Abstract
Recently, we isolated and characterized a new mouse mitochondrial RNA molecule containing the mitochondrial 16S RNA plus 121 nt joined to the 5′ end of the RNA. This fragment arises from the L strand of the same gene and we have named this transcript chimeric RNA. At position 121 of the RNA there is a C, which, according to the sequence of the mitochondrial 16S RNA gene, should be a U. We hypothesized that this RNA is synthesized having a U at position 121, which is later substituted to a C by a putative editing reaction. Based on the presence of sites for the restriction endonucleases RsaI and Fnu4HI around position 121, both forms of the RNA were detected in mouse tissues. To confirm the presence of the non-edited and putative edited RNA, a fragment containing the first 154 nt of the RNA was amplified by RT–PCR and cloned. The substitution of U for C was demonstrated by sequencing these clones. In vitro transcription experiments demonstrated that the substitution of U for C is not due to artifact of amplification or cloning. Moreover, in mitochondria from testis only the non-edited form was found. This, together with other experimental evidence, demonstrated that the base substitution was not due to polymorphism of the mitochondrial 16S RNA gene. This is the first demonstration of a substitution reaction from U to C in a mammalian mitochondrial transcript.
INTRODUCTION
Recently, we have described a novel mitochondrial transcript in mouse testis and sperm. The structure of this RNA comprises the 16S mitochondrial RNA plus a fragment of 121 nt joined to the 5′ end of the ribosomal RNA (1). The fragment of 121 nt arises from the transcript of the L strand of the mitochondrial DNA (mtDNA) corresponding to the 16S gene. Since the transcription of the 16S RNA and the transcript of the L strand depend on different promoters (2), we suggested naming this transcript chimeric RNA. This chimeric RNA was not codified in the mtDNA nor in the nuclear DNA, and therefore we suggested that the synthesis of this transcript is a post-transcriptional event that might involve a transplicing reaction (1).
The sequence of the first 120 nt of this RNA is fully complementary to the internal sequence of the 16S RNA from position 240 to 360, generating a long inverted repeat (1). At position 121 there is a C, just at the junction between the inverted repeat and the 16S mitochondrial RNA sequence (1). The sequence at the 5′ end of the 16S RNA component of the chimeric RNA is identical to the sequences reported before (1,3,4), and therefore, it is reasonable to propose that the C corresponds to the last nucleotide of the inverted repeat originated from the transcript of the L strand. This nucleotide should be complementary to the nucleotide in position 240 of the mouse 16S mitochondrial RNA. However, according to the sequence of the chimeric RNA (1) and the sequence of the mouse mitochondrial 16S RNA (3,4), at position 240 there is an A that will be copied as a U in the transcript of the L strand. A hypothetical explanation for the presence of a C at position 121 is that the transcript is synthesized having a U at position 121, which later is changed to or substituted by a C. In other words, we are proposing that following synthesis of the chimeric RNA a putative editing reaction takes place substituting the U at position 121 for a C (5,6). This is a provocative suggestion since this type of substitution reaction has not been reported in mammalian mitochondrial RNA. The U to C substitution has been described in plants (5–9) and in the Wilms’ tumor susceptibility gene WT1 RNA of rat and human (10). In this report we cloned and sequenced the non-edited (U at position 121) and the putative edited (C at position 121) forms of the RNA from mouse testis, sperm and somatic tissues, confirming for the first time the existence of a putative editing reaction from U to C in a mouse mitochondrial RNA.
MATERIALS AND METHODS
RNA and DNA isolation
Mouse testis mitochondria were isolated as described before (11) and purified by sucrose gradient centrifugation (12). The final purified fraction was resuspended in a solution containing 0.25 M sucrose, 10 mM KCl, 0.15 mM MgCl2 and 10 mM Tris–HCl, pH 7.4 and RNase A (Sigma) was added to a final concentration of 100 µg/ml and incubated at 25°C for 30 min. The mitochondria were recovered by centrifugation at 9000 g at 4°C and washed twice with the above mitochondria suspension buffer.
Total RNA from mouse sperm, testis, liver, brain, spleen, kidney and testis mitochondria was extracted as described before (1,13,14). mtDNA was isolated from mouse testis, sperm, liver, kidney and spleen using the Chellex 100 procedure (15).
RT–PCR
Reverse transcription was performed with 0.1–0.2 µg of total RNA. Briefly, the RNA was mixed with 50 ng of random hexamers and heated at 80°C for 10 min and then cooled in ice. The reaction mixture was completed with the enzyme buffer, 0.2 mM dATP, dGTP, dTTP and dCTP, 10 U ribonuclease inhibitor (RnaseOut, Gibco BRL) and 800 U reverse transcriptase (Superscript II or M-MVL, Gibco BRL). The reaction, in a final volume of 20 µl, was incubated first for 10 min at 25°C and then for 50 min at 42°C. The synthesis of cDNA was also carried out with either 300 U M-MVL reverse transcriptase (Gibco BRL) or 30 U AMV reverse transcriptase (Promega) (1).
Approximately 1–2 µl of the cDNA mix was added to 45 µl of the PCR reaction mixture containing 2.5 U Taq polymerase (Gibco BRL), 1.5 mM MgCl2, 200 µM dNTP, 50 pmol of each primer (primer 1 and 2; see Fig. 1) and buffer solution according to the manufacturer’s description (16). PCR was carried out for 30 cycles at 95°C for 1 min, at 58°C for 1 min and at 72°C for 1 min, and a final extension at 72°C for 10 min. In parallel, the same fragment was amplified using as template the DNA of clone λMS-134 (1). Using the same PCR protocol, a fragment of 84 bp of the mtDNA corresponding to the 16S gene (position 215–298; Fig. 1) was amplified using primers 3 and 4 (Fig. 1). The amplified fragments were analyzed by electrophoresis in 2% agarose gels and stained with ethidium bromide (17).
Figure 1.
Partial sequence of the 5′ region of the mouse mitochondrial chimeric RNA. The first 120 nt are complementary to the sequence of the 16S RNA from position 241 to 360 (both underlined). The C at position 121 is also underlined. The position and sequence of primers 1, 2, 3 and 4 used for amplification are shown. The sequence GCAC derived from the sequence GTAC (RsaI) in the putative non-edited RNA is shown in red.
Digestion with restriction enzymes
The amplified fragments obtained after PCR were digested with 5 U Fnu4HI (BioLabs Inc.) or with 10 U RsaI (Gibco) at 37°C for 2 h using the buffer and the reaction conditions recommended by the manufacturer. The digestion products were analyzed by electrophoresis in a 3% agarose gel and stained with ethidium bromide.
Cloning of PCR fragments
The RT–PCR product of 154 bp (Fig. 1) obtained with total RNA from testis or other tissues was purified using the Wizard system (Promega) and cloned in the linearized vector pCR 2.1 (InVitroGen) or pGEM-T (Promega) containing a single dT at the 5′ ends. Briefly, 6 µl of the purified fragment of 154 bp was mixed with a solution containing 60 mM Tris–HCl, pH 7.5, 6 mM MgCl2, 50 mM NaCl, 70 mM 2-mercaptoethanol, 1 mM ATP, 20 mM DTT, 10 mM spermidine, 1 mg/ml of BSA, 50 ng of the vector pCR 2.1 or pGEM-T and 4 U T4 DNA ligase in a final volume of 10 µl. The mixture was incubated overnight at 14°C.
Approximately 6 µl of the ligation product was used to transform 100 µl of competent HB101 or DH5α cells. The reaction was incubated for 30 min at 0°C followed by 30 s at 42°C and then cooled in ice for 2 min. The mixture was diluted with 400 µl of LB medium containing 100 µg/ml of ampicillin and incubated for 1 h at 37°C with shaking at 225 r.p.m. Between 100 and 200 µl of each transformation mixture was spread on LB agar plates containing 100 µg/ml of ampicillin, 160 µg/ml of X-Gal and 0.42 mM IPTG and incubated overnight at 37°C. Several colonies were picked and grown overnight in 5 ml of LB broth containing 100 µg/ml of ampicillin. The plasmid DNA of each culture was purified using the Concert Rapid Plasmid Miniprep System (Gibco BRL) and the presence of the insert was confirmed by PCR using primers 1 and 2 (Fig. 1).
In vitro transcription
The plasmids of non-edited and putative edited clones described in the previous section and with the sense or antisense orientation to the T7 promoter of the pGEM-T vector, were used as template for in vitro transcription. To linearize each plasmid, 1 µg of DNA was digested with 10 U DraI (Boehringer, Mannheim) under the conditions specified by the manufacturer in a final volume of 10 µl, at 37°C for 1 h. Then, 100 ng of DNA was mixed with 40 mM Tris–HCl, pH 8.0, 8 mM MgCl2, 2 mM spermidine, 25 mM NaCl, 0.4 mM NTPs, 5 mM DTT and 50 U T7 RNA polymerase (Gibco) in a final volume of 20 µl and incubated for 15 min at 37°C. To remove the template, 1 U RNase free-DNase I (Gibco) was added to the mixture and incubated for 15 min at 37°C. Then, 10 µl of the sense and antisense RNA were mixed, incubated at 80°C for 10 min, cooled in ice and 1 µl was used to amplify the fragment of 154 bp. The fragment was ligated to the vector pCR 2.1 or pGEM-T and the resulting construct was used to transform HB101 competent cells. The selection of the clones obtained from the non-edited or putative edited parental plasmids were carried out as described before.
Sequencing of DNA
The sequence of both strands of the insert in the recombinants clones was carried out using the forward and reverse M13 primers and the dideoxy-chain termination method (18). The sequence was performed using a Perkin-Elmer ABI Prism 310 Genetic Analyzer.
RESULTS
We have suggested that the chimeric RNA is a post-transcription product probably resulting from a transplicing reaction between the 16S mitochondrial RNA and a fragment of 121 nt of the transcript of the L strand of the mtDNA of the same gene (1). The first 400 bp of the sequence of clone λMS-134 corresponding to the chimeric RNA is shown in Figure 1, whereas the sequences of the inverted repeat complementary to the 16S RNA, are underlined. At position 121 is a C, just at the junction between the inverted repeat and the 16S mitochondrial RNA sequence. Since the sequence of the 16S RNA at the 5′ end is normal (1,3,4) it is reasonable to predict that the C corresponds to the last nucleotide of the inverted repeat originated from the transcript of the L strand. However, according to the sequence of clone λMS-134 (Fig. 1) and the sequence of the mouse mitochondrial 16S RNA (3,4), in position 240 (Fig. 1) there is an A that has become a U in the transcript of the L strand. Therefore, we propose that the chimeric transcript is synthesized having a U at position 121, which is later changed to a C. In other words, we postulate that following synthesis of the chimeric RNA, a putative editing reaction substituting the U for a C, takes place.
If this hypothesis is correct, we should find in testis, for example, both forms of the RNA. Amplification of testis RNA by RT–PCR using oligos 1 and 2 (Fig. 1) will generate a fragment of 154 bp. An interesting feature of the sequence of this fragment is that the last two residues of the inverted repeat are GC, which together with the two initial residues of the 16S RNA forms the sequence GCAC. If a T is present in a non-edited RNA, this sequence will be GTAC, which is the sequence recognized by the restriction enzyme RsaI. Notice also that there is another RsaI sequence beginning at position 60 (Fig. 1). Therefore, digestion of the PCR fragment of 154 bp with RsaI obtained from a non-edited transcript will generate fragments of 61, 60 and 33 bp. If C is at position 121 (Fig. 1), the site will become GCAC, which will not be digested by RsaI and only two fragments of 93 and 61 bp will be observed. Also, beginning at position 117 is the sequence GCGGC (Fig. 1), which corresponds to the sequence of the restriction enzyme Fnu4HI. Again, digestion of the PCR fragment of 154 bp obtained from a putative edited RNA will generate fragments of 118 and 36 bp, but no digestion will be observed in the amplicon of the non-edited RNA since this sequence will change to GCGGT.
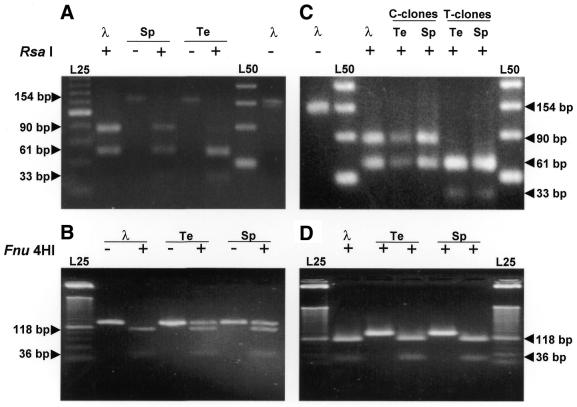
As shown in Figure 2, digestion of the 154 bp fragment amplified from testis RNA with RsaI generated fragments of 93, 60 and 33 bp (Fig. 2A). As expected, digestion with RsaI of the amplicon of 154 bp obtained from clone λMS-134 generated only fragments of 93 and 61 bp (Fig. 2A). Similarly, digestion of the same fragment with Fnu4HI generated fragments of 118 and 36 bp plus undigested product that should correspond to amplicons containing a T at position 121 (Fig. 2B). The same results were obtained with the 154 bp amplicons obtained with sperm RNA (Fig. 2A and B). These digestion patterns suggest that both the non-edited and the putative edited forms of the RNA are present in testis and sperm.
Figure 2.
Digestion patterns of the 154 bp fragment obtained with RsaI or with Fnu4HI. (A) The amplicon obtained by RT–PCR of sperm (Sp) and testis RNA (Te) were digested with (+) or without (–) RsaI and the products analyzed by agarose gel electrophoresis. The fragment of 154 bp obtained with clone λMS-134 (λ) was also digested with RsaI. (B) The same as in (A), but the digestion was carried out with or without Fnu4HI. (C) The plasmidial DNA from two representative C clones and T clones selected from the sperm and testis libraries were amplified by PCR and the fragment of 154 bp was digested with RsaI (+). The digestion pattern of the same fragment obtained from clone λMS-134 is also shown (λ). (D) The same as (C), but digested with Fnu4HI. The length of the digestion products is indicated in bp. L25 and L50 are ladders of 25 and 50 bp, respectively.
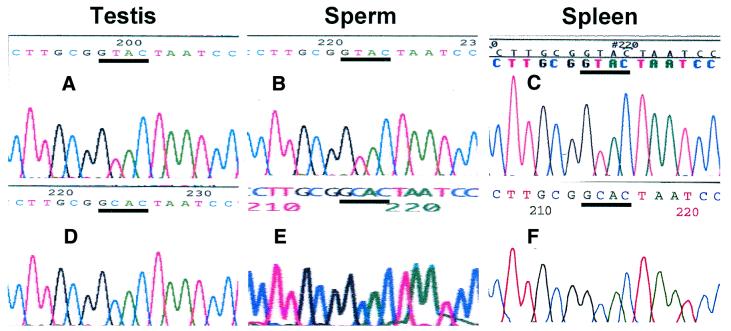
To corroborate these findings, the putative mixture of fragments of 154 bp obtained by RT–PCR were cloned in the vector pCR 2.1 or pGEM-T. Theoretically, the libraries should contain a mixture of clones corresponding to the putative edited and non-edited form of the RNA. Accordingly, colonies were picked randomly, grown in LB medium and the insert was amplified by PCR using primers 1 and 2 (Fig. 1). The digestion pattern of the amplicon of 154 bp obtained from a non-edited and putative edited clone from testis and sperm RNA is shown in Figure 2C and D. RsaI generated fragments of 93 and 60 bp with the putative edited clone of testis and sperm (C-clones; Fig. 2C), and fragments of 60 and 33 bp with the non-edited clones (T-clones; Fig. 2C). These results were confirmed by digestion with Fnu4HI (Fig. 2D). To confirm the substitution of the U for a C at position 121, both strands of the insert of each clone were sequenced. In the non-edited clones from testis and sperm, a T at position 121 was found (Fig. 3A and B), which changed to C in those clones defined as edited by the restriction endonuclease pattern (Fig. 3D and E).
Figure 3.
Sequence of the non-edited and the putative edited clones. Plasmids obtained from the libraries of testis, sperm and spleen were selected as non-edited or edited clones by digestion with RsaI and Fnu4HI, and sequenced. The sequence around position 121 of the non-edited clones (A–C) and the putative edited clones (D–F) are shown. The sequences GTAC and GCAC are underlined.
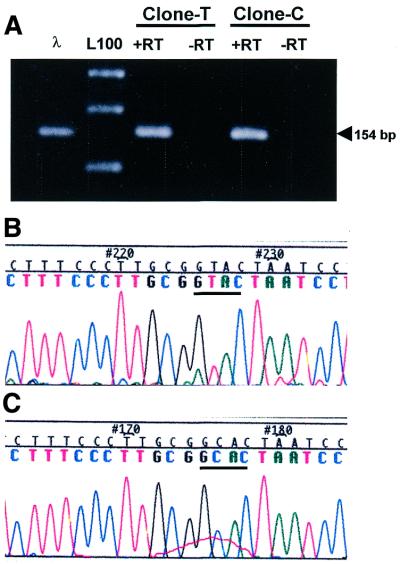
One way to explain these results is that the substitution of U for C at position 121 is an error arising during the synthesis of the cDNA, or during amplification with Taq polymerase or due to cloning artifacts. To investigate these possibilities, the purified plasmids from non-edited or putative edited clones were transcribed in vitro with T7 RNA polymerase (19). In both cases, a plasmid in the sense as well as in the antisense orientation respect the T7 promotor was used as described in Materials and Methods. After complete hydrolysis of the plasmid DNA template with DNase I, the sense and the antisense RNAs were mixed and amplified by RT–PCR using primers 1 and 2 (Fig. 1). As shown in Figure 4A, amplification of the mixture of sense and antisense RNAs transcribed in vitro using as template the plasmid of the non-edited (Clone-T) or putative edited clones of testis (Clone-C) yielded the expected fragment of 154 bp. No amplification product was obtained without reverse transcriptase (–RT; Fig. 4A), demonstrating the complete removal of the parental plasmid template.
Figure 4.
In vitro transcription of mouse testis clones. (A) A representative non-edited (Clone-T) and putative edited (Clone-C) parental clone of testis were linearized and transcribed in vitro with T7 RNA polymerase. After complete digestion of the plasmid template with DNase I, the fragment of 154 bp was amplified by RT–PCR with (+RT) or without (–RT) reverse transcriptase and using a mixture of the sense and antisense synthetic RNAs as template. The fragment of 154 bp obtained from clone λMS-134 is also shown (λ). L100, ladder of 100 bp. The amplicons were cloned in pGEM-T and sequenced. The partial sequences of a new non-edited (B) or a new putative edited (C) clone are shown.
The fragment of 154 bp from each reaction was cloned as described and the recombinant plasmids were sequenced. The sequence of the new non-edited clones was the same as that corresponding to the parental clone (Fig. 4B; GTAC underlined) and the same was true with the new putative edited clone (Fig. 4C; GCAC underlined). The same results were obtained when non-edited and putative edited clones from sperm were used for in vitro transcription (data not shown).
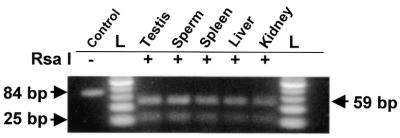
Polymorphism of the mtDNA at position 1212 (1,3,4) might be an alternative explanation of these results. It is possible that there are two sequences of this gene, the normal one carrying an A at this position and the other form carrying a G at the same position. Therefore, transcription of the L strand of these mtDNAs will yield two RNAs that in the corresponding fragment present in the chimeric RNA will carry a U or a C at position 121 (Fig. 1). To explore this possibility a fragment of 84 bp was amplified using primers 3 and 4 (Fig. 1) and mtDNA as templates. The sequence of this fragment (Fig. 1) has a site for the restriction endonuclease RsaI (GTAC), where the A is at position 240 of the chimeric RNA. As shown in Figure 5, the amplicon of 84 bp obtained from mtDNA of testis, sperm spleen, liver and kidney was completely digested by RsaI, generating the two expected fragments of 25 and 59 bp. If G was present at the same position of the putative polymorphic mtDNA, this site will become GTGC, which will not be digested by RsaI.
Figure 5.
Lack of polymorphism in the mouse mtDNA at position 1212. mtDNAs extracted from mouse testis, sperm, spleen, liver and kidney were amplified by PCR using primers 3 and 4, and the amplicon of 84 bp was digested with RsaI. In all cases, the amplicon was completely digested generating two fragment of 59 and 25 bp. The undigested fragment (–) and the ladder of 25 bp are also shown.
The chimeric RNA was also present in somatic tissues (1). Thus, it was reasonable to ask if both forms of the chimeric RNA coexist in these tissues. In the spleen, for example, the non-edited as well as the putative edited forms were found (Fig. 3E and F). To establish the proportion of non-edited to putative edited RNA present in each tissue, the amplicon of 154 bp was cloned and several recombinant clones from each library were analyzed according to the digestion pattern obtained with RsaI and Fnu4HI, and sequencing. A high proportion of clones carrying a C at position 121 were found in sperm and testis, in comparison with the low proportion observed in somatic tissues (Table 1). Using the same approach and total RNA from testis mitochondria, we were unable to detect C clones, although a large number of recombinant clones were analyzed (Table 1).
Table 1. Frequency of the putative edited clones in different tissues.
| Tissue | No. of recombinanta clones | T clones | C clones |
|---|---|---|---|
| Testis | 27 | 12 | 15 |
| Testis mitochondriab | 25 | 25 | – |
| Sperm | 12 | 3 | 9 |
| Spleen | 10 | 8 | 2 |
| Brain | 10 | 9 | 1 |
| Liver | 10 | 9 | 1 |
| Kidney | 8 | 7 | 1 |
aClones selected from each library and containing the insert of 154 bp.
bThe 25 recombinant clones were selected randomly from two libraries obtained with mitochondrial RNA. The presence of a T (T clones) or a C (C clones) at position 121 was determined by digestion with RsaI and Fnu4HI, and confirmed by sequencing.
DISCUSSION
The substitution of a U for a C found in the amplified fragment of 154 bp obtained from the chimeric RNA might be due to artifacts occurring during the synthesis of the cDNA by the reverse transcriptase or during the amplification reaction with Taq polymerase. Indeed, the inverted repeat of 120 nt will form a long hairpin (1) and the U at position 121 is just at the beginning of this stable secondary structure. Hence, this position could be a sort of hotspot for any of the enzymes used for RT–PCR inducing misincorporation of a C at position 121 instead of T. However, the results of the in vitro transcription studies (19) showed that, from a parental non-edited clone always non-edited new clones were obtained, and the same was true for the putative edited parental clones, demonstrating that the substitution reaction found in the chimeric RNA was not due to artifacts during amplification or cloning.
Polymorphism of the 16S RNA mitochondrial gene at position 1212 of the mouse mtDNA (1,3,4) was also investigated. However, the results presented in Figure 5 together with the absence of the chimeric RNA with a C at position 121 in testis mitochondria, rule out the possibility of polymorphism, at least at that position of the mtDNA.
It is also reasonable to argue that the form containing C at position 121 corresponds to a transcription product of a mitochondrial pseudogene (20–25). However, we were unable to find a pseudogene corresponding to the sequence of the 154 bp fragment in nuclear DNA from testis, sperm and somatic tissues (1).
The shift from U to C might be also due to misincorporation of a nucleotide by the mitochondrial RNA polymerase. For example, in the testis or sperm, where a high proportion of the putative edited RNA was found, the mitochondrial RNA polymerase should be doing a high rate of misincorporation of C instead of U. In contrast, we were unable to find in two testis mitochondrial libraries clones carrying a C at position 121, indicating that the organelle RNA polymerase transcribed correctly the expected sequence of the normal mtDNA 16S gene.
Another possibility is that the mechanism responsible for the joining or transplicing of the fragment of 121 nt to the 16S RNA might involve the addition of a U or a C at the 3′ end of that fragment. But this possibility also seems unlikely since, as discussed before, we were unable to find an RNA carrying a C at position 121 in testis mitochondria.
Altogether, these results strongly suggested that after the synthesis of the chimeric RNA in the mitochondria a substitution reaction from U to C takes place at position 121. Furthermore, since in testis mitochondria it is only possible to find the non-edited form of the RNA, the conversion from U to C seems to occur after the synthesis of the chimeric RNA and most probably outside the mitochondria.
RNA editing is found in diverse species of eukaryotes including mammals, trypanosomes, slime mold, plants and virus (6). Among animals, editing of mRNA includes the single substitution of C to U in the apoliprotein B and neurofibromatosis type 1 tumor suppressor mRNA, the A to I deaminations in the GluR channel mRNAs and in the 5HT2CR serotonin receptor mRNA (6). The change from A to I has been also described in the Kv2 K+ channel from squid, the Drosophila 4f-rnp mRNA and in the human hepatitis delta virus (5,6). Other changes include G to A in the mouse GlcNac-1 phosphate transferase mRNA and U to A in human α-galactosidase mRNA (5). It was reported that in rat and human the WT1 mRNA, the gene product of the Wilms’ tumor susceptibility gene, was edited at codon 280 (or 281 in human) from U to C (10). However, later, no editing of codon 281 was found in the WT1 mRNA obtained from 15 primary Wilms tumors (26).
As far as we know there is no previous description in mammalian mitochondria of a putative RNA editing reaction from U to C as reported here. Mitochondria and plastids of vascular and non-vascular plants exhibit examples of post-transcription substitution of C to U and U to C (9,27–31). Editing from C to U predominates greatly over U to C substitutions in organelles of dicots and monocots (9), although in certain lower plants, C to U as well as U to C editing events are both equally frequent (5,6,9).
Although there is experimental evidence supporting that cytidine deaminase catalyzes the editing reaction from C to U (32), much less clear is the mechanism that catalyzes the reverse process from U to C. Transamination as well as transglycosylation have been proposed as hypothetical mechanisms for the U to C conversion (33), but at present we have ignored the mechanism involved in the substitution reaction of the mouse chimeric RNA. The presence of a large double-strand structure in the chimeric RNA may suggest a mechanism of conversion from U to C similar to the transformation of A to I, where the reaction is catalyzed by adenosine deaminases (ADAR1 and ADAR2), the activity depending on the binding to a double-strand RNA (34). For example, one can envision a transaminase or transglycosydase that catalyzes the shift from U to C in which the activity will depend on the binding to the long double-strand region of the chimeric RNA.
At present, the function of the chimeric RNA is unknown (1). However, because of its unusual localization in the sperm nucleus, we have speculated that its function might be similar to the 16S mitochondrial RNA found outside the mitochondria in the egg of Drosophila and Xenopus (35–38). This RNA is localized in the polar cytoplasm in tight association with the polar granules, and the authors have proposed that this transcript plays an important role in pole cell formation (35–38). Therefore, one is tempted to speculate that the nuclear chimeric RNA is transferred to the oocyte after fertilization where it might play a similar role in the development of the germ line of the mouse. In this regard, the finding that a high proportion of the chimeric RNA in the sperm is present as the putative edited form (Table 1) is interesting and suggests that the modification of the transcript might be necessary for its function after fertilization.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Drs Axel Brennicke, Alejandro Araya, Simon Litvak and Pablo Valenzuela for their helpful discussion and suggestions. Grants 1990230 and 2960062 from FONDECYT, CHILE supported this work. J.A. was supported by a student fellowship from Fundación Ciencia Para La Vida.
REFERENCES
- 1.Villegas J., Zarraga,A.M., Muller,I., Montecinos,L., Werner,E., Brito,M., Meneses,A.M. and Burzio,L.O. (2000) A novel chimeric mitochondrial RNA localized in the nucleus of mouse sperm. DNA Cell Biol., 19, 579–588. [DOI] [PubMed] [Google Scholar]
- 2.Clayton D. (1992) Transcription and replication of animal mitochondrial DNAs. Int. Rev. Cytol., 141, 217–232. [DOI] [PubMed] [Google Scholar]
- 3.Bibb M.J., Van Etten,R.A., Wright,C.T., Walberg,M.W. and Clayton,D.A. (1981) Sequence and gene organization of mouse mitochondrial DNA. Cell, 26, 167–180. [DOI] [PubMed] [Google Scholar]
- 4.Mori M., Higuchi,K. and Tanaka,M. (2000) Mus musculus domesticus mitochondrial DNA, complete genome. Gene Bank, accession no. AB042432.
- 5.Smith H.C., Gott,J.M. and Hanson,M.R. (1997) A guide to RNA editing. RNA, 3, 1105–1123. [PMC free article] [PubMed] [Google Scholar]
- 6.Brennicke A., Marchfelder,A. and Binder,S. (1999) RNA editing. FEMS Microbiol. Rev., 23, 297–316. [DOI] [PubMed] [Google Scholar]
- 7.Schuster W., Hiesel,R., Wissinger,B. and Brennicke,A. (1990) RNA editing in the cytochrome b locus of the higher plant Oenothera berteriana includes a U-to-C transition. Mol. Cell. Biol., 10, 2428–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gualberto J.M., Weil,J.H. and Grienenberger,J.M. (1990) Editing of the wheat coxIII transcript: evidence for twelve C to U and one U to C conversions and for sequence similarities around editing sites. Nucleic Acids Res., 18, 3771–3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoshinaga K., Iinuma,H., Masuzawa,T. and Uedal,K. (1996) Extensive RNA editing of U to C in addition to C to U substitution in the rbcL transcripts of hornwort chloroplasts and the origin of RNA editing in green plants. Nucleic Acids Res., 24, 1008–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharma P.M., Browman,M., Madden,S.L., Rauscher,F.J.,III and Sukumar,S. (1994) RNA editing in the Wilms’ tumor susceptibility gene, WT1. Genes Dev., 8, 720–731. [DOI] [PubMed] [Google Scholar]
- 11.Kruse B., Murdter,N.N. and Attardi,G. (1995) Transcription system using a HeLa cell mitochondrial lysate. In Tymms,M.J. (ed.), Methods in Molecular Biology. Human Press Inc., Totowa, NJ, Vol. 37, pp. 179–197. [DOI] [PubMed]
- 12.Burzio L.O., Saez,L. and Cornejo,R. (1981) Poly(ADP-Ribose) synthetase activity in rat liver mitochondria. Biochem. Biophys. Res. Commun., 103, 369–375. [DOI] [PubMed] [Google Scholar]
- 13.Concha I.I., Urzua,U., Yañez,A., Schroeder,R., Pessot,C. and Burzio,L.O. (1993) U1 and U2 snRNA are localized in the sperm nucleus. Exp. Cell Res., 204, 378–381. [DOI] [PubMed] [Google Scholar]
- 14.Chomczynski P. and Sacchi,N. (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem., 162, 156–159. [DOI] [PubMed] [Google Scholar]
- 15.Walsh P., Metzger,D. and Higuchi,R. (1991) Chelex® 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques, 10, 506–513. [PubMed] [Google Scholar]
- 16.Saiki R.K., Gelfand,D.H., Stoffel,S., Scharf,S.J., Higuchi,R., Horn,G.T., Mullis,K.B. and Erlich,H.A. (1988) Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science, 239, 487–491. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook J., Fritsch,E.T. and Maniatis,T. (1989) Molecular Cloning. A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 18.Sanger F. and Coulson,A.R. (1975) A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J. Mol. Biol., 94, 441–457. [DOI] [PubMed] [Google Scholar]
- 19.Bourara K. Litvak,S. and Araya,A. (2000) Generation of G-to-A and C-to-U changes in HIV-1 transcripts by RNA editing. Science, 289, 1564–1566. [DOI] [PubMed] [Google Scholar]
- 20.Jacobs H.T. and Grimes,B. (1986) Complete nucleotide sequence of the nuclear pseudogenes for cytochrome oxidase subunit I and the large mitochondrial ribosomal RNA in the sea urchin Strongylocentrotus purpuratus. J. Mol. Biol., 187, 509–527. [DOI] [PubMed] [Google Scholar]
- 21.Arctander P. (1995) Comparison of a mitochondrial gene and a corresponding nuclear pseudogene. Proc. R. Soc. Lond. Biol. Sci., 262, 13–19. [DOI] [PubMed] [Google Scholar]
- 22.Smith M., Thomas,W. and Patton,W. (1992) Mitochondrial DNA-like sequence in the nuclear genome of an akodontine rodent. Mol. Biol. Evol., 9, 204–215. [DOI] [PubMed] [Google Scholar]
- 23.Zullo S. (1991) Mitochondrial D-loop sequences are integrated in the rat nuclear genome. J. Mol. Biol., 221, 1233–1235. [PubMed] [Google Scholar]
- 24.Wallace D., Stugard,C., Murdock,D., Schurr,T. and Brown,M. (1997) Ancient mtDNA sequences in the human nuclear genome: a potential source of errors in identifying pathogenic mutations. Proc. Natl Acad. Sci. USA, 94, 14900–14905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirano M., Shtilbans,A., Mayeux,R., Mercy,D., DiMauro,S., Knowles,J. and Schon,E. (1997) Apparent mtDNA heteroplasmy in Alzheimer’s disease patients and normals due to PCR amplification of nucleus-embedded mtDNA pseudogenes. Proc. Natl Acad. Sci. USA, 94, 14894–14899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gunning K.B., Cohn,S.L., Tomlinson,G.E., Strong,L.C. and Huff,V. (1996) Analysis of possible WT1 RNA processing in primary Wilms tumors. Oncogene, 13, 1179–1186. [PubMed] [Google Scholar]
- 27.Araya A., Bégu,D. and Litvak,S. (1994) RNA editing in plants. Biologia Plantarum, 91, 543–550. [Google Scholar]
- 28.Gray M.W. (1996) RNA editing in plant organelles: a fertile field. Proc. Natl Acad. Sci. USA, 93, 8157–8159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malek O., Lättig,K., Hiesel,R., Brennicke,A. and Knoop,V. (1996) RNA editing in bryophytes and a molecular phylogeny of land plants. EMBO J., 15, 1403–1411. [PMC free article] [PubMed] [Google Scholar]
- 30.Sper-Whitis G., Moody,J. and Vaughn,J. (1996) Universality of mitochondrial RNA editing in cytocrome-c oxidase subunit I (cox I) among the land plants. Biochim. Biophys. Acta, 1307, 301–308. [DOI] [PubMed] [Google Scholar]
- 31.Wakasagi T., Hirose,T., Horihata,M., Tsudzuki,T., Kössel,H. and Sigiura,M. (1996) Creation of novel protein-coding region at the RNA level in black pine chloroplasts: the pattern of RNA editing in the gymnosperm chloroplast is different from that in angiosperms. Proc. Natl Acad. Sci. USA, 93, 8766–8770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carter C.W. (1998) Nucleoside deaminases for cytidine and adenosine: comparison with deaminases acting on RNA. In Grosjean,H. and Benne,R. (eds), Modification and Editing of RNA. ASM Press, Washington, DC, pp. 363–375.
- 33.Marchfelder A., Binder,S., Brennicke,A. and Knoop,V. (1998) RNA editing by base conversion in plant organellar RNAs. In Grosjean,H. and Benne,R. (eds), Modification and Editing of RNA. ASM Press, Washington, DC, pp. 307–323.
- 34.Rueter S.M. and Emeson,R.B. (1998) Adenosine-to-inosine conversion in mRNA. In Grosjean,H. and Benne,R. (eds), Modification and Editing of RNA. ASM Press, Washington, DC, pp. 343–361.
- 35.Kobayashi S., Amikura,R. and Okada,M. (1993) Presence of mitochondrial large ribosomal RNA outside mitochondria in germ plasm of Drosophila melanogaster. Science, 260, 1521–1524. [DOI] [PubMed] [Google Scholar]
- 36.Iida T. and Kobayashi,S. (1998) Essential role of mitochondrially encoded large rRNA for germ-line formation in Drosophila embryos. Proc. Natl Acad. Sci. USA, 95, 11274–11278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kashikawa M., Amikura,R., Nakamura,A. and Kobayashi,S. (1999) Mitochondrial small ribosomal RNA is present on polar granules in early cleavage embryos of Drosophila melanogaster. Dev. Growth Differ., 41, 1–8. [DOI] [PubMed] [Google Scholar]
- 38.Amikura R., Kashikawa,M., Nakamura,A. and Kobayashi,S. (2001) Presence of mitochondria-type ribosomes outside mitochondria in germ plasm of Drosophila embryos. Proc. Natl Acad. Sci. USA, 98, 9133–9138. [DOI] [PMC free article] [PubMed] [Google Scholar]