerle C. T., Frieden C. Effect of pH on the mechanism of actin polymerization. Biochemistry. 1988 Oct 4;27(20):7766–7772. doi: 10.1021/bi00420a027. [DOI] [PubMed] [Google Scholar]
- Zimmerle C. T., Patane K., Frieden C. Divalent cation binding to the high- and low-affinity sites on G-actin. Biochemistry. 1987 Oct 6;26(20):6545–6552. doi: 10.1021/bi00394a039. [DOI] [PubMed] [Google Scholar]


