Abstract
The general principles of discrete, large self-assemblies composed of numerous components are not unveiled and the artificial formation of such entities is a challenging topic. In metal–organic cages, design strategies for tuning the coordination directions in multitopic ligands by the bend and twist angles were previously developed to solve this problem. In this study, the importance of remote geometric communications between components is emphasized, realizing several types of metal–organic assemblies based on dihedral angle control in multitopic ligands although they have the same coordination directions. Self-assembly of a tritopic ligand with dihedral angles θ = 36.5° and a cis-protected Pd(II) ion affords M9L6 and M12L8 cages as kinetic and thermodynamic products, respectively, whereas an M12L8 sheet is formed when θ = 90°. Geometric analyses of strains in the subcomponent rings reveals that remote geometric communications among neighboring multitopic ligands through coordination bonds are key for large assemblies.
Subject terms: Self-assembly, Metal-organic frameworks, Molecular self-assembly
General design principles for self-assembly of discrete, large metal-organic cages composed of numerous components are challenging to develop. Here, the authors use remote geometric communications between components to realize several types of metal–organic assemblies based on dihedral angle control in multitopic ligands.
Introduction
Molecular self-assembly is a powerful strategy for creating structurally well-defined large entities from small building blocks using reversible interactions between them and has been widely observed in nature, such as DNA origami1–3, protein assemblies4–6, and lipid bilayers7,8. Because of the reversibility of the interactions between the building blocks, the molecular self-assembly tends to reach thermodynamic equilibrium, where the products are determined by the Maxwell-Boltzmann distribution. Because the translational and rotational freedoms of the building blocks in self-assemblies composed of numerous building blocks are largely restricted compared with those in the disassembled state, the formation of such entities is entropically less favorable. Therefore, under the assumption that environmental effects such as solvation and templation are negligible, increasing the number of components in the self-assemblies enhances the entropic penalty. Consequently, when the enthalpic gains arising from the interactions between the building blocks (enthalpy change per component) are the same, assemblies composed of fewer components are thermodynamically advantageous, and the construction of self-assemblies with many building blocks under thermodynamic control is challenging. As observed in virus capsids9,10, general principles that enable the self-assembly of discrete structures composed of numerous building blocks must exist; however, no details are known beyond the geometric and electrostatic complementarity among the components.
Metal–organic cages11–13 are a well-established class of artificial self-assembled structures composed of multitopic ligands (L) and transition metal ions (M). The formation of M–L coordination bonds is the dominant enthalpic driving force of self-assembly. The geometric consistency between the coordination direction in the multitopic ligand and the coordination geometry of the metal centers determines the assembled structure. Metal–organic cages composed of many components were realized by precisely designing the bend angle of 4-pyridyl-based rigid ditopic ligands for a series of MnL2n assemblies (n = 2–48)14–31. Recently, it was reported that twisting a multitopic ligand, which alters the coordination direction of the ligand, enables the construction of a large molecular self-assembly32–34. In both cases, the change in the coordination directions in the multitopic ligands causes enthalpic destabilization of the self-assemblies consisting of fewer components, resulting in the thermodynamically most stable assemblies composed of more building blocks.
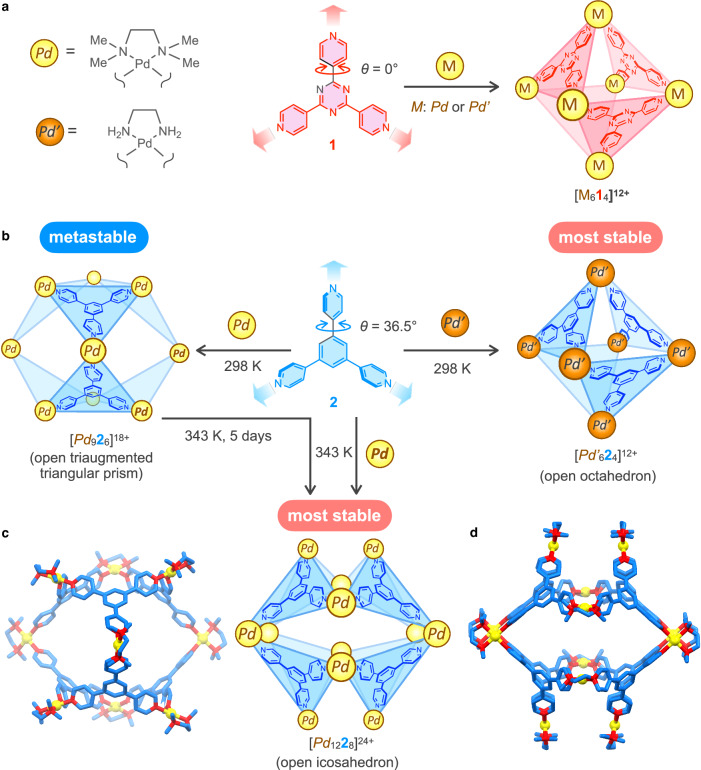
The self-assembly of the M6L4 open octahedron from the cis-protected square-planar Pd(II) ion (M: Pd2+ or Pd’2+) and tritopic ligand 1 was reported (Fig. 1a)35,36. Because the cis-protected Pd(II) ion has two binding sites, the assembled structures that satisfy the full occupations of the two building blocks have the general formula of M3nL2n. The minimum structure, M3L2, cannot be formed from Pd2+ and 1 because of the large strain. Such a strain is not found in the second minimum structure, M6L4. Thus, M6L4 was chosen in the thermodynamic selection process even if larger structures than M6L4, M3nL2n (n ≥ 3), also do not have large strains because of the above-mentioned entropic reason. The question is whether the assembled structures can be controlled by other changes in multitopic ligands while retaining the original coordination directions. Let us consider the self-assembly of Pd2+ and tritopic ligand 2 (Fig. 1b), whose structure is basically the same as 1 except for the dihedral angle (θ) between the 4-pyridyl rings and the central ring (θ for 1 is 0°, while θ for 2 is larger than 0°). Because the coordination directions in 2 are the same as those in 1, the assembly of the M6L4 open octahedron was expected from 2.
Fig. 1. Assembled structures from tritopic ligand 1 or 2 and cis-protected Pd(II) ions.
a Self-assembly of the [M614]12+ open octahedron from triazine-based tritopic ligand 135,36. b Self-assembly of the [Pd926]18+ open triaugmented triangular prism and the [Pd1228]24+ open icosahedron from benzene-based tritopic ligand 2 and Pd2+, whereas [Pd’624]12+ open octahedron was formed with Pd’2+ instead of Pd2+. c Energy-minimized structure of [Pd926]18+ by DFT calculations using B3LYP/def2-SV(P) level of theory. d Crystal structure of [Pd1228]24+. Color labels: Yellow = Pd; Blue = C; Red = N. Hydrogen atoms are omitted for clarity.
Herein, we report that the dihedral angle change in tritopic ligands with three 4-pyridyl rings attached to a central aromatic ring afforded different self-assemblies of M3nL2n (n = 3 and 4). Metal-complexation of tritopic ligand 2 with θ around 36° gave an M9L6 open triaugmented triangular prism37 as a kinetic product, and an M12L8 open icosahedron38 was produced as the thermodynamically most stable product (Fig. 1b–d). In contrast, the self-assembly of tritopic ligand 3 with a mesitylene moiety as the central ring to set θ > 70° afforded an M12L8 square sheet structure. Geometric analyses of MnLn (n = 2–4) subcomponent rings found in these assemblies revealed that a subcomponent ring(s) with small strain is(are) selected for each θ, resulting in the formation of self-assemblies composed of a larger number of building blocks (M9L6 and M12L8) by increasing θ. Remote geometric communication among the tritopic ligands through a cis-protected Pd(II) center connecting them, which is realized by fixing the relative orientation around the Pd(II) center, is key for reflecting the change in θ to the assembled structure, even though there is no contact among the tritopic ligands in the assemblies. These results imply that such a high level of design of building blocks is a hidden factor that enables the creation of large self-assemblies in nature.
Results
Self-assembly of an M12L8 cage
The self-assembly of tritopic ligand 2 and Pd2+ was conducted in CH3NO2 at 363 K in the presence of NO3– for 5 days, resulting in a broad 1H NMR spectrum at thermodynamic equilibrium (Fig. 2a). Downfield shift of Ha signals indicates the coordination of the 4-pyridyl groups in 2 to a Pd(II) center. 1H diffusion-ordered spectroscopy (DOSY) measurement indicated that all 1H signals of 2 have the same diffusion coefficient (2.0 × 10–10 m2 s–1), indicating the formation of a single assembly with a diameter of 35.6 Å, which is larger than the M6L4 open octahedron.
Fig. 2. 1H NMR spectra of the [Pd1228]24+ open icosahedron and the [Pd926]18+ open triaugmented triangular prism (500 MHz, CD3NO2).
a [Pd1228]24+ at 298 K. b 1H DOSY spectrum of [Pd1228]24+. c [Pd1228]24+ at 353 K. The dynamic motion in [Pd1228]24+, which caused the broadening of the 1H NMR spectrum at 298 K, is schematically shown on the right. d Free ligand 2 at 298 K. e [Pd926]18+ at 298 K. f 1H DOSY spectrum of [Pd926]18+.
Single crystals were obtained by slow diffusion of methanol into the reaction mixture for 8 days. Single-crystal X-ray analysis revealed the structure of an [Pd1228]24+ open icosahedron (Fig. 1d, Supplementary Fig. 1, and Methods section). Eight tritopic ligands 2 are connected by twelve Pd2+ and occupy eight faces of a distorted icosahedron. According to the crystal structure of [Pd1228]24+, all eight tritopic ligands 2 are chemically equivalent, and all Hc protons should be identical. In contrast, because of the distortion of the open icosahedron, the Ha and Hb protons in the 4-pyridyl rings of 2 are placed under two environments (denoted as aA, aB, bA, and bB in Fig. 2b). Thus, the observation of two types of Ha protons by 1H NMR spectroscopy is consistent with the crystal structure of [Pd1228]24+. Variable-temperature (VT) 1H NMR measurements of a solution of [Pd1228]24+ indicated that broad Ha and Hb signals at 298 K became sharp at 353 K, showing only one set of signals due to fast chemical exchange between aA and aB protons by dynamic motion in [Pd1228]24+ (Fig. 2c). Thermodynamic parameters for the dynamic motion were determined by the Eyring plot (Supplementary Fig. 2) to obtain ΔH‡ = 81.9 kJ mol–1 and ΔS‡ = 60.0 J mol–1 K–1. A large positive entropy change is probably due to the stabilization of the transition state by solvent and/or counter anions. Cold spray ionization Fourier transform ion cyclotron resonance (CSI FT-ICR) mass spectrometry showed the signals of ionic species for [Pd1228(BF4)x](24–x)+ (x = 13–19) (Supplementary Fig. 3). The [Pd1228]24+ open icosahedron was further characterized by 13C NMR, heteronuclear multiple quantum correlation (HMQC), and heteronuclear multiple bond coherence (HMBC) spectroscopy (Supplementary Figs. 4–6).
Self-assembly of an M9L6 cage
Careful monitoring of the self-assembly of 2 and Pd2+ in CH3NO2 without NO3– at 343 K indicated that a different structure was assembled as a kinetic species almost quantitatively (96%) in 2 days. The 1H NMR spectrum is much more complicated than expected from the highly symmetrical M6L4 open octahedron and M12L8 open icosahedron (Fig. 2e). 1H DOSY measurement indicated that all 1H NMR signals for 2 belong to a single species with a diffusion coefficient of 2.4 × 10–10 m2 s–1 (Fig. 2f), which is larger than that of the [Pd1228]24+ open icosahedron, indicating the formation of a discrete assembled structure smaller than [Pd1228]24+. An M9L6 open triaugmented triangular prism37 meets this requirement (Fig. 1b). All 1H NMR signals for 2 in the new assembly were assigned by (H,H)-COSY and (H,H)-NOESY spectroscopy (Supplementary Figs. 7, 8). A characteristic feature of the 1H NMR spectrum is that although three chemically inequivalent Ha protons were observed, Hc protons were observed as two signals in a 1:2 ratio. This signal pattern is consistent with a [Pd926]18+ open triaugmented triangular prism, in which all six chemically equivalent tritopic ligands 2 are desymmetrized into C2v symmetry. The Ha and Hb protons in the 4-pyridyl ring A were observed as two sets (a1–a2 and b1–b2) because of the slow rotation of ring A on the NMR time scale (Fig. 2e). The chemical exchange between these signals observed by (H,H)-exchange spectroscopy (EXSY) (Supplementary Fig. 8) is consistent with the two sets of protons belonging to the same pyridyl ring A. In contrast, only one set of Ha and Hb signals was observed for ring B because the exchange of the inner and outer protons in ring B is possible by tilting ring B instead of flipping (180° rotation of) it against the N4 plane of Pd2+, unlike flipping the ring required for the exchange between Ha and Hb protons in ring A. VT 1H NMR measurements of a solution of [Pd926]18+ showed that the a1, a2, b1, and b2 signals gradually broadened as the temperature increased and finally coalesced at 363 K because of the rotation of ring A on the NMR time scale (Supplementary Fig. 9). In contrast, the Hc protons were observed as two sets of signals in a 1:2 ratio even at high temperature, indicating that tritopic ligand 2 in the assembled structure maintained C2v symmetry at 363 K.
[Pd926]18+ is kinetically so stable that any decomposition of [Pd926]18+ was not observed at 298 K for 3 days and that heating at 363 K for 60 h caused only a 19% decrease of [Pd926]18+ without formation of [Pd1228]24+ (Supplementary Fig. 10). The conversion of [Pd926]18+ into [Pd1228]24+ proceeded in the presence of NO3– by heating at 363 K for 60 h (Supplementary Fig. 10). These results indicate that [Pd926]18+ is metastable and that NO3– promotes the equilibration of the system due to its coordination ability.
Good quality single crystals of [Pd926]18+ could not be obtained, and crystallization of a solution of [Pd926]18+ in the presence of NO3– afforded single crystals of the [Pd1228]24+ open icosahedron, preventing the characterization of [Pd926]18+ by X-ray analysis. The energy-minimized structure of [Pd926]18+ obtained by density functional theory (DFT) calculation shows that an M9L6 open triaugmented triangular prism can be constructed without significant distortion (Fig. 1c).
CSI FT-ICR mass spectrometry showed the signals of ionic species for [Pd926(BF4)x(NO3)yFz](18–x–y–z)+ (x, y, z) = (3,7,4) (4,6,4), (5,5,4), (8,4,2), (3,6,4), (7,4,2), (8,3,2) (Supplementary Fig. 11). The signal intensity of these mass signals was low, probably because of fragmentation of the metastable [Pd926]18+ during the ionization process. The [Pd926]18+ open triaugmented triangular prism was further characterized by 13C NMR, HMQC, and HMBC spectroscopy (Supplementary Figs. 12–14).
Geometric analysis
The self-assembly of the [Pd1228]24+ open icosahedron and the [Pd926]18+ open triaugmented triangular prism is contrary to the expectation that tritopic ligand 2, whose coordination directions are the same as those of 1, affords an M6L4 open octahedron. A question is why larger structures such as M9L6 and M12L8 were assembled with only the dihedral angles changed, retaining the same coordination directions as 1. DFT calculations of [Pd624]12+, [Pd926]18+, and [Pd1228]24+ showed that their total energies normalized as a Pd322 unit are in the order [Pd1228]24+ <[Pd926]18+ <[Pd624]12+ (Supplementary Table 1), which is consistent with the experimental results; however, the reason for this is unclear.
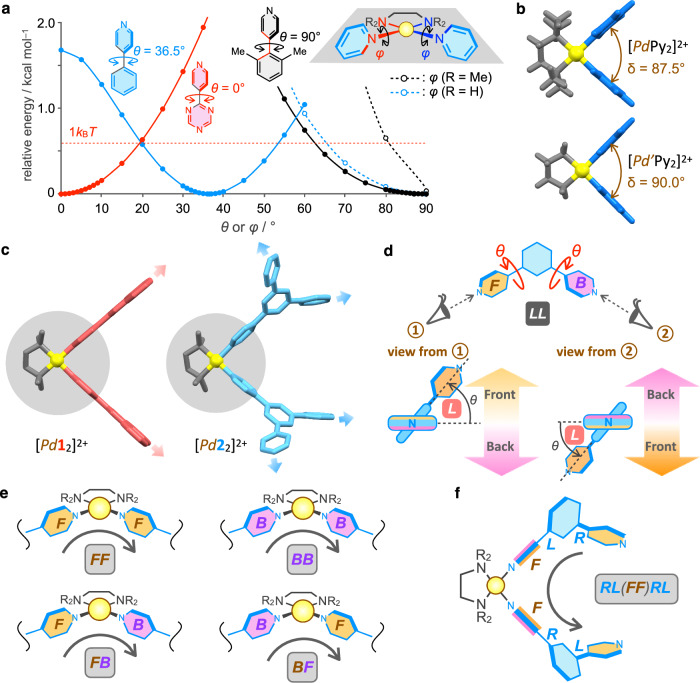
To understand how the dihedral change in the tritopic ligand affected self-assembly, first, the geometries of the building blocks (2 and Pd2+) were investigated by DFT calculations. The energy-minimized structure of 4-pyridylbenzene shows that the optimal dihedral angle (θ) is 36.5° (a blue solid line in Fig. 3a). The deviation from the optimal θ value destabilizes the structure. The red broken line in Fig. 3a indicates the energy level of 1 kBT at room temperature. Regarding the cis-protected square-planar geometry of the Pd(II) center, DFT calculations of [PdPy2]2+ were conducted. The optimal NTMEDA–Pd–NPy–C dihedral angle (φ) is 90°, and decreasing φ from 90° causes large destabilization (a black broken line in Fig. 3a), which indicates that the arrangement of the two pyridyl rings is sterically restricted by N,N,N’,N’-tetramethylethylenediamine (TMEDA) for Pd2+. The optimal NPy–Pd–NPy angle (δ) for [PdPy2]2+ is 87.5° (Fig. 3b), which is slightly smaller than the ideal 90° due to the steric effect of the methyl groups in TMEDA. The three angles (θ, φ, and δ) are the deterministic factors of the assembled structures that should be considered in addition to the coordination directions of 2.
Fig. 3. Conformation analysis of building blocks and precursory complexes.
a Change in the relative energy of 4-pyridyl-1,3,5-triazine, 4-phenylpyridine, and 4-(2,6-dimethylphenyl)pyridine by change of θ and change in the relative energies of [PdPy2]2+ and [Pd’Py2]2+ by change of φ. b Energy-minimized structures of [PdPy2]2+ and [Pd’Py2]2+ by DFT calculations using B3LYP/SDD (Pd), D95** (other atoms) level of theory. Color labels: Yellow = Pd; Gray = cis-protecting group; Blue = pyridine. c Energy-minimized structures of [Pd12]2+ and [Pd22]2+ by DFT calculations. Color labels: Yellow = Pd; Gray = TMEDA; Red = 1; Blue = 2. The structures highlighted by gray-filled circles are very similar. d Definition of L and R for the dihedral change (θ) and F (front) and B (back) faces of the pyridyl rings in the ligand. When θ = 0° and 90°, there is no discrimination between L and R. e Definition of the relative orientation of the ligands connected by a cis-protected Pd(II) ion. f Example of the notation of the conformation ID for [PdL2]2+. It is arbitrary from which side to read, so LR(FF)LR is the other notation of this conformation.
The effect of the dihedral angle (θ) in 2 was discussed by comparing the optimized structures of mononuclear [Pd12]2+ and [Pd22]2+ (Fig. 3c). The Pd(4-Py)2 moieties highlighted in filled gray circles are very similar between [Pd12]2+ and [Pd22]2+ and are essentially the same as the structure of [PdPy2]2+ (Fig. 3b). The structural difference between [Pd12]2+ and [Pd22]2+ is due to the arrangement of the 1,3-di(4-pyridyl)benzene moiety, which is caused by the difference in θ between 1 and 2. Therefore, although the coordination directions of the three nitrogen atoms in 1 and 2 are the same, the coordination directions of the four free pyridyl rings in [Pd12]2+ and [Pd22]2+ are different (red and blue bold arrows in Fig. 3c), which is caused by the fixation of the configuration in the Pd(4-Py)2 structure. In other words, the fixed value of φ enables remote geometric communications between the tritopic ligands through the connecting part (Pd2+), which energetically differentiates the assembled structures by a change in θ.
Reason why M6L4 was not formed from 2
The reason why the [Pd624]12+ open octahedron was not assembled is discussed on the basis of the structural change of a subcomponent M3L3 ring in the M6L4 open octahedron by change of θ (Fig. 4a). M6L4 is formed by connecting the three free coordination sites in the M3L3 ring with a free ligand L by three M. The optimized structure of [Pd313]6+ from 1 with θ = 0° shows that the three free pyridyl groups are properly positioned to bind the fourth L. Increasing θ value forces the three free pyridyl rings in [Pd3L3]6+ outward, and those in [Pd323]6+ are far from each other. Therefore, the large strain caused in the M6L4 open octahedron by increasing θ under the fixed φ value is the reason why [Pd624]12+ was not formed.
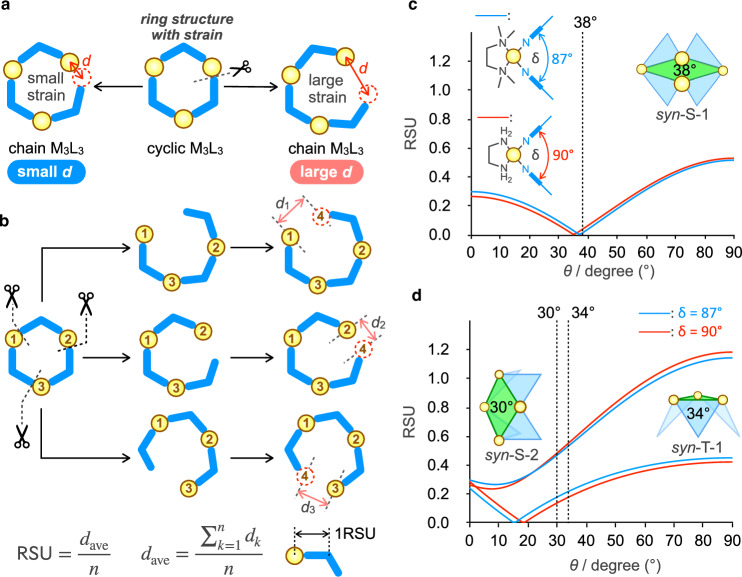
Fig. 4. Geometric analysis of the subcomponent rings in the [Pd3nL2n]6n+ (n = 2–4) cages.
a Structure change in the syn-T-1 conformation of [Pd3L3]6+ by change of θ. The energy-minimized structures of [Pd313]6+ and [Pd323]6+ are shown on the right. Color labels: Yellow = Pd; Blue = C; Red = N. Hydrogen atoms are omitted for clarity. b Subcomponent rings in the [Pd3nL2n]6n+ (n = 2–4) cages. The average θ values were obtained from the crystal structures or energy-minimized structures. The conformation IDs and RSU values of the subcomponent rings are listed in Table 1. The definition of RSU is shown in Fig. 5b.
Strain analysis of subcomponent rings
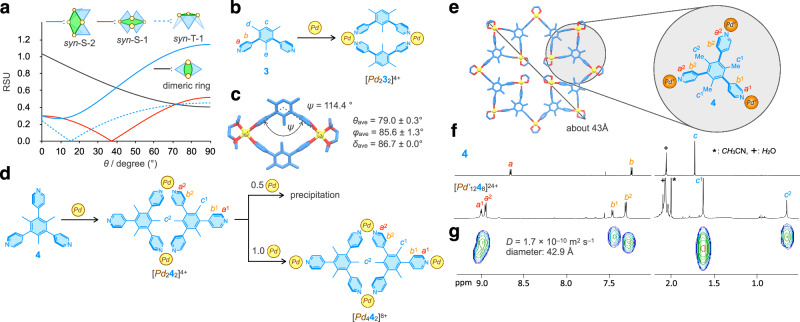
Next, we discuss why the [Pd1228]24+ open icosahedron is thermodynamically the most stable, whereas the [Pd926]18+ open triaugmented triangular prism is metastable. We focus on the subcomponent rings in the assembled structures (Fig. 4b). The M12L8 open icosahedron has only one type of conformation of M4L4 square in which all four free pyridyl groups are pointed toward the same side (syn-S-1). The M9L6 open triaugmented triangular prism has triangular and square subcomponents (syn-T-1 and syn-S-2). The M6L4 open octahedron contains triangles (syn-T-1) and squares (1,3-alt-S) as the subcomponent rings.
To analyze each subcomponent ring MnLn, the ditopic ligand L (1,3-di(4-pyridyl)benzene) was considered (Fig. 3d). The conformations of the ligand can be classified based on the screw sense concerning two dihedral angles (θ) (Fig. 3d). The clockwise rotation of θ indicates R, while the counterclockwise rotation of θ indicates L, and the conformation of the ligand can be indicated by two letters, such as LL. Next, the faces of the two pyridyl rings (F and B) are defined according to Fig. 3d. Four combinations of two pyridyl rings coordinating to a cis-protected Pd(II) center are possible (Fig. 3e). The conformational isomers of PdxLy can be expressed using four types of letters (R, L, B, and F). For example, the PdL2 structure shown in Fig. 3f is indicated as RL(FF)RL. According to this notation rule, the conformation IDs of the subcomponent rings in the crystal structures of [Pd614]12+ and [Pd1228]24+ and the optimized structure of [Pd926]18+ were determined (Table 1).
Table 1.
Ring strain per unit of subcomponent rings in Pd3nL2n cages
| cage | subcomponent ringa | θ / ° | RSUb |
|---|---|---|---|
| [Pd614]12+ |
syn-T-1 RL(FF)RL(FF)RL(FF)c |
0 | 0.243 |
|
1,3-alt-S RR(FF)RR(BB)RR(FF)RR(BB) |
0 | 0.284 | |
| [Pd926]18+ |
syn-T-1 RL(FF)RL(FF)RL(FF) |
34d | 0.220 |
|
syn-S-2 RR(FB)RL(BB)RR(FB)RL(BB) |
30d | 0.470 | |
| [Pd1228]24+ |
syn-S-1 RR(FF)LL(BB)RR(FF)LL(BB) |
38e | 0.010 |
aSchematic representation of the structures is shown in Fig. 4b.
bDefinition of the ring strain per unit (RSU) is shown in Fig. 5b.
cConformation ID of the subcomponent ring. The definitions of L and R for the counterclockwise and clockwise rotation of the dihedral change (θ) and F (front) and B (back) faces of the pyridyl rings in the ligand, as shown in Fig. 3d–f. When θ = 0° and 90°, there is no discrimination between L and R.
dAverage θ was obtained from the energy-minimized structure.
eAverage θ was obtained from the crystal structure38.
Strain in the subcomponent rings is purely estimated using the following method (Fig. 5a, b). Now, we consider the situation where an M–L coordination bond in an M3L3 ring is broken to form a chain structure without strain. When the ring strain is large, both terminals in the chain structure are largely separated from each other to relieve the strain. Thus, the distance between both ends in the M4L3 chain (d in Fig. 5a) can be used to evaluate the ring strain. In an M3L3 ring, there are three ways to form the M4L3 chain, and three d values are obtained from each M4L3 chain. The normalized ring strain in an MnLn ring per ML unit (RSU) is defined as dave/n (Fig. 5b). The effect of θ on the strain in the subcomponent rings MnLn was systematically analyzed by considering all possible mathematically generated conformations (see details in the Methods section).
Fig. 5. Strain analysis of MnLn rings.
a A concept of evaluating ring strain from the distance, d, between both ends in the chain structure obtained by breaking an M–L bond. The yellow circle and blue boomerang indicate a cis-protected Pd(II) ion and a ditopic ligand with a bend angle of 120°, respectively. b Definition of the ring strain per unit (RSU) for MnLn rings. The unit of RSU is the distance, and 1RSU is the distance between the centers of M and L. c Change in the RSU values of syn-S-1 in an M12L8 open icosahedron for Pd2+ (δ = 87°) and Pd’2+ (δ = 90°) by change of θ. A broken line indicates θave = 38° for syn-S-1 found in the crystal structure. d Change in the RSU values of syn-S-2 and syn-T-1 in an M9L6 open triaugmented triangular prism for Pd2+ (δ = 87°) and Pd’2+ (δ = 90°) by change of θ. Broken lines indicate θave = 30° and 34° for syn-S-2 and syn-T-1 in the energy-minimized structure of [Pd926]18+, respectively.
Strain analysis of Pd12L8
The change in RSU for the subcomponent square found in [Pd1228]24+, syn-S-1, is shown in Fig. 5c. The average θ value in the crystal structure of [Pd1228]24+ is 38°, which is slightly larger than the optimal value of θ for 4-pyridylbenzene (36.5°). It was found that the RSU of syn-S-1 for θ = 38° is 0.010 (Table 1), which is very close to the minimum RSU value at θ = 37°, and that syn-S-1 is the subcomponent square whose RSU is the smallest among the possible syn-M4L4 squares in the rage 37° ≤ θ ≤ 44°. Note that the RSU values of syn-T-1 and 1,3-alt-S in the [Pd614]12+ open octahedron (θ = 0°) are 0.243 and 0.284, respectively (Table 1). The strain in the [Pd1228]24+ open icosahedron is much smaller than that in [Pd614]12+. The small strain in [Pd1228]24+ is the reason why [Pd1228]24+ is thermodynamically the most stable, even though the number of components in [Pd1228]24+ is twice that of [Pd614]12+.
Strain analysis of Pd9L6
The changes in RSU of the subcomponent rings in [Pd926]18+ are shown in Fig. 5d. The average θ for syn-S-2 and syn-T-1 in the energy-minimized structure of [Pd926]18+ are 30° and 34°, respectively, and their RSU values are 0.470 and 0.220 (Table 1), which is much larger than RSU of syn-S-1 in [Pd1228]24+ at 38° (0.010). Therefore, a larger ring strain in [Pd926]18+ than in [Pd1228]24+ is the reason why [Pd926]18+ was obtained as a metastable assembly. The θ-dependence of RSU of syn-S-2 and syn-T-1 indicates that the ring strains are relieved by decreasing θ, and that approximately 20° is the best value to minimize the ring strains. The average θ values, 30° and 34°, are between 20° and the ideal θ for the free ligand, 36.5°. Considering that a total of six and eight rotatable pyridyl rings exist in syn-S-2 and syn-T-1, a large deviation of θ from the ideal 36.5° in all the rotatable parts significantly destabilizes the subcomponent rings, so these θ values would be taken to balance the two criteria. Smaller θ for syn-S-2, 30°, than θ for syn-T-1, 34°, would be due to a decrease in the large strain of syn-S-2.
Effect of cis-protecting group
The self-assembly of 2 and Pd’2+, in which ethylenediamine (en) is the cis-protecting group instead of TMEDA, produced neither M9L6 nor M12L8 but afforded a [Pd’624]12+ open octahedron (Fig. 1d). The [Pd’624]12+ open octahedron was characterized by 1H NMR, 1H DOSY, and mass spectroscopy (Supplementary Fig. 16). Unfortunately, single crystals of [Pd’624]12+ were not obtained. The assembly of the [Pd’624]12+ open octahedron indicates that not only θ but also φ is the important factor affecting the assembled structures. The cis-protecting group of a Pd(II) center alters φ and δ (Fig. 3b, c). DFT calculations indicate that the bite angle δ for [Pd’Py2]2+ is 90°, which is slightly larger than that for [PdPy2]2+, 87.5° (Fig. 3c). The effect of δ on RSU for the subcomponent rings of the assemblies is small (Fig. 5c, d), indicating that slight change in δ is not the reason for the formation of [Pd’624]12+.
Because the four N-methyl groups in TMEDA are replaced with hydrogens in en, the less steric effect of en allows a wider range of φ (Fig. 3a); therefore, the restriction of φ being 90° is relieved in the en-protected Pd(II) center, which enables [Pd’22]2+ to adopt a structure similar to that of [Pd12]2+ (Fig. 4a) by changing φ. In other words, in the case of en, variable φ values cancel the effect of θ, diminishing the geometric communication among the tritopic ligands. This idea was confirmed by comparing the DFT-optimized structures of the [Pd624]12+ and [Pd’624]12+ open octahedrons. Both cage structures have similar average θ values; 22.6 ± 1.9° for [Pd624]12+ and 27.5 ± 1.5° for [Pd’624]12+. In contrast, the φ values in [Pd624]12+ and [Pd’624]12+ show a larger difference; 82.0 ± 2.8° for [Pd624]12+ and 70.9 ± 7.2° for [Pd’624]12+ (Supplementary Fig. 16d), indicating that φ in [Pd’624]12+ is largely deviated from the ideal 90°. The small φ values in [Pd’624]12+ relieve the strain in subcomponent rings of [Pd’624]12+ caused by the dihedral angle θ being about 30°. Consequently, the fixed φ value is the key factor for differentiating the strain in the M3nL2n subcomponent rings caused by the θ change.
Strain analysis of Zn12L8
A similar M12L8 open icosahedron structure composed of 2 and a tetrahedral Zn(II) center, ZnCl2 (Zn), was reported in solid state (Fig. 6a)38,39. Because Zn(II) ions adopt tetrahedral coordinate, δ for Zn1228 is larger than 90°, and the average δ in the crystal structure is 102.8 ± 0.6°. The θ and φ in the crystal structure are 26.1 ± 9.0° and 84.1 ± 13.4°, respectively. The energy-minimized structure of the Zn323 ring shows that the three free pyridyl rings are separated (Fig. 6b). Note that the Zn313 ring also shows a similar spreading structure (Fig. 6b), indicating that the large ring strain in the Zn6L4 open octahedron (L = 1 or 2) is because of large δ regardless of θ.
Fig. 6. Strain analysis of the Zn1228 open icosahedron.
a Schematic representation of the Zn1228 open icosahedron. Zn indicates ZnCl2. θave, φave, and δave were obtained from the crystal structure38. b Energy-minimized structure of Zn323 by molecular mechanics calculations. Color labels: Purple = Zn; Blue = C; Red = N; Green = Cl. Hydrogen atoms are omitted for clarity. c Relative energy of mononuclear complexes by changing φ. φmin indicates φ in the energy-minimized structure. d Change in the RSU values of syn-S-1 and syn-S-3 in an M12L8 open triaugmented triangular prism for M (δ = 103°) by change of θ. syn-S-3 is the subcomponent ring found in the crystal structure of Zn122838. syn-S-1 is the same conformation as the subcomponent ring found in [Pd1228]24+. A broken line indicates θave = 26.1° found in the crystal structure of Zn1228. e Change in the RSU values of syn-S-2 and syn-T-1 in an M9L6 open triaugmented triangular prism for M (δ = 103°) by change of θ.
The subcomponent square in the crystal structure of Zn1228 is syn-S-3, in which one pyridyl ring rotates oppositely compared with syn-S-1 in [Pd1228]24+, so the conformation ID of syn-S-3 is different from that of syn-S-1 by two letters (Fig. 6d). The changes in the RSU values of syn-S-1 and syn-S-3 are shown in Fig. 6d. The ring strain in syn-S-3 is larger than that in syn-S-1 in the range 0° ≤ θ ≤ 70°. The reason why syn-S-3 with a larger strain was adopted in the crystal structure of Zn1228 is probably crystal packing. The RSU of syn-S-1 (δ = 103°) at the optimal θ (37°) is 0.15, which is larger than that at δ = 87° (RSU = 0.010, θ = 38°); therefore, the ring strain in Pd1228 is much smaller than that in Zn1228.
The changes in the RSU values of the subcomponent rings in M9L6 (δ = 103°) were calculated (Fig. 6e). The RSU of syn-S-2 was greater than 0.4 for θ > 20°, whereas the RSU of syn-T-1 was negligible around optimal θ = 36.5°. Therefore, Zn926 is largely destabilized by the strain in the square parts of Zn926 and is less stable than Zn1228. Because the complexation of 2 with ZnCl2 caused immediate precipitation38 and the resulting solid was not soluble in both polar and apolar solvents, thermodynamic discussion in the solution state was impossible. Thus, [Zn(TMEDA)]2+ (Zn’2+) was used as the metal source instead of ZnCl2 to solve the solubility problem. The complexation of 2 with Zn’2+ in CD3NO2 did not cause precipitation, and a slight downfield shift of the 1H NMR signals of 2 indicated the formation of coordination bonds between 2 with Zn’2+ (Supplementary Fig. 17a). 1H DOSY measurement of the resulting solution showed a diffusion coefficient of 6.7 × 10–10 m2 s–1 (Supplementary Fig. 17b), which is comparable to that of the free ligand 2 (6.7 × 10–10 m2 s–1) and larger than that of [Pd1228]24+ (2.0 × 10–10 m2 s–1). This result indicates that the equilibrium seldom shifts to the Zn’12L8 open icosahedron in solution, probably because of the weak Zn’–pyridine bond.
Note that φ in the crystal structure of Zn1228 shows a large deviation (84.1 ± 13.4°), which is likely due to the small steric effect of the two Cl atoms in the tetrahedral Zn(II) center, whereas θ values of 2 in the cage are close to the ideal value of 36.5°40. DFT calculations showed that destabilization by deviation of φ from the optimal φ for ZnCl2Py2 is much smaller than that for [PdPy2]2+ (Fig. 6c), which indicates that remote geometric communication among tritopic ligands 2 via the Zn–Py coordination bonds is weak. The Zn1228 open octahedron was assembled because of the destabilization of smaller assemblies (Zn624 and Zn926) by strain arising from large δ. This strategy is the same as that for the assembly of large [PdnL2n]2n+ assemblies by increasing the bend angle of the ditopic ligand; δ in Zn3n22n assemblies corresponds to the bend angle of L in [PdnL2n]2n+. [Pd1228]24+ and Zn1228 are structurally similar, but their formation origins are different.
Self-assembly of ligands with large dihedral angles
The next question is what assembly is produced when θ is greater than 40°. Methyl groups are introduced into the benzene ring to increase θ. DFT calculations indicate that the optimal θ for 4-(2,6-dimethylphenyl)pyridine is 90° (a black solid line in Fig. 3a). The RSU values of syn-T-1, syn-S-1, and syn-S-2 for the subcomponent rings in Pd9L6 and Pd12L8 increase with increasing θ, but the RSU of the Pd2L2 ring decreases as θ increases (Fig. 7a) and becomes the smallest among the four at θ > 74°. Thus, a Pd2L2 ring is expected when methyl groups are introduced in the central ring. To test this idea, the self-assembly of mesitylene-based ditopic ligand 3 was performed (Fig. 7b). A 1:1 mixture of Pd2+ and 3 afforded a clean 1H NMR spectrum, suggesting the formation of a single species (Supplementary Fig. 18). The [Pd222]4+ ring structure was unambiguously characterized by its single-crystal structure (Fig. 7c), where the bend angle of the ditopic ligands (ψ) was smaller than the ideal 120°, 114.4°, indicating a slight distortion, which is consistent with the large RSU of 0.416 at θ = 79°.
Fig. 7. Self-assembly of ligands with a large θ and cis-protected Pd(II) ions.
a Change in the RSU values of syn-S-1, syn-S-2, syn-T-1, and dimeric ring for Pd2+ (δ = 87°) by change of θ. The conformation of the dimeric ring whose RSU is the smallest at each θ is taken and their RSU values are plotted. b Formation of a [Pd232]4+ ring from the mesitylene-based ditopic ligand 3 and Pd2+. c Crystal structure of [Pd232]4+. Color labels: Yellow = Pd; Blue = C; Red = N. Hydrogen atoms are omitted for clarity. d Self-assembly of 4 and Pd2+ at different stoichiometry. e Energy-minimized structure of [Pd’1248]24+ composed of an M2L2 dimeric ring by molecular mechanics calculations. Color labels: Yellow = Pd; Blue = C; Red = N. Hydrogen atoms are omitted for clarity. f 1H NMR spectra of 4 and the [Pd’1248]24+ square sheet (500 MHz, CD3NO2, 298 K). g 1H DOSY spectrum of [Pd’1248]24+ (500 MHz, CD3NO2, 298 K).
Next, the self-assembly of tritopic ligand 4 and Pd2+ was investigated (Fig. 7d). The formation of the [Pd1248]24+ square sheet, where four molecules of the dimeric ring, [Pd242]4+, are connected by four Pd2+, was expected. 1:1 complexation of 4 and Pd2+ showed a characteristic 1H NMR spectrum of the [Pd242]4+ ring, where two sets of Ha, Hb, and Hc signals in a 1:2 integration ratio were observed (Supplementary Fig. 19a). The diffusion coefficients of these signals determined by 1H DOSY spectroscopy are the same (Supplementary Fig. 19b), indicating the formation of a single species with diameter of about 20.8 Å. Addition of 0.5 equiv. of Pd2+ to a solution of [Pd242]4+ to set the stoichiometry [Pd]/[4] = 1.5 caused precipitation, whereas addition of 1.0 equiv. of Pd2+ ([Pd]/[4] = 2) afforded a clear solution, whose 1H NMR spectrum showed a downfield shift of the a2 signal (Supplementary Fig. 19c), indicating the formation of Pd–N coordination bonds at the free pyridyl rings in [Pd242]4+ to form [Pd442]8+. [Pd442]8+ was characterized by 1H DOSY (Supplementary Fig. 19d), (H,H)-COSY, (H,H)-NOESY, 13C NMR, heteronuclear single quantum coherence (HSQC) and HMBC spectroscopy (Supplementary Figs. 20–24).
The precipitate produced at [Pd]/[4] = 1.5 may have oligomeric chain structures. Molecular modeling of the [Pd1248]24+ square sheet shows steric repulsion between the four TMEDA groups gathered in the center of the square (Supplementary Fig. 25), which may prevent the formation of the square sheet. When en is used as the cis-protecting group, such steric repulsion is not observed in the [Pd’1248]24+ square sheet by molecular modeling (Fig. 7e). Self-assembly of 4 and Pd’2+ in a 2:3 ratio gave a 1H NMR spectrum in which both a1 and a2 showed downfield shift, indicating that all of the pyridyl groups make a Pd(II)–N coordination bond (Fig. 7f). The 1H DOSY spectrum indicated the formation of a single species with a diameter of 42.9 Å (Fig. 7g), which is consistent with the size of the [Pd’1248]24+ square sheet (43 Å). According to the modeling structure of [Pd’1248]24+ square sheet, tritopic ligand 4 should be desymmetrized to C1; three 4-pyridyl groups in 4 are chemically inequivalent. Therefore, each Ha, Hb, and Hc proton is expected to be observed as three signals in a 1:1:1 ratio, which is not consistent with the observation of their signals in a 1:2 ratio (Fig. 7f). The environments of two types of 4-pyridyl groups of the [Pd’242]4+ ring parts (a2 protons) in the square sheet might be magnetically identical. CSI FT-ICR mass measurement of the solution detected the signals for [Pd’1248]24+ square sheet as several ionic species of [Pd’1248(BF4)x(NO3)yF6](18–x–y)+ (x, y) = (1,13), (2,12), (3,11), (0,13), (1,12), (2,11), (10,2), (11,1), (12,0), (11,0) (Supplementary Fig. 26). The [Pd’1248]24+ square sheet was further characterized by (H,H)-COSY, (H,H)-NOESY, 13C NMR, HSQC, and HMBC spectroscopy (Supplementary Figs. 27–31). Consequently, the introduction of three methyl groups in the central benzene ring to increase θ altered the assembled structure from the M12L8 open icosahedron to the M12L8 square sheet.
Narcissistic self-sorting behavior
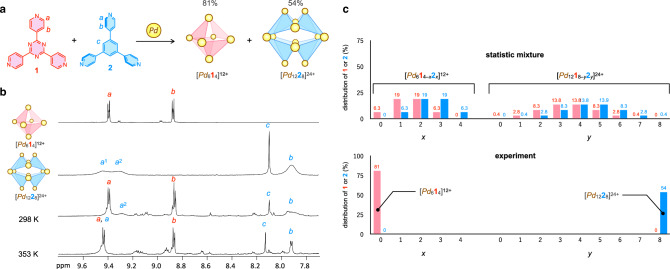
Finally, self-sorting of the tritopic ligands, 1 and 2, was investigated in the self-assembly of Pd3nL2n cages to evaluate the effect of the dihedral angle (θ) for the recognition between the two ligands with the same coordination direction. Although many examples of narcissistic self-sorting have been reported in molecular self-assembly41–53, the self-sorting of building blocks with high structural similarity is still challenging and such examples are scarce54. Tritopic ligands 1 and 2 are structurally the same except for a difference in the dihedral angle, θ (Fig. 1a, b). Tritopic ligands 1 and 2 were complexed with Pd2+ under thermodynamic control (heating at 363 K), resulting in intense 1H NMR signals of [Pd614]12+ and [Pd1228]24+ (Fig. 8b). The yields of [Pd614]12+ and [Pd1228]24+ determined by 1H NMR with the internal standard were 81% and 54%, respectively. The number of possible isomers of an M6L4 open octahedron composed of 1 and 2 is 5 and that of an M12L8 open icosahedron is 23, including an enantiomer (Supplementary Figs. 32 and 33). If there is no discrimination between the two ligands, the yields of [Pd614]12+ and [Pd1228]24+ based on 1 and 2 are calculated to be 6.25 and 0.39%, respectively (Fig. 8c and Supplementary Fig. 34). The difference in θ between 1 and 2 was well recognized in self-sorting, affording the homoleptic [Pd614]12+ and [Pd1228]24+ cages in much higher yields than those calculated for the statistical mixture. This indicates the power of remote geometric communications stemming from fixed φ.
Fig. 8. Narcissistic self-sorting of M6L4 and M12L8 cages from tritopic ligands 1 and 2 with the same coordination direction and Pd2+.
a A reaction scheme of the narcissistic self-sorting. The yields were determined based on tritopic ligands (1 or 2). b 1H NMR spectra of a mixture of 1, 2, Pd2+, and NO3– in a 1:1:3:1 ratio (500 MHz, CD3NO2/CDCl3/CD3OD = 7/3/2 (v/v/v)) after the convergence measured at 298 K and 353 K. NO3– was added to accelerate the equilibration of the system. 1H NMR spectra of [Pd614]12+ and [Pd1228]24+ at 298 K are shown for comparison. c Distribution of the tritopic ligands (1 and 2) in the [Pd6L4]12+ and [Pd12L8]24+ for a statistical mixture and experimental result (the yields of [Pd614]12+ and [Pd1228]24+). Only the distributions of 1 and 2 in the homoleptic assemblies are shown in the experimental result. Calculation of the distributions of 1 and 2 for a statistical mixture is shown in Supplementary Fig. 34.
Discussion
Although it has been recognized that the interactions among the building blocks in terms of shape and electrostatic complementarity are key for creating discrete, large self-assemblies in nature, a deeper understanding of the essence of the interactions is necessary to reveal the general principles. Simple artificial systems are a good toy model for abstracting the essential parts underlying complicated phenomena. It was found in this research that cis-protected Pd(II)-based self-assemblies are determined not only by the coordination direction of the multitopic ligand (L) and the coordination geometry of the metal ion (δ) but also by the dihedral angle (θ) in L. A narrow range of φ around 90° for Pd2+ enables propagation of the difference of θ in the multitopic ligand to neighboring multitopic ligands via cis-protected Pd(II) ions. There is no direct contact among the tritopic ligands in the Pd3nL2n (n = 2–4) assemblies, but remote geometric communications are realized through the connecting part (Pd2+) by fixing φ. This is the reason why larger assemblies, [Pd1228]24+ and [Pd926]18+, were assembled from the benzene-based tritopic ligand 2 (θ = 36.5°), whereas an M6L4 open octahedron was assembled from the triazine-based 1 (θ = 0°). The results obtained from a simple self-assembly system suggest that remote geometric communications among building blocks are a hidden important factor enabling giant assemblies in nature. Such a new perspective would allow us to rationally design large self-assemblies based on a higher level of geometric communication among the building blocks beyond the mere design of the shape of the building block itself.
Note that metastable [Pd926]18+ was produced in 96% yield, which must be realized by a pathway-dependent kinetic origin. Kinetic control can sometimes be involved in molecular self-assembly in nature55–57, but the understanding of kinetic control does not go beyond a kinetic trap. The advantage of kinetic control is that assemblies that are not thermodynamically most stable are accessible, which expands the possibility of molecular self-assembly. Understanding the self-assembly pathway to [Pd926]18+ would lead to the general principles of kinetic control in molecular self-assembly58, which would enable us to rationally design kinetic assemblies. We have just started working on this issue.
Methods
General information
1H NMR spectra were recorded using a Bruker AV-500 (500 MHz) spectrometer. All 1H NMR spectra were referenced using a residual solvent peak, CDCl3 (δ 7.26), CD3NO2 (δ 4.33). Electrospray ionization time-of-flight (ESI-TOF) mass spectra were obtained using a Waters Xevo G2-S Tof mass spectrometer and Bruker micrOTOF II-KE02. CSI FT-ICR Mass spectra were obtained using Bruker scimaX. Single-crystal X-ray analyses were conducted by Rigaku VariMax and SuperNova. Ligands 159,60, 261,62, and 463 were prepared according to the literature. Molecular mechanics calculations were performed using Materials Studio software (BIOVIA) with the universal force field.
Materials
Unless otherwise noted, all solvents and reagents were obtained from commercial suppliers (TCI Co., Ltd., WAKO Pure Chemical Industries Ltd., KANTO Chemical Co., Inc., and Sigma-Aldrich Co.) and were used as received. Deuterated solvents were used after dehydration with Molecular Sieves 4 Å.
Synthesis of 2,4,6-tri(4-pyridyl)-1,3,5-triazine (1)
18-Crown-6 (200 mg, 0.76 mmol) and KOH (45 mg, 0.8 mmol) were added to EtOH (4 mL), stirring at room temperature for 20 min. Then, 4-cyanopyrine (2.0 g, 19 mmol) was added to the reaction mixture, heating at 200 °C for 6 h. After cooling down to room temperature, pyridine (24 mL) was added to the reaction mixture, stirring for 5 min. The precipitation was obtained after filtration, washing with pyridine (10 mL) and toluene (10 mL). The solid was dissolved to 2 M. HCl aq. (24 mL) and collected into the filtrate. After the addition of NH3 aq. to the solution, until precipitation was generated. After filtration, tritopic ligand 1 was obtained as an off-white powder (0.51 g, 1.6 mmol, 25%). 1H NMR (500 MHz, CD3NO2/CDCl3/CD3OD = 7/3/2 (v/v/v), 298 K): δ 8.91 (dd, J = 1.5 and 4.3 Hz, 6 H), 8.71 (dd, J = 1.5 and 4.3 Hz, 6 H).
Synthesis of 1,3,5,-tri(4-pyridyl)-benzene (2)
In a 100-mL sealed tube flask, 1,3,5-tribromobenzene (0.3 g, 0.95 mmol, 1 eq.), 4-pyridyl-boronic acid (0.47 g, 3.8 mmol, 4 eq.), Na2CO3 (3.0 g, 29 mmol, 30 eq.), Pd(PPh3)4 (0.17 g, 0.14 mmol, 15 mol%) were mixed under inert atmosphere in dioxane (30 mL) and H2O (20 mL). The resulting mixture was stirred at 110 °C for 2 days. After evaporation of the mixture, the residue was added to H2O (50 mL), and extraction was performed using CHCl3 (40 mL, three times). The organic layer was dried over anhydrous MgSO4, and the solvent was removed by evaporation to obtain the crude product. After purification by silica gel column chromatography (Eluent: 5% CH3OH containing CHCl3 solution), tritopic ligand 2 was obtained as a colorless solid (0.22 g, 0.70 mmol, 73%). 1H NMR (500 MHz, CDCl3, 298 K): δ 8.76 (dd, J = 1.5 and 4.4 Hz, 6 H), 7.92 (s, 3 H), 7.61 (dd, J = 1.5 and 4.4 Hz, 6 H).
Synthesis of 4,4’,4”-(2,4,6-trimethylbenzene-1,3,5-triyl)tripyridine (4)
In a 100-mL sealed tube flask, 1,3,5-tribromo-2,4,6-trimethylbenzene (357 mg, 1.0 mmol, 1 eq.), 4-pyridyl-boronic acid (1.23 g, 10 mmol, 10 eq.), K3PO4 (2.55 g, 12 mmol, 12 eq.), Pd2(dba)3 (20.1 mg, 22 µmol, 0.022 eq.), and SPhos (18.1 mg, 44 µmol, 0.044 eq.) were mixed under inert atmosphere in n-butanol (40 mL). The resulting mixture was stirred at 80 °C for 3 days. After cooling to room temperature, the reaction mixture was filtered, and the filtrate was evaporated to remove the solvent. The residue was added to H2O (10 mL), and extraction was performed using CHCl3 (20 mL, three times). The organic layer was dried over anhydrous MgSO4, and the solvent was removed by evaporation to yield a yellow solid. After purification by silica gel column chromatography (Eluent: AcOEt), tritopic ligand 4 was obtained as a colorless solid (0.15 g, 0.42 mmol, 42%). 1H NMR (500 MHz, CD2Cl2, 298 K): δ 8.65 (dd, J = 1.5 and 4.3 Hz, 6 H), 7.14 (dd, J = 1.5 and 4.3 Hz, 4 H), 1.67 (s, 9 H).
Self-assembly of the [Pd1228]24+ open icosahedron
In a vial, [Pd(CH3CN)2](BF4)2 (12 mg, 24.2 µmol) was added to a solution of tritopic ligand 2 (5.0 mg, 16 µmol) in CH3NO2 in the presence of n-Bu4N·NO3 (2.5 mg, 8.1 µmol). The solution was heated at 363 K for 5 days. After cooling to room temperature, the solvent was removed by evaporation, and the residue was washed three times with CHCl3 (1.0 mL). The pale brown solid was dried in vacuo to obtain [Pd1228]24+ (15 mg, quant.). 1H NMR (500 MHz, CD3NO2, 353 K) δ 9.39 (d, J = 6.0 Hz, 48 H), 8.13 (s, 24 H), 7.94 (d, J = 6.0 Hz, 48 H), 3.28 (m, 48 H), 2.98 (s, 72 H), 2.84 (s, 72 H). 13C NMR (125 MHz, CD3NO2, 353 K): δ 153.2, 153.1, 140.2, 130.6, 127.4, 52.1, 51.9.
Self-assembly of the [Pd926]18+ open triaugmented triangular prism
In an NMR tube with [2.2]paracyclophane as an internal standard, [Pd(CH3CN)2](BF4)2 (0.86 mg, 1.8 µmol), tritopic ligand 2 (0.37 mg, 1.2 µmol), and CH3NO2 (480 µL) were added. The solution was then heated at 343 K for 2 days to obtain [Pd926]18+ in 96% NMR yield. 1H NMR (500 MHz, CD3NO2, 298 K): δ 9.21 (d, J = 6.0 Hz, 12 H), 9.15 (d, J = 6.0 Hz, 12 H), 9.10 (d, J = 6.0 Hz, 12 H), 8.08 (d, J = 1.5 Hz, 12 H), 7.95 (d, J = 6.5 Hz, 24 H), 7.66 (dd, J = 2.0, 6.0 Hz, 12 H), 7.58 (s, 6 H), 3.28 (m, 36 H), 3.03 (s, 36 H), 2.97 (s, 36 H), 2.73 (s, 36 H). 13C NMR (125 MHz, CD3NO2, 298 K): δ 153.0, 152.7, 152.5, 139.8, 139.6, 132.7, 128.8, 127.4, 127.0, 126.8, 105.5, 105.1, 51.9, 51.8, 51.5.
Self-assembly of the [Pd232]4+ ring
In an NMR tube with [2.2]paracyclophane as an internal standard, 1 equiv. of [Pd(CH3CN)2](BF4)2 (1.2 µmol) was added to a solution of 3 (1.2 µmol) in CD3NO2 (490 µL). After heating the solution at 343 K for 1 h, the [Pd232]4+ ring was obtained in 92% yield. 1H NMR (500 MHz, CD3NO2, 298 K): δ 9.14 (dd, J = 1.5, 5.5 Hz, 8 H), 7.40 (dd, J = 1.5 and 5.5 Hz, 8 H), 7.14 (s, 2 H), 3.20 (s, 8 H), 2.85 (s, 12 H), 2.06 (s, 12 H), 0.64 (s, 6 H). 13C NMR (125 MHz, CD3NO2, 298 K): δ 155.9, 152.6, 136.7, 136.4, 133.3, 130.3, 130.2, 51.7, 20.8, 20.3, 13.9.
Self-assembly of the [Pd242]4+ ring
In an NMR tube with [2.2]paracyclophane as an internal standard, 1 equiv. of [Pd(CH3CN)2](BF4)2 was added to a solution of 4 in CD3NO2 (500 µL). After heating at 343 K for 4 h, the [Pd242]4+ ring was obtained quantitatively. 1H NMR (500 MHz, CD3NO2, 298 K): δ 9.17 (dd, J = 1.5 and 5.5 Hz, 8 H), 8.65 (dd, J = 1.5 and 4.5 Hz, 4 H), 7.46 (d, J = 1.5 and 5.0 Hz, 8 H), 7.21 (d, J = 1.5 and 4.5 Hz,4 H), 3.18 (s, 8 H), 2.83 (s, 24 H), 1.77 (s, 12 H), 0.71 (s, 6 H). 13C NMR (125 MHz, CD3NO2, 298 K): δ 155.9, 152.8, 151.4, 139.3, 137.0, 134.1, 133.1, 130.2, 126.1, 79.2, 51.7, 21.0, 19.3, 13.9.
Self-assembly of the [Pd’1248]24+ square sheet
In an NMR tube with [2,2]paracyclophane as an internal standard, [Pd’(CH3CN)2](BF4)2 (0.76 mg, 1.8 μmol) was added to a solution of 4 (0.42 mg, 1.2 µmol) in CD3NO2 (500 µL) by titration at 343 K. After the convergence of the self-assembly, the [Pd’1248]24+ square sheet was obtained in 90% NMR yield. 1H NMR (500 MHz, CD3NO2, 298 K): δ 8.99 (d, J = 6.5 Hz, 16 H), 8.94 (d, J = 6.5 Hz, 32 H), 7.44 (d, J = 6.5 Hz, 16 H), 7.28 (d, J = 6.5 Hz, 32 H), 3.10 (br, 48 H), 1.62 (s, 48 H), 0.65 (s, 24 H). 13C NMR (125 MHz, CD3NO2, 298 K): δ 153.2, 137.4, 133.6, 132.6, 129.4, 129.2, 48.6, 47.8, 21.0, 19.1.
Single-crystal X-ray analyses
Single crystals of the [Pd1228]24+ open icosahedron suitable for X-ray crystallographic analysis were obtained by vapor diffusion of CH3OH in a CH3NO2 solution of [Pd1228]24+ at room temperature after 15 days. A crystal was immersed in and coated with FluorolubeⓇ (SIGMA-ALDRICH Corp.), and then mounted on a MicroMountTM (MiteGen LLC). The diffraction data of the single crystal were collected on a Rigaku VariMax with a Hybrid Photon Counting Detector at 93 K, using Mo Kα (λ = 0.71075 Å).
Single crystals of [Pd232]4+ were obtained at the interface between the two phases with CH3NO2 (100 µL) as a buffer solvent placed between them at room temperature for 15 days. One phase was a solution of 3 (5 mg, 18.2 µmol) and n-Bu4N·NO3 (2.8 mg, 9.1 µmol) in CH3NO2 (200 µL), and the other phase was a solution of [Pd(CH3CN)2](BF4)2 (8.7 mg, 18.2 µmol) in CH3NO2 (200 µL). A crystal was immersed in and coated with immersion oil (Cargille Corp.), and then mounted on a SuperNova single-crystal X-ray diffractometer with an Eos CCD detector (Rigaku Oxford Diffraction) at 180 K, using Cu Kα (λ = 1.54184 Å) radiation monochromated by multilayer mirror optics. Bragg spots were integrated using the CrysAlisPro program package (Rigaku Oxford Corporation). An empirical absorption correction based on the multi-scan method using spherical harmonics was implemented in the SCALE3 ABSPACK scaling algorithm. The structure was solved by an intrinsic phasing method on the SHELXT program64 and refined by a full-matrix least-squares minimization on F2 executed by the SHELXL program65 using the Olex2 software package (OlexSys Ltd.)66 and the ShelXle graphical user interface67. Thermal displacement parameters were refined anisotropically for all non-hydrogen atoms. The data were corrected for scattered electron density in large solvent voids using the PLATON SQUEEZE method68. The crystal structures are shown in Figs. 1d, 7d and Supplementary Fig. 35. The crystallographic data are summarized in Supplementary Table 2.
DFT calculations
The optimized structures of 2-(4-pyridyl)-1,3,5-triazine, 4-phenylpyridine, 4-(2,6-dimethylphenyl)pyridine, [PdPy2]2+,[Pd’Py2]2+, [Pd12]2+, [Pd22]2+ ZnPy2 were obtained using the B3LYP functional69,70 implemented in the Gaussian16 program package71. The SDD basis set72 was used for the Pd and Zn atoms, and the D95** basis set73 was used for the calculations of the H, C, N, and Cl atoms. The 6-31 G** basis set74,75 was employed for the calculations of 2-(4-pyridyl)-1,3,5-triazine, 4-phenylpyridine and 4-(2,6-dimethylphenyl)pyridine. To determine the relative energies depending on θ in the ligands and φ in [PdPy2]2+, [Pd’Py2]2+, and ZnPy2 (Figs. 3a and 6c), geometrical optimizations were performed by fixing the corresponding dihedral angle by inserting opt = modredundant keyword into the input file. Optimized structures and their energies were obtained using B3LYP/def2-SV(P) level of theory in [Pd624](BF4)12, [Pd926](BF4)18, [Pd1228](BF4)24, and [Pd’624](BF4)12. After optimization, single-point calculations were carried out with various functionals of LC-OLYP, M06, and ωB97X, taking into account an implicit solvent effect as a polarizable continuum model (PCM) of nitromethane (ε = 36.562). The cartesian coordinates of the optimized structures are listed in Supplementary Data 1.
Geometric analysis
For the calculation of RSU, it is necessary to determine the distance between the ends of the MnLn chain. Given that one end is situated at the origin of the global coordinate system, it suffices to ascertain the coordinates of the other end in the global coordinate system. When there exist coordinate systems A and B with bases and respectively, and the rotation matrix between them is denoted as i.e.,
| 1 |
the coordinates of a vector expressed as in coordinate system B can be calculated in coordinate system A as .
| 2 |
Note that vectors are quantities independent of coordinate systems. The vectors represented by here and later, such as , fall into this category. On the other hand, coordinates are representations of vectors that depend on the choice of coordinate systems and are expressed by arranging the components of the basis. Coordinates will be denoted by .
Each unit is assigned points A, B, and C, and coordinate systems A, B1, B2, and C, as illustrated in Fig. 9a–d. Because the positions of the coordinate systems do not affect the subsequent calculations, the coordinate systems can be located anywhere. Only the orientation of coordinate systems is crucial. Two dihedral angles between the rings in the ligand, denoted as θ1 and θ2, are defined as shown in Fig. 9a–d (0 ≤ θ1, θ2 ≤ 90°). To specify the rotation direction of the dihedral angles (R or L), integers j and k are introduced as follows.
| 3 |
| 4 |
Fig. 9. Definition of inner points and coordinates for ditopic ligands with cis-protected metal ions.
The ditopic ligand is depicted as a model of 1,3-di-4-pyridylbenzene. Two cis-protected metal ions with a bent ditopic ligand are shown (a–d). a RR, b RL, c LR, and d LL configurations. Points A and C are located at the center of the metal ions, whereas point B is located at the center of the middle hexagon in the ditopic ligand. The four coordinates are shown as the x, y, and z axes. The cis-protecting groups are omitted. Definitions of L and R are shown in the main text (Fig. 3d). A metal center with two coordinating rings is shown (e–h). e type FF, f type FB, g type BF, and h type BB. The point Ci is shown in the center of the Pd(II) ion, and the two coordinates are shown in the center of the coordinating rings. The coordinate is defined such that the front face of the coordinating ring is placed on the positive side of the z-axis. The cis-protecting ligand was omitted. Definitions of F and B are shown in the main text (Fig. 3d).
Definitions of L and R are shown in the main text (Fig. 3d).
What is ultimately required is only the transformation matrix between coordinate systems A and C, and the displacement vector between points A and C. Other coordinate systems, such as B1 and B2, are essentially not crucial. To determine the transformation matrix from A to C, the auxiliary coordinate systems B1 and B2 are introduced. Note that the z-axis of coordinate systems A and C always protrudes towards the front face.
Let the rotation matrix from A to B1 be denoted as , from B1 to B2 as , and from B2 to C as . Each matrix can be expressed as follows:
| 5 |
| 6 |
| 7 |
Since consecutive rotations can be expressed as the product of rotation matrices, the rotation matrix from A to C, denoted as , can be expressed as follows:
| 8 |
Let us now determine the components of the vector as seen in the local coordinate system A. As holds, it is necessary to express the right-hand side components and as coordinates seen from coordinate system A.
Since by definition, is expressed in coordinate system A as , and is expressed in the B2 coordinate system as . When converted to coordinates in the A system, is expressed as . Therefore, the components of as seen in the local coordinate system A are indicated as follows:
| 9 |
Consider two units, labeled as unit and unit . The two units are connected, and the unit numbers are indicated with superscripts in the alphabet (e.g., point A in unit is denoted as point , and the coordinate system of unit is represented as coordinate system ). Figure 9e–h shows an enlarged view of the junction part. φ was set to 90° throughout the calculation, which was demonstrated by DFT calculations (Fig. 3a) as the most stable for pyridyl groups coordinating to cis-protected Pd(II) center with TMEDA. While points and represent the same point, the coordinate systems and have different orientations.
Let the bite angle be denoted as δ (0° < δ < 180°). Additionally, to specify the relative orientations (F or B) of the connecting units, we defined integers and as follows:
| 10 |
| 11 |
Definitions of F and B are shown in the main text (Fig. 3d).
The rotation matrix can be expressed as follows:
| 12 |
The specific form of the coordinates in the coordinate system for is already known. The representation of in the coordinate system is obtained by multiplying from the left with the rotation matrix . Therefore, the calculation is given by .
Let us consider a chain consisting of n units (Unit 1, Unit 2,…, Unit n). The point A of Unit 1 (i.e., point ) is located at the origin of the global coordinate system, and the orientation of the local coordinate system of Unit 1 (i.e., coordinate system ) coincides with the global coordinate system.
Using the previously calculated , and , the position vector of the chain’s endpoint (i.e., point ) in the global coordinate system can be expressed in terms of components as follows.
| 13 |
Example
The method described above is employed to determine the distance between the endpoints of the M3L2 chain, illustrated through a specific example using the RR(FF)RL chain whose dihedral angles are all 30°. In this chain, the ditopic ligands with conformations RR and RL are connected to a cis-protected Pd(II) center with an FF relative orientation. Initially, the first unit (RR with both dihedral angles at 30°) is placed at the origin of the global coordinate system (i.e., ).
First, compute , and as follows.
| 14 |
| 15 |
| 16 |
Next, the components of the vector as viewed from the local coordinate system , denoted as , is calculated as follows:
| 17 |
Finally, the components of the position vector of point as seen from the global coordinate system, denoted as , is computed. Since the local coordinate system of Unit 1 is equivalent to the global coordinate system,
| 18 |
For calculations for subsequent units, the rotation matrix from the global coordinate system to the coordinate system is calculated as follows.
| 19 |
Next, consider connecting a new Unit 2 (RL with both dihedral angles at 30°) to the tip (point C) of Unit 1 (i.e., ). The relative orientation between the units is FF, and the bite angle is set at 90° (i.e., ).
First, calculate , , , and as follows.
| 20 |
| 21 |
| 22 |
| 23 |
Next, calculate the components of the vector as viewed from the local coordinate system , denoted as :
| 24 |
Subsequently, the coordinates of the vector as seen from the global coordinate system (i.e., the local coordinate system ) can be computed as follows.
| 25 |
Therefore, the components of the position vector of point as viewed from the global coordinate system (i.e., the local coordinate system ) are given by the following:
| 26 |
Therefore, the distance between the endpoints of the chain (tilt angle = 30°, bite angle = 90°, conformation id = RR(FF)RL is found to be
| 27 |
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
This work was supported by JSPS KAKENHI grant numbers 21K18974 and 23H01970 and the Asahi Glass Foundation. A part of single crystal X-ray measurement was supported by “Advanced Research Infrastructure for Materials and Nanotechnology in Japan (ARIM)” of the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Grant Number JPMXP1223UT0098. The authors thank Dr. Takashi Kikuchi (Rigaku corporation) for refining the single-crystal X-ray analysis, Dr. Akio Miyazato (JAIST) for the CSI FT-ICR MS measurements of [Pd926]18+, [Pd1228]24+, and [Pd’1248]24+, and Prof. Jun Terao, Dr. Hiroshi Masai, and Mr. Yudai Onda (Univ. of Tokyo) for the HRMS measurement of ditopic ligand 3. The computations were partly performed using the Research Center for Computational Science, Okazaki, Japan (Project: 24-IMS-C018).
Author contributions
S.H. conceived the project. T.A. performed all experiments. T.A., M.H., and H.S. performed DFT calculations. T.A. and K.T. performed geometric analyses. K.T. created a program to calculate RSU and strain analyses. S.H., T.A., and K.T. prepared the manuscript and the Supplementary Information, and all authors discussed the results and commented on the manuscript.
Peer review
Peer review information
Nature Communications thanks Javier Marti-Rujas and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The authors declare that all data supporting the findings of this study are available within the paper and its supplementary information files. Crystallographic data for the structures reported in this Article have been deposited at the Cambridge Crystallographic Data Centre (CCDC), under deposition numbers 2313022 (for the [Pd1228]24+ open icosahedron) and 2313023 (for the [Pd232]4+ ring). Copies of the data can be obtained free of charge via https://www.ccdc.cam.ac.uk/structures/. Calculated cartesian coordinates in this study are provided as Supplementary Data 1. All additional data were available from the corresponding author.
Code availability
All original code (RSU project) in this study has been deposited in Zenodo at the following: 10.5281/zenodo.1067730576 The data were available under the MIT license and is publicly accessible as of the date of publication.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-50972-z.
References
- 1.Dey, S. et al. DNA origami. Nat. Rev. Methods Prim.1, 13 (2021). 10.1038/s43586-020-00009-8 [DOI] [Google Scholar]
- 2.Nangreave, J., Han, D., Liu, Y. & Yan, H. DNA origami: a history and current perspective. Curr. Opin. Chem. Biol.14, 608–615 (2010). 10.1016/j.cbpa.2010.06.182 [DOI] [PubMed] [Google Scholar]
- 3.Saccà, B. & Niemeyer, C. M. DNA origami: the art of folding DNA. Angew. Chem. Int. Ed.51, 58–66 (2012). 10.1002/anie.201105846 [DOI] [PubMed] [Google Scholar]
- 4.Zhu, J. et al. Protein assembly by design. Chem. Rev.121, 13701–13796 (2021). 10.1021/acs.chemrev.1c00308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fegan, A., White, B., Carlson, J. C. T. & Wagner, C. R. Chemically controlled protein assembly: Techniques and applications. Chem. Rev.110, 3315–3336 (2010). 10.1021/cr8002888 [DOI] [PubMed] [Google Scholar]
- 6.Luo, Q., Hou, C., Bai, Y., Wang, R. & Liu, J. Protein assembly: versatile approaches to construct highly ordered nanostructures. Chem. Rev.116, 13571–13632 (2016). 10.1021/acs.chemrev.6b00228 [DOI] [PubMed] [Google Scholar]
- 7.Kulkarni, C. V. Lipid crystallization: from self-assembly to hierarchical and biological ordering. Nanoscale4, 5779–5791 (2012). 10.1039/c2nr31465g [DOI] [PubMed] [Google Scholar]
- 8.Pohorille, A. & Deamer, D. Self-assembly and function of primitive cell membranes. Res. Microbiol.160, 449–456 (2009). 10.1016/j.resmic.2009.06.004 [DOI] [PubMed] [Google Scholar]
- 9.Aumiller, W. M., Uchida, M. & Douglas, T. Protein cage assembly across multiple length scales. Chem. Soc. Rev.47, 3433–3469 (2018). 10.1039/C7CS00818J [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu, Z., Qiao, J., Niu, Z. & Wang, Q. Natural supramolecular building blocks: from virus coat proteins to viral nanoparticles. Chem. Soc. Rev.41, 6178–6194 (2012). 10.1039/c2cs35108k [DOI] [PubMed] [Google Scholar]
- 11.Tateishi, T. et al. Coordination/metal–organic cages inside out. Coord. Chem. Rev.467, 214612 (2022). 10.1016/j.ccr.2022.214612 [DOI] [Google Scholar]
- 12.Pilgrim, B. S. & Champness, N. R. Metal-organic frameworks and metal-organic cages – a perspective. ChemPlusChem85, 1842–1856 (2020). 10.1002/cplu.202000408 [DOI] [PubMed] [Google Scholar]
- 13.Chakrabarty, R., Mukherjee, P. S. & Stang, P. J. Supramolecular coordination: self-assembly of finite two- and three-dimensional ensembles. Chem. Rev.111, 6810–6918 (2011). 10.1021/cr200077m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujita, D. et al. Self-assembly of tetravalent Goldberg polyhedra from 144 small components. Nature540, 563–566 (2016). 10.1038/nature20771 [DOI] [PubMed] [Google Scholar]
- 15.Fujita, D. et al. Self-assembly of M30L60 icosidodecahedron. Chem1, 91–101 (2016). 10.1016/j.chempr.2016.06.007 [DOI] [Google Scholar]
- 16.Bunzen, J. et al. Self-assembly of M24L48 polyhedra based on empirical prediction. Angew. Chem. Int. Ed.51, 3161–3163 (2012). 10.1002/anie.201108731 [DOI] [PubMed] [Google Scholar]
- 17.Sun, S. et al. Self-assembled M24L48 polyhedra and their sharp structural switch upon subtle ligand variation. Science328, 1144–1147 (2010). 10.1126/science.1188605 [DOI] [PubMed] [Google Scholar]
- 18.Tominaga, M. et al. Finite, spherical coordination networks that self-organize from 36 small components. Angew. Chem. Int. Ed.43, 5621–5625 (2004). 10.1002/anie.200461422 [DOI] [PubMed] [Google Scholar]
- 19.Suzuki, K., Tominaga, M., Kawano, M. & Fujita, M. Self-assembly of an M6L12 coordination cube. Chem. Commun. 13, 1638–1640 (2009). [DOI] [PubMed]
- 20.Bloch, W. M. et al. Geometric complementarity in assembly and guest recognition of a bent heteroleptic cis-[Pd2LA2LB2] coordination cage. J. Am. Chem. Soc.138, 13750–13755 (2016). 10.1021/jacs.6b08694 [DOI] [PubMed] [Google Scholar]
- 21.Kumar Chand, D., Fujita, M., Biradha, K., Sakamoto, S. & Yamaguchi, K. Metal driven self-assembly of pyridine appended ligands with cis-protected/naked Pd(ii) ion: a comparative study. Dalton Trans. 13, 2750–2756 (2003).
- 22.Suzuki, K., Kawano, M. & Fujita, M. Solvato-controlled assembly of Pd3L6 and Pd4L8 coordination “Boxes”. Angew. Chem. Int. Ed46, 2819–2822 (2007). 10.1002/anie.200605084 [DOI] [PubMed] [Google Scholar]
- 23.Yu, H. et al. Elucidating anion-dependent formation and conversion of Pd2L4 and Pd3L6 metal–organic cages by complementary techniques. Eur. J. Inorg. Chem.2018, 80–85 (2018). 10.1002/ejic.201701319 [DOI] [Google Scholar]
- 24.Zhang, T., Zhou, L., Guo, X., Cai, L. & Sun, Q. Adaptive self-assembly and induced-fit transformations of anion-binding metal-organic macrocycles. Nat. Commun.8, 15898 (2017). 10.1038/ncomms15898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chand, D. et al. Dynamic self-assembly of an M3L6 molecular triangle and an M4L8 tetrahedron from naked PdII ions and bis(3-pyridyl)-substituted arenes. Chem. Asian J.1, 82–90 (2006). 10.1002/asia.200600029 [DOI] [PubMed] [Google Scholar]
- 26.Jurček, O. et al. Superchiral Pd3L6 coordination complex and its reversible structural conversion into Pd3L3Cl6 metallocycles. Angew. Chem. Int. Ed.54, 15462–15467 (2015). 10.1002/anie.201506539 [DOI] [PubMed] [Google Scholar]
- 27.Samanta, D. & Mukherjee, P. S. Component selection in the self-assembly of palladium(II) nanocages and cage-to-cage transformations. Chem. Eur. J.20, 12483–12492 (2014). 10.1002/chem.201402553 [DOI] [PubMed] [Google Scholar]
- 28.Engelhard, D. M., Freye, S., Grohe, K., John, M. & Clever, G. H. NMR-based structure determination of an intertwined coordination cage resembling a double trefoil knot. Angew. Chem. Int. Ed.51, 4747–4750 (2012). 10.1002/anie.201200611 [DOI] [PubMed] [Google Scholar]
- 29.Schmidt, A., Casini, A. & Kühn, F. E. Self-assembled M2L4 coordination cages: synthesis and potential applications. Coord. Chem. Rev.275, 19–36 (2014). 10.1016/j.ccr.2014.03.037 [DOI] [Google Scholar]
- 30.Liao, P. et al. Two-component control of guest binding in a self-assembled cage molecule. Chem. Commun.46, 4932–4934 (2010). 10.1039/c0cc00234h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McMorran, D. A. & Steel, P. J. The first coordinatively saturated, quadruply stranded helicate and its encapsulation of a hexafluorophosphate anion. Ange. Chem. Int. Ed.37, 3295–3297 (1998). [DOI] [PubMed] [Google Scholar]
- 32.Gao, C., Li, J., Yin, S., Sun, J. & Wang, C. Twist building blocks from planar to tetrahedral for the synthesis of covalent organic frameworks. J. Am. Chem. Soc.142, 3718–3723 (2020). 10.1021/jacs.9b13824 [DOI] [PubMed] [Google Scholar]
- 33.Pang, J. et al. Control the structure of Zr-tetracarboxylate frameworks through steric tuning. J. Am. Chem. Soc.139, 16939–16945 (2017). 10.1021/jacs.7b09973 [DOI] [PubMed] [Google Scholar]
- 34.Davies, J. A., Ronson, T. K. & Nitschke, J. R. Twisted rectangular subunits self-assemble into a ferritin-like capsule. Chem8, 1099–1106 (2022). 10.1016/j.chempr.2022.01.003 [DOI] [Google Scholar]
- 35.Stang, P. J., Olenyuk, B., Muddiman, D. C. & Smith, R. D. Transition-metal-mediated rational design and self-assembly of chiral, nanoscale supramolecular polyhedra with unique T symmetry. Organometallics16, 3094–3096 (1997). 10.1021/om9702993 [DOI] [Google Scholar]
- 36.Fujita, M. et al. Self-assembly of ten molecules into nanometre-sized organic host frameworks. Nature378, 469–471 (1995). 10.1038/378469a0 [DOI] [Google Scholar]
- 37.Lavendomme, R., Ronson, T. K. & Nitschke, J. R. Metal and organic templates together control the size of covalent macrocycles and cages. J. Am. Chem. Soc.141, 12147–12158 (2019). 10.1021/jacs.9b06182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Torresi, S., Famulari, A. & Martí-Rujas, J. Kinetically controlled fast crystallization of M12L8 poly-[n]-catenanes using the 2,4,6-tris(4-pyridyl)benzene ligand and ZnCl2 in an aromatic environment. J. Am. Chem. Soc.142, 9537–9543 (2020). 10.1021/jacs.0c03319 [DOI] [PubMed] [Google Scholar]
- 39.Martí-Rujas, J., Elli, S., Sacchetti, A. & Castiglione, F. Mechanochemical synthesis of mechanical bonds in M12L8 poly-[n]-catenanes. Dalton Trans.51, 53–58 (2022). 10.1039/D1DT03158A [DOI] [PubMed] [Google Scholar]
- 40.Martí-Rujas, J., Elli, S. & Famulari, A. Kinetic trapping of 2,4,6-tris(4-pyridyl)benzene and ZnI2 into M12L8 poly-[n]-catenanes using solution and solid-state processes. Sci. Rep.13, 5605 (2023). 10.1038/s41598-023-32661-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen, Q. et al. Narcissistic self-sorting and enhanced luminescence via catenation in water. Mater. Today Chem.23, 100679 (2022). 10.1016/j.mtchem.2021.100679 [DOI] [Google Scholar]
- 42.Ma, J. et al. Designing narcissistic self-sorting terpyridine moieties with high coordination selectivity for complex metallo-supramolecules. Commun. Chem.4, 136 (2021). 10.1038/s42004-021-00577-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao, X. et al. Narcissistic self-sorting in anion-coordination-driven assemblies. Chem. Commun.57, 6078–6081 (2021). 10.1039/D1CC01652K [DOI] [PubMed] [Google Scholar]
- 44.Rizzuto, F. J. & Nitschke, J. R. Narcissistic, Integrative, and kinetic self-sorting within a system of coordination cages. J. Am. Chem. Soc.142, 7749–7753 (2020). 10.1021/jacs.0c02444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tateishi, T., Kojima, T. & Hiraoka, S. Chiral self-sorting process in the self-assembly of homochiral coordination cages from axially chiral ligands. Commun. Chem.1, 20 (2018). 10.1038/s42004-018-0020-4 [DOI] [Google Scholar]
- 46.Wiley, C. A. et al. Electronic effects on narcissistic self-sorting in multicomponent self-assembly of Fe-iminopyridine meso-helicates. Inorg. Chem.55, 9805–9815 (2016). 10.1021/acs.inorgchem.6b01644 [DOI] [PubMed] [Google Scholar]
- 47.Sinha, N., Tan, T. T. Y., Peris, E. & Hahn, F. E. High-fidelity, narcissistic self-sorting in the synthesis of organometallic assemblies from Poly-NHC ligands. Angew. Chem. Int. Ed.56, 7393–7397 (2017). 10.1002/anie.201702637 [DOI] [PubMed] [Google Scholar]
- 48.Johnson, A. M. et al. Narcissistic self-sorting in self-assembled cages of rare earth metals and rigid Ligands. Angew. Chem. Int. Ed.54, 5641–5645 (2015). 10.1002/anie.201500400 [DOI] [PubMed] [Google Scholar]
- 49.Ayme, J., Beves, J. E., Campbell, C. J. & Leigh, D. A. The self-sorting behavior of circular helicates and molecular knots and links. Angew. Chem. Int. Ed.53, 7823–7827 (2014). 10.1002/anie.201404270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Singh, A. S. & Sun, S. Narcissistic self-sorting of hydrogen-bonded dimeric capsules formed through self-assembly of flexible tripodal receptors in polar solvents. Chem. Commun.48, 7392–7394 (2012). 10.1039/c2cc33645f [DOI] [PubMed] [Google Scholar]
- 51.Kramer, R., Lehn, J. M. & Marquis-Rigault, A. Self-recognition in helicate self-assembly: spontaneous formation of helical metal complexes from mixtures of ligands and metal ions. Proc. Nat. Acad. Sci. USA.90, 5394–5398 (1993). 10.1073/pnas.90.12.5394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lal Saha, M. & Schmittel, M. Degree of molecular self-sorting in multicomponent systems. Org. Biomol. Chem.10, 4651–4684 (2012). 10.1039/c2ob25098e [DOI] [PubMed] [Google Scholar]
- 53.Safont-Sempere, M., Fernández, G. & Würthner, F. Self-sorting phenomena in complex supramolecular systems. Chem. Rev.111, 5784–5814 (2011). 10.1021/cr100357h [DOI] [PubMed] [Google Scholar]
- 54.Abe, T., Horiuchi, S. & Hiraoka, S. Kinetically controlled narcissistic self-sorting of Pd(II)-linked self-assemblies from structurally similar tritopic ligands. Chem. Commun.58, 10829–10832 (2022). 10.1039/D2CC04496J [DOI] [PubMed] [Google Scholar]
- 55.Okazawa, A., Sanada, N., Takahashi, S., Sato, H. & Hiraoka, S. Pathway selection in the self-assembly of Rh4L4 coordination squares under kinetic control. Commun. Chem.6, 248 (2023). 10.1038/s42004-023-01053-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Foianesi-Takeshige, L. et al. Bifurcation of self-assembly pathways to sheet or cage controlled by kinetic template effect. Commun. Chem.2, 128 (2019). 10.1038/s42004-019-0232-2 [DOI] [Google Scholar]
- 57.ateishi, T. et al. Navigated Self-assembly of a Pd2L4 cage by modulation of an energy landscape under kinetic control. J. Am. Chem. Soc.141, 19669–19676 (2019). 10.1021/jacs.9b07779 [DOI] [PubMed] [Google Scholar]
- 58.Takahashi, S., Abe, T., Sato, H. & Hiraoka, S. Pathway bias and emergence of quasi-irreversibility in reversible reaction networks: extension of Curtin-Hammett principle. Chem9, 2971–2982 (2023). 10.1016/j.chempr.2023.06.015 [DOI] [Google Scholar]
- 59.Anderson, H. L., Anderson, S. & Sanders, J. K. M. Ligand binding by butadiyne-linked porphyrin dimers, trimers and tetramers. J. Chem. Soc. Perkin Trans.1, 2231–2245 (1995). 10.1039/p19950002231 [DOI] [Google Scholar]
- 60.He, Z., Li, M., Que, W. & Stang, P. J. Self-assembly of metal-ion-responsive supramolecular coordination complexes and their photophysical properties. Dalton Trans.46, 3120–3124 (2017). 10.1039/C7DT00174F [DOI] [PubMed] [Google Scholar]
- 61.Zheng, T. & Li, L. {[Ru(bda)]xLy}n cross-linked coordination polymers: toward efficient heterogeneous catalysis for water oxidation in an organic solvent-free system. New J. Chem.42, 2526–2536 (2018). 10.1039/C7NJ04330A [DOI] [Google Scholar]
- 62.Schmittel, M., He, B. & Mal, P. Supramolecular multicomponent self-assembly of shape-adaptive nanoprisms: wrapping up C60 with three porphyrin units. Org. Lett.10, 2513–2516 (2008). 10.1021/ol800796h [DOI] [PubMed] [Google Scholar]
- 63.Luisier, N. et al. Crystal engineering of polymeric structures with dative boron–nitrogen bonds: Design criteria and limitations. Crystal Growth Design16, 6600–6604 (2016). 10.1021/acs.cgd.6b01292 [DOI] [Google Scholar]
- 64.Sheldrick, G. SHELXT - Integrated space-group and crystal-structure determination. Acta Cryst. A71, 3–8 (2015). 10.1107/S2053273314026370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sheldrick, G. Crystal structure refinement with SHELXL. Acta Cryst. C71, 3–8 (2015). 10.1107/S2053229614024218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K. & Puschmann, H. OLEX2: a complete structure solution, refinement and analysis program. J. Appl. Cryst.42, 339–341 (2009). 10.1107/S0021889808042726 [DOI] [Google Scholar]
- 67.Hubschle, C. B., Sheldrick, G. M. & Dittrich, B. ShelXle: a Qt graphical user interface for SHELXL. J. Appl. Cryst.44, 1281–1284 (2011). 10.1107/S0021889811043202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Spek, A. Structure validation in chemical crystallography. Acta Cryst. D65, 148–155 (2009). 10.1107/S090744490804362X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee, C., Yang, W. & Parr, R. G. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B37, 785–789 (1988). 10.1103/PhysRevB.37.785 [DOI] [PubMed] [Google Scholar]
- 70.Becke, A. D. Density‐functional thermochemistry III. The role of exact exchange. J. Chem. Phys.98, 5648–5652 (1993). 10.1063/1.464913 [DOI] [Google Scholar]
- 71.Frisch, M. J. et al. Gaussian 16. Revision C.01 (2016).
- 72.Andrae, D., Häußermann, U., Dolg, M., Stoll, H. & Preuß, H. Energy-adjustedab initio pseudopotentials for the second and third row transition elements. Theor. Chim. Acta77, 123–141 (1990). 10.1007/BF01114537 [DOI] [Google Scholar]
- 73.Dunning, T. H. & Hay, P. J. in Methods of Electronic Structure Theory (ed. Schaefer, H. F.) (Springer, 1977).
- 74.Hehre, W. J., Ditchfield, R. & Pople, J. A. Self—consistent molecular orbital methods. XII. Further extensions of Gaussian—Type basis sets for use in molecular orbital studies of organic molecules. J. Chem. Phys.56, 2257–2261 (2003). 10.1063/1.1677527 [DOI] [Google Scholar]
- 75.Ditchfield, R., Hehre, W. J. & Pople, J. A. Self‐consistent molecular‐orbital methodS. IX. An extended Gaussian‐type basis for molecular‐orbital studies of organic moleculess. J. Chem. Phys.54, 724–728 (2003). 10.1063/1.1674902 [DOI] [Google Scholar]
- 76.Hiraoka, S. & Takeuchi, K. RSU project (v1.0.0). Zenodo.10.5281/zenodo.10677305 (2024).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The authors declare that all data supporting the findings of this study are available within the paper and its supplementary information files. Crystallographic data for the structures reported in this Article have been deposited at the Cambridge Crystallographic Data Centre (CCDC), under deposition numbers 2313022 (for the [Pd1228]24+ open icosahedron) and 2313023 (for the [Pd232]4+ ring). Copies of the data can be obtained free of charge via https://www.ccdc.cam.ac.uk/structures/. Calculated cartesian coordinates in this study are provided as Supplementary Data 1. All additional data were available from the corresponding author.
All original code (RSU project) in this study has been deposited in Zenodo at the following: 10.5281/zenodo.1067730576 The data were available under the MIT license and is publicly accessible as of the date of publication.