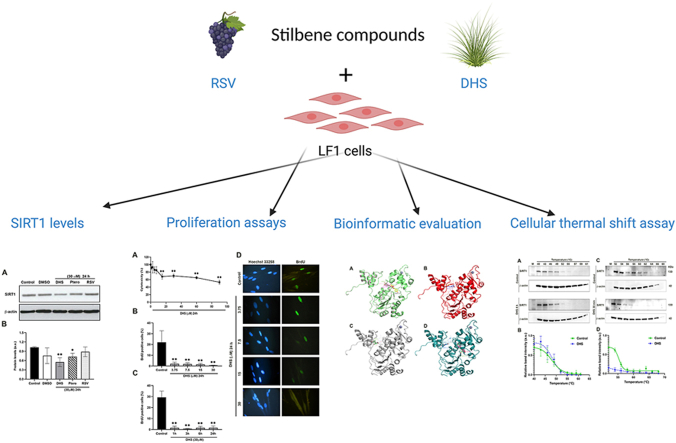
Abstract
Background and aim
Resveratrol (RSV), is a stilbene-based compound exerting wide biological properties. Its analogue 4,4′-dihydroxy-trans-stilbene (DHS) has shown improved bioavailability and antiproliferative activity in vitro and in vivo. One of the hypotheses on how resveratrol works is based on SIRT1 activation. Since their strict structural similarities, we have explored a potential interaction between DHS and SIRT1, in comparison with the parental molecule.
Experimental procedure
Timing of incubation and concentrations of DHS have been determined using MTT assay in normal human lung fibroblasts. Untreated, DHS- or RSV-treated cells were harvested and analysed by Western Blotting or RT-PCR, in order to evaluate SIRT1 levels/activity and expression, and by Cellular Thermal shift assay (CETSA) to check potential DHS or RSV-SIRT1 interaction. Transfection experiments have been performed with two SIRT1 mutants, based on the potential binding pockets identified by Molecular Docking analysis.
Results and conclusion
We unexpectedly found that DHS, but not RSV, exerted a time-dependent inhibitory effect on both SIRT1 protein levels and activity, the latter measured as p53 acetylation. At the mRNA level no significant changes were observed, whereas a proteasome-dependent mechanism was highlighted for the reduction of SIRT1 levels by DHS in experiments performed with the proteasome inhibitor MG132. Bioinformatics analysis suggested a higher affinity of RSV in binding all SIRT1 complexes compared to DHS, except comparable results for complex SIRT1-p53. Nevertheless, both CETSA and SIRT1 mutants transfected in cells did not confirm this interaction. In conclusion, DHS reduces SIRT1 protein level, thereby inhibiting its activity through a proteasome-mediated mechanism.
Keywords: 4,4′-dihydroxystilbene; Sirtuin-1; Resveratrol; Deacetylation
Graphical abstract
1. Introduction
Resveratrol (RSV) is a naturally occurring compound belonging to the stilbene family, which has gained extensive attention in recent years due to its potential health benefits.1, 2, 3, 4 Among the series of resveratrol derivatives designed and synthesized with the aim of enhancing the beneficial properties of the parental molecule,5, 6, 7 4, 4′-dihydroxy-trans-stilbene (DHS), known to possess in some model systems higher antioxidant8, 9, 10, 11 and anti-inflammatory properties,12 as well as antitumor activity,13, 14, 15, 16, 17 is one of the most promising. It shares with resveratrol several similarities, e.g. inhibiting cell proliferation by targeting specific cellular proteins, such as ribonucleotide reductase,18,19 DNA polymerase δ,20, 21, 22 or counteracting the expression of factors playing key roles in cancer invasion, like MMP-2/9, N/E-cadherin and survivin.17,23 This analogue demonstrates also specific characteristics that set it apart and contribute to its effects. Among these, DHS exhibits increased stability, thus contributing to its enhanced bioavailability and effectiveness, unlike resveratrol, which undergoes rapid metabolism and has a relatively short half-life.24, 25, 26 In addition, DHS appears to have, in normal human cells, additional molecular targets than RSV, thereby explaining its stronger antiproliferative activity.22 On the other hand, one key mechanism through which RSV exerts its effects is by activating Sirtuin 1 (SIRT1).27 In fact, numerous studies have shown that RSV can activate SIRT1, leading to a range of beneficial effects on cellular and molecular levels. It may improve mitochondrial function,28 enhance insulin sensitivity,29 and modulate inflammatory pathways,30 among other effects.31 SIRT1 is a member of the sirtuin family, a conserved family of NAD+ dependent histone deacetylases that regulate various physiological and pathological cellular processes, e.g. cellular response to stress factors, DNA repair, nutrient availability, metabolism, and aging.32 They have been extensively studied for their potential role in promoting beneficial health effects of caloric restriction and longevity in higher organisms.33,34 Resveratrol has been extensively investigated as well for its potential in promoting longevity and preventing age-related diseases and conditions.35 Moreover, dysregulation of histone deacetylases has been demonstrated to play a crucial role in carcinogenesis and tumor progression, providing a crucial attractive target against cancer.36 One of the mechanism by which SIRT1 can counteract cancer cell growth is by inducing autophagy as an alternative cell death.37 In fact, through its deacetylase activity, SIRT1 participates in the regulation of different steps of autophagy.38 We have recently provided the evidence that both the stilbenes are able to contrast tumor growth in mice by increasing autophagy, with DHS being more effective than RSV.15 These data in vivo strongly suggest that DHS effect could be dependent on a mechanism in which SIRT1 regulates autophagy.
In this paper, we aimed to explore a possible involvement of SIRT1 in the mechanism of action underlying the antiproliferative e antitumor activity of DHS, in comparison with its parental compound. No data are available in the literature about the potential DHS effect on SIRT1 function. In particular, this explorative work aimed to examine whether DHS could affect SIRT1 level and activity in primary cultures of human lung fibroblast (LF1). Understanding the interplay between DHS and SIRT1 will provide valuable insights into the biological effects of this resveratrol derivative.
2. Materials and methods
2.1. Reagents and cell culture
4,4′-dihydroxy-trans-stilbene (DHS) and resveratrol (RSV) were purchased from Santa Cruz Biotechnology. Pterostilbene (PTS) was synthetized by L. Forti (University of Modena and Reggio Emilia).10 All other chemicals of reagent grade, if not specified, were obtained from Sigma (St. Louis, MO). Normal human lung embryonic fibroblasts (LF1) cells (gift from J. Sedivy, Brown University Providence, RI), were cultured in E-MEM supplemented with 10% FBS, 1% Sodium pyruvate, 200 mM l-glutamine, 100 IU/mL penicillin, and 100 μg/mL streptomycin, all obtained from Thermo Fisher Scientific, Waltham, MA, USA. Etoposide (Eto) and MG132 were provided by MERCK and used at the 20 and 25 μM working solution respectively.
2.2. Cytotoxicity and proliferation assay
Cell toxicity was determined by the MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] assay. LF1 cells, seeded at a density of 1 × 105 in 24 wells plate, were treated for 24 h with 1, 2.5, 5, 7.5, 15, 30, 60 and 90 μM DHS, then processed as previously described.13 Cell proliferation was assessed by 5-bromo-2′-deoxyuridine (BrdU) immunofluorescence assay, after incubation of cells with 30 μM BrdU during the last hour of treatment. All samples were then harvested, washed in PBS, fixed in 70% ethanol for at least 2 h at −20 °C and processed as previously reported.22 Dose-response and time-course experiments have been performed, treating LF1 cells with DHS or RSV at the different concentrations of 3.75, 7.5, 15, 30 μM for 24 h, and 30 μM for 1, 3, 6, 24 h, before performing BrdU proliferation assay with anti BrdU antibody (Becton Dickinson, 1:50).
2.3. Western blotting and antibodies
Proteins involved in cell cycle progression were investigated by Western blotting, as described previously.22 The primary antibodies used for Western blotting and Cellular Thermal shift assay (CETSA) were the following: anti-SIRT1 mouse monoclonal antibody (Cell Signaling Technology 1:1000), anti-actin (Sigma Aldrich – FLUKA 1:5000), anti-vimentin (Sigma Aldrich - FLUKA 1:1000), anti p53 (DO7, Santa Cruz Biothechnology 1:500), anti p53 acetylated (Cell Signaling Technology, 1:1000), anti-FLAG (Sigma Aldrich 1: 500). Densitometric analyses were performed with the Fiji software.39
2.4. Quantitative real-time RT-PCR
Total RNA was extracted from cultured cells using the RNeasy Mini Kit purification system (Qiagen, Valencia, CA) according to the manufacturer's instructions. The concentration was measured by the Nanodrop 2000 (Thermo Fisher Scientific, Wilmington, DE, USA) and samples were diluted to a final concentration of 100 ng/μl. Quality was assessed by the Bioanalyzer 2100 RNA Nano kit (Agilent Tech, Santa Clara, CA, USA) in order to estimate RNA Integrity Number (RIN).
For each sample, 1 μg RNA was reverse transcribed using the specific High Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA, USA), following manufacturer's recommendation. The yield of retro-transcription was checked by the Bioanalyzer and Nanodrop and samples were diluted to 100 ng cDNA/μl.
For TaqMan-based qPCR expression profiling, 1 μl of each cDNA sample was amplified using TaqMan Gene Expression Assays (20X) for human SIRT1 (Hs0109006_m1, Applied Biosystems) and TaqMan Advanced Master Mix (2X) (Thermofisher Scientific) in 10 μl reactions. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as internal reference and co-amplified with target samples using identical experimental conditions. Real time fluorescence monitoring was performed with the Real Time PCR 7500 Fast (Applied Biosystems). Expression reactions were performed in triplicate.
2.5. Cell transfections and treatments
Plasmid and siRNA transfections were performed using Lipofectamine 2000 (Thermo Fisher Scientific); for SIRT1 overexpression, 4 different plasmids mock (empty plasmid), SIRT1 full length, Δ5 (296–377) and Δ6 (378–458) were used. The plasmids were kindly provided to LZ by Ja-Eun Kim.40 Transfections were carried out following the manufacturers’ instructions. DNA damage was induced by treating cells with Eto (20 μM), MG132 was added 20 min before treatment, at the concentration of 25 μM.
2.6. In silico docking
SIRT1 structure in complex with p53AMC was obtained from rcsb (PDB ID: 5BTR).27 We consider X-ray structure as complex SIRT1-AMC (AMC peptide: ACERHKFDL), and starting from this we obtain the SIRT1 free structure (SIRT1), and model with Pymol mutagenesis tool, the complexes SIRT1-p53 (p53 peptide: RHKALYLMFKT) and SIRT1-p53W (p53W peptide: ACERHKW), as indicated by Hou et al.41 Ligands structures were obtained from PubChem database (Resveratrol (RSV) CID: 445154; 4,4′-Dihydroxystilbene (DHS): 5282363). Docking simulations were performed with AutoDock4,42 using Lamarckian Genetic Algorithm and 100 individuals as population size.
2.7. Cellular thermal shift assay
CETSA was performed as previously described.43 Briefly, cells were treated with 30 μM DHS or RSV for 30 min and 4 h; untreated cells were used as control group. After washing with ice-cold PBS (supplied with Protease Inhibitor Cocktail, Sigma-Aldrich-FLUKA), cells were then aliquoted into PCR tubes (100 μl each) and then incubated at different temperatures, in particular 40–61 °C (for the 4-h treatments) and 52–68 °C (for the 30-min treatments) for 4 min. The cells were then frozen and thawed twice using liquid nitrogen, proteins were isolated after centrifugation and incubated at 70 °C for 10 min for the analysis by Western blot; Biorad system was used (7.5% precasting gel and Trans-Blot Turbo Transfer System). Densitometric analyses were performed with the Fiji software8 using β-actin as internal standard.
2.8. Statistical analysis
At least three biological replicates (unless otherwise stated) were performed for each experiment. Statistical analysis was carried out to calculate significance with the Student t-test (two-tailed), with p values ≤ 0.05 considered to be significant.
3. Results
3.1. Sirt1 protein levels with different stilbenes
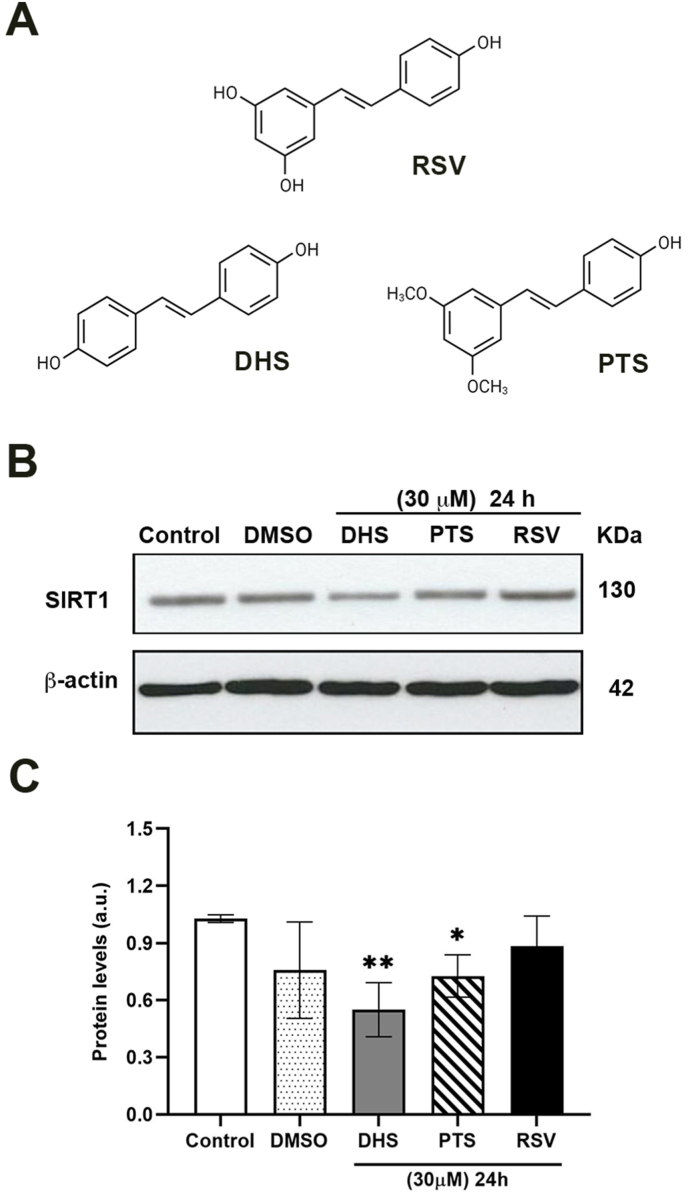
The expression levels of SIRT1 were investigated in LF1 cells after treatment with three different stilbenes, RSV, DHS and PTS (Fig. 1A) at the concentration of 30 μM for 24 h. Unexpectedly, both DHS and PTS are able to reduce SIRT1 (50 and 30%, respectively) as compared to RSV-treated sample, in which the protein levels remain similar to the control and DMSO-treated cells (Fig. 1B and C).
Fig. 1.
(A) Chemical structure of the studied stilbenes. (B) Effects of 24 h-incubation of DHS, PTS and RSV (30 μM) on the SIRT1 protein levels in LF1 cells. (C) Quantification of SIRT1 protein by densitometric analysis of the Western blot and normalized to the internal loading control (β-actin). Data are the mean ± SD from at least three independent experiments; values are expressed as arbitrary units (a.u.). (*p ≤ 0.05 and **p ≤ 0.01 compared to control cells).
3.2. Cytotoxic and antiproliferative effect of DHS
The strongest and significant reduction of SIRT1 induced by DHS prompted us to focus our attention on this compound, evaluating the effects of increasing concentrations and time of incubation on LF1 viability and proliferation. The MTT-based colorimetric assay after 24 h of treatment, with increasing concentrations (in the 1–90 μM range) of DHS, was performed to analyse its cytotoxicity, whereas the quantitative analysis of BrdU incorporation was carried out to study cell proliferation both at different concentrations and different time of treatments.
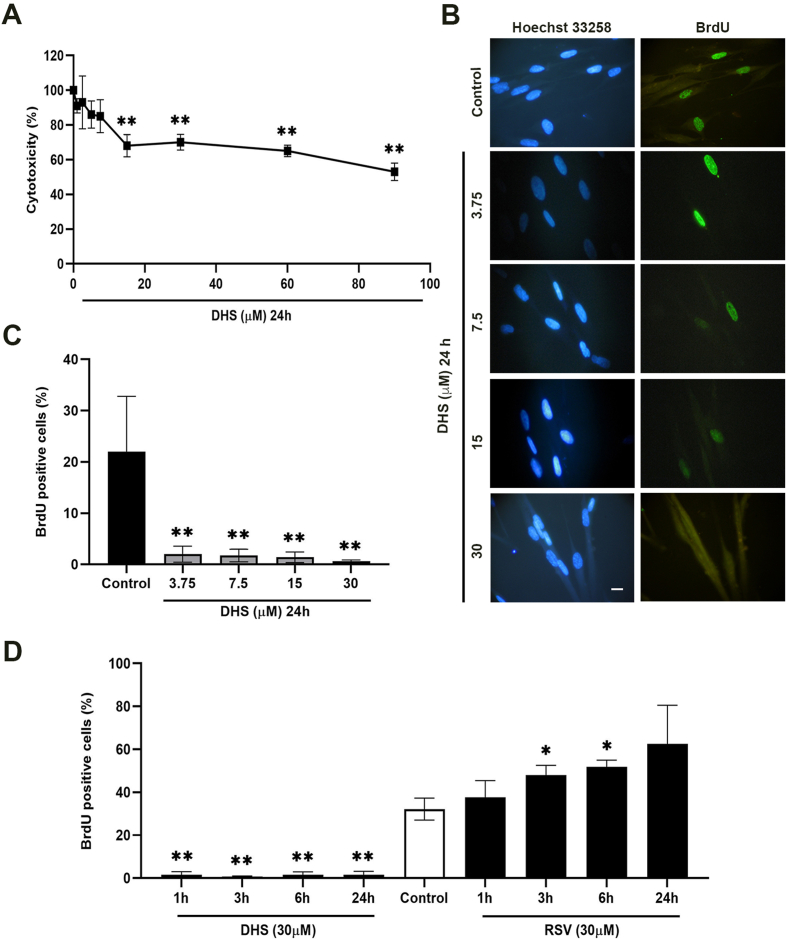
All the concentrations used exert a reduction of viability compared to control cells, from the lowest (10% at 1 μM) to the highest concentration (47% at 90 μM) (Fig. 2A), although data start to be statistically significant at 15 μM concentration (32%, p ≤ 0.01); similar results were previously obtained treating LF1 with RSV, although this stilbene appeared less cytotoxic than DHS.22 We decided to keep the 30 μM dose as the maximum in DHS concentration-effect experiments (range 3.75–30 μM) or as single one in time-course kinetics. Representative images of BrdU immunofluorescence were shown in Fig. 2B, and the corresponding positively labelled cell counts, at different concentrations of treatments, reported in Fig. 2C. This quantitative analysis demonstrated that, already at the lowest concentration (3.75 μM), DHS induces a high significant reduction of BrdU positive cells (90%) compared to the control cells. This inhibition was dose-independent since at 30 μM concentrations a similar percentage (98%) of cells were BrdU-negative (Fig. 2C). In contrast, a dose-dependent increase of RSV-treated LF1 in the S-phase was already demonstrated by Savio et al..22 In the time-response experiment, after 1 h of 30 μM DHS-treatment, BrdU incorporation was highly significant reduced (80%, p ≤ 0.01), and this strong inhibition was maintained until 24 h of treatment (Fig. 2D). RSV confirmed an increase in BrdU positive cells starting from 1 h of 30 μM treatment; this increase is time-dependent, reaching a 55% of BrdU positive cells at 24 h (Fig. 2D).
Fig. 2.
(A) Cell toxicity determined by the MTT assay after 24 h of treatment with different concentrations of DHS (1, 2.5, 5, 7.5, 10, 15, 30, 60 and 90 μM) in LF1 cells. (B) Effect on DNA synthesis evaluated by BrdU incorporation in LF1 cells seeded on coverslips, and treated with increasing concentration of DHS as indicated. Representative images of BrdU-positive cells are showed in the panel (Scale bar = 10 μm). (C) Analysis of BrdU incorporation in LF1 cells treated with different concentration of DHS as indicated. (D) Analysis of BrdU incorporation of 30 μM DHS or RSV at different time (1, 3, 6, 24 h). All results are expressed as mean ± SD from three independent experiments; values are expressed as percentage of BrdU-stained positive cells (*p ≤ 0.05; **p ≤ 0.01 compared to control cells).
3.3. Protein and expression levels of SIRT1 after treatment with DHS
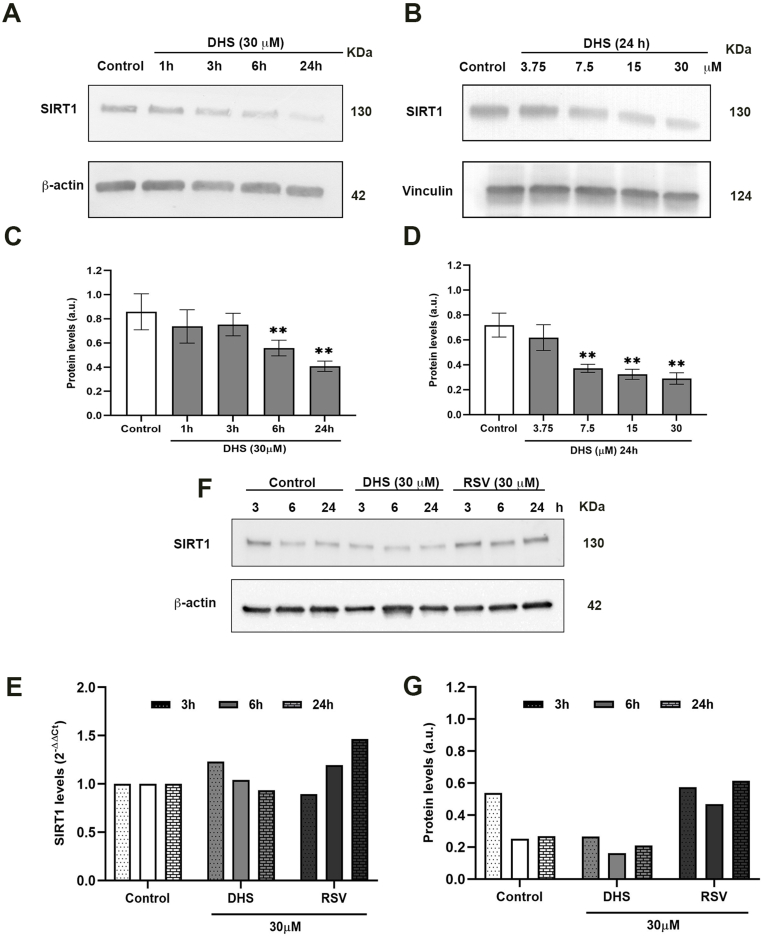
A time-dependent reduction of SIRT1 protein levels was demonstrated in Fig. 3A. Increasing the incubation times with DHS (30 μM), a significant reduction in SIRT1 of about 40% and 60% was observed at 6 and 24 h (p ≤ 0.01), respectively (Fig. 3A and C). Similar trend was observed after incubation with the different concentration of DHS treatment (Fig. 3B and D).
Fig. 3.
(A) Effects of 30 μM of DHS in LF1 cells at different incubation times and (B) at different concentrations for 24 h on SIRT1 protein levels. (C and D) Quantification of SIRT1 protein by densitometric analysis of the Western blot and normalization to the internal loading control (β-actin and vinculin, respectively). Data are the mean ± SD from at least three independent experiments; values are expressed as arbitrary units (a.u.). (**p ≤ 0.01 compared with control cells). (E) Time-dependent analysis of SIRT1transcripts by quantitative real-time PCR (RT-PCR) in LF1 cells treated with 30 μM DHS or RSV at the indicated times. (F) Time dependent analysis of SIRT1 protein levels by Western blot in LF1 cells treated with 30 μM DHS or RSV at the indicated time and (G) the relative densitometric analysis. Both RT-PCR and Western Blot were performed on samples derived from the same experiment.
At the mRNA levels (Fig. 3E), DHS treatment did not significantly modify the SIRT1 expression in comparison to the control cells. Since it is known from the literature that RSV induces the expression and activity of SIRT1,27 we performed PCR experiments also with this compound. As shown, RSV confirms its effect on SIRT1, which is different with that exerted by DHS. These results were also confirmed at protein level, as demonstrated in Fig. 3F and G, in which RSV clearly induces the deacetylase.
3.4. Cell cycle regulatory proteins modification after treatment with DHS
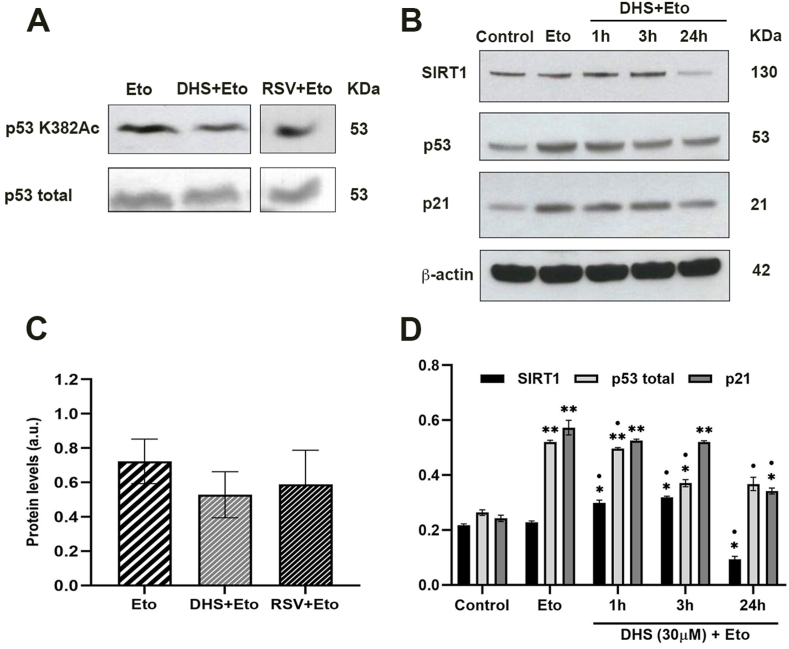
SIRT1 is involved in post translational modifications of p53, such as acetylation which is indispensable for p53 activation. To assess the influence of DHS or RSV treatment on SIRT1 activity, 20 μM Etoposide for 3 h was used to cause DNA damage, which promotes p53 activation through its acetylation by SIRT1. A trend towards a reduction of acetylated p53 was observed only after treatment with DHS for 1 h in comparison with cells treated with Eto (Fig. 4A and C). To confirm and better understand this decrease in p53 protein levels, a kinetic of treatment (1-3-24 h) with DHS 30 μM in presence of Eto was performed. Fig. 4B shows representative image of Western blot of SIRT1, p53 and p21 and their related quantitative analysis (Fig. 4D). A significant induction of SIRT1 was observed after Eto treatment as well as at 1 and 3 h of DHS + Eto (p ≤ 0.05), whereas at 24 h the protein was significantly reduced (p ≤ 0.05). Similarly, p53 was significantly augmented until 3 h in comparison to the control cells, while a time-dependent reduction versus the Eto-sample was clearly detectable. The levels of p21 clearly overlap with those of p53, increasing after DHS + Eto treatment in comparison to control cells, and slightly decreasing when compared to Eto-sample. This is not surprising since it is largely known that p21 transcription is p53-dependent.
Fig. 4.
(A) p53 acetylation levels after treatment with 20 μM Eto (3 h) and in combination 30 μM DHS or RSV (1 h) + 20 μM Eto (3 h) and (C) the relative densitometric analysis. (B) Kinetic experiments on SIRT1, p53 and p21 protein levels in untreated or treated with 20 μM Eto plus 30 μM DHS LF1 cells, and (D) the relative densitometric analysis of the correspondent protein levels. Data are the mean ± SD from at least three independent experiments; values are expressed as arbitrary units (a.u.). (*p ≤ 0.05, **p ≤ 0.01 compared with control cells; • p ≤ 0.05 compared with Eto-treated cells).
3.5. DHS and proteasome inhibition
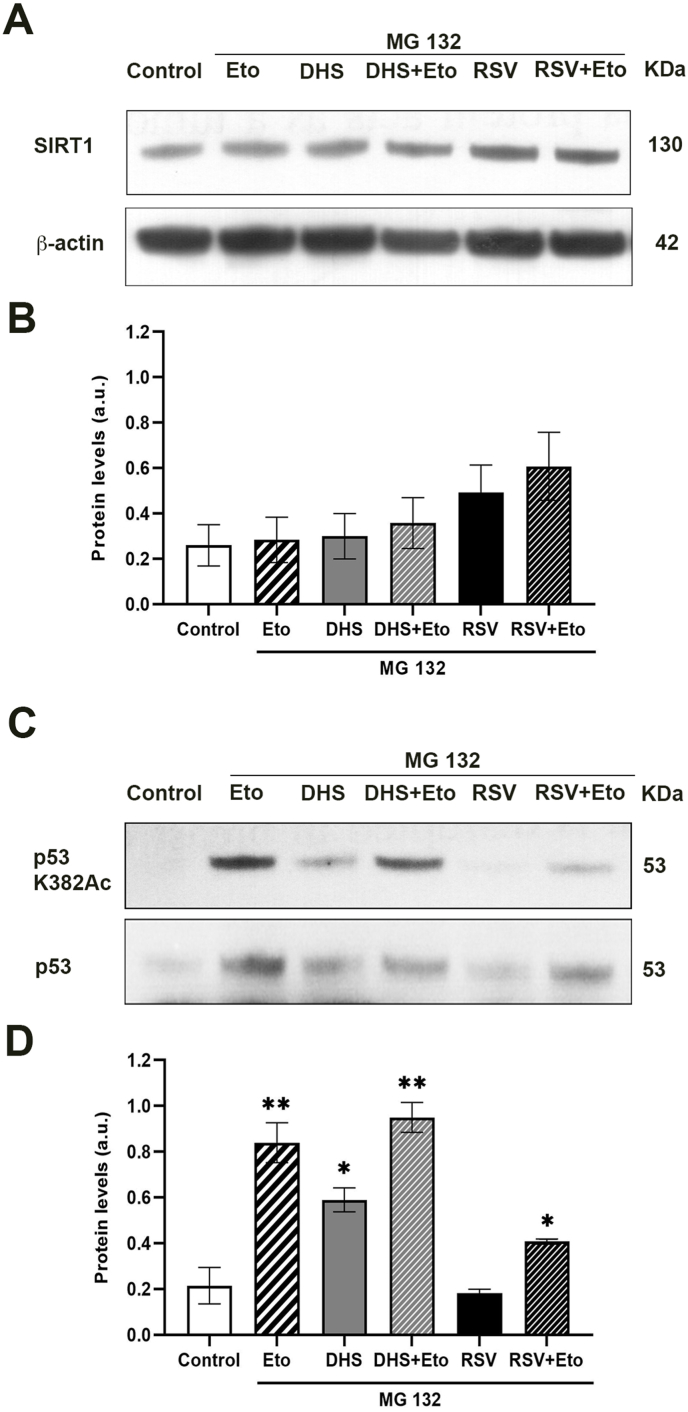
To dig into the mechanism of action underlying SIRT1 reduction and p53 acetylation status in the presence of DHS or RSV, we determined SIRT1 stability by examining the effect of the proteasome inhibitor MG132. LF1 cells were treated with 25 μM MG132 for 30 min before Eto (for 3 h), then with each stilbene (for 24 h). Western blot of SIRT1 protein levels, as shown in Fig. 5A and B, did not change indicating that the reduction of the protein observed with DHS was dependent on its degradation by the proteasome pathway; on the contrary, RSV maintained high levels of the protein. In parallel, in the same experimental conditions, p53-Ac-K382 and the relative quantitative analysis (Fig. 5C and D) revealed, as expected, a significant increase (p ≤ 0.01) of p53 acetylation after treatment with Eto in relation to untreated cells. This increase persisted in cells treated with DHS plus Eto, while the only DHS-treated cells showed well detectable levels of acetylated p53, significantly higher than those of control, suggesting a possible inhibition of SIRT1 activity. RSV did not show any influence in itself p53 acetylation, whereas a significant (p ≤ 0.05) increase at p53-Ac-k382 was measured in the presence of Eto.
Fig. 5.
(A) SIRT1 protein levels obtained by Western blotting and (B) the correspondent quantitative analysis. Data are the mean ± SD from at least three independent experiments; values are expressed as arbitrary units (a.u.). (C) p53 acetylation levels in untreated or pre-treated with MG132 (25 μM) samples in the presence of Eto, DHS, DHS + Eto, RSV and RSV + Eto and (D) the relative densitometric analysis. (*p ≤ 0.05, **p ≤ 0.01 compared with control cells).
3.6. Docking studies
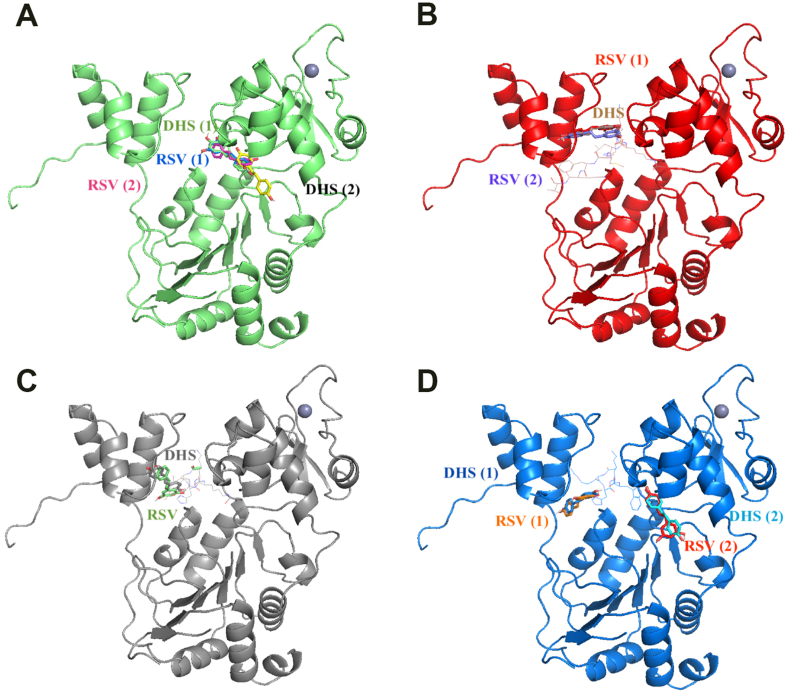
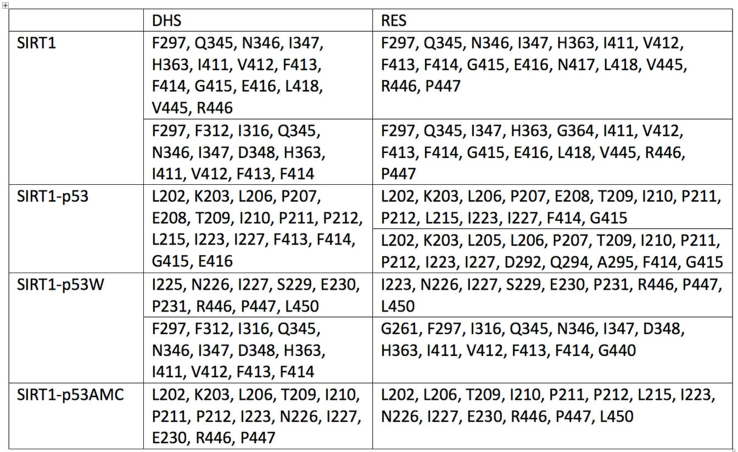
In order to provide further insights into the activity exerted on SIRT1 level by DHS compared to RSV, whose direct interaction with SIRT1 has been published,44 a docking simulation run was performed to evaluate DHS binding and the possible differences compared with the known ligand RSV. We report the docking best results in Table 1. Computational analysis suggests the effect of modified p53 (p53AMC) to increase binding energy of both ligands, while overall resveratrol bind with a higher affinity all SIRT1 complexes compared to DHS, except comparable results for complex SIRT1-p53. Moreover, docked RSV and DHS occupy the same binding cavity in SIRT1, while as SIRT1 is bound to p53, the main binding site is in the near of p53, suggesting a role of this molecule in ligand binding (Fig. 6).
Table 1.
Binding energy and cluster numerosity, in brackets, of docking best results. Results of the second cluster with numerosity comparable to the first one and energy difference lower than 1 kcal/mol, are considered.
| DHS | RES | |
|---|---|---|
| SIRT1 | −7.17 (41), −6.93 (59) | −7.82 (56), −7.49 (22) |
| SIRT1-p53 | −7.88 (87) | −7.79 (64), −7.49 (23) |
| SIRT1-p53W | −7.29 (36), −7.00 (57) | −7.36 (31), −7.17 (29) |
| SIRT1-p53AMC | −7.87 (77) | −8.47 (85) |
Fig. 6.
SIRT1 – RSV/DHS docking pose. SIRT1 is represented in cartoon, RSV and DHS ligands are in licorice and p53 is in lines. (A) SIRT1, (B) SIRT1-p53, (C) SIRT1-p53AMC, (D) SIRT1-p53W.
3.7. DHS and cellular thermal shift assay
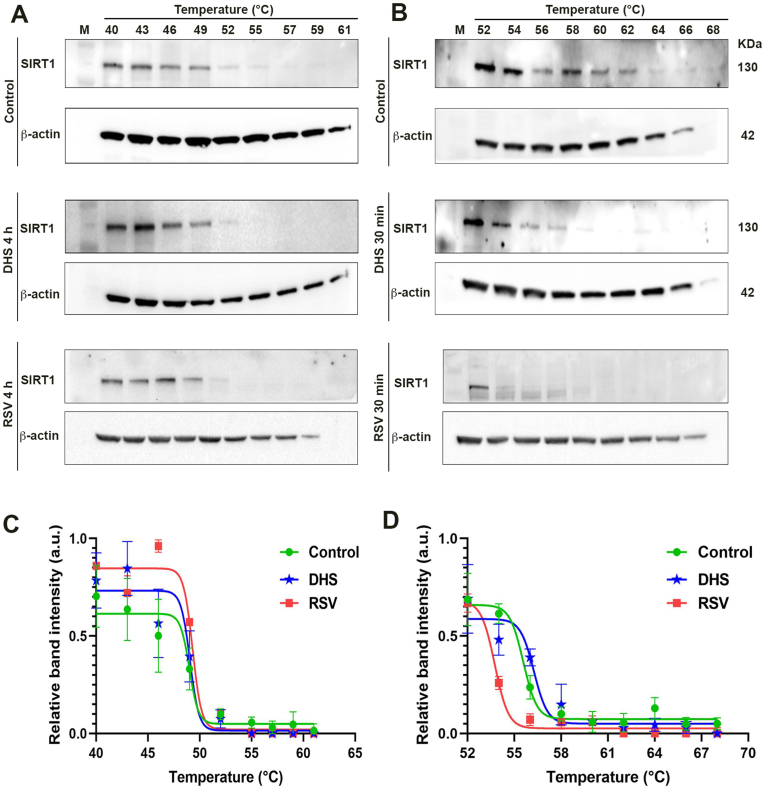
In order to confirm the in silico studies, we carried out the CETSA. This assay is used to study thermal stabilization of proteins upon ligand binding. LF1 cells were treated with 30 μM DHS or RSV for 4 h and processed as reported in material and methods. The results have been obtained as Western blot of SIRT1 using β-actin as internal standard (Fig. 7A). After 4 h of treatment with DHS or RSV, in the range of 40 and 61 °C temperatures, the bands of SIRT1 are detectable until 52 °C for both compounds, as demonstrated in the quantitative analysis in Fig. 7C. In control cells, the SIRT1 levels decreased with the increase of the temperature, but are still evident at 61 °C. These results demonstrated that both stilbenes are not able to stabilize the SIRT1, thereby excluding their direct interaction with the protein. Considering data from the literature according to which DHS is able to stabilize the ribonucleotide reductase after an exposure of 30 min, we decided to repeat these experiments by reducing the incubation time to 30 min. SIRT1 levels (Fig. 7B and D) remain weakly detectable until the 64 °C of temperature in DHS-treated cells, whereas in control sample the signal persists until 66 °C. Compared to DHS, SIRT1 degradation of RSV-treated sample is detectable at lower temperature, in which some bands reactive to the antibody are also evident, probably due to the presence of degraded forms of the protein. These results confirm that also with short time of treatment, both stilbenes are not able to interact directly with the protein.
Fig. 7.
(A and B) DHS or RSV and SIRT1 interaction evaluated by CETSA in untreated LF1 cells or treated with 30 μM DHS or RSV at the indicated times. The levels of non-denaturated protein fractions are normalized to β-actin, and (C and D) the relative densitometric analysis of the correspondent bands. Data are the mean ± SD from at least three independent experiments; values are expressed as arbitrary units (a.u.).
3.8. Analysis of exogenous SIRT1 and its mutant levels
To further investigate DHS-SIRT1 direct interaction, LF1 cells have been transfected with plasmids containing FLAG-tagged full length SIRT1, Δ5 (296−377) and Δ6 (378−458) mutants, which are deleted of the putative binding pockets according to docking analyses (Fig. S1).
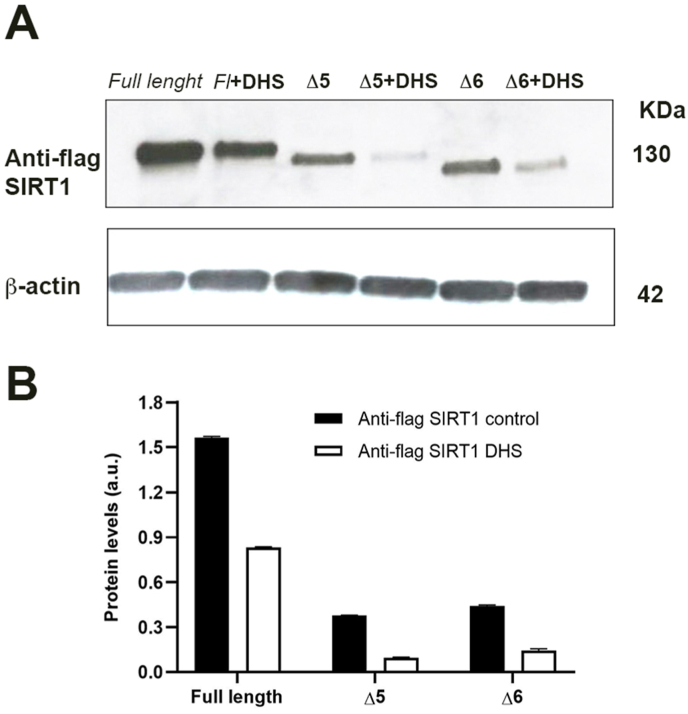
The Flag tag allows to distinguish the exogenous and endogenous target protein, hypothesizing that DHS could interact with the SIRT1 forms. The Flag SIRT1 levels were assessed by Western blot and representative image are shown in Fig. 8A, with the relative quantitative analysis in Fig. 8B. After treatment with 30 μM DHS for 24 h, the full-length protein and its deletions seems to be reduced by about 50%, thereby excluding the involvement of these regions as a potential actor in SIRT1-DHS interaction.
Fig. 8.
(A) Full length (Fl) SIRT1 and its mutant Δ5 e Δ6 in the absence or presence of 30 μM DHS for 24 h and (B) the relative densitometric analysis. Data are the mean ± SD from at least three independent experiments; values are expressed as arbitrary units (a.u.).
4. Discussion
In this study, we have obtained data indicating that the resveratrol analogue DHS is a novel natural inhibitor of SIRT1, downregulating the protein level through a proteasomal dependent degradation. Being RSV a well-known activator of SIRT1, this result is somewhat unexpected since the initial hypothesis, considering the strong similarities between the two stilbenes, was that it could act as an inducer. Sirtuins, NAD+ dependent histone deacetylases, are stress responsive proteins that are considered master regulators of different biological processes including lifespan regulation, cell proliferation, apoptosis and cancer. Recently, emerging evidence suggests an important role of SIRT1 in cancer development, progression and therapeutic resistance45 but its role is still controversial and contradictory; in part it seems to be depend on the tumor type, in part on the stage of cancer (early/advanced disease).45,46 The SIRT1 levels are increased in myeloid acute leukemia and melanoma, whereas are reduced in breast and liver cancers. Interestingly, it has been suggested that SIRT1 activation may be beneficial during the early stage of the cancer growth, whereas its inhibition may be useful for treatment of advanced disease.45
It has been reported that resveratrol is able to activate SIRT127 with an allosteric mechanism,44,47 leading to a conformational change in N-terminal domain that has significant implications for the rational design of new substrate-specific SIRT1 modulators. Other RSV-related compounds, such as piceatannol and polydatin, were demonstrated to be comparable activator of SIRT148, 49, 50; many additional synthetic compounds from different compound families have also been described to have effects on Sirtuins or in biological systems through positively or negatively modulation of Sirtuins. Their effect may involve multiple downstream signalling pathways, which are still debated, involving signal molecules such as Nrf2, p38MAPK, NLPR3 inflammasome, and p53.50 Importantly, acting on SIRT1 means to regulate p53 functions, since it is one of the several deacetylation targets of this enzyme; the tight regulation of p53 allows for p53 to react appropriately to different cellular contexts, including its tumor suppressor activity.51 From our research and several other studies in the literature, DHS has shown a stronger antiproliferative/antitumor activity than that of RSV in both in vitro and in vivo models.15,16,22 Starting from this observation we have investigated the involvement of the histone deacetylase SIRT1 in the mechanism of action underlying the stronger antiproliferative activity of DHS, as compared to RSV. To this end, we used primary cultures of LF1, without p53 alterations, in which the well-established interplay between SIRT1 and this tumor suppressor is supposed to be maintained.52,53
Both resveratrol analogues DHS and PTS efficiently reduced the SIRT1 levels (about 50%), as compared to RSV, but DHS was the more potent one. Thus, we decided to examine in depth this last molecule and its potential interaction with SIRT1 since, despite different studies reported the antiproliferative/anticancer activity of DHS,19,54 no data are available in the literature about the potential involvement of SIRT1 in its activities. In this work, LF1 cells treated with increasing concentrations of DHS (1–90 μM) for 24 h, showed a dose-dependent increase in cytotoxicity, achieving a percentage of 50% at the highest concentration. These results are in agreement with those reported in the literature.19 Whereas, in time course experiments, DHS negatively affected cell proliferation, in a dose-independent manner, being the BrdU incorporation levels already very low after 1 h of treatment; similarly, at 3.75 μM it already induced about 90% of reduction in BrdU incorporation, with very few BrdU staining-positive cells counted at 30 μM. Kinetic experiments from 1 to 24 h of DHS- or RSV-treatment showed different behaviour in cell cycle. As already reported in Savio et al.,22 RSV increases BrdU-positive cells indicating a block of the cells in a different cell-cycle phase compared to DHS.
Kinetic treatments for identifying the best incubation time and concentration of DHS for the analysis of SIRT1, demonstrated an efficient and significant reduction in SIRT1 levels at 6 and 24 h compared to the control sample; while the lowest level of the protein was detected at 30 μM. This reduction was independent on the expression of the protein since no significative influence of DHS was found at the mRNA levels. Whereas, in RSV-treated samples, we still detect an increase of SIRT1 at both mRNA and protein levels.
The analysis of acetylated p53 in LF1 cells, under basal conditions, showed undetectable levels (data not shown), for this reason cells were pre-incubated with Eto for 3 h to promote DNA damage and activation of the p53 pathway. A weak reduction, although not significant, of the acetylated band was observed in the DHS sample, probably due to a minor reduction of the total p53 at 1 h of treatment, as compared to the Eto sample. Adding the proteasome inhibitor MG132, we observed that DHS in itself, but not RSV, induce an increase in the acetylated form of p53, thereby supporting a decrease in SIRT1 activity, according to other SIRT1 inhibitors.55,56 Concomitantly, no reduction in SIRT1 protein level was detected, thus demonstrating that DHS causes SIRT1 reduction by a mechanism dependent on proteasome activity. This result is in agreement from the mechanistic point of view to the inhibition of ribonucleotide reductase regulatory subunit M2 (RRM2) by DHS.19
Considering a potential interaction between DHS and SIRT1, using three distinct p53 derived substrates, p53AMC, p53W and p53, as previously reported for RSV,41 we found that RSV seems to be once again more efficient to bind all SIRT1 complexes compared to DHS, which in turn appears to bind SIRT1 at the same binding cavity of p53. To verify the potential interaction between SIRT1 and DHS, a double approach has been adopted such as the cellular thermal shift assay, and cell transfection with SIRT1 protein mutated in the regions indicated as potential binding pockets by docking.
In our experimental setting, we obtained results at first sight conflicting to our proposed hypothesis of the interaction between DHS and SIRT1. In fact, both after 30 min and 4 h-treatments, DHS was not able to thermal stabilize SIRT1 in the range of the temperature tested, thus confirming no direct interaction among them. A similar result was obtained for the parental compound. However, we cannot rule out the possibility that DHS-SIRT1 or RSV-SIRT1binding is weak, thus non-detectable by CETSA. As second approach, we used the two SIRT1 mutants, in which the amino acids involved in the hypothetical binding only to DHS at the catalytic site of the protein were deleted. By using the antibody (anti-FLAG) discriminating the endogenous protein from the exogenous SIRT1-full length (wt) protein, and the two mutants Δ5 and Δ6, we highlighted how both full-length protein and Δ5 and Δ6 mutant levels decreased by about 50% after treatment with DHS, thus allowing to exclude once again this potential interaction at the defined amino acid sites identified by docking simulation. These data are in agreement with those obtained by CETSA, despite the performed docking analysis. At now, only the SIRT1 catalytic domain has been solved, therefore it cannot be excluded that there are other crucial interactions in different sites of the enzyme which confer the same activity. Finally, it cannot be ruled out an interference given by the presence of the endogenous SIRT1 on the transfected samples.
In conclusion, our work further demonstrates that the mechanism of action of the two stilbenes involves different targets, not only in inhibiting cell proliferation but also in modulating SIRT1 level and activity. In fact, it can be stated that DHS, but not RSV, reduces SIRT1 protein level, thereby inhibiting its activity through a proteasome-mediated degradation. Further studies are needed to better clarify the involvement of SIRT1 in the antiproliferative mechanism of DHS.
Funding
This research was funded by a grant from the Ministero dell’Università e della Ricerca (MUR) to the Department of Molecular Medicine of the University of Pavia under the initiative ‘Dipartimenti di Eccellenza (2018–2022)’.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgments
We are very grateful to Prof. Cristian Turato for his help in performing RT-PCR analysis.
Footnotes
Peer review under responsibility of The Center for Food and Biomolecules, National Taiwan University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jtcme.2024.03.001.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Fig. S1.
Binding pockets as resulted by the in silico docking.
References
- 1.Bradamante S., Barenghi L., Villa A. Cardiovascular protective effects of resveratrol. Cardiovasc Drug Rev. 2004;22:169–188. doi: 10.1111/j.1527-3466.2004.tb00139.x. [DOI] [PubMed] [Google Scholar]
- 2.Soleas G.J., Diamandis E.P., Goldberg D.M. The world of resveratrol. Adv Exp Med Biol. 2001;492:159–182. doi: 10.1007/978-1-4615-1283-7_13. [DOI] [PubMed] [Google Scholar]
- 3.Yadav E., Yadav P., Khan M.M.U., Singh H., Verma A. Resveratrol: a potential therapeutic natural polyphenol for neurodegenerative diseases associated with mitochondrial dysfunction. Front Pharmacol. 2022;13 doi: 10.3389/fphar.2022.922232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hazafa A., Iqbal M.O., Javaid U., et al. Inhibitory effect of polyphenols (phenolic acids, lignans, and stilbenes) on cancer by regulating signal transduction pathways: a review. Clin Transl Oncol. 2022;24:432–445. doi: 10.1007/s12094-021-02709-3. [DOI] [PubMed] [Google Scholar]
- 5.Kang J.H., Park Y.H., Choi S.W., Yang E.K., Lee W.J. Resveratrol derivatives potently induce apoptosis in human promyelocytic leukemia cells. Exp Mol Med. 2003;35:467–474. doi: 10.1038/emm.2003.61. [DOI] [PubMed] [Google Scholar]
- 6.Lappano R., Rosano C., Madeo A., et al. Structure-activity relationships of resveratrol and derivatives in breast cancer cells. Mol Nutr Food Res. 2009;53:845–858. doi: 10.1002/mnfr.200800331. [DOI] [PubMed] [Google Scholar]
- 7.Ciccone L., Piragine E., Brogi S., et al. Resveratrol-like compounds as SIRT1 activators. Int J Mol Sci. 2022;23 doi: 10.3390/ijms232315105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fang J.G., Lu M., Chen Z.H., et al. Antioxidant effects of resveratrol and its analogues against the free-radical-induced peroxidation of linoleic acid in micelles. Chemistry. 2002;8:4191–4198. doi: 10.1002/1521-3765(20020916)8:18<4191::AID-CHEM4191>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 9.Cai Y.J., Fang J.G., Ma L.P., Yang L., Liu Z.L. Inhibition of free radical-induced peroxidation of rat liver microsomes by resveratrol and its analogues. Biochim Biophys Acta. 2003;1637:31–38. doi: 10.1016/s0925-4439(02)00174-6. [DOI] [PubMed] [Google Scholar]
- 10.Stivala L.A., Savio M., Carafoli F., et al. Specific structural determinants are responsible for the antioxidant activity and the cell cycle effects of resveratrol. J Biol Chem. 2001;276:22586–22594. doi: 10.1074/jbc.M101846200. [DOI] [PubMed] [Google Scholar]
- 11.Fan G.J., Liu X.D., Qian Y.P., et al. 4,4'-Dihydroxy-trans-stilbene, a resveratrol analogue, exhibited enhanced antioxidant activity and cytotoxicity. Bioorg Med Chem. 2009;17:2360–2365. doi: 10.1016/j.bmc.2009.02.014. [DOI] [PubMed] [Google Scholar]
- 12.Wang T., Dai F., Li G.H., et al. Trans-4,4'-dihydroxystilbene ameliorates cigarette smoke-induced progression of chronic obstructive pulmonary disease via inhibiting oxidative stress and inflammatory response. Free Radic Biol Med. 2020;152:525–539. doi: 10.1016/j.freeradbiomed.2019.11.026. [DOI] [PubMed] [Google Scholar]
- 13.Maccario C., Savio M., Ferraro D., et al. The resveratrol analog 4,4'-dihydroxy-trans-stilbene suppresses transformation in normal mouse fibroblasts and inhibits proliferation and invasion of human breast cancer cells. Carcinogenesis. 2012;33:2172–2180. doi: 10.1093/carcin/bgs244. [DOI] [PubMed] [Google Scholar]
- 14.Savio M., Ferraro D., Maccario C., et al. Resveratrol analogue 4,4'-dihydroxy-trans-stilbene potently inhibits cancer invasion and metastasis. Sci Rep. 2016;6 doi: 10.1038/srep19973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Savio M., Ferraresi A., Corpina C., et al. Resveratrol and its analogue 4,4'-Dihydroxy-trans-stilbene inhibit lewis lung carcinoma growth in vivo through apoptosis, autophagy and modulation of the tumour microenvironment in a murine model. Biomedicines. 2022;10 doi: 10.3390/biomedicines10081784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kimura Y., Sumiyoshi M., Baba K. Antitumor activities of synthetic and natural stilbenes through antiangiogenic action. Cancer Sci. 2008;99:2083–2096. doi: 10.1111/j.1349-7006.2008.00948.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saha B., Patro B.S., Koli M., et al. -4,4'-Dihydroxystilbene (DHS) inhibits human neuroblastoma tumor growth and induces mitochondrial and lysosomal damages in neuroblastoma cell lines. Oncotarget. 2017;8:73905–73924. doi: 10.18632/oncotarget.17879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fontecave M., Lepoivre M., Elleingand E., Gerez C., Guittet O. Resveratrol, a remarkable inhibitor of ribonucleotide reductase. FEBS Lett. 1998;421:277–279. doi: 10.1016/s0014-5793(97)01572-x. [DOI] [PubMed] [Google Scholar]
- 19.Chen C.W., Li Y., Hu S., et al. DHS (trans-4,4'-dihydroxystilbene) suppresses DNA replication and tumor growth by inhibiting RRM2 (ribonucleotide reductase regulatory subunit M2) Oncogene. 2019;38:2364–2379. doi: 10.1038/s41388-018-0584-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun N.J., Woo S.H., Cassady J.M., Snapka R.M. DNA polymerase and topoisomerase II inhibitors from Psoralea corylifolia. J Nat Prod. 1998;61:362–366. doi: 10.1021/np970488q. [DOI] [PubMed] [Google Scholar]
- 21.Locatelli G.A., Savio M., Forti L., et al. Inhibition of mammalian DNA polymerases by resveratrol: mechanism and structural determinants. Biochem J. 2005;389:259–268. doi: 10.1042/BJ20050094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Savio M., Coppa T., Bianchi L., et al. The resveratrol analogue 4,4'-dihydroxy-trans-stilbene inhibits cell proliferation with higher efficiency but different mechanism from resveratrol. Int J Biochem Cell Biol. 2009;41:2493–2502. doi: 10.1016/j.biocel.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 23.Li W., Ma J., Ma Q., et al. Resveratrol inhibits the epithelial-mesenchymal transition of pancreatic cancer cells via suppression of the PI-3K/Akt/NF-κB pathway. Curr Med Chem. 2013;20:4185–4194. doi: 10.2174/09298673113209990251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nawaz W., Zhou Z., Deng S., Ma X., Li C., Shu X. Therapeutic versatility of resveratrol derivatives. Nutrients. 2017;9 doi: 10.3390/nu9111188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walle T., Hsieh F., DeLegge M.H., Oatis J.E., Walle U.K. High absorption but very low bioavailability of oral resveratrol in humans. Drug Metab Dispos. 2004;32:1377–1382. doi: 10.1124/dmd.104.000885. [DOI] [PubMed] [Google Scholar]
- 26.Chen W., Yeo S.C., Elhennawy M.G., Xiang X., Lin H.S. Determination of naturally occurring resveratrol analog trans-4,4'-dihydroxystilbene in rat plasma by liquid chromatography-tandem mass spectrometry: application to a pharmacokinetic study. Anal Bioanal Chem. 2015;407:5793–5801. doi: 10.1007/s00216-015-8762-7. [DOI] [PubMed] [Google Scholar]
- 27.Howitz K.T., Bitterman K.J., Cohen H.Y., et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 28.Lagouge M., Argmann C., Gerhart-Hines Z., et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 29.Milne J.C., Lambert P.D., Schenk S., et al. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature. 2007;450:712–716. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malaguarnera L. Influence of resveratrol on the immune response. Nutrients. 2019;11 doi: 10.3390/nu11050946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iside C., Scafuro M., Nebbioso A., Altucci L. SIRT1 activation by natural phytochemicals: an overview. Front Pharmacol. 2020;11:1225. doi: 10.3389/fphar.2020.01225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu Q.J., Zhang T.N., Chen H.H., et al. The sirtuin family in health and disease. Signal Transduct Targeted Ther. 2022;7:402. doi: 10.1038/s41392-022-01257-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guarente L. Calorie restriction and SIR2 genes--towards a mechanism. Mech Ageing Dev. 2005;126:923–928. doi: 10.1016/j.mad.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 34.Haigis M.C., Sinclair D.A. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ungurianu A., Zanfirescu A., Margină D. Sirtuins, resveratrol and the intertwining cellular pathways connecting them. Ageing Res Rev. 2023;88 doi: 10.1016/j.arr.2023.101936. [DOI] [PubMed] [Google Scholar]
- 36.Patra S., Panigrahi D.P., Praharaj P.P., et al. Dysregulation of histone deacetylases in carcinogenesis and tumor progression: a possible link to apoptosis and autophagy. Cell Mol Life Sci. 2019;76:3263–3282. doi: 10.1007/s00018-019-03098-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patra S., Praharaj P.P., Singh A., Bhutia S.K. Targeting SIRT1-regulated autophagic cell death as a novel therapeutic avenue for cancer prevention. Drug Discov Today. 2023 doi: 10.1016/j.drudis.2023.103692. [DOI] [PubMed] [Google Scholar]
- 38.Lee I.H., Cao L., Mostoslavsky R., et al. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci U S A. 2008;105:3374–3379. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schindelin J., Arganda-Carreras I., Frise E., et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim J.E., Chen J., Lou Z. DBC1 is a negative regulator of SIRT1. Nature. 2008;451:583–586. doi: 10.1038/nature06500. [DOI] [PubMed] [Google Scholar]
- 41.Hou X., Rooklin D., Fang H., Zhang Y. Resveratrol serves as a protein-substrate interaction stabilizer in human SIRT1 activation. Sci Rep. 2016;6 doi: 10.1038/srep38186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morris G.M., Huey R., Lindstrom W., et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jafari R., Almqvist H., Axelsson H., et al. The cellular thermal shift assay for evaluating drug target interactions in cells. Nat Protoc. 2014;9:2100–2122. doi: 10.1038/nprot.2014.138. [DOI] [PubMed] [Google Scholar]
- 44.Cao D., Wang M., Qiu X., et al. Structural basis for allosteric, substrate-dependent stimulation of SIRT1 activity by resveratrol. Genes Dev. 2015;29:1316–1325. doi: 10.1101/gad.265462.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang S.B., Rivas P., Yang X., et al. SIRT1 inhibition-induced senescence as a strategy to prevent prostate cancer progression. Mol Carcinog. 2022;61:702–716. doi: 10.1002/mc.23412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang R.H., Zheng Y., Kim H.S., et al. Interplay among BRCA1, SIRT1, and Survivin during BRCA1-associated tumorigenesis. Mol Cell. 2008;32:11–20. doi: 10.1016/j.molcel.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hubbard B.P., Gomes A.P., Dai H., et al. Evidence for a common mechanism of SIRT1 regulation by allosteric activators. Science. 2013;339:1216–1219. doi: 10.1126/science.1231097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang H., Baur J.A., Chen A., et al. Design and synthesis of compounds that extend yeast replicative lifespan. Aging Cell. 2007;6:35–43. doi: 10.1111/j.1474-9726.2006.00259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dai H., Sinclair D.A., Ellis J.L., Steegborn C. Sirtuin activators and inhibitors: promises, achievements, and challenges. Pharmacol Ther. 2018;188:140–154. doi: 10.1016/j.pharmthera.2018.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun Z., Wang X., Xu Z. SIRT1 provides new pharmacological targets for polydatin through its role as a metabolic sensor. Biomed Pharmacother. 2021;139 doi: 10.1016/j.biopha.2021.111549. [DOI] [PubMed] [Google Scholar]
- 51.Lee J.T., Gu W. SIRT1: regulator of p53 deacetylation. Genes Cancer. 2013;4:112–117. doi: 10.1177/1947601913484496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gonfloni S., Iannizzotto V., Maiani E., Bellusci G., Ciccone S., Diederich M. P53 and Sirt1: routes of metabolism and genome stability. Biochem Pharmacol. 2014;92:149–156. doi: 10.1016/j.bcp.2014.08.034. [DOI] [PubMed] [Google Scholar]
- 53.Yi J., Luo J. SIRT1 and p53, effect on cancer, senescence and beyond. Biochim Biophys Acta. 2010;1804:1684–1689. doi: 10.1016/j.bbapap.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saha B., Pai G.B., Subramanian M., et al. Resveratrol analogue, trans-4,4'-dihydroxystilbene (DHS), inhibits melanoma tumor growth and suppresses its metastatic colonization in lungs. Biomed Pharmacother. 2018;107:1104–1114. doi: 10.1016/j.biopha.2018.08.085. [DOI] [PubMed] [Google Scholar]
- 55.Magni M., Buscemi G., Maita L., et al. TSPYL2 is a novel regulator of SIRT1 and p300 activity in response to DNA damage. Cell Death Differ. 2019;26:918–931. doi: 10.1038/s41418-018-0168-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chao S.C., Chen Y.J., Huang K.H., et al. Induction of sirtuin-1 signaling by resveratrol induces human chondrosarcoma cell apoptosis and exhibits antitumor activity. Sci Rep. 2017;7:3180. doi: 10.1038/s41598-017-03635-7. [DOI] [PMC free article] [PubMed] [Google Scholar]