Abstract
We present a genome assembly from a Linaria vulgaris specimen (common toadflax; Streptophyta; Magnoliopsida; Lamiales; Plantaginaceae). The genome sequence is 760.5 megabases in span. Most of the assembly is scaffolded into six chromosomal pseudomolecules. Two mitochondrial genomes were assembled, which were 330.8 and 144.0 kilobases long. The plastid genome was also assembled and is 156.7 kilobases in length.
Keywords: Linaria vulgaris, common toadflax, genome sequence, chromosomal, Lamiales
Species taxonomy
Eukaryota; Viridiplantae; Streptophyta; Embryophyta; Tracheophyta; Spermatophyta; Magnoliopsida; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Plantaginaceae; Antirrhineae; Linaria; Linaria vulgaris Mill. (NCBI:txid43171).
Background
Common toadflax, Linaria vulgaris Mill. (Plantaginaceae; 2 n = 2 x = 12 ( Henniges et al., 2022), is a short-lived herbaceous perennial plant. It is native to most of Europe to North and Central Asia, the Himalayas and eastern China ( POWO, 2022), and was introduced to South Africa, northeast Asia and the temperate Americas, where it can be highly invasive ( Sing & Peterson, 2011). It is widely distributed throughout Britain and Ireland, although it is less common in northern Scotland and western Ireland ( Preston et al., 2002; Stace et al., 2019). Linaria vulgaris avoids acid soils but tolerates a wide range of habitats, such as open meadows and disturbed roadsides ( Sing & Peterson, 2011; Stace et al., 2019). This species readily hybridises with other members of its genus ( Stace et al., 2015; Ward et al., 2009). One of the best-studied Linaria hybrids in Britain and Ireland is L. vulgaris × repens, which is highly fertile and able to backcross with both parental species ( Druce, 1896; Stace et al., 2015).
The foliage superficially resembles that of the flax ( Linum L.), whence the names ‘toadflax’ and ‘ Linaria’ derive ( Gledhill, 2008). The zygomorphic flowers, borne on terminal racemes, are pale-yellow with orange centres ( Figure 1A, B). Each flower contains a tubular nectar spur, which holds a reservoir of nectar that is accessible to long-tongued bumblebee pollinators ( Newman & Thomson, 2005) ( Figure 1C). Linaria vulgaris reproduces sexually, via bee-mediated pollination, and asexually, via vegetative propagation from adventitious buds which develop on the roots ( Bakshi & Coupland, 1960). Seeds are dust-like and dispersed by wind.
Figure 1. Flowers of Linaria vulgaris (not the sampled specimen).

A. Flowers are borne on terminal racemes. B. Flowers are lobed and bilaterally symmetrical. C. Each flower possesses a single nectar spur. D Photograph of Linaria vulgaris expressing YFP following Agrobacterium-mediated transformation.
Linaria vulgaris has been of particular interest to botanists for over 250 years, since Linnaeus described mutant toadflax flowers that displayed radial, instead of the typical bilateral, symmetry ( Gustafsson, 1979; Linneaus, 1749). Later characterisation of these mutant flowers led to the first description of naturally occurring epigenetic mutations (epimutations) ( Cubas et al., 1999). It is currently being developed as a model system for studying nectar spur development and floral evolution ( Cullen et al., 2018), and has recently been genetically transformed via Agrobacterium-mediated transformation ( Figure 1D).
We hope that the sequence provided here will contribute to the study of Linaria vulgaris as an emerging model for understanding the genetics of floral evolution and flower development. This whole genome sequence will also facilitate population genetic studies of natural toadflax populations.
Genome sequence report
The genome was sequenced from a specimen of Linaria vulgaris ( Figure 1) collected from along the River Thames towpath in Kingston upon Thames, Surrey, UK (latitude 51.43, longitude –0.31). Using flow cytometry, the genome size (1C-value) was estimated to be 0.96 pg, equivalent to 940 Mb. A total of 28-fold coverage in Pacific Biosciences single-molecule HiFi long reads was generated. Primary assembly contigs were scaffolded with chromosome conformation Hi-C data. Manual assembly curation corrected 69 missing joins or mis-joins and removed 58 haplotypic duplications, reducing the assembly length by 3.2% and the scaffold number by 59.8%, and decreasing the scaffold N50 by 17.75%.
The final assembly has a total length of 760.5 Mb in 41 sequence scaffolds with a scaffold N50 of 127.5 Mb ( Table 1). Most (99.62%) of the assembly sequence was assigned to six chromosomal-level scaffolds. Chromosome-scale scaffolds confirmed by the Hi-C data are named in order of size ( Figure 2– Figure 5; Table 2). While not fully phased, the assembly deposited is of one haplotype. Contigs corresponding to the second haplotype have also been deposited.
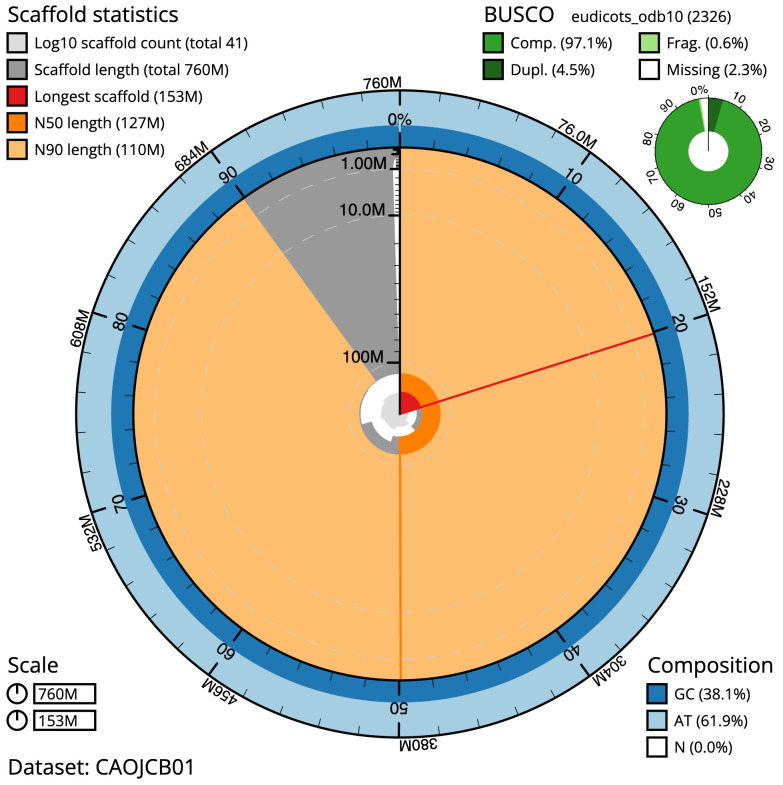
Figure 2. Genome assembly of Linaria vulgaris, daLinVulg1.1: metrics.
The BlobToolKit Snailplot shows N50 metrics and BUSCO gene completeness. The main plot is divided into 1,000 size-ordered bins around the circumference with each bin representing 0.1% of the 760,456,579 bp assembly. The distribution of scaffold lengths is shown in dark grey with the plot radius scaled to the longest scaffold present in the assembly (153,338,251 bp, shown in red). Orange and pale-orange arcs show the N50 and N90 scaffold lengths (127,462,361 and 110,092,459 bp), respectively. The pale grey spiral shows the cumulative scaffold count on a log scale with white scale lines showing successive orders of magnitude. The blue and pale-blue area around the outside of the plot shows the distribution of GC, AT and N percentages in the same bins as the inner plot. A summary of complete, fragmented, duplicated and missing BUSCO genes in the eudicots_odb10 set is shown in the top right. An interactive version of this figure is available at https://blobtoolkit.genomehubs.org/view/daLinVulg1.1/dataset/CAOJCB01/snail.
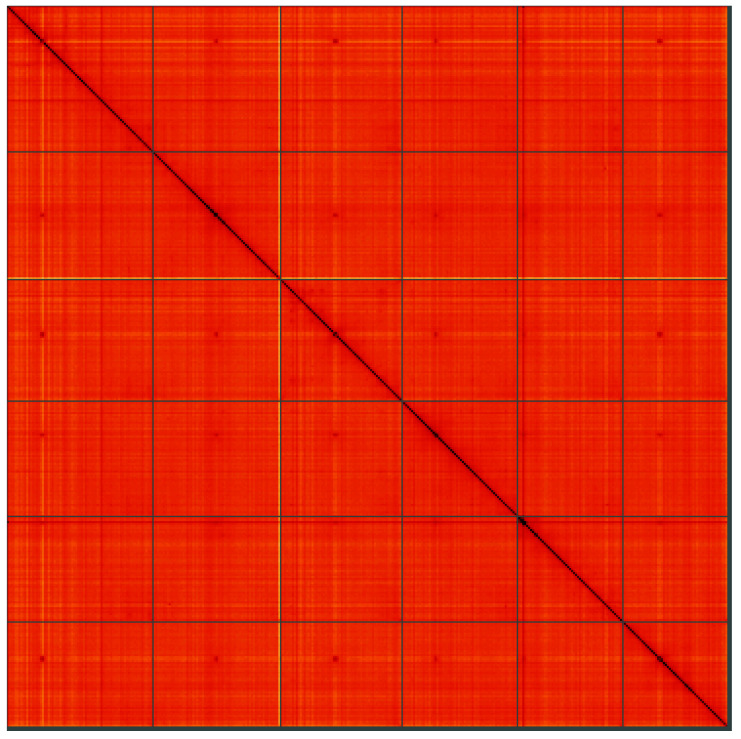
Figure 5. Genome assembly of Linaria vulgaris, daLinVulg1.1: Hi-C contact map.
Hi-C contact map of the daLinVulg1.1 assembly, visualised using HiGlass. Chromosomes are shown in order of size from left to right and top to bottom. An interactive version of this figure may be viewed at https://genome-note-higlass.tol.sanger.ac.uk/l/?d=Lroe3SmHRWOiVBKAhWt8bQ.
Table 1. Genome data for Linaria vulgaris, daLinVulg1.1.
| Project accession data | ||
|---|---|---|
| Assembly identifier | daLinVulg1.1 | |
| Species | Linaria vulgaris | |
| Specimen | daLinVulg1 | |
| NCBI taxonomy ID | 43171 | |
| BioProject | PRJEB50878 | |
| BioSample ID | SAMEA7522288 | |
| Isolate information | daLinVulg1; leaf (genome sequencing and Hi-C scaffolding); flower
(RNA sequencing) |
|
| Assembly metrics * | Benchmark | |
| Consensus quality (QV) | 61.8 | ≥ 50 |
| k-mer completeness | 100% | ≥ 95% |
| BUSCO ** | C:97.1%[S:92.6%,D:4.5%],
F:0.6%,M:2.3%,n:2,326 |
C ≥ 95% |
| Percentage of assembly mapped to
chromosomes |
99.62% | ≥ 95% |
| Sex chromosomes | Not applicable | localised homologous pairs |
| Organelles | Two mitochondrial scaffolds, one
chloroplast scaffold |
complete single alleles |
| Raw data accessions | ||
| PacificBiosciences SEQUEL II | ERR8705859, ERR8705860 | |
| Hi-C Illumina | ERR8702786 | |
| PolyA RNA-Seq Illumina | ERR10378004, ERR10378005 | |
| Genome assembly | ||
| Assembly accession | GCA_948329865.1 | |
| Accession of alternate haplotype | GCA_948329855.1 | |
| Span (Mb) | 760.5 | |
| Number of contigs | 132 | |
| Contig N50 length (Mb) | 12.7 | |
| Number of scaffolds | 41 | |
| Scaffold N50 length (Mb) | 127.5 | |
| Longest scaffold (Mb) | 153.3 | |
* Assembly metric benchmarks are adapted from column VGP-2020 of “Table 1: Proposed standards and metrics for defining genome assembly quality” from ( Rhie et al., 2021).
** BUSCO scores based on the eudicots_odb10 BUSCO set using v5.3.2. C = complete [S = single copy, D = duplicated], F = fragmented, M = missing, n = number of orthologues in comparison. A full set of BUSCO scores is available at https://blobtoolkit.genomehubs.org/view/daLinVulg1.1/dataset/CAOJCB01/busco.
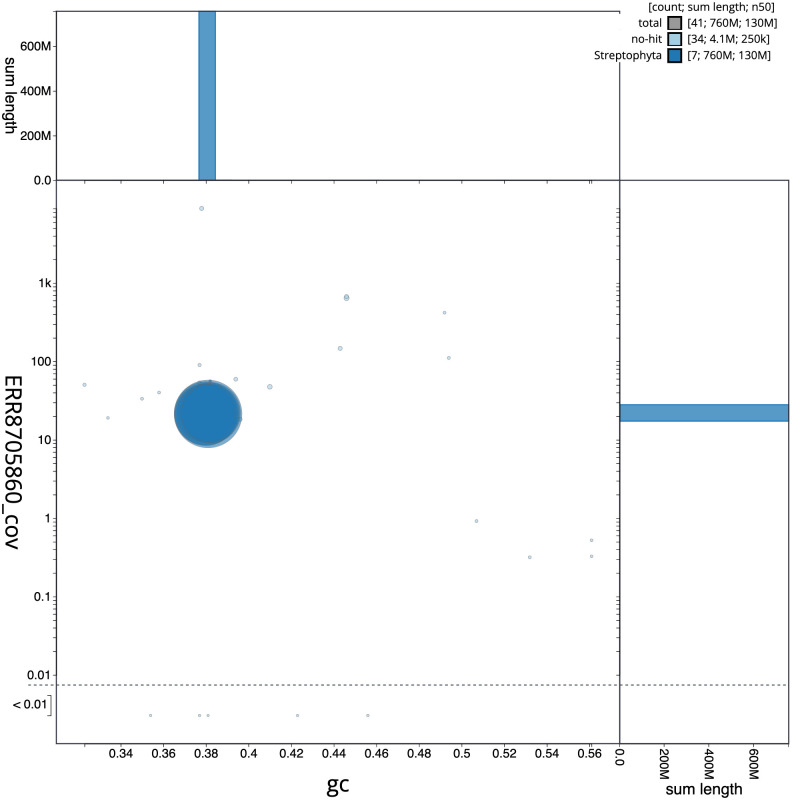
Figure 3. Genome assembly of Linaria vulgaris, daLinVulg1.1: GC coverage.
BlobToolKit GC-coverage plot. Scaffolds are coloured by phylum. Circles are sized in proportion to scaffold length. Histograms show the distribution of scaffold length sum along each axis. An interactive version of this figure is available at https://blobtoolkit.genomehubs.org/view/daLinVulg1.1/dataset/CAOJCB01/blob.
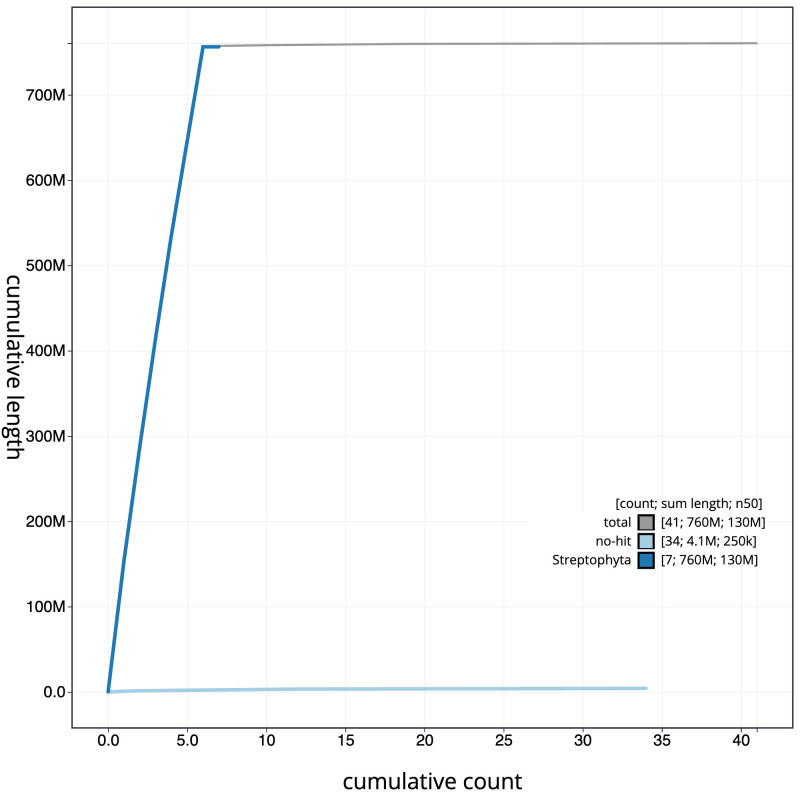
Figure 4. Genome assembly of Linaria vulgaris, daLinVulg1.1: cumulative sequence.
BlobToolKit cumulative sequence plot. The grey line shows cumulative length for all scaffolds. Coloured lines show cumulative lengths of scaffolds assigned to each phylum using the buscogenes taxrule. An interactive version of this figure is available at https://blobtoolkit.genomehubs.org/view/daLinVulg1.1/dataset/CAOJCB01/cumulative.
Table 2. Chromosomal pseudomolecules in the genome assembly of Linaria vulgaris, daLinVulg1.
| INSDC accession | Chromosome | Size (Mb) | GC% |
|---|---|---|---|
| OX415249.1 | 1 | 153.34 | 38.1 |
| OX415250.1 | 2 | 133.81 | 38 |
| OX415251.1 | 3 | 127.46 | 38 |
| OX415252.1 | 4 | 120.92 | 38.2 |
| OX415253.1 | 5 | 110.69 | 38 |
| OX415254.1 | 6 | 110.09 | 38.2 |
| OX415255.1 | MT1 | 0.33 | 44.5 |
| OX415256.1 | MT2 | 0.14 | 44.6 |
| OX415257.1 | Pltd | 0.16 | 37.8 |
| - | unplaced | 3.51 | 39.6 |
The estimated Quality Value (QV) of the final assembly is 61.8 with k-mer completeness of 100%, and the assembly has a BUSCO v5.3.2 completeness of 97.1% (single = 92.6%, duplicated = 4.5%), using the eudicots_odb10 reference set ( n = 2,326).
Metadata for specimens, spectral estimates, sequencing runs, contaminants and pre-curation assembly statistics can be found at https://links.tol.sanger.ac.uk/species/43171.
Methods
Sample acquisition, genome size estimation and nucleic acid extraction
A Linaria vulgaris specimen (daLinVulg1) was collected from Kingston upon Thames, Surrey, UK (latitude 51.43, longitude –0.31) on 1 September 2020. The specimen was picked by hand by Maarten Christenhusz (Royal Botanic Gardens, Kew) from riparian vegetation on sand along the River Thames towpath. The specimen was identified based on its morphology by Maarten Christenhusz, and was preserved by freezing at –80°C.
The genome size was estimated by flow cytometry using the fluorochrome propidium iodide and following the ‘one-step’ method as outlined in Pellicer et al. (2021). Specifically for this species, the General Purpose Buffer (GPB) supplemented with 3% PVP and 0.08% (v/v) beta-mercaptoethanol was used for isolation of nuclei ( Loureiro et al., 2007), and the internal calibration standard was Petroselinum crispum ‘Champion Moss Curled’ with an assumed 1C-value of 2,200 Mb ( Obermayer et al., 2002).
DNA was extracted at the Tree of Life laboratory, Wellcome Sanger Institute (WSI). The daLinVulg1 sample was weighed and dissected on dry ice with tissue set aside for Hi-C sequencing. Leaf tissue was cryogenically disrupted to a fine powder using a Covaris cryoPREP Automated Dry Pulveriser, receiving multiple impacts. High molecular weight (HMW) DNA was extracted using the Illustra Nucleon PhytoPure HMW DNA extraction kit. HMW DNA was sheared into an average fragment size of 12–20 kb in a Megaruptor 3 system with speed setting 30. Sheared DNA was purified by solid-phase reversible immobilisation using AMPure PB beads with a 1.8× ratio of beads to sample to remove the shorter fragments and concentrate the DNA sample. The concentration of the sheared and purified DNA was assessed using a Nanodrop spectrophotometer and Qubit Fluorometer and Qubit dsDNA High Sensitivity Assay kit. Fragment size distribution was evaluated by running the sample on the FemtoPulse system.
RNA was extracted from flower tissue of daLinVulg1 in the Tree of Life Laboratory at the WSI using TRIzol, according to the manufacturer’s instructions. RNA was then eluted in 50 μl RNAse-free water and its concentration assessed using a Nanodrop spectrophotometer and Qubit Fluorometer using the Qubit RNA Broad-Range (BR) Assay kit. Analysis of the integrity of the RNA was done using the Agilent RNA 6000 Pico Kit and Eukaryotic Total RNA assay.
Sequencing
Pacific Biosciences HiFi circular consensus DNA sequencing libraries were constructed according to the manufacturers’ instructions. Poly(A) RNA-Seq libraries were constructed using the NEB Ultra II RNA Library Prep kit. DNA and RNA sequencing was performed by the Scientific Operations core at the WSI on Pacific Biosciences SEQUEL II (HiFi) and Illumina NovaSeq 6000 (RNA-Seq) instruments. Hi-C data were also generated from leaf tissue of daLinVulg1 using the Arima2 kit and sequenced on the Illumina NovaSeq 6000 instrument.
Genome assembly, curation and evaluation
Assembly was carried out with Hifiasm ( Cheng et al., 2021) and haplotypic duplication was identified and removed with purge_dups ( Guan et al., 2020). One round of polishing was performed by aligning 10X Genomics read data to the assembly with Long Ranger ALIGN, calling variants with FreeBayes ( Garrison & Marth, 2012). The assembly was then scaffolded with Hi-C data ( Rao et al., 2014) using YaHS ( Zhou et al., 2023). The assembly was checked for contamination and corrected using the gEVAL system ( Chow et al., 2016) as described previously ( Howe et al., 2021). Manual curation was performed using gEVAL, HiGlass ( Kerpedjiev et al., 2018) and Pretext ( Harry, 2022). The mitochondrial and chloroplast genomes were assembled using MBG ( Rautiainen & Marschall, 2021) from PacBio HiFi reads mapping to related genomes: a representative circular sequence was selected for each from the graph based on read coverage.
A Hi-C map for the final assembly was produced using bwa-mem2 ( Vasimuddin et al., 2019) in the Cooler file format ( Abdennur & Mirny, 2020). To assess the assembly metrics, the k-mer completeness and QV consensus quality values were calculated in Merqury ( Rhie et al., 2020). This work was done using Nextflow ( Di Tommaso et al., 2017) DSL2 pipelines “sanger-tol/readmapping” ( Surana et al., 2023a) and “sanger-tol/genomenote” ( Surana et al., 2023b). The genome was analysed within the BlobToolKit environment ( Challis et al., 2020) and BUSCO scores ( Manni et al., 2021; Simão et al., 2015) were calculated.
Table 3 contains a list of relevant software tool versions and sources.
Table 3. Software tools: versions and sources.
| Software tool | Version | Source |
|---|---|---|
| BlobToolKit | 4.0.7 | https://github.com/blobtoolkit/blobtoolkit |
| BUSCO | 5.3.2 | https://gitlab.com/ezlab/busco |
| FreeBayes | 1.3.1-17-gaa2ace8 | https://github.com/freebayes/freebayes |
| gEVAL | N/A | https://geval.org.uk/ |
| Hifiasm | 0.15.3 | https://github.com/chhylp123/hifiasm |
| HiGlass | 1.11.6 | https://github.com/higlass/higlass |
| Long Ranger
ALIGN |
2.2.2 |
https://support.10xgenomics.com/genome-
exome/software/pipelines/latest/advanced/other- pipelines |
| MBG | - | https://github.com/maickrau/MBG |
| Merqury | MerquryFK | https://github.com/thegenemyers/MERQURY.FK |
| PretextView | 0.2 | https://github.com/wtsi-hpag/PretextView |
| purge_dups | 1.2.3 | https://github.com/dfguan/purge_dups |
| YaHS | yahs-1.1.91eebc2 | https://github.com/c-zhou/yahs |
Wellcome Sanger Institute – Legal and Governance
The materials that have contributed to this genome note have been supplied by a Darwin Tree of Life Partner. The submission of materials by a Darwin Tree of Life Partner is subject to the ‘Darwin Tree of Life Project Sampling Code of Practice’, which can be found in full on the Darwin Tree of Life website here. By agreeing with and signing up to the Sampling Code of Practice, the Darwin Tree of Life Partner agrees they will meet the legal and ethical requirements and standards set out within this document in respect of all samples acquired for, and supplied to, the Darwin Tree of Life Project.
Further, the Wellcome Sanger Institute employs a process whereby due diligence is carried out proportionate to the nature of the materials themselves, and the circumstances under which they have been/are to be collected and provided for use. The purpose of this is to address and mitigate any potential legal and/or ethical implications of receipt and use of the materials as part of the research project, and to ensure that in doing so we align with best practice wherever possible. The overarching areas of consideration are:
• Ethical review of provenance and sourcing of the material
• Legality of collection, transfer and use (national and international)
Each transfer of samples is further undertaken according to a Research Collaboration Agreement or Material Transfer Agreement entered into by the Darwin Tree of Life Partner, Genome Research Limited (operating as the Wellcome Sanger Institute), and in some circumstances other Darwin Tree of Life collaborators.
Acknowledgements
We thank Alex D. Twyford for helpful comments on the introduction section.
Funding Statement
This work was supported by Wellcome through core funding to the Wellcome Sanger Institute (206194, <a href=https://doi.org/10.35802/206194>https://doi.org/10.35802/206194</a>) and the Darwin Tree of Life Discretionary Award (218328, <a href=https://doi.org/10.35802/218328>https://doi.org/10.35802/218328</a>).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; peer review: 2 approved]
Data availability
European Nucleotide Archive: Linaria vulgaris. Accession number PRJEB50878; https://identifiers.org/ena.embl/PRJEB50878 ( Wellcome Sanger Institute, 2022)
The genome sequence is released openly for reuse. The Linaria vulgaris genome sequencing initiative is part of the Darwin Tree of Life (DToL) project. All raw sequence data and the assembly have been deposited in INSDC databases. The genome will be annotated using available RNA-Seq data and presented through the Ensembl pipeline at the European Bioinformatics Institute. Raw data and assembly accession identifiers are reported in Table 1.
Author information
Members of the Royal Botanic Gardens Kew Genome Acquisition Lab are listed here: https://doi.org/10.5281/zenodo.4786680.
Members of the Royal Botanic Garden Edinburgh Genome Acquisition Lab are listed here: https://doi.org/10.5281/zenodo.4786682.
Members of the Plant Genome Sizing collective are listed here: https://doi.org/10.5281/zenodo.7994306.
Members of the Darwin Tree of Life Barcoding collective are listed here: https://doi.org/10.5281/zenodo.4893703.
Members of the Wellcome Sanger Institute Tree of Life programme are listed here: https://doi.org/10.5281/zenodo.4783585.
Members of Wellcome Sanger Institute Scientific Operations: DNA Pipelines collective are listed here: https://doi.org/10.5281/zenodo.4790455.
Members of the Tree of Life Core Informatics collective are listed here: https://doi.org/10.5281/zenodo.5013541.
Members of the Darwin Tree of Life Consortium are listed here: https://doi.org/10.5281/zenodo.4783558.
References
- Abdennur N, Mirny LA: Cooler: Scalable storage for Hi-C data and other genomically labeled arrays. Bioinformatics. 2020;36(1):311–316. 10.1093/bioinformatics/btz540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakshi TS, Coupland RT: Vegetative Propagation in Linaria vulgaris. Can J Bot. 1960;38(2):243–249. 10.1139/b60-022 [DOI] [Google Scholar]
- Challis R, Richards E, Rajan J, et al. : BlobToolKit - interactive quality assessment of genome assemblies. G3 (Bethesda). 2020;10(4):1361–1374. 10.1534/g3.119.400908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Concepcion GT, Feng X, et al. : Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat Methods. 2021;18(2):170–175. 10.1038/s41592-020-01056-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow W, Brugger K, Caccamo M, et al. : gEVAL — a web-based browser for evaluating genome assemblies. Bioinformatics. 2016;32(16):2508–2510. 10.1093/bioinformatics/btw159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubas P, Vincent C, Coen E: An epigenetic mutation responsible for natural variation in floral symmetry. Nature. 1999;401(6749):157–161. 10.1038/43657 [DOI] [PubMed] [Google Scholar]
- Cullen E, Fernández-Mazuecos M, Glover BJ: Evolution of nectar spur length in a clade of Linaria reflects changes in cell division rather than in cell expansion. Ann Bot. 2018;122(5):801–809. 10.1093/aob/mcx213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Tommaso P, Chatzou M, Floden EW, et al. : Nextflow enables reproducible computational workflows. Nat Biotechnol. 2017;35(4):316–319. 10.1038/nbt.3820 [DOI] [PubMed] [Google Scholar]
- Druce GC: The hybrids of Linaria repens and L. vulgaris in Britain. Ann Bot. 1896;10(40):622–524. 10.1093/oxfordjournals.aob.a088630 [DOI] [Google Scholar]
- Garrison E, Marth G: Haplotype-based variant detection from short-read sequencing.2012. 10.48550/arXiv.1207.3907 [DOI] [Google Scholar]
- Gledhill D: The names of plants.Cambridge; New York: Cambridge University Press,2008. Reference Source [Google Scholar]
- Guan D, McCarthy SA, Wood J, et al. : Identifying and removing haplotypic duplication in primary genome assemblies. Bioinformatics. 2020;36(9):2896–2898. 10.1093/bioinformatics/btaa025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson Å: Linnaeus’ Peloria: The history of a monster. Theor Appl Genet. 1979;54(6):241–248. 10.1007/BF00281206 [DOI] [PubMed] [Google Scholar]
- Harry E: PretextView (Paired REadTEXTure Viewer): A desktop application for viewing pretext contact maps. 2022; [Accessed 19 October 2022]. Reference Source
- Henniges MC, Powell RF, Mian S, et al. : A taxonomic, genetic and ecological data resource for the vascular plants of Britain and Ireland. Sci Data. 2022;9(1):1. 10.1038/s41597-021-01104-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe K, Chow W, Collins J, et al. : Significantly improving the quality of genome assemblies through curation. Gigascience. Oxford University Press,2021;10(1): giaa153. 10.1093/gigascience/giaa153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerpedjiev P, Abdennur N, Lekschas F, et al. : HiGlass: web-based visual exploration and analysis of genome interaction maps. Genome Biol. 2018;19(1): 125. 10.1186/s13059-018-1486-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linneaus C: Peloria.In: Amoenitates Academicae. Apud Godofredum Kiesewetter.1749;1:55–73. [Google Scholar]
- Loureiro J, Rodriguez E, Dolezel J, et al. : Two new nuclear isolation buffers for plant DNA flow cytometry: A test with 37 species. Ann Bot. 2007;100(4):875–888. 10.1093/aob/mcm152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manni M, Berkeley MR, Seppey M, et al. : BUSCO update: Novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol Biol Evol. 2021;38(10):4647–4654. 10.1093/molbev/msab199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman DA, Thomson JD: Effects of nectar robbing on nectar dynamics and bumblebee foraging strategies in Linaria vulgaris (Scrophulariaceae). Oikos. 2005;110(2):309–320. 10.1111/j.0030-1299.2005.13884.x [DOI] [Google Scholar]
- Obermayer R, Leitch IJ, Hanson L, et al. : Nuclear DNA C-values in 30 species double the familial representation in pteridophytes. Ann Bot. 2002;90(2):209–217. 10.1093/aob/mcf167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicer J, Powell RF, Leitch IJ: The application of flow cytometry for estimating genome Size, Ploidy Level Endopolyploidy, and Reproductive Modes in Plants. In: Besse, P. (ed.) Methods in Molecular Biology (Clifton, N.J.).New York, NY: Humana,2021;2222:325–361. 10.1007/978-1-0716-0997-2_17 [DOI] [PubMed] [Google Scholar]
- POWO: Linaria linaria (L.). H.Karst., Plants of the World Online.2022; [Accessed 3 April 2023]. Reference Source
- Preston CD, Pearman D, Trevor DD: New atlas of the British & Irish flora.Oxford: Oxford University Press,2002. Reference Source [Google Scholar]
- Rao SSP, Huntley MH, Durand NC, et al. : A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159(7):1665–1680. 10.1016/j.cell.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rautiainen M, Marschall T: MBG: Minimizer-based sparse de Bruijn Graph construction. Bioinformatics. 2021;37(16):2476–2478. 10.1093/bioinformatics/btab004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhie A, Walenz BP, Koren S, et al. : Merqury: Reference-free quality, completeness, and phasing assessment for genome assemblies. Genome Biol. 2020;21(1): 245. 10.1186/s13059-020-02134-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhie A, McCarthy SA, Fedrigo O, et al. : Towards complete and error-free genome assemblies of all vertebrate species. Nature. 2021;592(7856):737–746. 10.1038/s41586-021-03451-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, et al. : BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31(19):3210–3212. 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]
- Sing SE, Peterson RKD: Assessing Environmental Risks for Established Invasive Weeds: Dalmatian ( Linaria dalmatica) and Yellow ( L. vulgaris) toadflax in North America. Int J Environ Res Public Health. 2011;8(7):2828–2853. 10.3390/ijerph8072828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stace CA, Preston CD, Pearman DA: Hybrid flora of the British Isles.Bristol: Botanical Society of Britain and Ireland,2019. Reference Source [Google Scholar]
- Stace CA, Thompson H, Stace M: New flora of the British Isles.4th ed. C&M Floristics,2015. [Google Scholar]
- Surana P, Muffato M, Qi G: sanger-tol/readmapping: sanger-tol/readmapping v1.1.0 - Hebridean Black (1.1.0). Zenodo. 2023a; [Accessed 21 July 2023]. 10.5281/zenodo.7755665 [DOI] [Google Scholar]
- Surana P, Muffato M, Sadasivan Baby C: sanger-tol/genomenote (v1.0.dev). Zenodo. 2023b; [Accessed 21 July 2023]. 10.5281/zenodo.6785935 [DOI] [Google Scholar]
- Vasimuddin Md, Misra S, Li H, et al. : Efficient Architecture-Aware Acceleration of BWA-MEM for Multicore Systems. In: 2019 IEEE International Parallel and Distributed Processing Symposium (IPDPS). IEEE,2019;314–324. 10.1109/IPDPS.2019.00041 [DOI] [Google Scholar]
- Ward SM, Fleischmann CE, Turner MF, et al. : Hybridization between Invasive Populations of Dalmatian Toadflax ( Linaria dalmatica) and Yellow Toadflax ( Linaria vulgaris). Invasive Plant Sci Manag. 2009;2(4):369– 378. 10.1614/IPSM-09-031.1 [DOI] [Google Scholar]
- Wellcome Sanger Institute: The genome sequence of common toadflax, Linaria vulgaris Mill., 1768. European Nucleotide Archive.[dataset], accession number PRJEB50878,2022.
- Zhou C, McCarthy SA, Durbin R: YaHS: yet another Hi-C scaffolding tool. Bioinformatics. 2023;39(1): btac808. 10.1093/bioinformatics/btac808 [DOI] [PMC free article] [PubMed] [Google Scholar]